Abstract
Background.
Allergic skin inflammation elicited in mice by epicutaneous (EC) sensitization with antigen shares characteristics with human atopic dermatitis (AD).
Objective.
To characterize gene expression by single cells in mouse skin undergoing antigen-driven allergic inflammation and compare the results with findings in AD skin lesions.
Methods.
Mice were EC sensitized by application of ovalbumin (OVA) or saline to tape stripped skin. Single-cell RNA-Seq (scRNA-Seq) was performed on skin cells twelve days later. Flow cytometry analysis was performed to validate results.
Results.
scRNA-Seq identified seven nonhematopoietic and six hematopoietic cell subsets in EC sensitized mouse skin. OVA sensitization resulted in the expansion in the skin of T cells, dendritic cells (DCs), macrophages, mast cell/basophils, fibroblasts and myocytes cell clusters, and in upregulation of Th2 cytokine gene expression in CD4+ T cells and mast/cell basophils. Genes differentially expressed in OVA sensitized skin included genes important for inflammation in DCs and macrophages, collagen deposition and leukocyte migration in fibroblasts, chemotaxis in endothelial cells and skin barrier integrity and differentiation in KCs, findings that recapitulate those in AD skin lesions. Unexpectedly, mast/cell basophils, rather than T cells, were the major source of Il4 and ll13 in OVA sensitized mouse skin. In addition, our results suggest novel pathways in fibroblast and endothelial cells that may contribute to allergic skin inflammation.
Conclusion.
The gene expression profile of single cells in mouse skin undergoing antigen-driven shares many features with that in AD skin lesions, and unveils novel pathways that may be involved in allergic skin inflammation.
Keywords: Atopic dermatitis, single cell RNA-seq, allergic skin inflammation
CAPSULE SUMMARY.
Analysis of gene expression at the single cell level reveals similarities between antigen-driven allergic skin inflammation in mice and AD skin lesions and unveils novel pathways that may contribute to allergic skin inflammation.
INTRODUCTION.
Atopic dermatitis (AD) is the most common skin inflammatory disease in infants and young children. It is characterized by a defective skin barrier, epidermal hyperplasia, dermal infiltration by T cells and eosinophils, and type 2 dominated local and systemic immune responses evidenced by increased cutaneous expression of type 2 cytokines, eosinophilia, elevated levels of serum IgE and the presence of IgE antibodies to environmental and food allergens1, 2.
The type 2 cytokines IL-4 and IL-13 are responsible for many of the pathological features of AD 1–3. IL-4 and IL-13 drive the expression of chemoattractants for eosinophils, basophils and Th2 cells, inhibit keratinocyte differentiation and production of antimicrobial peptides, impair skin barrier function, promote lipid abnormalities in the epidermis, increase endogenous protease activity, and drive skin remodeling 2, 4–7. A key role for IL-4 and IL-13 in allergic skin inflammation has been documented in several mouse models 8–10, and has been demonstrated in patients with AD by the beneficial effect of monoclonal antibody against the IL4Rα chain, shared by the IL-4 and IL-13 receptors11–13.
Our understanding of the physiopathology of AD has benefited enormously from the use of animal models 8–10. Our laboratory has developed a mouse model of allergic skin inflammation induced by repeated epicutaneous sensitization (EC) of tape stripped skin with ovalbumin (OVA) or peanut antigens 8, 9, 14–16. This model shares several characteristics with human AD. These include skin barrier disruption, scratching behavior, local and systemic type 2 immune response, epidermal thickening and dermal infiltration by CD4+ T cells and eosinophils 8, 9, 15. In addition, mice EC sensitized with OVA develop food allergy following oral antigen challenge and airway inflammation following intranasal antigen challenge 14, 16–18, thus mimicking the atopic march. These shared features have made our mouse model widely used in investigating allergic skin inflammation in AD 8, 9, 19–23.
Single cell RNA-Seq (scRNA-Seq) allows measuring gene expression profiles at the single cell level in tissues by combining barcoding technology, sequencing and computational analysis 24–26. Due to the complex cellular composition of the skin and technically challenging disruption of this tissue into single cell suspension, most scRNA-Seq studies performed on skin to date have focused on limited cell types27–34, with very few studies having evaluated the global cell populations in the skin 35–37. These studies have been instrumental in determining the diversity of different skin cell types, such as epidermal keratinocytes, fibroblasts, endothelial cells and immune cells. Recently, two studies reported scRNA-Seq of lesional and non-lesional skin from AD patients. They have shown that T cell subsets in AD lesional skin upregulate markers for Th2/Th22 cells, such as Il13 and Il22, and demonstrated increased infiltration of myeloid cells with inflammatory phenotypes, including dendritic cells, macrophages and Langerhans cells 36, 37. One of the studies further identified a population of COL6A5+COL18A1+ fibroblasts in lesional AD skin that express chemokines, cytokines and extracellular matrix products induced in type 2 inflammatory conditions36.
We performed scRNA-Seq analysis of full thickness mouse skin that has been subjected to EC sensitization with OVA or saline as control. The results show striking similarities with the results of scRNA-Seq studies and other studies of human AD lesions, providing further support for the utility of this mouse model in studying disease mechanisms in AD. Unexpectedly, mast/cell basophils, rather than T cells, were the major source of Il4 and ll13 in OVA sensitized skin. In addition, fibroblasts in OVA sensitized mouse skin expressed genes involved in mesenchymal/epidermal transition and regulation of NF-κβ activation, and endothelial cells expressed genes that stimulate angiogenesis, suggesting novel pathways that may contribute to allergic skin inflammation.
METHODS.
Epicutaneous (EC) sensitization.
Female BALB/c mice were purchased from Charles River Laboratories. Il13eGFP/+ mice in Balb/c background were obtained from Dr. Andrew N.J. McKenzie. Foxp3eyfp-cre (C57BL/6) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). 6–8-weeks old mice were epicutaneously sensitized for 10 days as described previously 15. Briefly, mice were anesthetized, and their back skin was shaved and tape-stripped with a film dressing (TegadermTM, 3M) followed by the application of 200 μg OVA (Sigma-Aldrich) or saline every other day. Analyses were done at day 12. All mice were kept in a pathogen-free environment and fed an OVA-free diet. All procedures were performed in accordance with the Animal Care and Use Committee of the Children’s Hospital Boston.
Skin cell preparation.
1 cm2 skin pieces from EC sensitized mice were obtained. Skin pieces were finely chopped using scissors after fat removal and digested for 90 minutes in the media containing Liberase (0.2 mg/mlRoche) and DNAse II (Sigma), with continuous shaking at 37° C. Digested skin homogenates were passed through a 100 μm cell strainer, washed and resuspended in 0.05% fetal calf serum in PBS.
Droplet-based scRNA-Seq.
Experiments were performed on the 10x Genomics Chromium platform, with the Chromium Single Cell 3′ Library & Gel Bead Kit v2 and Chromium Single Cell 3′ Chip kit v2 according to the manufacturer’s instructions in the Chromium Single Cell 3′ Reagents Kits V2 User Guide. Briefly, ~6,000 cells re-suspended in PBS supplemented with 0.05% FCS were loaded in each channel. Cells were then partitioned into Gel Beads in Emulsion in the Chromium controller, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Barcoded single cell libraries were sequenced with 38bp paired end reads on an Illumina NextSeq 500 instrument.
Single-cell transcriptome analysis.
The analysis was performed with the computational resources provided by Research Computing Group at Boston Children’s Hospital and Harvard Medical School (Boston, USA), including High-Performance Computing Cluster Enkefalos 2 (E2) and BioGrids scientific software made available for data analysis. The raw sequencing data were preprocessed to generate the read count per gene per cell matrices using CellRanger (10x Genomics, version 3.1.0), including BCL to FASTQ file conversion, data multiplexing according to cell barcodes, genome alignment to mouse genome mm10 with STAR, and reads counting per gene per cell into matrices. A pipeline for the single cell profiling was built based on R toolkit, Seurat 38, 39 (R version 3.6.2; Seurat version 3.1.2), including cell filtering, clustering, annotation, differential gene expression, and visualization. Cells expressing 200–2500 genes that were detected in at least three cells and a percentage of mitochondrial genes below 5% in droplets were considered viable and non-multiplets and were included for analysis. Following this quality control criteria to extract cells of good quality, the three replicates per group were merged and the two experimental groups were integrated for analysis by identifying common sources of variation using canonical correlation analysis 38, 39. Cell clustering was performed with a shared nearest neighbor graph-based method followed by the original Louvain algorithm for modularity optimization (resolution = 2.5) after data dimension reduction using principal component analysis (number of dimensions = 35). After the cell clusters were determined, their marker genes were identified with FindMarkers function. For cluster annotation, the top marker genes based on the adjusted p value were manually curated to match canonical cell types and their marker genes based on literature research and the resources in ImmGen (The Immunological Genome Project). Lastly, the differential gene expression between groups was calculated by likelihood-ratio test 40 as well as Wilcoxon rank-sum test with Bonferroni correction implemented in FindMarkers function in Seurat for further study. Raw and processed scRNA-seq data for all samples have been deposited in the GEO database under accession code GSE194254.
Flow cytometry.
Skin cells were preincubated with FcγR-specific blocking mAb (2.4G2) and washed before staining with the following monoclonal antibodies (mAbs): CD3 (17A2), CD4 (GK1.5), CD45 (30F11) and δ TCR (ebioGL3) from eBioscience, CD11b (M1/70), and CD117 (2B8) from Biolegend and anti-IgE (R35–72) from BD Biosciences. For cytokine staining, cells were stimulated with Ionomycin (0.5ug/ml; Sigma), Phorbol 12,13-dibutyrate (1ug/ml; Sigma), Brefeldin A (eBioscience), Monensin (eBioscience) in complete RPMI for 3 hours before surface staining. Then, cells were fixed and permeabilized (BD Biosciences Cytofix/Cytoperm) and stained in permeabilization solution with IL-22 (1H8PWSR), from Ebiosciences. Cells were analyzed by flow cytometry using an LSRFortessa machine (BD Biosciences). The data was analyzed with FlowJo software.
Statistical analysis.
The differential gene expression between groups was calculated by likelihood-ratio test 40 as well as Wilcoxon rank-sum test with Bonferroni correction. Statistical significance in the proportion of cell types was determined by the two-tailed Student’s t test. A p value <0.05 was considered statistically significant.
RESULTS
Cell composition of EC sensitized mouse skin by single cell RNA-Seq.
We previously showed that EC sensitization of mouse skin with OVA for 12 days results in allergic skin inflammation that recapitulates many pathological features of AD lesional skin15, 22. As previously reported, mouse skin EC sensitized with OVA showed increased transepidermal water loss, increased epidermal thickness and increased infiltration with mononuclear cells compared to mouse skin EC sensitized with saline (Fig. E1A and E1B). In addition, OVA sensitized skin exhibited increased infiltration of CD45+ T cells, including CD4+ T cells and eosinophils, and upregulation of Il4 and Il13 mRNA levels compared to saline sensitized skin (Fig. E1C–G). To perform scRNA-Seq, the skin of Balb/C mice EC sensitized with OVA or saline (n=3/goup, total 6 samples) was dissociated into a single cell suspension by enzymatic digestion, followed immediately by bar coding the cells and scRNA sequencing. We profiled 6,200±1,010 cells per skin sample with ~18,000±2,940 reads per cell and 1,250±52 unique genes detected per cell. Following quality control (QC) to remove cells with low quality profiles and suspected doublets 14,862 high quality single cell profiles from the six samples were retained for subsequent analysis.
Unsupervised clustering and post-hoc annotation integrating saline- and OVA-sensitized skin identified 47 clusters spanning 13 distinct cell types (Fig. 1A, Fig. E2 and Table E1). Fibroblasts, identified by the expression of Lum, Dcn and Col1a2, were the most frequent cell type (~46%) followed by keratinocytes (~26%), identified by the expression of Krt1, Krt5, Krt14 and Krt15. Endothelial cells (~10%) were identified by the expression of Cldn5 and Vwf, macrophages (~4%) by the expression of Lyz2 and Cd14, pericytes (~3.7%) by the expression of Rgs4, Rgs5 and Kcnj8, dendritic cells (DCs) (~3.0%) by the expression of Cd74 and H2-Eb1, Schwann cells (~2.3%) by the expression of Mbp and Kcna1, T cells (~2.3%) by the expression of Trbc1, Trbc2 and Cxcr6, neutrophils(~0.6%) by the expression of S100a9 and S100a8, Langerhans cells (LCs) (~0.4%) by the expression of Cd207 and Ltc4s, melanocytes by the expression of Oca2, Sox10 and Pax3, myocytes (~0.2%) by the expression of Csrp3, Apobec2 and Ckmt2 and mast cells (MCs)/basophils (~0.1%) by the expression of Fcer1a, Cpa3 and Mcpt8 (Fig. 1B–C and Table E1).
Figure 1. Cell composition of EC sensitized mouse skin by scRNA-Seq.
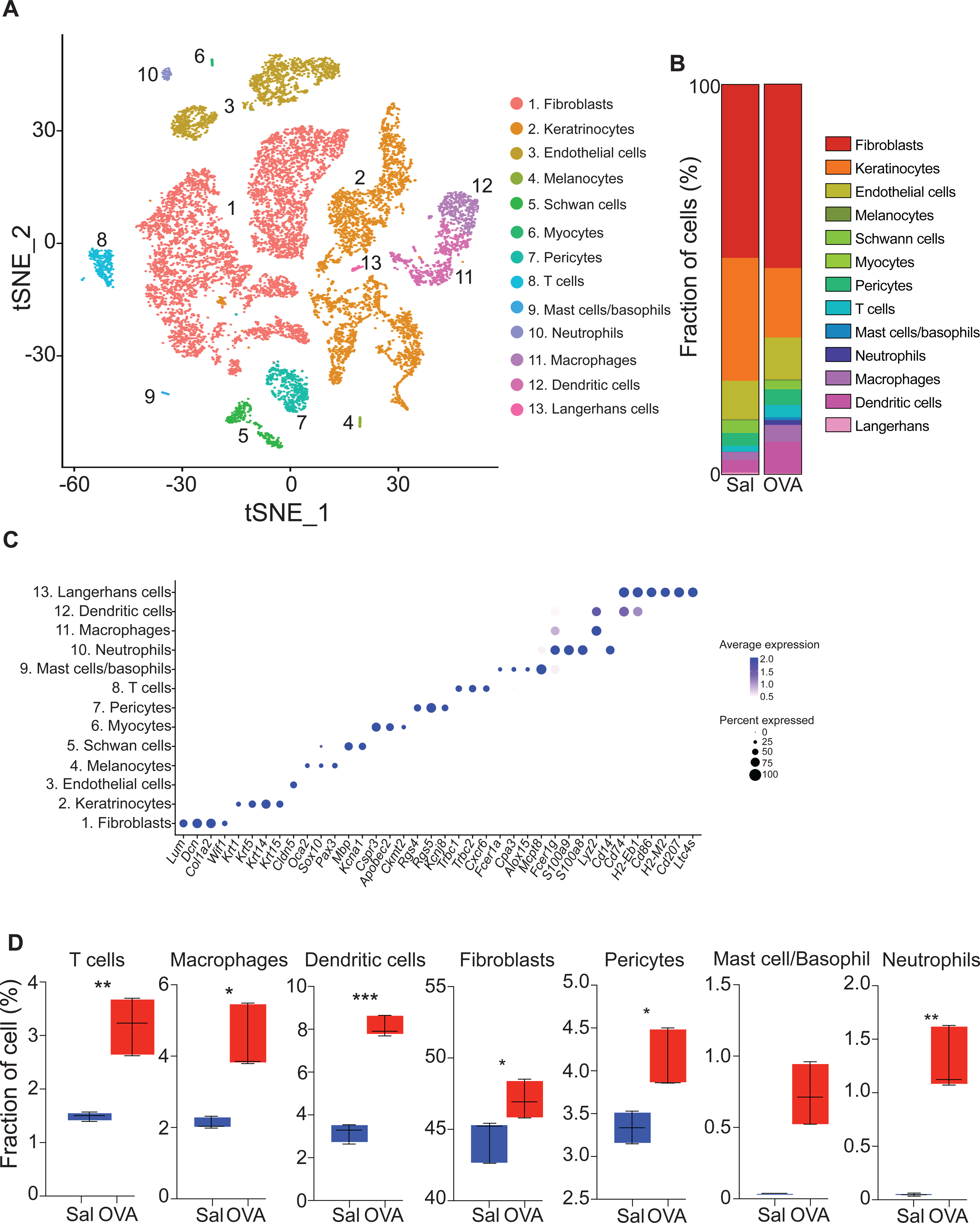
A. t-Distributed stochastic neighbor embedding (tSNE) plot for 14,211 high quality single cell transcriptomes from mouse skin EC sensitized with saline (n=3) and OVA (n=3), revealing 13 different cell populations. B. Fractions of cell populations identified in EC sensitized skin with saline (Sal) and OVA. C. Dot plot displaying top expressed genes in each cell population compared to the other cell populations identified in EC sensitized skin. D. Boxplot of the distribution of seven selected cell populations in mouse skin EC sensitized with OVA or saline. *p< 0.05, ** p< 0.005 and *** p< 0.001.
Consistent with the development of allergic skin inflammation, OVA sensitized skin demonstrated an increase in the total number of cells recovered compared to saline sensitized skin. This was caused by expansion of both immune cells, including T cells, macrophages, and DCs, and non-hematopoietic cells, including fibroblasts and pericytes, as well as by the appearance of neutrophils and MC/basophil populations (Fig. 1D, Table 1 and Fig. E2).
Table 1.
Cell number and percentage of the different cell populations identified in our analysis.
| Cluster | Cell number | % of total | ||
|---|---|---|---|---|
| EC Sal | EC OVA | EC Sal | EC OVA | |
| 1. Fibroblasts | 2885 | 3895 | 44.3 | 46.25 |
| 2. Keratinocytes | 2053 | 1739 | 31.62 | 20.78 |
| 3. Endothelial cells | 656 | 864 | 10.10 | 10.32 |
| 4. Melanocytes | 22 | 32 | 0.34 | 0.38 |
| 5. Schwann cells | 188 | 173 | 2.90 | 2.07 |
| 6. Myocytes | 11 | 31 | 0.17 | 0.37 |
| 7. Pericytes | 216 | 328 | 3.33 | 3.92 |
| 8. T cells | 95 | 233 | 1.46 | 2.78 |
| 9. Mast cells / Basophils | 1 | 48 | 0.02 | 0.57 |
| 10. Neutrophils | 3 | 81 | 0.05 | 0.97 |
| 11. Macrophages | 133 | 322 | 2.05 | 3.85 |
| 12. Dendritic cells | 197 | 595 | 3.03 | 7.11 |
| 13. Langerhans cells | 33 | 28 | 0.51 | 0.33 |
| Total | 6,493 | 8,369 | 100 | 100 |
Characterization of fibroblast populations in mouse EC sensitized skin.
Fibroblast spanned 16 clusters (Fig. 2A and Table E2). Cells in clusters 1 to 6 were collagen I-producing fibroblasts characterized by the expression of Col1a1 and Col1a2 and comprised ~44% of the fibroblast population. Clusters 7 to 10 were collagen VIII/IX-producing fibroblasts characterized by the expression of Col8a1, Col8a2, Col9a1, Col9a2 and Col9a3 and comprised ~37% of the fibroblast population. Cells in clusters 11 and 12 expressed Sst, Hhip and Sox2 characteristic of dermal papilla fibroblasts and comprised ~7% of the fibroblast population. Cells in cluster 13 expressed Myod1 and Lypd2 characteristic of myofibroblasts comprised ~1% of the fibroblast population. Cells in cluster 14 to 16 expressed Mfap5, Cygb and Apod characteristic of lipofibroblasts comprised ~11% of the fibroblast population (Fig. 2B).
Figure 2. Characterization of the fibroblast population in mouse EC sensitized skin by scRNA-Seq.
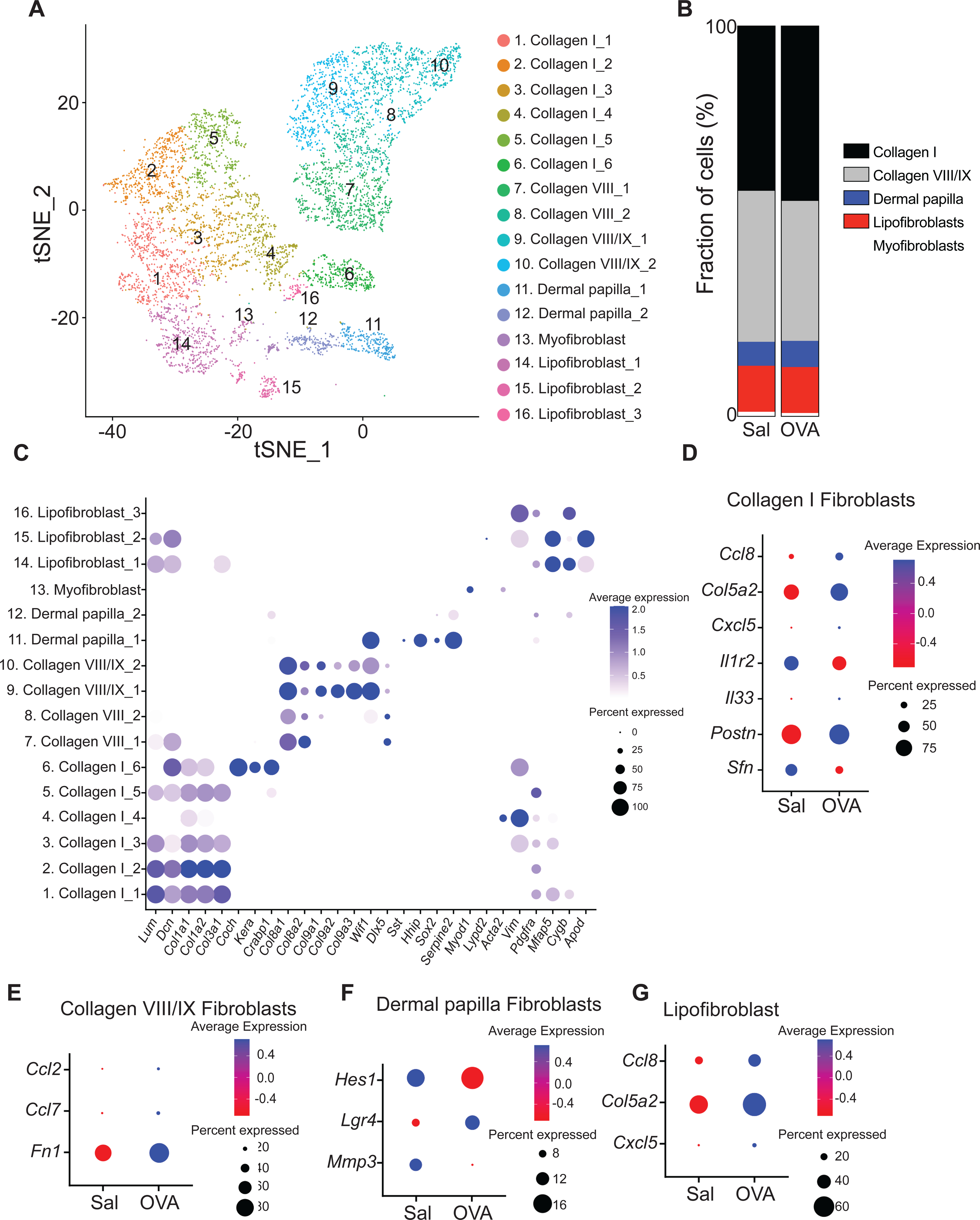
A. t-Distributed stochastic neighbor embedding plot for fibroblast cluster from EC sensitized mouse skin, revealing 16 clusters. B. Distribution of fibroblast populations identified in mouse skin EC sensitized with OVA or saline. C. Dot plot displaying top expressed genes in each fibroblast cluster. D-G. Dot plot of differentially expressed genes in the collagen I-producing fibroblasts (D), collagen VIII/IX-producing fibroblasts (E), Dermal papilla fibroblast (F), and lipofibroblast (G) clusters from skin EC sensitized with saline or OVA.
There were important differences between OVA sensitized and saline sensitized skin in the expression of key genes in fibroblast subsets. Collagen I-producing fibroblasts from OVA sensitized skin showed increased Ccl8, Col5a2, Cxcl5, Il33 and Postn expression and diminished Sfn and Il1r2 expression compared to those in saline sensitized skin (Fig. 2C and Table E3). Lipofibroblasts from OVA sensitized skin showed increased Ccl8, Col5a2 and Cxcl5 expression (Fig. 2D and Table E3). In addition, collagen VIII/IX-producing fibroblasts from OVA sensitized skin showed increased Ccl2, Ccl7 and Fn1 expression (Fig. 2D and Table E3). Following OVA sensitization, dermal papilla fibroblasts showed increased Lgr4 expression and diminished Hes1 and Mmp3 expression (Fig. 2E and Table E3). These results suggest that fibroblasts play an important role in skin remodeling and the attraction of immune cell into allergic inflamed skin.
Characterization of the keratinocyte population in mouse EC sensitized skin.
The keratinocyte (KC) population spanned 13 clusters, tracking with the different layers of the epidermis, the various parts of the hair follicle and the sebaceous gland (Fig. 3A and Table E2). Specifically, cells in clusters 1, 2 and 3 comprised ~26% of the KC population and showed high expression of Krt14, Krt5 and Krt15, characteristic of the basal layer of the epidermis. Cells in cluster 4 comprised ~13% of KCs and expressed Krt1 and Krt10 characteristic of the spinous layer. Cells in cluster 5 comprised ~9% of KCs and expressed Flg and Lor characteristic of the granular layer (Fig. 3B). Cells in cluster 6 and 7 comprised ~14% of the population and expressed Krt17 and Lrg5 characteristic of the hair follicle outer bulge. Cells in cluster 8 expressed the hair follicle inner bulge gene group 1 (Krt25, Krt27 and Krt28) and clusters 9 to 11 expressed Plet1 characteristic of hair follicle inner bulge gene group 2 (Fig. 3B) comprised ~26% of the population. Cells in cluster 12 comprised ~4% of KCs and expressed Awat2, Far2, Acot5 and Pnpla5 characteristic of the sebaceous gland (Fig. 3B). Cells in cluster 13 comprised ~8% of KCs and expressed Defb6, and Cst6 characteristic of the upper hair follicle (Fig. 3B).
Figure 3. Characterization of the keratinocyte population in mouse EC sensitized skin by scRNA-Seq.
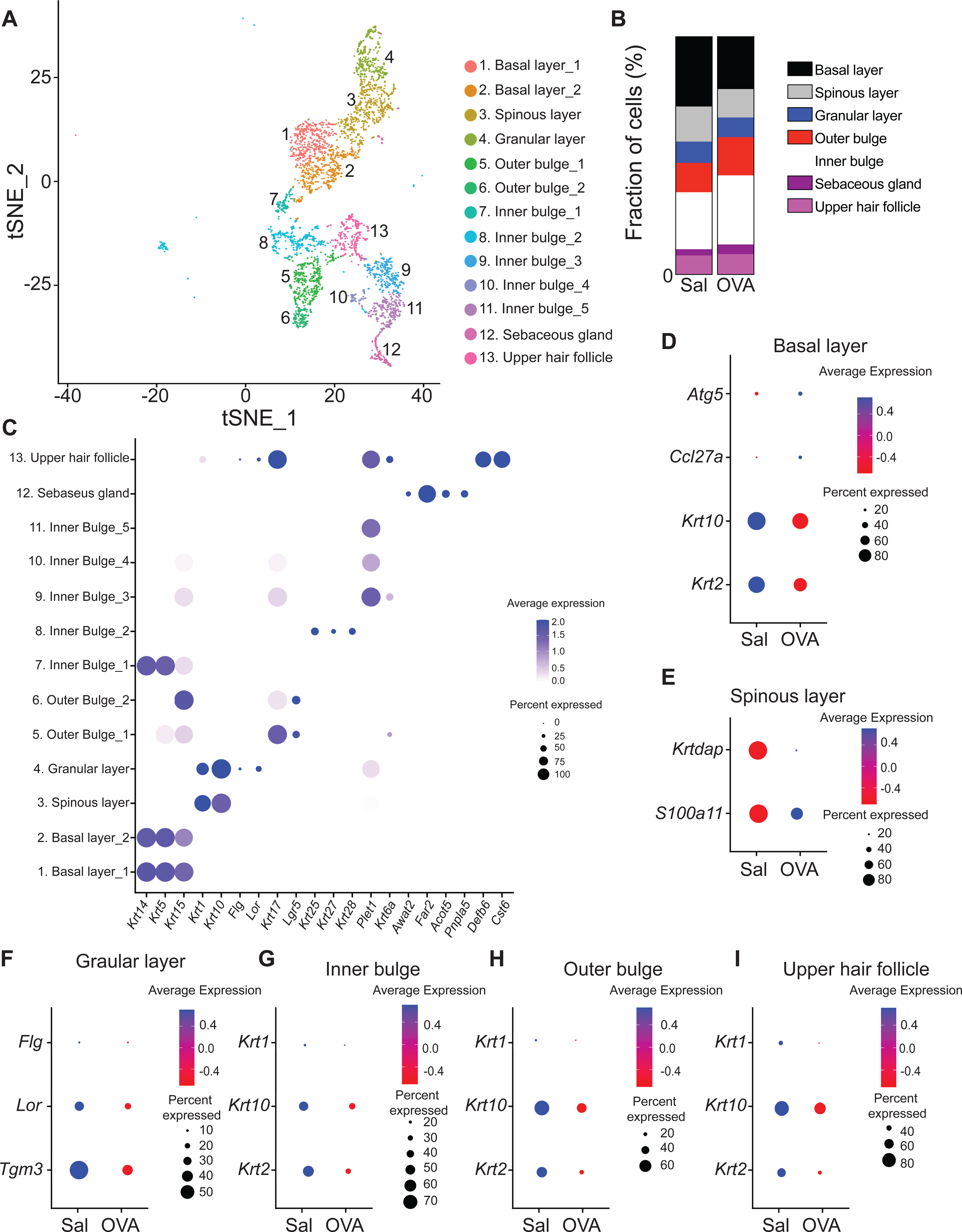
A. t-Distributed stochastic neighbor embedding plot for keratinocyte cluster from EC sensitized mouse skin, revealing 13 clusters. B. Distribution of keratinocyte populations identified in mouse skin EC sensitized with OVA or saline. C. Dot plot displaying top expressed genes in each keratinocyte cluster. D-I. Dot plot of differentially expressed genes in the keratinocyte basal layer (D), spinous layer (E), granular layer (F), inner bulge (G), outer bulge (H) and upper hair follicle (I) clusters from skin EC sensitized with saline or OVA.
As with fibroblasts, keratinocytes showed cell intrinsic differences between OVA and saline sensitization. Keratinocytes from OVA sensitized skin showed diminished Krt10 and Krt2 and increased Ccl27a and Atg5 expression in the basal layer, increased S100a11 and Krtdap expression in the spinous layer and decreased expression of Flg, Lor, and Tgm3 in the granular layer compared to those in saline sensitized skin (Fig. 3C–E and Table E3). Furthermore, hair follicle inner and outer bulge, as well as upper hair follicle keratinocytes from OVA sensitized skin showed diminished Krt1, Krt10 and Krt2 expression (Fig. 3F–H and Table E3). Together, these results suggest that allergic skin inflammation impairs keratinocyte differentiation.
Characterization of endothelial cell populations in mouse EC sensitized skin.
The endothelial cell population spanned four clusters (Fig. 4A and Table E2). Cells in cluster 1 comprised ~32% of endothelial cells and was characterized by the expression of the lymphatic endothelium genes Mmrn1, Flt4 and Lyve1. Cells in clusters 2 to 4 comprised ~68 % of endothelial cells and were characterized by the expression of the vascular endothelium genes Vwf, Esam and Ly6c1 (Fig. 4B). Vascular endothelial cells from OVA sensitized skin showed increased Selp and Cxcl2 expression (Fig. 4C and Table E3) compared with those from saline sensitized skin, whereas lymphatic endothelial cells from OVA sensitized skin showed increased expression of Selp and S100a6 (Fig. 4D and Table E3), which encode for a protein essential for cell cycle progression in endothelial cells. These results suggest that endothelial cells play an important role in the attraction of immune cells into allergic inflamed skin.
Figure 4. Characterization of the endothelial cell population in mouse EC sensitized skin by scRNA-Seq.
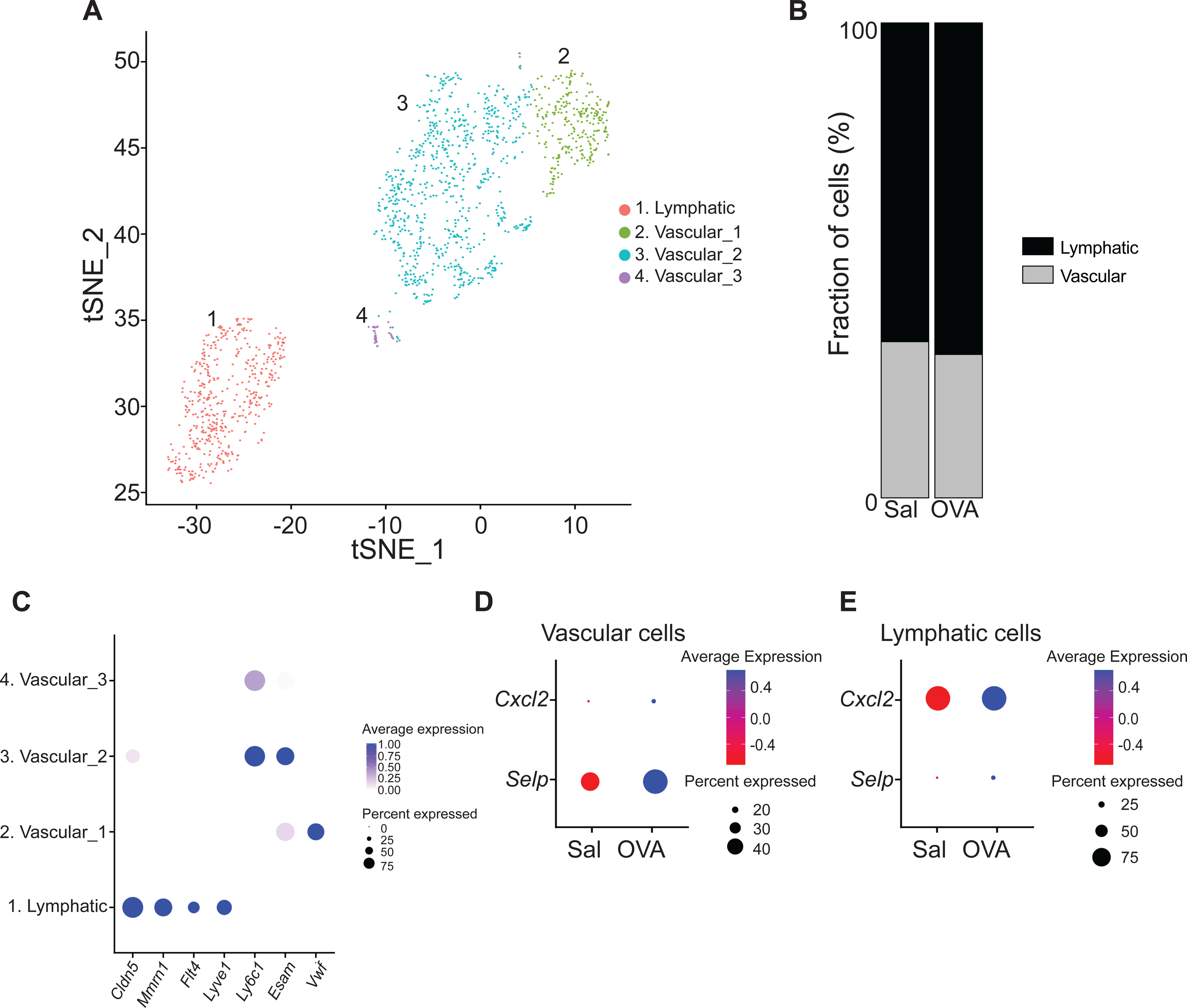
A. t-Distributed stochastic neighbor embedding plot for endothelial cell cluster from EC sensitized mouse skin, revealing 4 clusters. B. Distribution of endothelial cell populations identified in mouse skin EC sensitized with OVA or saline. C. Dot plot displaying top expressed genes in each endothelial cell cluster. D-E. Dot plot of differentially expressed genes in the endothelial cell vascular (D) and lymphatic (E) clusters from skin EC sensitized with saline or OVA.
Characterization of immune cell populations in mouse EC sensitized skin.
The T cell cluster in OVA sensitized skin demonstrated increased expression of Cd4 and decreased expression of Tcrd compared to saline sensitized skin (Fig. 5A and Table E3). Flow cytometry analysis of CD3+ cells revealed an increased percentage of CD4+ T cells at the expense of the percentage of TCR γδ + T cells in OVA sensitized skin compared to saline sensitized control (Fig. 5B). These results indicate a shift in CD4+/TCR γδ+ T cell ratio at sites of allergic skin inflammation.
Figure 5. Characterization of immune cell populations in mouse EC sensitized skin by scRNA-Seq.
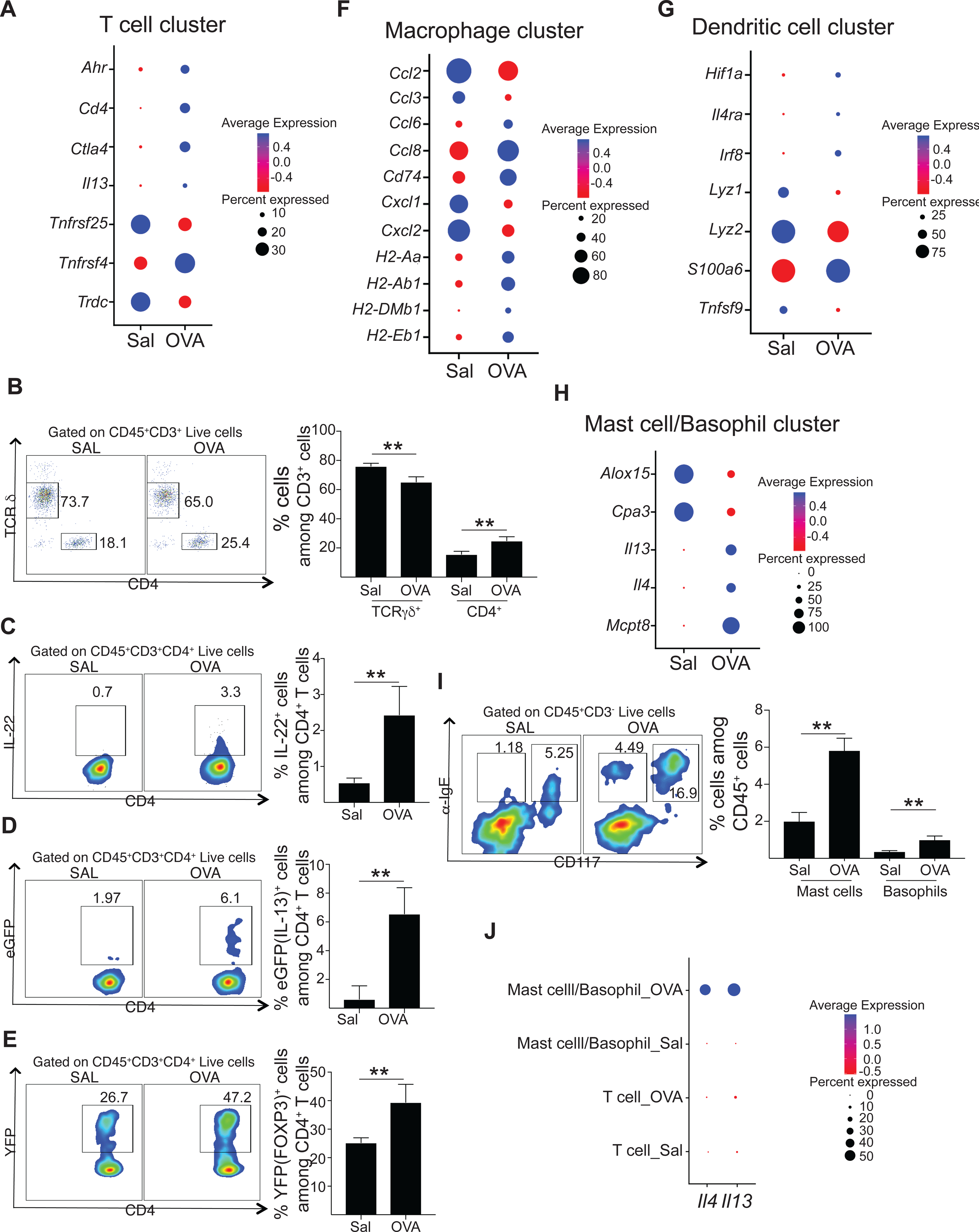
A. Dot plot of differentially expressed genes in T cell cluster of skin EC sensitized with saline or OVA. B-E. Representative flow cytometry plots (left) and percentages of CD4+ T cells and TCR γδ + T cells (B), CD4+ IL-22+ T cells (C), CD4+eGFP(IL-13)+ T cells (D) and CD4+YFP(FOXP3)+ T cells (E) of skin EC sensitized with saline or OVA. F-H. Dot plot of differentially expressed genes in DC (F), macrophages (G) and mast cell/basophil (H) clusters of skin EC sensitized with saline or OVA. H. Representative flow cytometry plots (left) and percentages of mast cells and basophils of skin EC sensitized with saline or OVA. I. Dot plot displaying the relative Il4 and Il13 expression in the T cell cluster and the mast cell/basophil cluster from skin EC sensitized with saline or OVA.
T cells from OVA sensitized skin further showed increased expression of genes associated with Th22 (Ahr), Th2 (Il13 and Tnfrs4) and regulatory T cells (Ctla4 and Tnfrs25) compared to saline sensitized skin (Fig. 5A and Table E3), suggesting accumulation of these cell populations in skin undergoing allergic inflammation. Consistent with these results, intracellular staining showed an increase in the percentage of IL-22 producing CD4+ T cells in OVA sensitized skin compared to saline sensitized skin (Fig. 5C). Furthermore, analysis of EC sensitized skin from Il13eGFP/+ and Foxp3Cre-YFP showed respectively increased percentages of CD4+ T cells expressing IL-13 and FOXP3 in OVA sensitized skin compared with saline sensitized skin (Fig. 5D and E).
Macrophages in OVA sensitized skin demonstrated increased expression of antigen presentation genes (H2-Ab1, H2-Aa, H2-Eb1, H2-DMb1, H2-DMa and Cd74), the chemokines Ccl8 and Ccl6 and the fatty acid transporter Fabp5, but decreased expression of inflammatory macrophage markers such as the chemokines Ccl2, Ccl3, Cxcl1 and Cxcl2 and the inflammatory cytokine Tnfa compared with saline sensitized skin (Fig. 5F and Table E3). These results suggest that allergic skin inflammation promotes macrophages with an alternative activation phenotype.
DCs in OVA sensitized skin demonstrated increased expression of Irf8, Il12b, Fscn1, Cts2, and Cd24a, but decreased expression of Areg, Cd14, Ctss, Fcer1g and Fcgr3 compared to saline sensitized skin (Fig. 5G and Table E3). These data suggest that allergic skin inflammation promotes an inflammatory phenotype in skin DCs.
Cells in the mast cell/basophil cluster in OVA sensitized skin exhibited expression of Il4, Il13, Mcpt8, and Ccrl2 (Fig. 5H and Table E3), suggesting increased infiltration of mast cells and basophils in allergic inflamed skin. Flow cytometry analysis of skin cells confirmed the increase in basophils (identified as CD3−IgE+CD117− cells) and mast cells (identified as CD3−IgE+CD117+ cells) in OVA sensitized skin compared to saline sensitized skin (Fig. 5I).
Finally, the Th2 cytokine Il4 was detected only in OVA sensitized skin with mast cells/basophils being the only contributors. More Il13 was detected in OVA-sensitized than in saline-sensitized skin. In both samples mast cells/basophils, rather than T cells, were the major 1l13 contributors (Fig. 5J).
DISCUSSION.
We describe the single cell RNA-Seq profile of mouse skin undergoing acute allergic inflammation elicited by EC sensitization with antigen.
Our unsupervised clustering analysis revealed seven non-hematopoietic and six hematopoietic cell types in mouse skin. The identification in our study of these thirteen different cell types in mouse skin is consistent with the recently published scRNA-Seq analysis of full-thickness unmanipulated mouse skin 35. In addition, our analysis identifies mast cell/basophil and neutrophil populations in OVA sensitized skin that were not detected in saline sensitized skin and that were not previously reported in unmanipulated mouse skin35. This likely reflects the cutaneous recruitment and/or or expansion of mast cell/basophil and neutrophil populations in allergic skin inflammation.
We detected expansion of immune cells, fibroblasts and pericytes following in mouse skin undergoing antigen driven acute allergic inflammation. We identified sixteen different clusters of fibroblasts in mouse skin, consistent with the heterogeneity of dermal fibroblasts 31, 35, 41, as well as four subsets of endothelial cells. Analysis of differentially expressed genes in dermal fibroblasts from OVA sensitized skin reveals increased expression of genes coding for chemokines (Ccl8, Ccl2, Cxcl5), and molecules involved in tissue remodeling (Tnc, Postn, Col5a2 and Col6a3). In addition, fibroblasts from OVA sensitized skin expressed higher levels of Il33 and Prg4, molecules implicated in the suppression of NF-κB activation induced by IL-1β and TLR activation 42, 43. Our results suggest that in addition to their known role in attracting immune cells and skin remodeling 44, fibroblasts may play a role in the susceptibility of cutaneous infection observed in AD patients.
Endothelial cells from OVA sensitized skin showed increased expression of Cxcl2, Sele and Selp, consistent with the role of endothelial cells in the cutaneous attraction of immune cells 45 In addition, they exhibited differential expression of genes regulating endothelial cell growth and sprouting, suggesting that allergic skin inflammation activates an autocrine loop that stimulates angiogenesis in endothelial cells.
AD is characterized by a defective skin barrier due to impaired keratinocyte differentiation, lipid abnormalities and increased protease activity in the epidermis 46–49. Our analysis reveals that keratinocytes from OVA sensitized skin exhibit dysregulated expression of keratins (Krt1, Krt2, Krt10 and Krtdap) in multiple layers, increased expression of Atg5 in the basal layer and decreased expression of genes important for barrier integrity (Flg, Lor and Tgm3), changes that have been reported in skin from AD patients and in our mouse model of antigen driven allergic skin inflammation5, 47, 50–56.
OVA sensitized mouse skin exhibited increased numbers of T cells that expressed Cd4 as well as genes associated with Th2 cells, including Il13 and Tnfrs4, and demonstrated increased expression of genes associated with Tregs, including Tnfrsf25 and Ctla4. These findings are in line with the previously reported increase in CD4+ T cells and in Th2 cytokine expression in OVA sensitized skin in our model and in AD skin lesions 4, 14, 15, 57, 58, as well as with results of scRNA-Seq analysis of AD skin lesions 36, 37. IL-4 and IL-13 play a major role in AD pathophysiology. Importantly, our analysis identifies the mast cell/basophil population, rather than T cells, as the major source of Il4 and ll13 in acute allergic skin inflammation 59. This is in line with our recent findings using flow cytometry analysis of cells from OVA sensitized mouse skin 15.
DCs and macrophages play an important role in AD 60, 61. Our results show that skin acutely sensitized with OVA exhibits an increased number of macrophages and DCs, but not LCs. This is consistent with the increase in inflammatory DCs and macrophages in AD skin lesions previously demonstrated by immunohistochemistry and digital imaging 62 and by scRNA-Seq 36, 37. The macrophage cluster from OVA sensitized skin shows an increased expression of genes involved in antigen presentation and alternative macrophage activation. This is consistent with the increased number of CD163+ cells, a marker for alternatively activated macrophages, and the presence of cells expressing markers for both macrophages and dendritic cells in AD lesional skin 59, 62. In addition, DCs from OVA sensitized skin downregulated genes coding for proteins highly expressed in monocytes (Cd14, Fcer1g and Fcgr3). Taken together, these results suggest that the type 2 cytokine environment promotes in situ differentiation of monocytes to dendritic cells at sites of allergic skin inflammation.
Our study has limitations. First, we were unable to identify cell populations, such as eosinophils and type 2 innate lymphoid cells, and detect cytokines, such as Il17a and Il22, previously described in OVA sensitized mouse skin and in AD skin lesions 63–66, possibly because of the limited number of hematopoietic cells recovered and some limitations of droplet based scRNA-Seq at capturing eosinophils and neutrophils. Second, the limited number of hematopoietic cells analyzed did not allow us to perform a more granular analysis of their different states. Third, we were unable to distinguish between T cell subsets with clustering, such that both Treg cells and Th2 are members of a single T cell cluster or split mast cell and basophil populations. Fourth, our analysis is limited to a single time point providing only a snapshot of the disease, and thus does not inform on the dynamics of the cutaneous response during the course of allergic skin inflammation. Future studies are needed to overcome these limitations.
Importantly, our study demonstrates that our mouse model of antigen-driven allergic skin inflammation elicited by EC sensitization shares at the level of gene expression by single cells many features found in AD lesional skin by scRNA-Seq and demonstrate general, although not total, agreement with the results of published scRNA-Seq and global gene expression studies of AD lesional skin (Table 2, Table E4, Fig. E3 and E4). Our comparative analysis of our data and data on gene expression in AD lesional skin is based on the publicly available datasets in human AD, with the limitations that skin samples were collected and processed under different conditions in different laboratories, and analyzed using different bioinformatic pipelines.
Table 2.
scRNAseq findings in mouse allergic skin inflammation shared with AD lesional skin
| scRNAseq of EC sensitized mouse skin | AD | Reference | |
|---|---|---|---|
| scRNAseq | Other | ||
| Increased T cell cluster | IHC | 68 | |
| Increased DC cluster | IF | 69 | |
| Increased macrophages cluster | X | IHC | 36,37,62 |
| Increased mast cell/basophils cluster | FACS, IF, Histology | 70,71 | |
| Increased neutrophil cluster | IHC | 72 | |
| Upregulation of Il33 in collagen I-producing fibroblasts | IHC | 73 | |
| Upregulation of Ccl2 in fibroblasts | X | 36 | |
| Upregulation of Postn in fibroblasts | X | 36 | |
| Upregulation of Ccl2 in fibroblasts | X | 36 | |
| Upregulation of Ccl27 in KC basal layer cluster | X | 36 | |
| Downregulation of Krt1 and Krt10 in KC clusters | IF | 74 | |
| Downregulation of Flg in KC basal layer cluster | PCR, IHC | 75 | |
| Downregulation of Lor in KC basal layer cluster | PCR, IHC | 50 | |
| Upregulation of Selp expression in endothelial cell clusters | IHC | 76,77 | |
| Upregulation of Cd4 in T cell cluster | IHC | 68,78 | |
| Upregulation of Ctla4 andTnfrs25 in T cell cluster | X | IHC | 36,37, 79, 80 |
| Upregulation of Il13 in T cell cluster | X | FACS | 36,37,57 |
| Upregulation of Tnfrs4 in T cell cluster | X | 36, 37 | |
| Upregulation of Ahr expression in T cell cluster | X | FACS | 36, 37, 66 |
| Upregulation of Fab5, Ccl8 and ccl6 in macrophage cluster | IHC | 59 | |
| Upregulation of antigen presentation genes in macrophage cluster | IHC | 62 | |
X: scRNAseq data, IHC: Immunohistochemistry, IF: Immunofluorescence, FACS: Fluorescence-Activated Cell Sorting, PCR: Polymerase Chain Reaction.
In summary, the application of scRNA-Seq analysis to our mouse model of antigen-driven allergic skin inflammation further enhances the utility of this model in investigating potential disease mechanisms, and identifies potentially novel therapeutic targets in AD.
Supplementary Material
CLINICAL IMPLICATIONS.
scRNA-Seq validates the use of the mouse model of antigen-driven allergic skin inflammation to investigate the pathogenesis of AD and unveils novel pathways in allergic skin inflammation.
Acknowledgements.
We thank Dr. Aviv Regev for her help in the cell barcoding and generation of scRNA-Seq libraries and Dr. Hans Oettgen and Dr. Aviv Regev for reading the manuscript and useful discussions.
Funding:
This work was funded by the National Institutes of Health/National Institute of Allergy and Infectious Diseases Atopic Dermatitis Research Network grant U19AI117673, NIH grant AI113294-01A1. J.M.L.C. was supported by Boston Children’s Hospital OFD/BTREC/CTREC Faculty Career Development Fellowship, NIAID T32 training grant 5T32AI007512-32 and Dermatology Foundation Research Career Development Award.
ABBREVIATIONS
- AD
Atopic dermatitis
- DCs
Dendritic cells
- EC
Epicutaneous
- KCs
Keratinocytes
- MCs
Mast cells
- OVA
Ovalbumin
- scRNA-Seq
Single-cell RNA sequencing
Footnotes
Declaration of interests.
The rest of the authors declare that they have no relevant conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES.
- 1.Bieber T. Atopic dermatitis. N Engl J Med 2008; 358:1483–94. [DOI] [PubMed] [Google Scholar]
- 2.Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers 2018; 4:1. [DOI] [PubMed] [Google Scholar]
- 3.May RD, Fung M. Strategies targeting the IL-4/IL-13 axes in disease. Cytokine 2015; 75:89–116. [DOI] [PubMed] [Google Scholar]
- 4.Brunner PM, Guttman-Yassky E, Leung DY. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol 2017; 139:S65–S76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omori-Miyake M, Yamashita M, Tsunemi Y, Kawashima M, Yagi J. In vitro assessment of IL-4- or IL-13-mediated changes in the structural components of keratinocytes in mice and humans. J Invest Dermatol 2014; 134:1342–50. [DOI] [PubMed] [Google Scholar]
- 6.Callewaert C, Nakatsuji T, Knight R, Kosciolek T, Vrbanac A, Kotol P, et al. IL-4Ralpha Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J Invest Dermatol 2020; 140:191–202 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werfel T, Allam JP, Biedermann T, Eyerich K, Gilles S, Guttman-Yassky E, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol 2016; 138:336–49. [DOI] [PubMed] [Google Scholar]
- 8.Jin H, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol 2009; 129:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyoshi MK, He R, Kumar L, Yoon J, Geha RS. Cellular and molecular mechanisms in atopic dermatitis. Adv Immunol 2009; 102:135–226. [DOI] [PubMed] [Google Scholar]
- 10.Kim D, Kobayashi T, Nagao K. Research Techniques Made Simple: Mouse Models of Atopic Dermatitis. J Invest Dermatol 2019; 139:984–90 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman-Yassky E, Bissonnette R, Ungar B, Suarez-Farinas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol 2019; 143:155–72. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton JD, Suarez-Farinas M, Dhingra N, Cardinale I, Li X, Kostic A, et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 2014; 134:1293–300. [DOI] [PubMed] [Google Scholar]
- 13.Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 2014; 371:130–9. [DOI] [PubMed] [Google Scholar]
- 14.Spergel JM, Mizoguchi E, Brewer JP, Martin TR, Bhan AK, Geha RS. Epicutaneous sensitization with protein antigen induces localized allergic dermatitis and hyperresponsiveness to methacholine after single exposure to aerosolized antigen in mice. J Clin Invest 1998; 101:1614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leyva-Castillo JM, Galand C, Mashiko S, Bissonnette R, McGurk A, Ziegler SF, et al. ILC2 activation by keratinocyte-derived IL-25 drives IL-13 production at sites of allergic skin inflammation. J Allergy Clin Immunol 2020; 145:1606–14 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartnikas LM, Gurish MF, Burton OT, Leisten S, Janssen E, Oettgen HC, et al. Epicutaneous sensitization results in IgE-dependent intestinal mast cell expansion and food-induced anaphylaxis. J Allergy Clin Immunol 2013; 131:451–60 e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leyva-Castillo JM, Yoon J, Geha RS. IL-22 promotes allergic airway inflammation in epicutaneously sensitized mice. J Allergy Clin Immunol 2019; 143:619–30 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galand C, Leyva-Castillo JM, Yoon J, Han A, Lee MS, McKenzie ANJ, et al. IL-33 promotes food anaphylaxis in epicutaneously sensitized mice by targeting mast cells. J Allergy Clin Immunol 2016; 138:1356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamari M, Orimo K, Motomura K, Arae K, Matsuda A, Nakae S, et al. The optimal age for epicutaneous sensitization following tape-stripping in BALB/c mice. Allergol Int 2018; 67:380–7. [DOI] [PubMed] [Google Scholar]
- 20.Kypriotou M, Rivero D, Haller S, Mariotto A, Huber M, Acha-Orbea H, et al. Activin a inhibits antigen-induced allergy in murine epicutaneous sensitization. Front Immunol 2013; 4:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Oh MH, Park JU, Myers AC, Dong C, Zhu Z, et al. Epicutaneous exposure to staphylococcal superantigen enterotoxin B enhances allergic lung inflammation via an IL-17A dependent mechanism. PLoS One 2012; 7:e39032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J Invest Dermatol 2013; 133:154–63. [DOI] [PubMed] [Google Scholar]
- 23.Fyhrquist N, Lehtimaki S, Lahl K, Savinko T, Lappetelainen AM, Sparwasser T, et al. Foxp3+ cells control Th2 responses in a murine model of atopic dermatitis. J Invest Dermatol 2012; 132:1672–80. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez Conde C, Teichmann SA. Deciphering immunity at high plexity and resolution. Nat Rev Immunol 2020; 20:77–8. [DOI] [PubMed] [Google Scholar]
- 25.Wen T, Rothenberg ME. Cell-by-cell deciphering of T cells in allergic inflammation. J Allergy Clin Immunol 2019; 144:1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Yang B, Udo-Inyang I, Ji S, Ozog D, Zhou L, et al. Research Techniques Made Simple: Single-Cell RNA Sequencing and its Applications in Dermatology. J Invest Dermatol 2018; 138:1004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, et al. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep 2018; 25:871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 2019; 20:915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghahramani A, Donati G, Luscombe NM, Watt FM. Epidermal Wnt signalling regulates transcriptome heterogeneity and proliferative fate in neighbouring cells. Genome Biol 2018; 19:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lonnerberg P, et al. Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 2016; 3:221–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philippeos C, Telerman SB, Oules B, Pisco AO, Shaw TJ, Elgueta R, et al. Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J Invest Dermatol 2018; 138:811–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzer MC, Lafzi A, Berenguer-Llergo A, Youssif C, Castellanos A, Solanas G, et al. Identity Noise and Adipogenic Traits Characterize Dermal Fibroblast Aging. Cell 2018; 175:1575–90 e22. [DOI] [PubMed] [Google Scholar]
- 33.Tabib T, Morse C, Wang T, Chen W, Lafyatis R. SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin. J Invest Dermatol 2018; 138:802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E. Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 2017; 169:483–96 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, et al. The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 2020; 26:441–57 e7. [DOI] [PubMed] [Google Scholar]
- 36.He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol 2020; 145:1615–28. [DOI] [PubMed] [Google Scholar]
- 37.Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J Allergy Clin Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 38.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell 2019; 177:1888–902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018; 36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 2013; 29:461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sole-Boldo L, Raddatz G, Schutz S, Mallm JP, Rippe K, Lonsdorf AS, et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol 2020; 3:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, et al. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol 2011; 187:1609–16. [DOI] [PubMed] [Google Scholar]
- 43.Iqbal SM, Leonard C, Regmi SC, De Rantere D, Tailor P, Ren G, et al. Lubricin/Proteoglycan 4 binds to and regulates the activity of Toll-Like Receptors In Vitro. Sci Rep 2016; 6:18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berroth A, Kuhnl J, Kurschat N, Schwarz A, Stab F, Schwarz T, et al. Role of fibroblasts in the pathogenesis of atopic dermatitis. J Allergy Clin Immunol 2013; 131:1547–54. [DOI] [PubMed] [Google Scholar]
- 45.Steinhoff M, Steinhoff A, Homey B, Luger TA, Schneider SW. Role of vasculature in atopic dermatitis. J Allergy Clin Immunol 2006; 118:190–7. [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Farinas M, Tintle SJ, Shemer A, Chiricozzi A, Nograles K, Cardinale I, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol 2011; 127:954–64 e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berdyshev E, Goleva E, Bronova I, Dyjack N, Rios C, Jung J, et al. Lipid abnormalities in atopic skin are driven by type 2 cytokines. JCI Insight 2018; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voegeli R, Rawlings AV, Breternitz M, Doppler S, Schreier T, Fluhr JW. Increased stratum corneum serine protease activity in acute eczematous atopic skin. Br J Dermatol 2009; 161:70–7. [DOI] [PubMed] [Google Scholar]
- 49.Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol 2009; 124:1235–44 e58. [DOI] [PubMed] [Google Scholar]
- 50.Kim BE, Leung DY, Boguniewicz M, Howell MD. Loricrin and involucrin expression is down-regulated by Th2 cytokines through STAT-6. Clin Immunol 2008; 126:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howell MD, Fairchild HR, Kim BE, Bin L, Boguniewicz M, Redzic JS, et al. Th2 cytokines act on S100/A11 to downregulate keratinocyte differentiation. J Invest Dermatol 2008; 128:2248–58. [DOI] [PubMed] [Google Scholar]
- 52.Morizane S, Yamasaki K, Kajita A, Ikeda K, Zhan M, Aoyama Y, et al. TH2 cytokines increase kallikrein 7 expression and function in patients with atopic dermatitis. J Allergy Clin Immunol 2012; 130:259–61 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danso MO, van Drongelen V, Mulder A, van Esch J, Scott H, van Smeden J, et al. TNF-alpha and Th2 cytokines induce atopic dermatitis-like features on epidermal differentiation proteins and stratum corneum lipids in human skin equivalents. J Invest Dermatol 2014; 134:1941–50. [DOI] [PubMed] [Google Scholar]
- 54.Bernard FX, Morel F, Camus M, Pedretti N, Barrault C, Garnier J, et al. Keratinocytes under Fire of Proinflammatory Cytokines: Bona Fide Innate Immune Cells Involved in the Physiopathology of Chronic Atopic Dermatitis and Psoriasis. J Allergy (Cairo) 2012; 2012:718725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di ZH, Ma L, Qi RQ, Sun XD, Huo W, Zhang L, et al. T Helper 1 and T Helper 2 Cytokines Differentially Modulate Expression of Filaggrin and its Processing Proteases in Human Keratinocytes. Chin Med J (Engl) 2016; 129:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honzke S, Wallmeyer L, Ostrowski A, Radbruch M, Mundhenk L, Schafer-Korting M, et al. Influence of Th2 Cytokines on the Cornified Envelope, Tight Junction Proteins, and ss-Defensins in Filaggrin-Deficient Skin Equivalents. J Invest Dermatol 2016; 136:631–9. [DOI] [PubMed] [Google Scholar]
- 57.Mashiko S, Mehta H, Bissonnette R, Sarfati M. Increased frequencies of basophils, type 2 innate lymphoid cells and Th2 cells in skin of patients with atopic dermatitis but not psoriasis. J Dermatol Sci 2017; 88:167–74. [DOI] [PubMed] [Google Scholar]
- 58.Leung DY. Atopic dermatitis: the skin as a window into the pathogenesis of chronic allergic diseases. J Allergy Clin Immunol 1995; 96:302–18; quiz 19. [DOI] [PubMed] [Google Scholar]
- 59.Sugaya M, Miyagaki T, Ohmatsu H, Suga H, Kai H, Kamata M, et al. Association of the numbers of CD163(+) cells in lesional skin and serum levels of soluble CD163 with disease progression of cutaneous T cell lymphoma. J Dermatol Sci 2012; 68:45–51. [DOI] [PubMed] [Google Scholar]
- 60.Chu CC, Di Meglio P, Nestle FO. Harnessing dendritic cells in inflammatory skin diseases. Semin Immunol 2011; 23:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kasraie S, Werfel T. Role of macrophages in the pathogenesis of atopic dermatitis. Mediators Inflamm 2013; 2013:942375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiekens RC, Thepen T, Oosting AJ, Bihari IC, Van De Winkel JG, Bruijnzeel-Koomen CA, et al. Heterogeneity within tissue-specific macrophage and dendritic cell populations during cutaneous inflammation in atopic dermatitis. Br J Dermatol 2001; 145:957–65. [DOI] [PubMed] [Google Scholar]
- 63.He R, Oyoshi MK, Jin H, Geha RS. Epicutaneous antigen exposure induces a Th17 response that drives airway inflammation after inhalation challenge. Proc Natl Acad Sci U S A 2007; 104:15817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon J, Leyva-Castillo JM, Wang G, Galand C, Oyoshi MK, Kumar L, et al. IL-23 induced in keratinocytes by endogenous TLR4 ligands polarizes dendritic cells to drive IL-22 responses to skin immunization. J Exp Med 2016; 213:2147–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol 2008; 128:2625–30. [DOI] [PubMed] [Google Scholar]
- 66.Nograles KE, Zaba LC, Shemer A, Fuentes-Duculan J, Cardinale I, Kikuchi T, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol 2009; 123:1244–52 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arkwright PD, Chase JM, Babbage S, Pravica V, David TJ, Hutchinson IV. Atopic dermatitis is associated with a low-producer transforming growth factor beta(1) cytokine genotype. J Allergy Clin Immunol 2001; 108:281–4. [DOI] [PubMed] [Google Scholar]
- 68.Bos JD, Hagenaars C, Das PK, Krieg SR, Voorn WJ, Kapsenberg ML. Predominance of “memory” T cells (CD4+, CDw29+) over “naive” T cells (CD4+, CD45R+) in both normal and diseased human skin. Arch Dermatol Res 1989; 281:24–30. [DOI] [PubMed] [Google Scholar]
- 69.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Whynot J, Novitskaya I, Cardinale I, et al. Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. J Allergy Clin Immunol 2007; 119:1210–7. [DOI] [PubMed] [Google Scholar]
- 70.Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, et al. Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol 2014; 193:3717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mihm MC Jr., Soter NA, Dvorak HF, Austen KF. The structure of normal skin and the morphology of atopic eczema. J Invest Dermatol 1976; 67:305–12. [DOI] [PubMed] [Google Scholar]
- 72.Choy DF, Hsu DK, Seshasayee D, Fung MA, Modrusan Z, Martin F, et al. Comparative transcriptomic analyses of atopic dermatitis and psoriasis reveal shared neutrophilic inflammation. J Allergy Clin Immunol 2012; 130:1335–43 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savinko T, Matikainen S, Saarialho-Kere U, Lehto M, Wang G, Lehtimaki S, et al. IL-33 and ST2 in atopic dermatitis: expression profiles and modulation by triggering factors. J Invest Dermatol 2012; 132:1392–400. [DOI] [PubMed] [Google Scholar]
- 74.Totsuka A, Omori-Miyake M, Kawashima M, Yagi J, Tsunemi Y. Expression of keratin 1, keratin 10, desmoglein 1 and desmocollin 1 in the epidermis: possible downregulation by interleukin-4 and interleukin-13 in atopic dermatitis. Eur J Dermatol 2017; 27:247–53. [DOI] [PubMed] [Google Scholar]
- 75.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, Debenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol 2007; 120:150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyazaki Y, Satoh T, Nishioka K, Yokozeki H. STAT-6-mediated control of P-selectin by substance P and interleukin-4 in human dermal endothelial cells. Am J Pathol 2006; 169:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sigurdsson V, de Vries IJ, Toonstra J, Bihari IC, Thepen T, Bruijnzeel-Koomen CA, et al. Expression of VCAM-1, ICAM-1, E-selectin, and P-selectin on endothelium in situ in patients with erythroderma, mycosis fungoides and atopic dermatitis. J Cutan Pathol 2000; 27:436–40. [DOI] [PubMed] [Google Scholar]
- 78.Laberge S, Ghaffar O, Boguniewicz M, Center DM, Leung DY, Hamid Q. Association of increased CD4+ T-cell infiltration with increased IL-16 gene expression in atopic dermatitis. J Allergy Clin Immunol 1998; 102:645–50. [DOI] [PubMed] [Google Scholar]
- 79.Fujimura T, Okuyama R, Ito Y, Aiba S. Profiles of Foxp3+ regulatory T cells in eczematous dermatitis, psoriasis vulgaris and mycosis fungoides. Br J Dermatol 2008; 158:1256–63. [DOI] [PubMed] [Google Scholar]
- 80.Szegedi A, Barath S, Nagy G, Szodoray P, Gal M, Sipka S, et al. Regulatory T cells in atopic dermatitis: epidermal dendritic cell clusters may contribute to their local expansion. Br J Dermatol 2009; 160:984–93. [DOI] [PubMed] [Google Scholar]
- 81.Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, et al. Developmental cell programs are co-opted in inflammatory skin disease. Science 2021; 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bangert C, Rindler K, Krausgruber T, Alkon N, Thaler FM, Kurz H, et al. Persistence of mature dendritic cells, TH2A, and Tc2 cells characterize clinically resolved atopic dermatitis under IL-4Ralpha blockade. Sci Immunol 2021; 6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


