Abstract
Protein-based vaccines are playing an increasingly important role in the COVID-19 pandemic. As late-stage clinical data are finalized and released, the number of protein-based vaccines expected to enter the market will increase significantly. Most protein-based COVID-19 vaccines are based on the SARS-CoV-2 spike protein (S-protein), which plays a major role in viral attachment to human cells and infection. As a result, in order to develop and manufacture quality vaccines consistently, it is imperative to have access to selective and efficient methods for the bioanalytical assessment of S-protein. In this study, samples of recombinant S-protein (hexS-protein) and commercial S-protein were used to develop a selective reversed-phase HPLC (RP-HPLC) method that enabled elution of the intact S-protein monomer as a single peak on a wide pore, C8-bonded chromatographic column. The S-protein subunits, S1 and S2 subunits, were clearly separated from intact S-protein and identified. The results of this study set the foundation for reversed-phase HPLC method development and analysis for selective and efficient separation of S-protein monomer from its subunits.
Keywords: Covid-19, Spike protein, RP-HPLC, bioanalytical characterization
1. Introduction
Within a year of the first COVID-19 cases, vaccines were developed rapidly to combat the public health crisis. First-generation vaccines were based on nucleic acids and non-replicating viral vectors, which allowed novel technologies to debut effective vaccine candidates at the global scale [1,2]. However, as the SARS-CoV-2 virus spread rapidly, the need for global immunization coverage increased dramatically [3]. As a result, conventional protein-based vaccines are expected to find a strong foothold in logistically difficult regions as the pandemic develops [4]. The advantages of these vaccines include a strong history of triggering safe and robust immune responses [5,6] along with significant flexibility in storage and transportation.
The vast majority of protein-based COVID-19 vaccines use full-length or truncated forms of the SARS-CoV-2 virus spike protein (S-protein) as the primary antigenic component [5,7]. The S-protein is a heavily glycosylated fusion protein (140 kDa by amino acid sequence) that homotrimerizes on the virus surface and is critical for membrane fusion and host cell receptor binding [8]. During infection, the S-protein is activated by proteolytic cleavage into its S1 and S2 subunits [9] by host-cell proteases. Importantly, viral surveillance show that mutations in the S-protein have the highest impact on transmissibility and immune escape [10,11].
To date, a growing number of protein-based COVID-19 vaccines are at the late stages of clinical trials and seeking approval [7]. One highly anticipated candidate is the NVX-CoV2373 protein subunit vaccine, which contains a cleavage-resistant full-length S-protein stabilized in the prefusion conformation [12]. Another vaccine in late-stage development, SCB-2019, also uses the full-length S-protein but maintains the wild-type protease cleavage site, making it prone to cleavage by host-cell proteases during manufacturing [13]. Given the diversity of protein-based COVID-19 vaccines, the complexity of the S-protein, and the potential for variants in future vaccine formulations, robust analytical techniques will be needed to separate S-protein species and assess the critical quality attributes which ensure product consistency and comparability throughout its lifecycle.
Reversed-phase (RP) high-performance liquid chromatography (HPLC) is one such technique well-established for the separation and quality assessment of proteins. Compared to other techniques such as SDS-PAGE, the advantages of RP-HPLC in resolving power, selectivity, and sensitivity are significant, especially for complex and heterogeneous biotherapeutics. Not only can antigens be separated by sequence but also post-translational modifications. As a result, RP-HPLC is a highly reproducible analytical tool widely used for a variety of biotherapeutics, such as purified protein, virus-like particles, and whole viruses [14], [15], [16]. However, as of this writing, only one brief application note with limited detail on S-protein analysis by RP-HPLC exists [17] and no published methods have characterized intact S-protein with its major subunits, S1 and S2.
To address this gap, we developed a selective, repeatable, and versatile RP-HPLC method to characterize S-protein. Using an in-house purified, full-length S-protein variant called HexaPro S-protein (hexS-protein) and a variety of commercial S-protein products, we identified clear and reproducible peak patterns associated with full-length S-protein as well as its S1 and S2 subunit cleavage products. Finally, a preliminary method qualification is presented for additional direction in biotherapeutic applications.
2. Material and methods
2.1. Spike protein purification
The plasmid encoding hexS-protein was purchased from Addgene (plasmid #154,754) [18]. Cells were transfected according to previously developed methods [19]. Briefly, FreeStyle 293-F cells (Invitrogen) were cultured at 37 °C, 8% CO2 with Gibco™ FreeStyle™ 293 Expression Medium (Fisher Scientific) and seeded at 1.12×106 cells/mL for transfection. The hexS-protein plasmid and polyethylenimine (25 kDa linear PEI, Polysciences) were diluted and sterile filtered (0.22 µm) in Opti-MEM™ I Reduced Serum Medium (Life Technologies). The plasmid/PEI mixture was added to cells at a final density of 1.0 × 106 cells/mL (final concentrations: 0.5 µg/mL plasmid, 1.5 µg/mL PEI).
For protein purification, cell suspensions were centrifuged 4 days post-transfection, supplemented with 1 mM benzamidine HCl, and purified on Ni-NTA Superflow resin (Qiagen) by FPLC (Bio-Rad NGC Chromatography System). Resin was equilibrated with Equilibration Buffer (10 mM Tris–HCl, 100 mM sodium phosphate, pH 8.0). After loading the clarified supernatant, the column was washed with Wash 1 (10 mM Tris–HCl, 100 mM sodium phosphate, 30 mM imidazole), Wash 2 (10 mM Tris–HCl, 100 mM sodium phosphate, 60 mM imidazole), and eluted with Elution Buffer (10 mM Tris–HCl, 100 mM sodium phosphate, 250 mM imidazole). Protein size and purity were confirmed by gel electrophoresis. Final samples were dialyzed against buffer (1.0 M Phosphate Buffer Solution pH 7.4 from Sigma-Aldrich diluted to 5 mM phosphate, supplemented with 50 mM sodium chloride) at room temperature and stored at 4 °C or −80 °C.
2.2. HPLC instrumentation and chemicals
All chemicals used for HPLC assays were analytical reagent grade. Sodium chloride, sodium phosphate dibasic, polysorbate-80, and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN) and 2-propanol were purchased from Merck KGaA (Darmstadt, Germany). Distilled water (dH2O) was deionized on a Nanopure Diamond™ system (Barnstead International, Dubuque, IA, USA). Commercial reference S-proteins and related subunits were purchased from Sino Biological Inc., summarized in Table 1 .
Table 1.
Commercial S-proteins used in this manuscript.
| catalog no. | manufacturer protein name | study protein name | expression host |
|---|---|---|---|
| 40589-V27B-B | S1+S2 ECD-Avi-His | comS-protein A | Baculovirus-Insect Cells |
| 40589-V08B1 | S1+S2 ECD-His | comS-protein B | Baculovirus-Insect Cells |
| 40590-V08B | S2-His | comS2 | Baculovirus-Insect Cells |
| 40591-V08H | S1-His | comS1 | HEK293 Cells |
The HPLC system consisted of a Waters Alliance 2695 chromatograph equipped with a column heater and an auto-sampler with a sample cooling device. Fluorescence and UV spectroscopy detection was accomplished by coupling in-series a Waters 2475 Multichannel Fluorescence Detector (8 µL flow cell, excitation wavelength of λex 280 nm and emission wavelength at λem 335 nm) and a Waters 2998 UV–VIS photodiode array detector (Waters, QC, Canada). Data acquisition and integration were performed with Empower 3 Chromatography Data Software.
Additional details on LC-MS/MS analysis are provided in Supplemental Information.
2.3. RP-HPLC analysis
The chromatographic column used for the optimized protocol was an Aeris Widepore XB-C8, 150 mm x 4.6 mm, 3.6 µm particles, 200 Å pore size (Phenomenex, Torrance, CA, USA). The optimized separation method were carried out at 40ºC with an AB gradient elution of 45 min at a flow rate of 1.0 mL/min (Gradient I, Table 2 ). Eluent A was 0.1% (v/v) aqueous TFA and eluent B was 0.08% (v/v) TFA in 50% ACN and 50% 2-propanol. Prior to analysis, two zero injections (no sample injected) are performed with Gradient I. Injection volume for full-length S-protein material was 20 µL at 50 µg/mL and 10 µL at 50 µg/mL for comS1 and comS2.
Table 2.
Gradient I conditions used in RP-HPLC method.
| Time(min) | Flow rate(mL/min) | %A | %B |
|---|---|---|---|
| 0 | 1.0 | 95 | 5 |
| 2 | 1.0 | 95 | 5 |
| 4 | 1.0 | 65 | 35 |
| 34 | 1.0 | 35 | 65 |
| 37 | 1.0 | 5 | 95 |
| 39 | 1.0 | 5 | 95 |
| 40 | 1.0 | 95 | 5 |
| 45 | 1.0 | 95 | 5 |
For linearity assessment with polysorbate-80, final solutions were diluted to 0.05 w/v polysorbate-80 using a 1% w/v stock solution.
2.4. Size-exclusion (SE) HPLC
The chromatographic column used was a TSKgel G4000SWXL, 300 mm x 7.8 mm, 8 µm particles, 450 Å pore size (Tosoh Bioscience, Philadelphia, PA, USA). Separations were carried out at room temperature with isocratic elution at 0.5 mL/min phosphate buffer (0.1 M sodium phosphate, 0.2 M sodium chloride, pH 7). The mobile phase was filtered through a 0.45 µm nylon filter (GVS, Sanford, ME, USA). The injection volume for hexS-protein was 20 µL at 50 µg/mL. Molecular weight (MW) standards used were as follows: Thyroglobulin (Sigma T1001), Apoferritin (Sigma A3660), Ovalbumin (Sigma A2512), 4-Aminobenzoic acid (Sigma A9878).
3. Results and discussion
3.1. Spike protein material used for method development
To generate vaccine-relevant protein material for RP-HPLC method development, a modified full-length S-protein variant called hexS-protein was purified from FreeStyle293-F cells [18]. As one of the earliest published S-protein constructs available, many manufacturers have incorporated the features of hexS-protein into protein-based COVID-19 vaccines such as NVX-CoV2373 (Supplemental Figure S1) [5,12]. Some features enhance immunogenicity, originally established for a related betacoronavirus [20], while other features improve production yields [18].
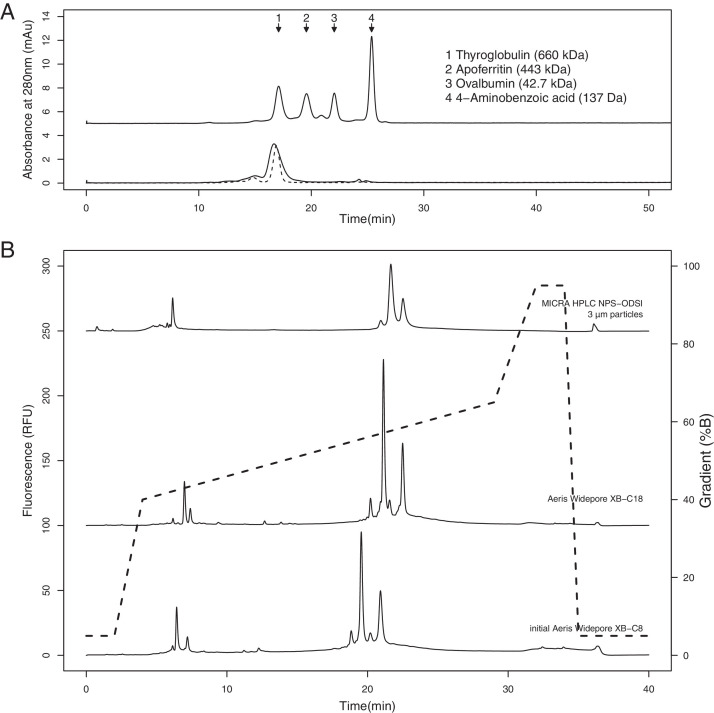
As a quality check to confirm that in-house purified glycosylated hexS-protein formed the expected trimeric structure, samples were analyzed by SE-HPLC using a TSKgel G4000SWXL column, appropriate for fractionating biomolecules up to 7000 kDa. A comparison to MW standards showed that the main hexS-protein peak eluted at a MWapp of 700 kDa (Fig. 1A), consistent with a previous SE-HPLC report on the size of trimeric glycosylated S-protein determined by an Xbridge BEH200 column [21]. It is not clear whether the small group of unresolved peaks eluting before the major peak correspond to higher order aggregates or alternate S-protein conformers, since both have been observed as earlier-eluting species by SE-HPLC [21,22]. However, it's noted that column parameters in this study, such as particle size, could contribute to resolution loss between species of similar hydrodynamic radius. Overall, the observed size of the main hexS-protein was consistent with the trimeric form.
Fig. 2.
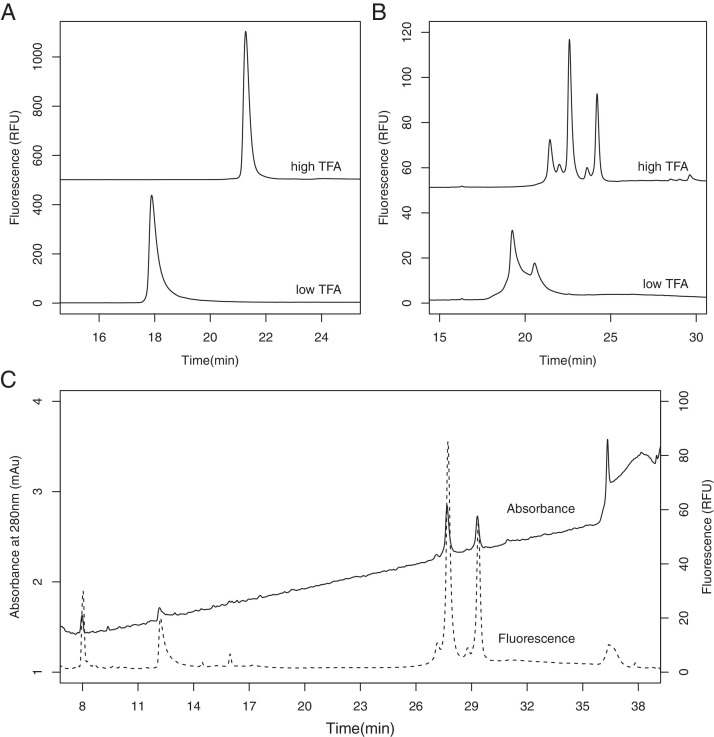
RP-HPLC performance in high and low TFA% for (A) purified hexS-protein and (B) comS-protein A. (C) Comparison of fluorescence (dashed line) and absorbance detection in RP-HPLC analysis of comS-protein A.
In addition to in-house purified hexS-protein, commercial products available at the time of this study were used to represent vaccines closer to the wild-type S-protein sequence. These include two full-length wild-type S-proteins (comS-protein A and comS-protein B) along with the S1 and S2 subunits alone (comS1 and comS2, respectively) (Table 1).
3.2. Method development and optimization
Under the organic and acidic solvent conditions of RP-HPLC, proteins are typically denatured and higher-order species dissociated. As a result, RP-HPLC analysis will characterize the physiochemical features of monomers (140 kDa for hexS-protein).
Method development using hexS-protein and commercial S-protein products began with a number of RP-HPLC columns established for bioanalytical characterization of viral membrane proteins, such as hemagglutinin (Fig. 1B) [16]. Under low TFA conditions, 0.04% TFA Eluent A and 0.03% TFA Eluent B, two MICRA® HPLC NPS-ODSI columns (33 mm × 4.6 mm with 1.5 µm particles, and 100 mm × 4.6 mm with 3 µm particles) were first tested with a separation gradient from 40% to 65% Eluent B over 25 min. Both showed some peak resolution and acceptable back pressure, with the 3 µm particle column allowing for higher sample loading (Fig. 1B, top trace). Later, these columns were replaced with the Aeris Widepore XB-C8 core-shell silica column (150 mm x 4.6 mm, 3.6 µm particles, 200 Å), also suitable for separating large hydrophobic proteins. This provided even lower back pressure, significantly improved near-baseline resolution between peaks, and comparable sample loading capacity (Fig. 1B, bottom trace). An Aeris Widepore XB-C18 column was also tested and showed similar performance to the C8 version, except for the co-elution of an apparent S-protein peak with TFA impurities at 6 min (Fig. 1B, middle trace). Due to this minor difference in performance, the Aeris Widepore XB-C8 column was chosen for further evaluation. Adjusting column temperatures from 55 °C to 40 °C had no notable impact on sample recovery or resolution.
Fig. 1.
(A) SE-HPLC analysis of purified hexS-protein (bottom, black trace) superimposed with a thyroglobulin reference (bottom, dashed line). MW standards were used to estimate hexS-protein size (top trace). (B) RP-HPLC chromatograms showing the performance of different columns on comS-protein A under low TFA conditions. The solvent gradient used in development for all columns is shown by the dashed line.
Interestingly, with a newly purchased Aeris Widepore XB-C8 column and the gradient elution profile used in Fig. 1B (40% to 65% B over 25 min), TFA concentrations needed to be increased to 0.1% in Eluent A and 0.08% in Eluent B for similarly good peak resolution (Fig. 2A and B, high TFA traces). In contrast, the lower TFA concentrations used in development (0.04% TFA Eluent A and 0.03% TFA Eluent B) produced significant peak tailing and reduced resolution (Fig. 2A and B, low TFA traces). Later, the gradient was extended (35% to 65% Eluent B over 30 min) to prevent the co-elution of S1 peaks in comS-proteins with TFA impurity peaks (Table 2, Gradient I). These results were reproducible across multiple recently (2021) manufactured columns. The difference in performance between these newly purchased columns and the older columns used in method development could be due to unknown impurities introduced in the column's usage or manufacturing history, which emphasizes the importance of interlaboratory studies for robustness assessment across equipment and environments.
In all studies, both native fluorescence (ex/em: 280 nm/335 nm) and absorbance (210 – 400 nm) were monitored, which showed comparable peak retention patterns using Gradient I (Fig. 2C). Native fluorescence detection of aromatic residues, primarily tryptophan, is advantageous for low-abundance samples due to its sensitivity [23], [24], [25]. Since the hexS-protein sequence contains 12 tryptophan residues, fluorescence detection allowed for significantly lower injection volumes and improved resolution.
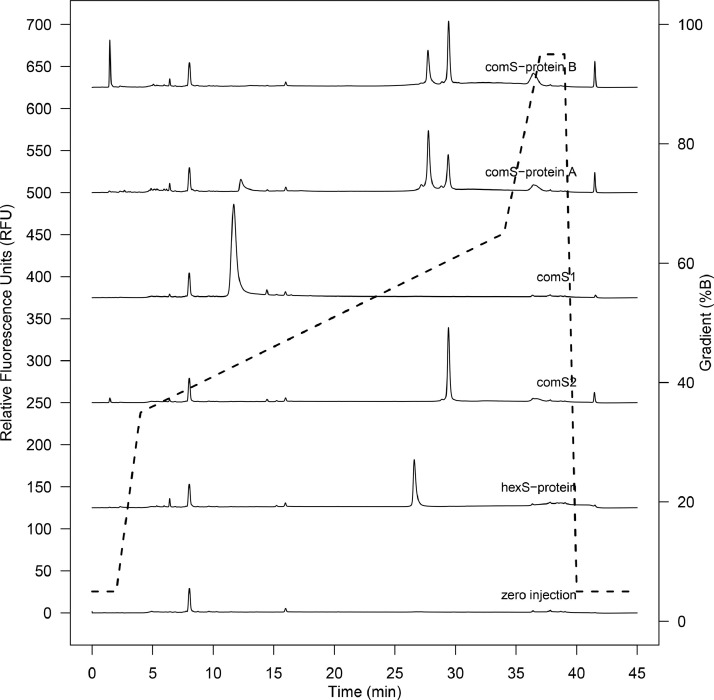
The final method (Gradient I, using the Aeris Widepore XB-C8 column) showed good peak resolution and was highly reproducible across independent sample preparations (Supplemental Figure S2). Analysis of purified hexS-protein showed elution of a single major peak at a retention time of 26.7 min (Fig. 3 , second trace from bottom), with a corresponding absorbance signal at 280 nm. Confirmation of the S-protein identity (60% sequence coverage with 46 unique peptide sequences identified) was obtained by MS/MS analysis of the collected HPLC peak (Supplemental Figure S3).
Fig. 3.
RP-HPLC chromatograms with Gradient I comparing hexS-protein to commercial products: full-length S-protein (comS-protein A and comS-protein B), the S1 subunit (comS1), and the S2 subunit (comS2). No sample injected is in the zero injection and solvent gradient is shown by the dashed line.
3.3. Separation of S1, S2, and full-length S-protein
A potential quality issue for COVID-19 vaccines, such as SCB-2019, is uncontrolled cleavage during production [26]. Since both comS-protein A and comS-protein B retain the native S1/S2 protease cleavage site, the susceptibility to cleavage is increased [9], which can be assessed by RP-HPLC analysis with Gradient I. Indeed, chromatograms of comS-protein A and comS-protein B showed the presence of multiple peaks (Fig. 3, top two traces). These were identified by comparison to comS1 and comS2 subunit products (Fig. 3, third and fourth traces from the top respectively). Both comS-protein A and comS-protein B showed a peak eluting at around 29.4 min, which corresponded to the comS2 subunit retention time of 29.4 min. However, only comS-protein A showed a significant peak at 12.3 min, which corresponded roughly to the comS1 subunit retention time of 11.7 min. In contrast, no S1 cleavage product could be detected in the comS-protein B product, despite significant S2 presence. While not a quantitative analysis, which would require a calibration curve with a carefully selected reference standard, this relative difference in S1/S2 fluorescence between comS-protein A and B cannot be solely explained by aromatic residue content. These results suggest significant differences in subunit content between products, possibly caused by expression or purification conditions impacting degradation or recovery.
It was noted that the full-length species of comS-protein A and comS-protein B both eluted at 27.7 min, which was shifted relative to full-length hexS-protein at 26.7 min (Fig. 3, second trace from bottom) and likely due to sequence or glycosylation differences. An extraneous peak at 1.5 min in comS-protein B (Fig. 3, top trace) was determined non-proteinaceous from lack of absorbance. All other S-protein and subunit peak identities were confirmed by MS/MS analysis of the corresponding fractions collected from HPLC (Supplemental Figure S4).
3.4. Preliminary method qualification
In a preliminary qualification assessment, the method's linearity, limit of detection (LOD), limit of quantification (LOQ), and intra-assay precision was assessed.
Using hexS-protein, we confirmed linearity between 50 – 100 µg/mL using fluorescence (R2 = 0.995, 20 µL injections). This range was significantly improved with the addition of 0.05% w/v polysorbate-80, a common ingredient in the final formulation of vaccines such as NVX-CoV2373, which likely prevented non-specific protein adsorption to vial surfaces. With the polysorbate-80 detergent, method linearity was determined between 2.5 – 100 µg/mL using fluorescence (R2 = 1.000, 20 µL injections) with no impact on the retention time or shape of the hexS-protein peak. In both linearity assessments, with and without polysorbate-80, 3 injections were analysed for each of 6 concentrations.
The LOD, defined as the lowest protein amount injected to give a signal-to-noise ratio of 3:1, was determined with hexS-protein at 25 ng for fluorescence detection and 500 ng for absorbance detection at 280 nm. The LOQ, defined as the lowest protein amount injected to give a signal-to-noise ratio of 10:1, was determined with hexS-protein at 50 ng for fluorescence detection and 1 µg for absorbance detection at 280 nm. These results show that fluorescence detection is significantly advantageous for low-concentration S-protein samples.
Intra-assay precision was measured by 8 injections at 20 µL of 50 µg/mL hexS-protein. For peak area, RSD% was 3.34%. For retention time, RSD% was 0.05%. These results show acceptable intra-assay precision.
4. Conclusions
Protein-based vaccines, including those based on whole viruses and recombinant protein, are some of the most well-established and robust vaccine types [5]. Overall, these advantages in efficacy and stability are significant, especially for areas without access to the stable cold-chain distribution system required for first-generation mRNA vaccines [27] and for future endemic scenarios where addressing local needs will be priority.
In this work, we show that RP-HPLC is a useful tool for assessing vaccine-relevant S-protein quality and consistency. The separation and identification of full-length S-protein from its major cleavage subunits, S1 and S2, was consistently demonstrated for a variety of products. Preliminary method qualification suggests that the RP-HPLC method could be easily adapted to meet the stringent quality requirements of drug assessment. The observations reported here, such as the impact of TFA conditions and polysorbate-80 addition, will be informative for adapting the method to different systems.
Historically, vaccines and their quality assays were developed and standardized in parallel, slowly over decades [28]. However, in the COVID-19 era, a variety of modern techniques will need to be established rapidly. As a bioanalytical technique, RP-HPLC is one such tool that provides the rapid results, reagent flexibility, and method adaptability well-suited for vaccine analysis.
CRediT authorship contribution statement
Barry Lorbetskie: Conceptualization, Methodology, Investigation. Taryn White: Investigation. Marybeth Creskey: Investigation, Formal analysis, Visualization. Xu Zhang: Conceptualization, Formal analysis, Writing – review & editing. Michel Girard: Conceptualization, Writing – review & editing. Roger Y. Tam: Investigation, Writing – review & editing, Visualization. Simon Sauvé: Conceptualization, Writing – review & editing. Huixin Lu: Conceptualization, Methodology, Writing – original draft, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Jason McLellan for the generous gift of S-protein constructs and protocols, Dr. Sean Li and Dr. Yves Aubin for their expertise on S-protein biology and production, and Geneviève Gingras for the expertise in protein purification.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.chroma.2022.463424.
Appendix. Supplementary materials
References
- 1.Jackson N.A.C., et al. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rauch S., et al. New Vaccine TECHNOLOGIES to Combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchy P., et al. COVID-19 pandemic: lessons learned from more than a century of pandemics and current vaccine development for pandemic control. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolgin E. How protein-based COVID vaccines could change the pandemic. Nature. 2021;599(7885):359–360. doi: 10.1038/d41586-021-03025-0. [DOI] [PubMed] [Google Scholar]
- 5.Pollet J., Chen W.H., Strych U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug. Deliv. Rev. 2021;170:71–82. doi: 10.1016/j.addr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin S. History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 2014;111(34):12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.COVID-19 vaccine tracker and landscape. 2022 [cited 2022 March 14, 2022]; Available from: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 8.Duan L., et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peacock T.P., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 10.Harvey W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinde V., et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richmond P., et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet. 2021;397(10275):682–694. doi: 10.1016/S0140-6736(21)00241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dillon T.M., et al. Optimization of a reversed-phase high-performance liquid chromatography/mass spectrometry method for characterizing recombinant antibody heterogeneity and stability. J. Chromatogr. A. 2006;1120(1–2):112–120. doi: 10.1016/j.chroma.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Shytuhina A., et al. Development and application of a reversed-phase high-performance liquid chromatographic method for quantitation and characterization of a Chikungunya virus-like particle vaccine. J. Chromatogr. A. 2014;1364:192–197. doi: 10.1016/j.chroma.2014.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorbetskie B., et al. Fast and highly selective determination of hemagglutinin content in quadrivalent influenza vaccine by reversed-phase high-performance liquid chromatography method. J. Chromatogr. A. 2017;1528:18–24. doi: 10.1016/j.chroma.2017.10.041. [DOI] [PubMed] [Google Scholar]
- 17.Jennifer M. Nguyen, M.A.L.Comprehending COVID-19: reversed-phase liquid chromatography (RPLC) of intact SARS-CoV-2 spike protein. 2020; Available from: https://www.waters.com/nextgen/us/en/library/application-notes/2020/comprehending-covid-19-reversed-phase-liquid-chromatography-rplc-of-intact-sars-cov-2-spike-protein.html.
- 18.Hsieh C.L., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369(6510):1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallesen J., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U.S.A, 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro R., et al. Production of high-quality SARS-CoV-2 antigens: impact of bioprocess and storage on glycosylation, biophysical attributes, and ELISA serologic tests performance. Biotechnol. Bioeng. 2021;118(6):2202–2219. doi: 10.1002/bit.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera N.G., et al. Characterization of the SARS-CoV-2 S protein: biophysical, biochemical, structural, and antigenic analysis. ACS Omega. 2021;6(1):85–102. doi: 10.1021/acsomega.0c03512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K.C., Veenstra T.D., Issaq H.J. Comparison of fluorescence, laser-induced fluorescence, and ultraviolet absorbance detection for measuring HPLC fractionated protein/peptide mixtures. Anal. Chem. 2011;83(6):2394–2396. doi: 10.1021/ac1032462. [DOI] [PubMed] [Google Scholar]
- 24.Joshi P.R.H., et al. Development and validation of an anion exchange high-performance liquid chromatography method for analysis of empty capsids and capsids encapsidating genetic material in a purified preparation of recombinant adeno-associated virus serotype 5. Hum. Gene Ther. 2021;32(21–22):1390–1402. doi: 10.1089/hum.2020.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Canas V., et al. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Anal. Chem. 2007;79(8):3164–3172. doi: 10.1021/ac0621120. [DOI] [PubMed] [Google Scholar]
- 26.Ma J., et al. Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19. J. Virol. 2021 doi: 10.1128/JVI.00194-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crommelin D.J.A., et al. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 2021;110(3):997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood J.M., Weir J.P. Standardisation of inactivated influenza vaccines-learning from history. Influenza Other Respir. Viruses. 2018;12(2):195–201. doi: 10.1111/irv.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.