Abstract
Integration host factor (IHF) is a bacterial protein that binds and severely bends a specific DNA target. IHF binding sites are approximately 30 to 35 bp long and are apparently divided into two domains. While the 3′ domain is conserved, the 5′ domain is degenerate but is typically AT rich. As a result of physical constraints that IHF must impose on DNA in order to bind, it is believed that this 5′ domain must possess structural characteristics conducive for both binding and bending with little regard for specific contacts between the protein and the DNA. We have examined the sequence requirements of the 5′ binding domain of the IHF binding target. Using a SELEX procedure, we randomized and selected variants of a natural IHF site. We then analyzed these variants to determine how the 5′ binding domain affects the structure, affinity, and function of an IHF-DNA complex in a native system. Despite finding individual sequences that varied over 100-fold in affinity for IHF, we found no apparent correlation between affinity and function.
In solution, B-form DNA is isotropically flexible only when it vastly exceeds its persistence length (150 bp; reviewed in reference 8). Nevertheless, DNA binding proteins often impose an anomalous structure onto their DNA targets despite typically contacting fewer than 30 bp. These deformations can be dramatic, however, such that the generated bend itself has a functional consequence. Since these DNA bending proteins generally require no coupled energy source for binding, the deformations are probably derived from the favorable thermodynamics of protein-DNA interactions. Proteins accomplish this feat by using various mechanisms, for example, charge neutralization of phosphates and/or destabilization of DNA base-stacking interactions (25, 30).
What role does the DNA sequence play in its own distortion? While specific protein-DNA interactions are most often mediated via specific hydrogen bonding between DNA bases and amino acid residues, there is an ever-growing class of proteins that select their target site based on indirect readout, i.e., a structure or subset of structures that are specifically recognized by a protein (34).
The bacterial protein integration host factor (IHF) epitomizes such proteins. It is a small heterodimeric protein consisting of homologous subunits, α and β, that binds and bends DNA specifically. Although known DNA targets are selected over bulk DNA by at least 1,000-fold (40), there is an inherent degeneracy of sequence in the target selection. IHF was originally discovered as a protein required for efficient integration of the bacteriophage lambda into the Escherichia coli chromosome (36). Subsequently, IHF has been shown to participate in virtually every type of nucleoprotein system (e.g., transcription, replication, and recombination). While its role is always that of an accessory factor, its involvement can be anything from a nominal 2-fold influence, as in promoter activation (23), to as great as a 10,000-fold effect, as in lambda integrative recombination.
It is the complex of IHF with DNA that makes it unique among bending proteins; the recently solved cocrystal structure of IHF bound to one of its natural binding sites shows that the DNA is bent by 180° into a virtual U-turn (24, 25). IHF typically binds to a 30- to 35-bp sequence which can be divided into at least two domains. The 3′ region is significantly conserved, and sites share the consensus WATCAANNNNTTR (where W is A or T, R is purine, and N is any base). The lone cytosine base is conserved in every known natural site. Unlike the 3′ region, the 5′ region seems almost random; natural sites are typically AT rich, but no obvious patterns of sequence emerge as conserved.
How does the IHF binding site accommodate specific binding and bending? Of particular interest is the 5′ binding domain, since it appears variant in each sequence. Recent evidence suggests that this domain is most successful at binding when an appropriately positioned run of adenines, or an A-tract, is present (11). A-tracts typically consist of three to six consecutive adenines that, in the proper sequence context, create an intrinsically rigid structure with a narrow minor groove; adjacent sequences typically possess an anisotropic bend. What facets of A-tracts, if any, attract IHF for preferred binding?
We have attempted to answer these questions by performing a systematic evolution of ligands by exponential enrichment (SELEX) analysis. Selective pressure in vitro for high-affinity binding was applied to a population of IHF binding sites where the wild-type 3′ domain was held constant and the 5′ domain was randomized. Individual sequences were then compared for binding affinity, gross structure of nucleoprotein complexes, and the ability to function in bacteriophage lambda site-specific recombination. We found that the 5′ region can vary the affinity for IHF at least 100-fold. Although comprehensive rules for these sequence determinants could not be deduced, this region contributes significantly to the structure but little to the function of these nucleoprotein complexes. Finally, we found that while the base composition of this region is skewed in native sites, there appears to be no gross base composition advantage for either affinity or function.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5 was the host for all the plasmids used in this work. pHN868, pHN872, and pHN873 were derived from pBR322 and have been described previously (4). pHN868 possesses a 1.0-kb HindIII-BamHI attR-containing insert. pHN872 and pHN873 contain two inserts, a 1.2-kb PstI segment from pUC4K, which confers kanamycin resistance, and a 1.0-kb PstI-BamHI attL-containing segment. pHN872 contains the wild-type attL, and pHN873 contains a mutant attL (QH′) possessing four missense mutations (A37C, A38C, T43G, and T44G) at the H′ site.
SELEX.
Oligonucleotides were purchased from the University of Southern California Microchemical Core Facility. Our randomized H′ pool of attL was constructed from a starting template, oSG42L, with the sequence 5′ GCC TGC TTT TTT ATA CTA AGT TGG CAN NNN NNN NNN NNN NNN NNN NNC AAT TTG TTG CAA CGA ACA GGT CAC TA 3′. This sequence corresponds to the top-strand bases −11 to +62 of attL. We found that we could amplify an intact attL (119 bp) by using this single-stranded template and two additional oligonucleotides. oSG47 (5′ GGA ATT CAA ATA ATG ATT TTA TTT TGA CTG ATA GTG ACC TGT TCG TTG CAA CAA ATT G 3′) possesses 27 bases of complementary DNA sequence to oSG42L in addition to sequence that encompasses the entire 3′ end of the attL locus (+88) and five additional bases which create an EcoRI restriction site. The third oligonucleotide, oSG46 (5′ GGA ATT CCG TTG AAG CCT GCT TTT TTA TAC TAA GTT GGC 3′), corresponds to bases −20 to +13 of attL and also possesses an EcoRI linker. Exponential PCR amplification of a population of attLs was then initiated under the following conditions: strands were melted at 95°C for 3 min, and Taq polymerase (Promega) was added. Subsequent temperature cycling was performed by heating to 94°C for 40 s, annealing at 47°C for 30 s, and extending at 72°C for 40 s. This first amplification to create duplex starting material was performed for only five cycles.
Once the attL population was prepared, it was used as a substrate in an electrophoretic mobility shift assay (EMSA). Twenty-one replicates of 300 ng (80 pmol total) of SELEX-derived attL were each incubated with 60 nM IHF in 50 mM Tris-Cl–50 mM KCl–50 μg of bovine serum albumin (BSA) per ml–3.75 μg of salmon sperm DNA per ml–10% glycerol–1 mM EDTA at pH 7.8 for 20 min at room temperature in a final volume of 20 μl. Each reaction mixture was loaded onto an 8% nondenaturing polyacrylamide gel (29:1 acrylamide-to-bisacrylamide ratio) and electrophoresed (6 to 10 V/cm) for 2 to 3 h. The gels were then stained with ethidium bromide solution (0.5 μg/ml) for 1 h and visualized under long-wave UV light (365 nm). All DNA that was chased into the vicinity (± 0.5 cm) of the wild-type shifted product was excised and purified from the gel fragment by the crush-and-soak method (26). DNA concentrations were estimated by measurement of absorbance at 260 nm.
The amplification portion of the SELEX strategy was then used. Gel-purified IHF-shifted DNA was used as a template for PCR with oligonucleotides oSG46 and oSG49 (5′GGA ATT CAA ATA ATG ATT TTA TTT TGA CTG ATA GTG ACC TGT TCG 3′, a shortened version of oSG47 and one with a melting temperature more similar to that of oSG46) at a final concentration of 400 nM. Shifted DNA from the first round was added to 3 μg/ml. The cycling conditions for amplification were identical to that of the original SELEX substrate reaction, except that nine cycles were performed. The small number of cycles was crucial since we found that under our conditions the polymerizing reaction started to fail significantly beyond nine cycles. This results in increasing formation of a heteroduplex, which does not seem to be a substrate for IHF binding (data not shown). In addition, we found that the highest yield was attained by keeping the reaction volume to 100 μl but performing 20 reactions in parallel. PCR products were pooled, and unincorporated oligonucleotides were removed by using the QIAquick PCR purification kit (Qiagen Inc.). All subsequent rounds of SELEX EMSA were performed the same way.
Cloning attL.
attL PCR products were cloned into the EcoRI site of pBR322 by using the ligation express kit (Clontech) under conditions described by the manufacturer and transformed into E. coli DH5. Plasmid DNA from transformants was analyzed by restriction and DNA sequencing. Once a unique attL-containing plasmid was isolated, the attL portion was amplified by PCR as described above with oligonucleotides oSG46 and oSG49. QIAquick-purified attLs were eluted in 10 mM Tris–1 mM EDTA (TE), pH 8.0.
Isotopic labeling of DNA.
Linear DNA was isotopically labeled with 32P in either of two ways. For PCR amplicons, oligonucleotides were first labeled isotopically with [γ-32P]ATP by using the RTS T4 kinase-labeling system (Life Technologies Inc.). These labeled oligonucleotides were then used in subsequent PCRs (see below). For restriction fragments with 5′ protruding ends, a fill-in reaction was used with Sequenase version 2.0 and an α-32P-labeled deoxynucleoside triphosphate. For either labeling method, free nucleotides were removed with the QIAquick nucleotide removal kit (Qiagen Inc.) or on Sephadex G-50 spin columns (26).
Quantitative EMSA.
Substrates were synthesized by PCR with oSG46 and oSG49, using wild-type attL-containing plasmid (pHN872) as a template. Quantitative EMSAs with our wild-type attL were performed similarly to the experiments described by Yang and Nash (40). Labeled attL PCR amplicon (119 bp) and IHF to 10 nM were added simultaneously to 50 mM Tris-HCl (pH 7.8)–60 mM KCl–50 μg of BSA per ml–10% glycerol to a final volume of 20 μl. The reaction was allowed to reach equilibrium by incubation at 25°C for 40 min. For competition experiments, EMSA mixtures were similarly assembled except that IHF was limiting at 50 pM. The DNA substrate consisted of 10 fmol of 32P-labeled attL with a variable amount of unlabeled competitor DNA.
In all cases, the reaction mixtures were loaded onto an 8% polyacrylamide gel immediately after incubation. IHF nucleoprotein complexes were separated from free DNA by electrophoresis (10 V/cm) in 0.5× Tris-borate-EDTA (TBE) (26). Dried gels were then exposed to a PhosphorImager screen (Molecular Dynamics) and quantitated with ImageQuant software. The fraction of DNA shifted into complex was estimated by dividing the amount of complexed DNA by the total amount. For each competition experiment, a range of competitor concentrations were used. Curve fitting with Cricket software was used to estimate the 50% inhibitory concentration (IC50).
Quantitative in vitro recombination.
All recombination reactions were performed in 25 to 50 mM Tris-Cl (pH 7.8)–60 to 70 mM KCl–250 μg of BSA–5 mM spermidine–0.5 mM EDTA–10% glycerol in a final volume of 15 μl. Purified Int, IHF, and Xis were gifts from Howard Nash. In all the reactions, Int and IHF were added to a final concentration of 70 to 140 nM and 50 nM respectively, unless specified. In excisive recombination reactions, Xis was added to 30 to 60 nM. Plasmid DNA substrates were extracted and purified with the QIAprep spin miniprep kit (Qiagen). Substrates derived from PCR amplicons (typically 100 to 200 bp) were purified of free nucleotides and unincorporated oligonucleotides via the QIAquick PCR purification kit. The concentration of DNA substrates was typically 1 to 2 nM. Salmon sperm DNA was added to 20 μg/ml in reactions with PCR-derived substrates. The reaction mixtures were always incubated at 25°C for the times indicated. In some cases, reactions were terminated by incubation at 60°C for 5 min and MgCl2 was added to 10 mM along with select restriction enzymes. Digests of the DNA followed by gel electrophoresis were performed to distinguish substrates from products.
Construction of attPs.
Amplicons (119 bp) of select attLs along with the wild-type attR-containing plasmid, pHN868, were used as substrates for in vitro excisive recombination. Recombination reactions were performed as described above, except that the incubations were extended to 3 h and the reaction mixture sizes were doubled. Recombination yielded an integrated linear-form DNA molecule the sum of the sizes of the two constituent substrates and featured an attB and an attP (5.1 kb). Following the incubations, the reaction products were digested with BamHI, which separated the attB (0.8 kb)- from the attP (4.3 kb)-containing fragments. The ends of the DNA were filled in with T4 DNA polymerase. The restriction fragments were gel purified, self-ligated, and transformed into E. coli. Plasmid DNA isolated from transformants was mapped by restriction enzyme analysis to verify that the attPs contained a 350-bp EcoRI fragment indicative of the recombinant attP.
Hydroxyl radical footprinting.
Footprinting reactions were performed on PCR-amplified attL with oligonucleotides oSG46 and oSG49. To examine the top-strand protections, oSG46 was labeled isotopically with [γ-32P]ATP, and 32P-labeled and unlabeled oSG46 and unlabeled oSG49 were used to PCR amplify the designated attLs. Hydroxyl radical footprinting was carried out as described by Yang and Nash (38).
RESULTS
Experimental strategy.
We created a population of randomized IHF binding sites and used IHF to select for high-affinity DNA targets. We chose the H′ site of bacteriophage lambda attL/attP, which is required for efficient phage recombination, as the parental target DNA. IHF requires a region comprising 32 bp for maximum binding (+15 to +46 of the attL/attP locus of bacteriophage lambda). The H′ site was chosen because it has been extensively mutagenized and studied (2, 4, 6, 9–11, 15–18, 21, 24, 27, 28, 35, 38–40) and because its structure bound to IHF was recently solved by X-ray crystallography (24). IHF bound to the H′ site has been shown to act as an architectural element; similar deformations of DNA at this site replace IHF for function (6, 27).
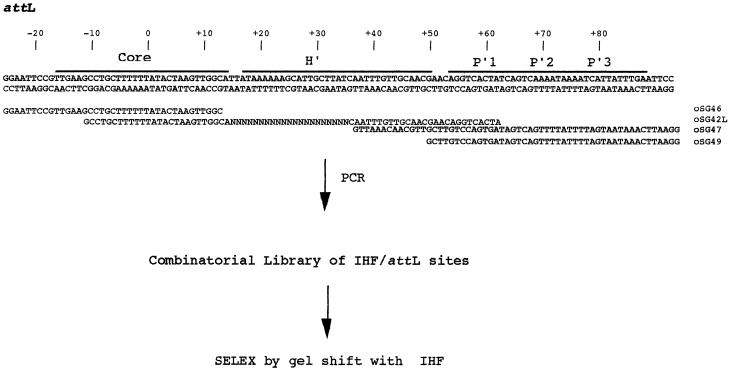
To identify high-affinity targets, we used a version of the SELEX protocol (32). Briefly, this entails synthesizing a DNA template in which the ends have a defined sequence and surround a region of random sequence. The selection is based on binding IHF and partitions high-affinity members from low-affinity members. High-affinity members are amplified by PCR, and the selection-amplification cycle is repeated until the selection has enriched the population to a suitable percentage of high-affinity members. Although we had desired to randomize an entire 32-base site, this turned out to be impractical for sites in excess of 22 bases due to the large quantity of DNA and protein required to ensure a completely random population. Instead, we focused on the 21 bp comprising the 5′ region of H′, +15 to +35 of attL-attP, next to the conserved cytosine at +36. The ends of the amplifying oligonucleotides were designed to intentionally exceed the template so that the final amplicons would be 119 bp and comprise the entire length of the attL locus (Fig. 1).
FIG. 1.
SELEX strategy. The SELEX template, oSG42L, was synthesized and mixed with amplification oligonucleotides oSG46 and oSG47. The template was made duplex, extended, and amplified by PCR. The full-length product, a complete attL site, acts as a combinatorial library for selection of high-affinity IHF binders. Members selected on the basis of high affinity were assessed as substrates for in vitro site-specific recombination. The region designated the core represents a site for Int binding and DNA crossover. Sites designated as P′ are also Int type binding sites.
Since this strategy enabled us to assess selected candidates individually for both affinity for IHF and utility as substrates for excisive recombination, we could determine the relationship between binding affinity and function for each selected attL. We speculated at the outset that the function of the IHF binding site, in our case measured as the recombination competence of the attL site, may be more dependent on the final structure of the IHF-DNA complex than on how readily the complex forms (as indicated by the affinity of IHF for its target site). The case for attL has been well defined; although the attL site needs to assume a very specific structure for efficient recombination to occur, the structure can be achieved in a variety of ways (6, 11, 15, 18, 21, 27, 35).
SELEX of the IHF site of attL of bacteriophage lambda.
Perhaps the most important consideration for SELEX is the affinity difference between strong binding members and the bulk population. The larger the difference in affinity, the fewer rounds of selection are required. Since our original goal was to find IHF binding sites with 5′ domains of equal or better binding with respect to the H′ site of lambda, we chose the wild-type H′ site as our lower limit for a winning affinity. The dissociation constant (Kd) for IHF binding to H′ has been previously measured in the nanomolar range (40), while IHF binding to nonspecific DNA (e.g., salmon sperm DNA) has been measured to micromolar Kd values. Thus, we expected our SELEX DNA substrate with an intact 3′ binding domain to bind with intermediate affinity.
We picked conditions where sufficient IHF (60 nM) could shift 20% of the wild-type attL site while failing to shift (<1.0%) an attL with a mutated H′ site (QH′ [4]; this site fails to yield a DNase I footprint) and the bulk attL population. As expected, the first gel shift of our attL population failed to yield a visible shifted product as assessed by ethidium bromide staining. Nevertheless, gel fragments corresponding to the position where a shifted complex would migrate were excised, and whatever DNA was present was extracted and purified.
This DNA was amplified by PCR to create an enriched attL population. Three additional cycles of gel shift, DNA extraction, and PCR again yielded no observable shifted complex. Each time, we recovered between 25 and 35% of the substrate DNA. Although we originally expected the enrichment of a discrete shifted complex with each successive round of selection, we observed only a faint smear. The smear did not increase in intensity or extent with each successive round, and the amount of DNA extracted from the gel in the vicinity of the wild-type attL-IHF complex never changed significantly. In retrospect, an altered strategy during these subsequent SELEX cycles under more stringent conditions (e.g., with lower IHF concentrations) could have been used for a stronger selection of high-affinity sites.
Since sequence variations in the 5′ domain of H′ yielded alternative migration during EMSA, smearing of the IHF-DNA complex was not surprising (11, 28). This observation is consistent with the 5′ domain acting as a sequence-dependent modulator of the final structure of the IHF-DNA complex. Therefore, we took the smear to represent a population of attLs that, when bound to IHF, formed complexes where migration was specifically dependent on the 5′ domain. Indeed, we observed no smearing or blurring of the uncomplexed population of attL fragments during gel electrophoresis.
Preliminary sequence and binding analysis.
To characterize our selected population, individual members were cloned. Seventy randomly selected attLs were isolated and sequenced. Each was found to be unique, indicating that a large number of sites are capable of, at least grossly, creating high-affinity complexes with IHF. This also suggests that our original conditions for selection were suboptimal and that enrichment of the highest-affinity members was incomplete. Of the 70, 33 suffered from either insertions or deletions, probably an artifact of PCR. Thus, we chose to further analyze only the 37 attLs possessing wild-type spacing.
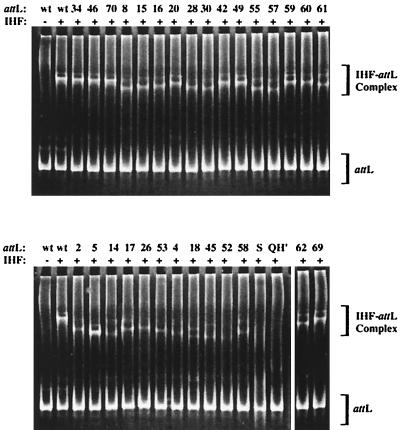
PCR amplicons of each of the 37 attLs were then analyzed in an EMSA with IHF under the same conditions as used for the original selection. Only 28 of the attLs were capable, to different degrees, of forming discrete complexes. We refer to these attLs as class I. The remaining nine that failed to shift in the presence of IHF (data not shown) have consequently been designated class II. As shown in Fig. 2, class I members yielded complexes that migrated to different degrees. Only three attLs, 1-34, 1-46, and 1-70, had gel shifts indistinguishable from that of wild-type attL. No attL had a greater shift than the wild type.
FIG. 2.
EMSA of attL with IHF. Each attL (200 nM) was incubated in the presence of IHF (60 nM) for 40 min at 25°C. Each reaction mixture was then resolved by polyacrylamide gel electrophoresis. The gels were stained in ethidium bromide and visualized under 302-nm light. Complexed DNA has a retarded migration pattern relative to free DNA. Numbers atop each lane indicate the attL. Lanes marked wt contain wild-type attL. Lane S contains the original attL population from which selection was performed. Lane QH′ contains attL with four base mutations in the 3′ binding domain (see the text).
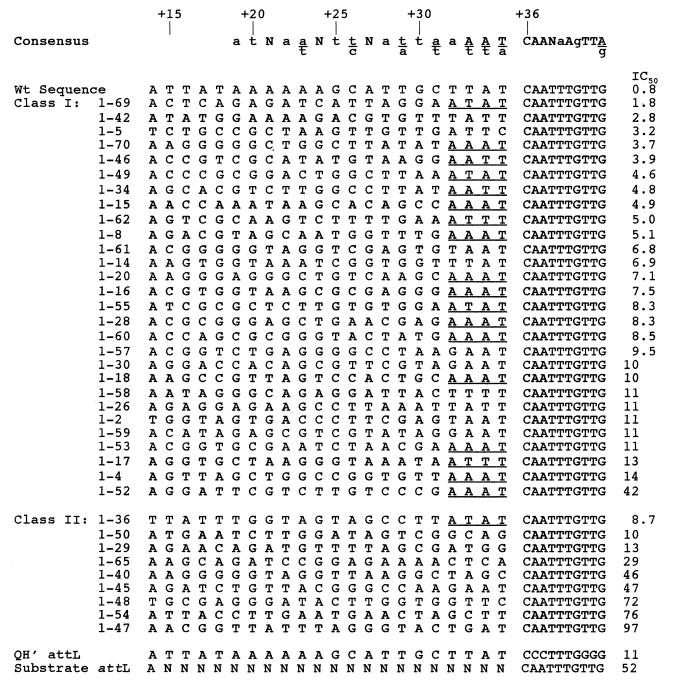
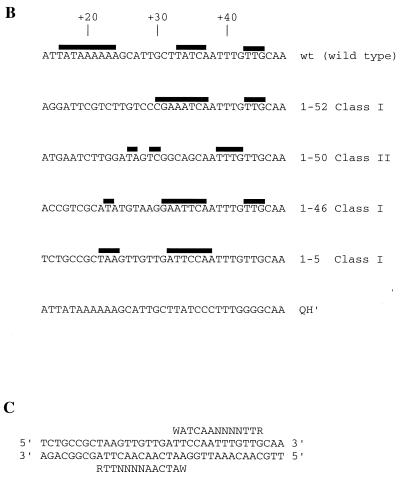
A cursory inspection of the class I attLs showed that specific portions of each sequence were conserved (Fig. 3 and 4). Based on the original design of the IHF site population, 21 bases 5′ to the conserved cytosine (+36) were randomized. Others have identified the 3 bases 5′ from this cytosine as being highly conserved in natural sites (7). In fact, Craig and Nash (2) originally defined the consensus as WATCAANNNNTTR (where W is A or T, R is purine, and N is any base). We found that all class I attLs except one (attL 1-5) possessed the sequence WWT 5′ to the +36 cytosine. In addition, 17 of these 28 members possessed an adenine directly 5′ to this sequence, yielding a conserved sequence of AWWT 5′ to the +36 cytosine (bases +32 to +35). These three bases were much less highly conserved among class II members, with only 1-36 yielding an exact match. Thus, selection for affinity and the screening for discrete complexes during EMSA mostly coincided with the canonical 3′ consensus.
FIG. 3.
DNA sequences of SELEX-derived attLs. The consensus sequence is derived from a compilation of 27 natural IHF binding sites (7). All sequences are aligned with the numbering of the H′ site of bacteriophage lambda. Bases +14 to +35 constitute the varied region. Sequences 5′ (+13 and lower [not shown]) and 3′ (+36 and higher [+36 to +45 are shown]) were not altered. IC50 is the concentration (nanomolar) of competitor ligand which reduces the quantity of shifted complex by 50%; it represents an estimate of DNA binding affinity. Class I sites form stable complexes with IHF in EMSA under the initial selection conditions. Class II sites fail to form complexes with IHF in EMSA under the initial selection conditions. The substrate attL represents the SELEX starting material and was originally designed to possess an adenine nucleotide in the first position (as shown). Underlined regions possess the common sequence AWWT, where W is A or T.
FIG. 4.
Compilation of DNA sequence of class I attLs. The sum of bases at each position for 27 of our attL class I clones is shown (attL 1-5 was excluded from this analysis [see Discussion]). Consensus #1 is from reference 7. Consensus #2 was derived from class I attL sequences. Capital letters represent strong conservation, while lowercase letters represent weaker conservation. Positions labeled N fail to show significant conservation. Bases found beneath the line represent a significant conservation among the remaining three bases. The significance of each conservation was determined as in reference 7.
Despite these matches to published consensus sequences, a plethora of other sequence motifs were observed in our populations, demonstrating that high affinity and gross structure could be accommodated by multiple sequences. In fact, for the 21 randomized bases, none of the 37 attLs possessed more than 12 identical bases and two contained as few as 3 identical bases compared to the wild-type attL sequence.
Within the wild-type H′ sequence, the most conspicuous sequence motif is an A tract between bases +19 and +24 that others have speculated enhances DNA binding via an accommodating sequence-directed architecture (11, 28). Several groups have demonstrated that point mutations within the A tract cause a significant and sometimes severe binding defect (9, 11, 16, 18, 28). To the best of our knowledge, no mutation in this region has ever enhanced DNA affinity over the wild type. Only three class I attLs (1-42, 1-15, and 1-14) possessed an A tract at these coordinates. In each case, the length of the A tract was 4 adenines or less. Whether these adenine residues are important for binding in these variants is not known, but the lack of A tracts in the other high-affinity class I members precludes this motif from being essential.
As shown in Fig. 4, additional conservations among 27 class I members occurred at bases +14, +16, +18, +19, +22, and +23. By oligonucleotide design, base +14 should have been exclusively adenine. The fact that it varied within our population indicates that our starting oligonucleotides were not homogeneous and/or there were additional PCR artifacts. Indeed, several sequenced isolates that were not further analyzed possessed either additions or deletions within the attL fragment (data not shown). Since we did not examine the proportion of adenines at +14 in the unselected starting material, variability at +14 may not be relevant. Among the most highly conserved bases were those at +18 and +19, where at least half of the bases were guanine, and base +22, which was mostly adenine (16 of 27 bases). These conservations were absent in the class II members. With the exception of the adenine at base +22, these conservations were also absent in a published alignment of 27 natural sequences (7).
Quantitative binding analysis of attL-IHF interactions.
To further characterize and classify our SELEX-derived attL populations, we determined the apparent equilibrium dissociation constants (Kd) by a competition assay for all members of each class as well as the original randomized SELEX starting material. To start, a binding isotherm was generated by using an EMSA with a constant amount of IHF and increasing concentrations of our wild-type attL substrate (119 bp). Based on the amounts of free and bound DNA, we were able to estimate the concentration of free attL which yields half the maximal concentration of bound complex, the Kd, for the wild-type substrate at 0.7 nM (40). We measured the Kd of all variant sites by a competition method: isotopically labeled wild-type attL (0.5 nM) was incubated with limiting amounts of purified IHF (50 pM) in the presence or absence of different amounts of a specific competitor attL. Dissociation constants were based on the measured IC50 (the concentration of the competitor that yielded 50% inhibition of the wild-type shifted complex in the absence of competitor [see Materials and Methods]). With the wild-type attL as the competitor, the average IC50 in three trials was 0.8 nM, very similar to the value we measured by using the more direct approach of a binding isotherm. The IC50s determined for each SELEX-derived attL are shown in Fig. 3.
Each class had distinctive affinity characteristics. Consistent with our preliminary designations, class I members had a higher average affinity than class II members did. No SELEX-derived site had a higher affinity than the wild-type sequence, although site 1-69 had a binding affinity within a factor of 3 of the wild type. Although the average affinity of each class was distinct, there was overlap between individual members of class I and class II. The worst binder, 1-47, had only a threefold-lower affinity than did the worst class I binder. It is noteworthy, however, that discrete complexes were formed with class I variants at IC50s as high as 42 nM yet the highest-affinity class II member had an IC50 of 8.7 nM. Thus, binding affinity is not an absolute measure of stable complex formation in our gel system. This may indicate that some class II members are capable of more than the canonical mode of binding, implying multiple metastable interactions to yield a net affinity.
The worst class II competitors had affinities over 100-fold lower than the wild type. This is the first indication of the severe effect of randomly altering this region of the DNA ligand binding surface. Since we did not originally select for poor binders, this value represents a higher limit. It is not clear whether the sequences with the lowest affinities are poor due to limited positive interactions with IHF or to the presence of destructive interactions with IHF. Finally, we have also measured the affinity of IHF to another mutant target (QH′ [see Table 1]). In this case, the altered bases are in the highly conserved 3′ domain. As others have shown, the affinity of IHF for this target is at least 10-fold lower than for the wild type (40). Thus, IHF interactions which contribute to the binding affinity are distributed over both the 3′ and 5′ domains of the site (35).
TABLE 1.
Quantitative recombination
| attL | % Excisive recombinationa | % Integrative recombinationb |
|---|---|---|
| Wild type | 100 | 100 |
| QH′ | 34 | 7 |
| SELEX substrate | 4 | NDd |
| Class Ic | ||
| 1-69 | 28 | ND |
| 1-42 | 58 | 100 |
| 1-5 | <2 | ND |
| 1-70 | 25 | 33 |
| 1-46 | 50 | 35 |
| 1-49 | 30 | ND |
| 1-34 | 44 | ND |
| 1-15 | 31 | ND |
| 1-62 | 34 | ND |
| 1-8 | 40 | ND |
| 1-61 | 46 | 67 |
| 1-14 | 32 | ND |
| 1-20 | 57 | ND |
| 1-16 | 18 | 67 |
| 1-55 | 45 | 39 |
| 1-28 | 42 | 31 |
| 1-60 | 52 | ND |
| 1-57 | 30 | ND |
| 1-30 | 30 | ND |
| 1-18 | 23 | ND |
| 1-58 | 26 | ND |
| 1-26 | 50 | ND |
| 1-2 | 28 | 15 |
| 1-59 | 42 | 80 |
| 1-53 | 56 | 35 |
| 1-17 | 13 | 75 |
| 1-4 | 39 | ND |
| 1-52 | 26 | 12 |
| Class IIc | ||
| 1-36 | 50 | 110 |
| 1-50 | 13 | 8 |
| 1-29 | 15 | ND |
| 1-65 | 15 | 9 |
| 1-40 | 16 | ND |
| 1-45 | 43 | ND |
| 1-48 | 28 | ND |
| 1-54 | 23 | ND |
| 1-47 | 42 | 85 |
Recombination reaction mixtures were incubated at 25°C for 20 min. All recombination efficiencies are the average of two trials and were normalized against the wild-type attL. 100% is equivalent to 19% product formation.
Recombination reaction mixtures were incubated at 25°C for 60 min. All recombination efficiencies are the average of two trials and were normalized against the wild-type attP. 100% is equivalent to 19% product formation.
Columns are listed in decreasing order of IHF binding affinity within each class.
ND, not determined.
Quantitative excisive recombination.
Since we chose the H′ IHF binding site of bacteriophage lambda attL, we were able to test the SELEX-derived IHF sites in site-specific recombination. Lambda excisive recombination occurs between the attR and attL loci found at the junction of the phage and bacterial genomes in a lambda lysogen. attL has a single IHF site, which is required for recombination. We measured the efficiency of each SELEX-derived H′ site in supporting excisive recombination between attLs with a wild-type attR. We also tested the efficiency of the SELEX starting material as a substrate for recombination.
The results for all excision reactions are shown in Table 1. Although no selected attL was superior to the native site in recombination efficiency, the general recombination defect of most of the tested attLs was within fourfold of the wild type. With only one exception, attL 1-5, the recombination efficiency was within a factor of 10 for any of the attLs, independent of their individual affinity for IHF. The QH′ attL also showed little effect, yielding about 50% of the wild-type recombination efficiency. In contrast, the SELEX starting material (the initial random attL population) was particularly poor as a substrate, indicating that most members of the population were not functional in recombination. This may be due to the high percentage of attLs that did not have native spacing between the consensus elements (21). As for attL 1-5, although it binds with high affinity (within a factor of 4 of the wild-type sequence), it yielded no recombinant products. This may be due to the inadvertent creation of a second, competing IHF binding site within the randomized region (see Discussion). In summary, there is no obvious relationship between the recombination efficiency of these attLs and either their affinity for IHF or their ability to form discrete complexes with IHF in our gel system.
Quantitative integrative recombination.
To further investigate the ability of IHF to function at these SELEX-selected sites, we tested their proficiency in integrative recombination. In this case, the phage recombination locus, attP, integrates site specifically into the bacterial site attB. Prior results show that while the general phenotypes of H′ variants in attL and in attP are similar, the phenotypes of H′ mutations in attP are more severe than those of mutations in attL. To test individual H′ variants in integrative recombination, we created attPs with the altered H′ sequence by in vitro excisive recombination with a wild-type attR (this was possible since the effect of the H′ variants on excision was modest). In all, 16 attPs were constructed.
As shown in Table 1, the phenotypes of our SELEX-derived attPs are relatively small for integrative recombination, despite the apparent binding affinity, which is within 10-fold of that of the wild-type att site. In fact, attPs derived from sites 1-42 and 1-36 gave recombination efficiencies at least as good as the wild-type attP. In addition, the severities of excisive and integrative recombination defects of sites possessing a common sequence were similar, with less than threefold variation. There were two exceptions: the 1-16 site and 1-17 site had more severe phenotypes in excisive than in integrative recombination. A converse phenotype where a SELEX-derived site was more strongly proficient in excisive than in integrative recombination was not observed; this, however, may not be statistically significant. In contrast, the QH′ mutations in the 3′ consensus had a much stronger defect in an attP context than in an attL context. These examples show that the 5′ and 3′ regions of the H′ site contribute both to binding of IHF and to the function of the resulting complex. The fact that H′ variants affect excisive and integrative recombination differently suggests that the higher-order complexes assembled by Int and IHF on attL and attP have distinct conformations.
Hydroxyl radical footprinting.
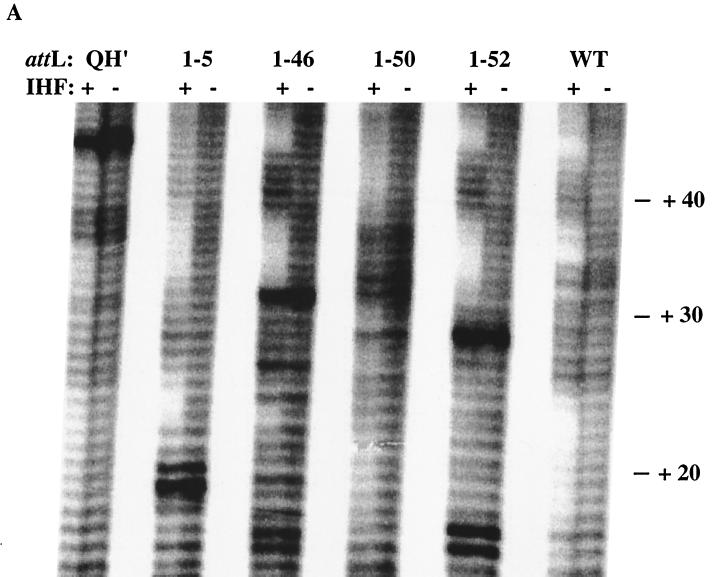
We compared the nature of the binding interactions of IHF with some of our SELEX-derived attLs with the interactions with the wild-type H′ by using hydroxyl radical footprinting. In the presence of hydroxyl radicals, IHF-DNA complexes on H′ yield a characteristic pattern of three protected patches (38): in the 3′ TTR portion of the DNA binding site, in the WATC region of the 3′ consensus, and in the 5′ region at the A tract. This footprinting method is very precise and mostly sequence independent (although there are apparent contextual effects). In addition to the wild type and the QH′ mutant, we assessed the top-strand interactions of four different attLs: 1-46 (high-affinity class I site, proficient in excisive recombination and modest in integrative recombination), 1-5 (high-affinity class I site but recombination deficient), 1-52 (lowest-affinity class I site, modest in both excisive and integrative recombination and with the most accelerated migration in complexes with IHF), and 1-50 (class II site, modest affinity for IHF but no formation of a discrete complex, poor in both excisive and integrative recombination).
As shown in Fig. 5, we examined each attL site with an IHF concentration of 1 μM. At this concentration of IHF, we reproduced a protection pattern on the wild-type site similar to that observed by Yang and Nash (38). Little or no protection was observed on the QH′ mutant. The footprint of 1-46 and 1-52 most resembled that of the wild type, with the only obvious differences being a less protected third patch which coincides with the A tract in the wild type. This suggests that IHF makes a more intimate contact with this region in the wild type than in 1-46 and 1-52. Site 1-50 yielded several uncharacteristic but weak protections near the middle of the 3′ consensus between conserved regions. Site 1-5 also had a very complicated and extensive protection pattern. At first glance, the protection patterns appear similar to those of the wild type except at the TTR region, which was not protected. This is atypical, since this region is well protected in other sites. At a fivefold-lower IHF concentration (200 nM [data not shown]), only the protection pattern of 1-5 and the wild type persisted. This indicates that despite the novel interactions with IHF, 1-5 still bound well, consistent with our estimate of high-affinity binding. We propose that the noncanonical binding pattern for 1-5 is due to the presence of a new IHF site out of register with the original wild-type H′ site (see below). This easily explains the complete failure of this site in excisive recombination.
FIG. 5.
Hydroxyl radical footprinting. Preformed complexes of IHF (1 μM) and top strand-labeled attL DNA (0.5 nM) were exposed to hydroxyl radicals generated by the Fenton reaction. The conditions were adjusted to create about one cleavage per DNA molecule. The reaction mixtures were separated by denaturing gel electrophoresis. (A) Autoradiography of six attL top strands in the absence and presence of IHF. Base positions are indicated in the right-hand margin. (B) Summary of protection patterns. Each solid bar represents bases that are moderately to strongly protected from hydroxyl-radical attack. (C) Alignment of the pertinent region of the H′ locus of attL 1-5 with the IHF 3′ consensus (2) for the top and bottom strands.
DISCUSSION
The IHF protein is highly conserved among gram-negative eubacteria (19, 22). This conservation extends beyond apparent amino acid sequence homology to a specific recognition sequence and subsequent structural deformation of the bound DNA target. Despite DNA site specificity, DNA targets possess significant amounts of sequence degeneracy between sites. This degeneracy is most apparent in the 5′ region of the DNA target. In this work, we studied variants of the lone natural site from the bacteriophage lambda recombination locus attL. Using a minimal attL locus (119 bp) as a template, we generated DNA targets for IHF de novo via the SELEX strategy. By this approach, we randomized the 5′ region of the IHF site while keeping the 3′ region and the remainder of the attL locus unaltered. Specific members of the population were initially selected for affinity with IHF by EMSA. Selected attLs were subsequently assessed individually in EMSA for affinity and gross structure and as substrates in recombination assays. Although the affinity of IHF for individual members of our selected sequences varied over 100-fold, we could find no relationship to recombination function.
Our SELEX selection yielded IHF binding species that, as a group in complex with IHF, produced a smear during EMSA. We found that the smear consisted of complexes with the original population of SELEX-derived attLs and was, at least in part, the result of a population of distinctly shifted IHF-attL complexes with different migrations. No members, complexed with IHF, produced shifts even more retarded than the wild-type attL-IHF complex. In addition, none of the 37 members had an altered migration on polyacrylamide gel electrophoresis in the absence of IHF, indicating that the altered migration was indeed due to the nucleoprotein complex. We conclude that the 5′ region of the binding site plays a critical role in IHF binding and in nucleoprotein complex structure.
The affinity of each SELEX-derived attL substrate was measured by competition with the wild-type attL for IHF. This obviates the necessity for the mutant sites to form an electrophoretically stable complex. We found that the ability to compete for binding was not absolutely related to the ability to form a stable complex with IHF during gel electrophoresis. No member of our population bound with greater affinity than the wild-type site, although the strongest binders had IC50s within a factor of 3. Interestingly, when the wild-type 5′ binding domain of H′ was left intact and the conserved 3′ region was mutated at the most conserved residues (QH′), binding was reduced to a level 14-fold lower than the wild type and no shifted bands were observed during EMSA. In addition, the IC50s of the starting material and the lowest-affinity attLs were 50- and 100-fold higher, respectively, demonstrating the critical contribution of the 5′ region to affinity. These results are consistent with binding affinity being distributed between the 5′ and 3′ domains.
In general, the functionality of the SELEX-derived sites in excisive or integrative lambda site-specific recombination does not correlate with their affinity for IHF. It is very likely that the recombination defect was ameliorated in the presence of Int. We have previously shown that the nucleoprotein intermediates on various att sites are codependent on Int and IHF to facilitate the formation of the appropriate structure (27); Int cooperates even with the nonspecific bending protein HU to make a functional nucleoprotein complex. Since the nucleoprotein complex between Int, IHF, and attL or attP is the functional substrate for recombination, it seems more likely that the variant sequences are applying their effect on recombination via these nucleoprotein complexes rather than any intermediate complex between either att site and IHF. We are beginning to investigate the role that Int may play in mitigating the defects of our H′ variants in recombination.
Of the 40 attLs tested, only attL 1-5 was inactive as a substrate for excisive recombination. IHF binds to this site with high affinity, suggesting that complex formation per se does not cause the recombination defect. Based on our footprinting analysis, we propose that the true failure of this site lies in the presence of an alternative IHF site out of register with the wild-type H′, resulting in a nucleoprotein complex with the wrong geometry for recombination. To identify potential alternative and overlapping IHF sites, we used the MacTargsearch program, which was developed to identify possible IHF sites based on similarity to a set of 27 known IHF sites (7). It is particularly useful in identifying possible sites based on the conserved 3′ region. It is much less effective in identifying sites based on the unconserved 5′ region, a primary reason for this investigation. Of all the SELEX-derived sites, only attL 1-5 was distinguished as possessing a second possible site. One of these, which has a weaker similarity to bona fide IHF sites, is in register with the H′ site, while the second, with a much stronger similarity to IHF sites, is out of register with the H′ site. The misalignment of the IHF site with respect to the loci of Int-mediated catalysis is known to interfere strongly with lambda recombination (21).
While many reports, including the solution of the cocrystal structure of IHF bound to this very site (H′), have described the interactions of IHF with its DNA site, the specifics of this nucleoprotein association are still not fully defined. There appear to be no unique contacts between the protein and the DNA bases, although specific components may aid IHF in recognizing both regions of its binding site. Conventional protein-DNA binding specificity is thought to be driven by unique hydrogen bond donor and acceptors; in contrast, IHF appears to rely on indirect readout (34). Structural configurations associated with both the protein and target DNAs (e.g., ionic interactions, stacking interactions, etc.) limit the degrees of freedom of interactions which restrict and impose a high-affinity binding surface even when the protein-DNA base contacts are degenerate. The E. coli bacterial DNA binding proteins Fis and H-NS, which also modulate nucleoprotein functions, also share these characteristics. Fis binds and bends DNA site specifically and, like IHF, has a highly degenerate consensus sequence, making site identification difficult (1, 12). The DNA binding protein H-NS is even more problematic, since DNA sites are bound with high affinity but without any sequence conservation. H-NS has been shown to bind preferentially to existing DNA deformations (29, 37). Thus, the phenomenon of indirect readout coupled to DNA deformation is not unique to IHF.
Evidence of contacts or at least intimate proximity between IHF and the 5′ region at various IHF sites has been demonstrated at virtually every base pair in the 5′ domain from +35 to +19 (Fig. 1) (reviewed in references 19 and 25). Beyond +19, the number of demonstrable interactions falls off dramatically, although some investigators have shown biochemical evidence of interaction as distal as base +14 (31). In our selected sequences, we have found that strong conservation between positions +35 and +32 is associated with stability of the IHF-DNA complex during EMSA. This patch of four bases coincides with the 5′ boundary of the 3′ region observed in the great majority of natural IHF sites. By selecting IHF sites de novo, we demonstrate that this 4-base stretch is essential for forming at least one type of complex. Cocrystal analysis did not find unique hydrogen bonding of amino acid side chains to the canonical H′ sequence in this region.
While it is beyond the scope of this paper to interpret the contribution of specific motifs, there appear to be patterns of conservation. Data from other studies demonstrate the importance and the proximity of IHF to the region from +19 to +29 (reviewed in reference 19). Indeed, the six consecutive adenines of H′ are found from +19 to +24 and one of two kinks induced by IHF is found between bases +28 and +29. The A-tract figures prominently in the cocrystal structure of IHF bound to H′ and demonstrates that, at least in this complex, IHF forms a clamp around a particularly narrow minor groove. One possibility is that all high-affinity members of class I have or are predisposed to form narrow minor grooves in the vicinity of +19 to +24 and/or sequences that readily kink about bases +28 and +29. While there is no obvious conservation among our class I attLs at coordinates +28 and +29, there is conservation at bases +18, +19, and +22. Conservation of these three bases persists over a 20-fold range of affinities and suggests that they specify important binding features that at least stabilize IHF-DNA interactions.
Evidence of a noncanonical structure associated with these sequences (+19 to +29) can be seen in our preliminary footprinting analysis. With the exception of the wild-type attL sequence, all of the attLs tested here displayed strong cleavage enhancements upon exposure to hydroxyl radicals in the absence of IHF. These enhancements result from increasing solvent accessibility of the hydroxyl radical target within the ribose ring through sequence-directed distortion. It is not clear whether these distortions play a role in enhancing IHF binding, although it is interesting that in attL 1-52 the enhancements coincide with the IHF-induced kink between +28 and +29 in the wild-type H′ site. A more detailed biochemical analysis, focused on this region of class I attLs of similar IHF binding affinity, would clarify the nature and extent of these contacts.
In addition to base conservation (or lack thereof), we observed an intriguing phenomenon concerning the nucleotide content of the IHF-selected sequences. We compared the nucleotide content at bases +14 to +35 for both class I and class II with the corresponding region of the 27 natural IHF sites cited by Goodrich et al. (7), including the wild-type H′ site. Remarkably, the base content differed dramatically between the selected sites and the native sequences (Table 2). These 22 bp in the native sequences are extremely AT rich, while they are only slightly enriched for A · T base pairs in the selected sequences regardless of class. Since natural sites and class I selected sequences conserve A · T base pairs from +35 to +32, this 4-base stretch weighs significantly in the overall base composition of the region. If we examine only bases +31 to +14, the result becomes even more striking: while 72% of the base pairs in this region are A · T among the native sequences, there is virtually no bias in base composition among our SELEX-derived sequences. We interpret this to mean that IHF sites, while typically possessing AT-rich 5′ regions, are not dependent on this base composition. This implies that, on average, many motifs of various base compositions can accommodate high-affinity binding. The biological role of IHF may be a more significant determinant in target site selection and conservation than a specific binding motif that is independent of base composition. It is also possible that there are alternative binding modes of IHF, particularly within the 5′ domain, as indicated in our preliminary footprint analysis.
TABLE 2.
Base composition of the 5′ domain of selected IHF binding sites
| H′ DNA sequence | % A+T (no. in region/total no.)a in:
|
|
|---|---|---|
| +35 to +14 | +31 to +14 | |
| Class I | 57 (340/594) | 49 (236/486) |
| Class II | 58 (114/198) | 57 (93/162) |
| McClure consensusb | 76 (454/594) | 72 (352/486) |
| Wild-type H′ | 82 (18/22) | 78 (14/18) |
The percentage is the fraction of the sum of A · T base pairs in the designated region of each group of H′ sequences divided by the total number of bases and multiplied by 100.
The McClure consensus (7) was derived from 27 natural sequences aligned with the native H′ site.
We are currently attempting to identify critical determinants in the 3′ region of the IHF site by using a SELEX approach. Despite its greater conservation, the 3′ region is still mystifying, since IHF contacts appear degenerate in the cocrystal structure. Finally, to understand the essential determinants of IHF function, a similar selection will be used with recombination as the partition selector in order to ask how efficient recombination is associated with DNA affinity.
ACKNOWLEDGMENTS
We are particularly indebted to Richard Deonier for his many helpful suggestions and for comments on the manuscript. We also thank Bruce Teter and Howard Nash for comments on the manuscript.
This work was supported by Public Health Service grants GM55392 (to S.D.G.) and GM52847 (to A.M.S.) and by a grant from Pioneer Hi-Bred International Inc. (to S.D.G.).
REFERENCES
- 1.Betermier M, Galas D J, Chandler M. Interactions of Fis protein with DNA: bending and specificity. Biochimie. 1994;76:958–967. doi: 10.1016/0300-9084(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 2.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 3.Cui Y, Wang Q, Stormo G D, Calvo J M. A consensus sequence for binding of LRP to DNA. J Bacteriol. 1995;177:4872–4880. doi: 10.1128/jb.177.17.4872-4880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner J F, Nash H A. Role of Escherichia coli IHF protein in lambda site-specific recombination: a mutational analysis of binding sites. J Mol Biol. 1986;191:181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- 5.Goodman S D, Nash H A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989;341:251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- 6.Goodman S D, Nicholson S C, Nash H A. Deformation of DNA during site-specific recombination of bacteriophage λ: replacement of IHF protein by HU protein or sequence-directed bends. Proc Natl Acad Sci USA. 1992;89:11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagerman P J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 9.Hales L M, Gumport R I, Gardner J F. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J Bacteriol. 1994;176:2999–3006. doi: 10.1128/jb.176.10.2999-3006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hales L M, Gumport R I, Gardner J F. Mutants of Escherichia coli integration host factor: DNA-binding and recombination properties. Biochimie. 1994;76:1030–1040. doi: 10.1016/0300-9084(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 11.Hales L M, Gumport R I, Gardner J F. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 1996;24:1780–1786. doi: 10.1093/nar/24.9.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengen P N, Bartram S L, Stewart L E, Schneider T D. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvine D, Tuerk C, Gold L. SELEXION, systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991;222:739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Moitoso de Vargas L, Nunes-Duby S E, Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in λ attL. Cell. 1990;63:773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- 16.Lee E C, MacWilliams M P, Gumport R I, Gardner J F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage λ H′ recognition site. J Bacteriol. 1991;173:609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E C, Hales L M, Gumport R I, Gardner J F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992;11:305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacWilliams M, Gumport R I, Gardner J F. Mutational analysis of protein binding sites involved in formation of the bacteriophage λ attL complex. J Bacteriol. 1997;179:1059–1067. doi: 10.1128/jb.179.4.1059-1067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash H A. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 149–179. [Google Scholar]
- 20.Numrych T E, Gumport R I, Gardner J F. A comparison of the effects of single-base and triple-base changes in the integrase arm-type binding sites on the site-specific recombination of bacteriophage lambda. Nucleic Acids Res. 1990;18:3953–3959. doi: 10.1093/nar/18.13.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunes-Duby S E, Smith-Mungo L I, Landy A. Single base-pair precision and structural rigidity in a small IHF-induced DNA loop. J Mol Biol. 1995;253:228–242. doi: 10.1006/jmbi.1995.0548. [DOI] [PubMed] [Google Scholar]
- 22.Oberto J, Drlica K, Rouviere-Yaniv J. Histones, HMG, HU, IHF: meme combat. Biochimie. 1994;76:901–908. doi: 10.1016/0300-9084(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 23.Peacock S, Weissbach H, Nash H A. In vitro regulation of phage λ cII gene expression by Escherichia coli integration host factor. Proc Natl Acad Sci USA. 1984;81:6009–6013. doi: 10.1073/pnas.81.19.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice P A, Yang S, Mizuuchi K, Nash H A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 25.Rice P A. Making DNA do a U-turn: IHF and related proteins. Curr Opin Struct Biol. 1997;7:86–93. doi: 10.1016/s0959-440x(97)80011-5. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Segall A M, Goodman S D, Nash H A. Architectural elements in nucleoprotein complexes: interchangeability of specific and non-specific DNA binding proteins. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shindo H, Kanke F, Miyake M, Matsumoto U, Shimizu M. The binding specificity and affinity of E. coli integration host factor (IHF) are influenced by the flexibility of flanking regions of its recognition sites. Biol Pharm Bull. 1995;18:1328–1334. doi: 10.1248/bpb.18.1328. [DOI] [PubMed] [Google Scholar]
- 29.Spurio R, Falconi M, Brandi A, Pon C L, Gualerzi C O. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss J K, Maher L J. DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1833. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, Hurley L H, Harshey R M. Structural distortions induced by integration host factor (IHF) at the H′ site of phage λ probed by (+)-CC-1065, pluramycin, and KMnO4 and by DNA cyclization studies. Biochemistry. 1996;35:10815–10827. doi: 10.1021/bi952786x. [DOI] [PubMed] [Google Scholar]
- 32.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 33.Vant-Hull B, Payano-Baez A, Davis R H, Gold L. The mathematics of SELEX against complex targets. J Mol Biol. 1998;278:579–597. doi: 10.1006/jmbi.1998.1727. [DOI] [PubMed] [Google Scholar]
- 34.von Hippel P H. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994;263:769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]
- 35.Werner M H, Clore G M, Gronenborn A M, Nash H A. Symmetry and asymmetry in the function of Escherichia coli integration host factor: implications for target identification by DNA-binding proteins. Curr Biol. 1994;4:477–487. doi: 10.1016/s0960-9822(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 36.Williams J G K, Wulff D L, Nash H A. A mutant of Escherichia coli deficient in a host function required for phage lambda integration and excision. In: Bukhari A, Shapiro J, Adhya S, editors. DNA insertion elements, plasmids and episomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1977. pp. 357–361. [Google Scholar]
- 37.Yamada H, Muramatsu S, Mizuno T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J Biochem. 1990;108:420–425. doi: 10.1093/oxfordjournals.jbchem.a123216. [DOI] [PubMed] [Google Scholar]
- 38.Yang C-C, Nash H A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989;57:869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- 39.Yang S-W, Nash H A. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its DNA target. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S-W, Nash H A. Comparison of protein binding to DNA in vivo and in vitro: defining an effective intracellular target. EMBO J. 1995;14:6292–6300. doi: 10.1002/j.1460-2075.1995.tb00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]