Abstract
In this article, a review of a series of applications of atomic force microscopy (AFM) and fluidic Atomic Force Microscopy (fluidic AFM, hereafter fluidFM) in single-cell studies is presented. AFM applications involving single-cell and extracellular vesicle studies, colloidal force spectroscopy, and single-cell adhesion measurements are discussed. FluidFM is an offshoot of AFM that combines a microfluidic cantilever with AFM and has enabled the research community to conduct biological, pathological, and pharmacological studies on cells at the single-cell level in a liquid environment. In this review, capacities of fluidFM are discussed to illustrate (1) the speed with which sequential measurements of adhesion using coated colloid beads can be done, (2) the ability to assess lateral binding forces (LBFs) of endothelial or epithelial cells in a confluent cell monolayer in an appropriate physiological environment, and (3) the ease of measurement of vertical binding forces (VBFs) of intercellular adhesion between heterogeneous cells. Furthermore, key applications of fluidFM are reviewed regarding to extracellular vesicle absorption, manipulation of a single living cell by intracellular injection, sampling of cellular fluid from a single living cell, patch clamping, and mass measurements of a single living cell.
Keywords: Fluidic atomic force microscopy, atomic force microscopy, single living cell, extracellular vesicle
1. Introduction
Cell biomechanical properties play an important role in regulating cellular activities, such as cell adhesion, migration, and barrier functions, and are related to intracellular structures, signal transductions, biochemical pathways, and metabolic functions. Modeling host-pathogen interactions in the cell surface microenvironment requires a platform in which to explore single cells if we are to gain deeper insights into the mechanisms that underlie infection and inflammation at the nanoscale level(Mager, LaPointe, & Stevens, 2011).
Invented by Binnig et al. in 1986(Binnig, Quate, & Gerber, 1986), atomic force microscopy (AFM) has emerged as an indispensable technique for the study of the biomechanical features of the cell surface in real time(Bhat, Price, & Dahms, 2021). AFM measures the force dynamics between a probe tip and a sample at the nanoscale level. AFM is capable of probing surface biophysical aspects of single cells, including those of microbes(Alsteens, Beaussart, El-Kirat-Chatel, Sullan, & Dufrêne, 2013), by measuring the interacting force between the probe and cell surface. Unlike other microscopy techniques, AFM captures high-resolution, three-dimensional (3-D) images of the cell surface without special sample preparation such as fixation, staining, or labeling, and therefore can be applied to models of living cells(Alsteens et al., 2013). Furthermore, the probe tips can be functionalized with various biochemical materials, including recombinant proteins(Dufrêne, 2008), antibodies(Dufrêne, 2008), and cultured cells(Friedrichs, Helenius, & Muller, 2010). This affords AFM the unique capacity to directly measure protein-protein and protein-cell interacting forces at the single molecule or single-cell level(Dufrêne, 2008; Friedrichs et al., 2010), thus allowing for the biomechanical characterization of target-specific cell surfaces.
In 2009, Zambelli’s group introduced the fluidic force microscope (fluidic AFM, hereafter fluidFM)(André Meister et al., 2009) that employed a micromachined fluidFM cantilever to the conventional AFM system. They proposed its use in biological applications and demonstrated the capacity of intracellular injections. Afterwards, such applications have been extended to various cell biological studies, including single-cell injection, agent and biomolecule delivery, single-cell content extractions, and patch clamping, and single-cell adhesion measurements, etc.
In this review, we first explain a transferring from AFM to the fluidFM for the application in cell biology studies. Then, we focus on the unique applications of fluidFM, which are designed to investigate the biomechanical mechanisms underlying the initiation of intracellular pathogen infection and host responses, biophysical characterization of single living leukocytes or bacterial adhesion to endothelial cells, and single living cell-virion binding force measurements, and the biomechanical nature of endothelial or epithelial cell barrier function. Multiple novel technologies using fluidFM in the cell biology have been summarized, including manipulation of single living cells by intracellular injection, sampling of cellular fluid from a single living cell, and single living cell mass measurements(Aebersold et al., 2015; Amarouch, El Hilaly, & Mazouzi, 2018; Cohen, Sarkar, Hondroulis, Sabhachandani, & Konry, 2017; Pablo Dörig et al., 2010; Guillaume-Gentil et al., 2014; A. Meister et al., 2009; Nagy et al., 2019). Meanwhile, a faster method to sequentially measure adhesion with coated colloid beads using fluidFM is mentioned(Gerecsei et al., 2019). Assessment of lateral binding forces (LBFs) of endothelial cells (ECs) on a mature monolayer, intercellular vertical binding forces (VBFs) between heterogeneous cells, and the potentials of the applications of AFM and fluidFM in the field of extracellular vesicle (EV) are explained.
2. From AFM to fluidFM
2.1. Fundamentals of fluidFM
The principle of fluidFM is shown in Fig. 1. The basic components of the fluidFM unit are the same as those of an ordinary AFM system, which is commonly composed of a piezoelectric tube driver that is connected to a probe, a laser beam, a four-quadrant, position-sensitive detector (PSD), a signal processing module, and the feedback control electronics. The fluidic part is composed of a fluidic microcantilever, connecting tubing, and a fluid pressure controller. Thus, a fluidFM system has a continuous and closed fluidic channel that can be filled with an arbitrarily chosen liquid that can be locally dispensed through an aperture at the end of the cantilever(A. Meister et al., 2009).
Fig. 1:
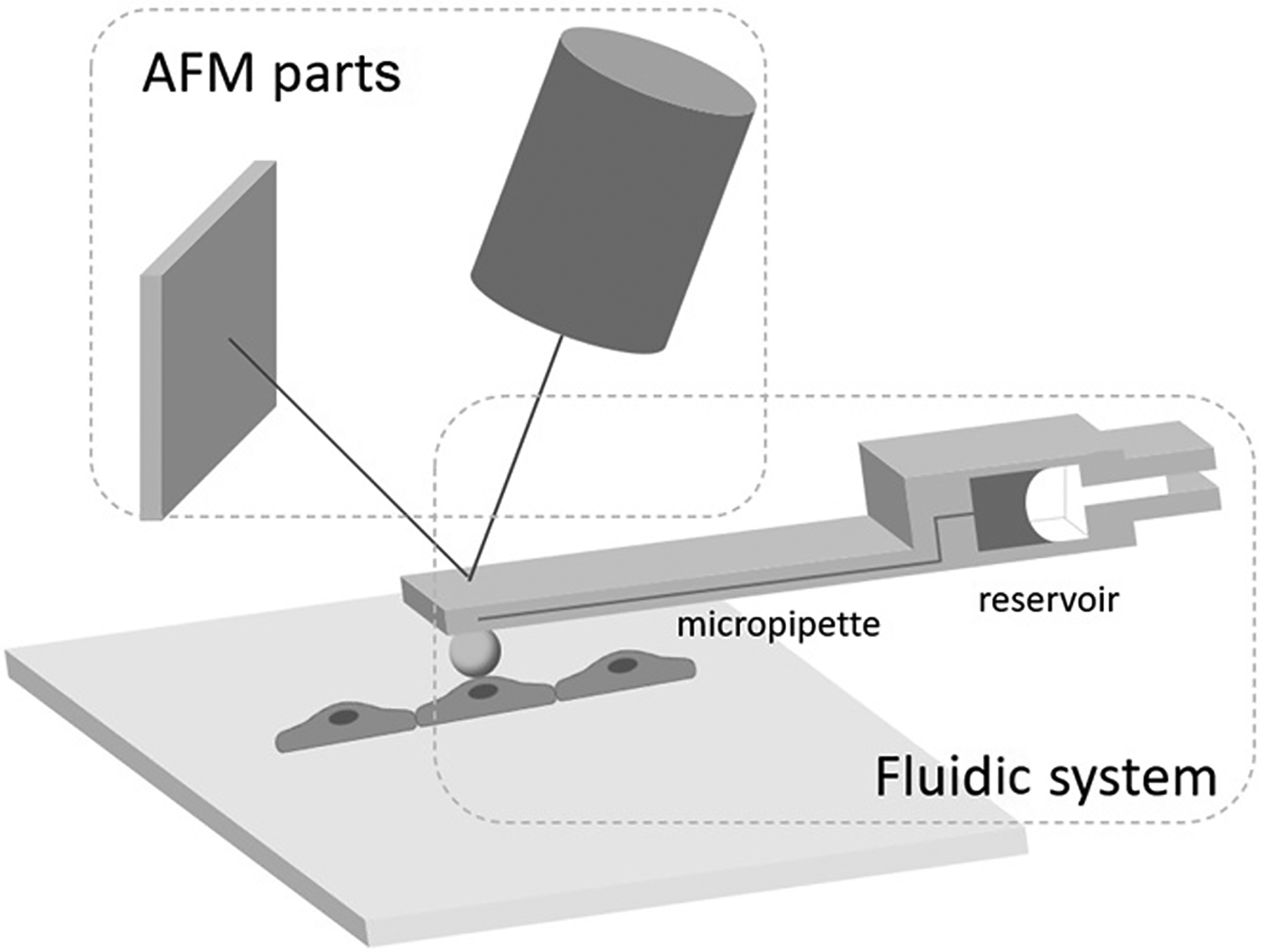
A graphic representation of a fluidFM system designed to measure the adhesion force between a coated microsphere and a target cell. The coated microsphere can be replaced with another coated microbead or a cell depending on the objective of the experiment.
2.2. Fluidic microcantilever - the key component of fluidFM
Glass micropipettes have been used in a number of applications in biology, such as intracellular injection(Graessmann & Graessman, 1976) and patch clamping for electrophysiology measurements(Hamill, Marty, Neher, Sakmann, & Sigworth, 1981). Near-field scanning optical microscopy (NSOM), first reported in 1983(Betzig, Lewis, Harootunian, Isaacson, & Kratschmer, 1986; A Lewis, Isaacson, Muray, & Harootunian, 1983), made use of micropipettes in a scanning arrangement. The pipettes were later combined with scanning probe microscopes such as AFM(Lieberman et al., 1994) and scanning tunneling microscopes(Lieberman & Lewis, 1993). Glass micropipette thermocouples were constructed and used with AFM for nanoscale temperature measurements(Fish et al., 1995). Glass micropipettes of nanomolar size apertures combined with AFM were also used for nanoscale liquid and gaseous material delivery(Aaron Lewis et al., 1999) and protein printing (Taha et al., 2003). Francis et al. in 1987(Francis, Fisher, Gamble, & Gingell, 1987) measured single cell adhesion by applying suction with a glass micropipette and tracking the movement with an interference reflection microscope (which is similar to an AFM). Other detection schemes (non-AFM based) with glass micropipettes have been used for cell adhesion measurements(Bowers, Fisher, Francis, & Williams, 1989; Chesla, Selvaraj, & Zhu, 1998; Moussy, Neumann, & Zingg, 1990; Palmer et al., 2008). Glass micropipettes are pulled individually using a mechanical pipette puller and heated in a serial process that is relatively cumbersome and often does not produce uniform results.
Computer-controlled micropipette (CCMP) can manipulate and sort cells in a Petri dish, individually(Lomakina & Waugh, 2004). Cells are selected on the basis of their phase contrast and/or fluorescent images(Rita Ungai-Salánki et al., 2019). Sorting is performed by a micropipette with an aperture of 10–70 μm with a sorting speed of 3–4 cell/min(Rita Ungai-Salánki et al., 2019). After sorting, single cells are deposited into another Petri dish/multiwall plate/PCR tube or glass cover slip in the nL to μL range(Környei et al., 2013). Individual cells inside the drops on the glass cover slip can be studied with high resolution, immediately after sorting(Salánki et al., 2014). The technique is suitable for high throughput single-cell adhesion force measurements by repeating the pick-up process with an increasing vacuum(Jani et al., 2016). Furthermore, the adhesion force of cells attached to specific molecular surfaces can be probed when cells grow on the molecules-coated surface(Jani et al., 2016).
Micromachined (micro-electromechanical system, MEMS) fluidic cantilevers can be fabricated for fluidFM applications. Compared to glass pipettes that are individually produced by pipette pullers, MEMS cantilevers are mass-produced using microfabrication techniques with minimal variations between devices, overcoming throughput and non-uniformity issues. In addition, micromachining offers greater control over the size and location of the aperture(Deladi et al., 2005; Deladi et al., 2004; Hug, Biss, De Rooij, & Staufer, 2005; Kim, Ke, Moldovan, & Espinosa, 2003).
The use of fluidFM cantilevers overcomes many issues encountered with conventional AFM cantilevers. For example, in conventional AFM single-cell force spectroscopy, cells are irreversibly attached to an AFM cantilever via biochemical functionalization(Friedrichs et al., 2010), which is labor-intensive and time consuming. In addition, single-cell force spectroscopy requires that the cell-cantilever coupling force is stronger than the adhesion interaction force to be detected. The biochemical adhesive-mediated attachment of cells to the conventional AFM cantilever may not be strong enough(Guillaume-Gentil et al., 2014). Consequently, cell-to-cell binding force measurements become challenging, particularly when probing the lateral forces between cells in a monolayer(Mathieu, Martin-Jaular, Lavieu, & Théry, 2019). Furthermore, biochemical-mediated immobilization techniques may perturb cells(Friedrichs et al., 2010). Using the fluidFM technique(A. Meister et al., 2009), it is relatively easy to attach a cell by applying negative pressure in the microchannel of the micropipette and bring it into contact with other cells or functionalized surfaces to measure adhesion(Wysotzki, Sancho, Gimsa, & Groll, 2020). Importantly, fluidFM cantilevers can be reused.
Available fluidic cantilevers are either tipless (with a micron-size aperture) or have a pyramid-shaped tip (with an aperture of a few hundred nanometers in diameter located on the side or the apex of the tip)(Guillaume-Gentil et al., 2014). Typically the nanometer-sized opening is fabricated using focused ion beam technology(Álvarez-Asencio, Thormann, & Rutland, 2013). The channel height ranges from 0.2 to 1 μm, and the fluidic channel on the cantilever chip is connected to a reservoir. Tipless cantilevers are utilized in applications such as exchangeable colloidal force spectroscopy(Dehullu et al., 2019; Su et al., 2021), spatial manipulation of a targeted cell(V. Martinez et al., 2016), and single-cell or cell-to-cell binding force measurements(Vincent Martinez, Pascal Behr, et al., 2016). The cantilevers with pyramid-shaped tips are mainly employed in applications that require delivery of biomolecules and sampling(Guillaume-Gentil et al., 2016; Guillaume-Gentil et al., 2013). For dispensing material onto the apical surface of a cell, a tip with an aperture at the apex is commonly used. For intracellular experiments, an aperture on the side of the pyramidal-shaped tips(Guillaume-Gentil et al., 2014) is preferred to deliver or extract loads from a cell’s plasma membrane or the cell’s nucleus.
Various research groups have reported on the development of novel devices that have not been commercialized. Inspired by NSOM and fountain-pen lithography, Meister et al.(Meister et al., 2003) developed a nanoscale dispensing fluidic probe that includes a hollow SixNy tip on a Si–SixNy cantilever. Hug et al. reported a fluidic cantilever entirely made of silicon oxide(Hug et al., 2005); a version of this device was later used in fluidic force microscope work(André Meister et al., 2009). A hollow silicon nitride (Si3N4) tip on a silicon dioxide (SiO2) fluidic cantilever and an array of these devices was developed for high throughput applications(van Oorschot, Garza, Derks, Staufer, & Ghatkesar, 2015). A silicon nitride fluidic cantilever without a tip was reported by Schön et al.(Schön, Geerlings, Tas, & Sarajlic, 2013) and in a later effort, a tip with a submicron aperture was included(Verlinden et al., 2020). Other efforts included the use of flexible materials like SU-8 as a cantilever material(Angelo Gaitas & Hower, 2014; Han et al., 2018; Vincent Martinez, Pascal Behr, et al., 2016; Vincent Martinez, Csaba Forró, et al., 2016). Several of the fluidic cantilevers also included embedded sensing elements, eliminating the need for an AFM laser for deflection detection(Angelo Gaitas & Hower, 2014; Han et al., 2020). In a recent effort, a fluidFM cantilever was fabricated using 3-D printing(Kramer et al., 2020).
2.3. FluidFM printing, colloidal, and cell adhesion
Micropatterning of living single cells and cell clusters is reported in the field of cell-based biosensors(Hynes et al., 2014). This approach requires both a suitable biomimetic support and a printing technology. Saftics et al. presented the micropatterning of living mammalian cells on carboxymethyl dextran (CMD) hydrogel layers using the robotic fluidFM (FluidFM BOT) technology, developing CMD layers in order to provide support for the adhesion of living cells(Saftics et al., 2019).
Polymer or protein adsorption are often measured by colloidal particle deposition. Force measurements on microbeads by AFM or optical tweezers are conventional options in molecular biophysics, although throughput is low(Gerecsei et al., 2019). Washing and centrifuge assays with (bio)chemically decorated microbeads provide better statistics, but only qualitative results to be collected(Gerecsei et al., 2019). Gerecsei et al. demonstrated that a CCMP is a straightforward and high-throughput alternative to quantify the surface adhesion of functionalized microparticles, measuring the binding forces of the microbeads with both FluidFM BOT and CCMP(Gerecsei et al., 2019).
Adhesion of tumor cells is a potential therapeutic target(R. Ungai-Salánki et al., 2021). Ungai-Salánki et al. investigated the integrin-mediated adhesion between HeLa cells and the tripeptide Arg-Gly-Asp motif, using CCMP and a high spatial resolution, label-free resonant waveguide grating-based optical sensor at the single-cell level(R. Ungai-Salánki et al., 2021). They found that the overall binding force of single cells is approximately constant in all phases except the mitotic phase with a significantly lower adhesion and the cell material mass per unit area inside the cell-substratum contact zone is significantly less at this phase, which leads to the conclusion that the weaker mitotic adhesion is not simply a direct consequence of the measured smaller contact area(R. Ungai-Salánki et al., 2021). Sztilkovics et al. presented a high spatial and temporal resolution resonant waveguide grating based label-free optical biosensor which was combined with FluidFM BOT to measure the adhesion of living cells(Sztilkovics et al., 2020). In contrast to traditional fluidFM, the FluidFM BOT can address single cells over mm-cm scale areas(Sztilkovics et al., 2020). Thus, this feature significantly increased measurement throughput and the capacities to couple other technologies using microplate-based and large area biosensor(Sztilkovics et al., 2020).
3. AFM and fluidic AFM for EV analysis
Cell-to-cell communication is critical for maintaining mammalian homeostasis and responding quickly to environmental stimuli, including pathogens. Besides direct intercellular contact, this communication is often mediated by soluble factors that can convey signals to a large repertoire of responding cells, either locally or remotely. Extracellular vesicles (EVs) are membrane-enveloped vesicles and are naturally released from almost all mammalian cells(Schorey & Harding, 2016). They ferry various biological cargos, including proteins, multiple RNA species, and DNAs, and transfer these potential functional mediators to neighboring and distant recipient cells, forming a novel mode of cell-to-cell communication and contributing to changes in cellular function in health and disease(Bhatnagar et al., 2007; Coelho et al., 2019; Davis et al., 2019; Garcia-Martin et al., 2022; Hui et al., 2018; Jones et al., 2018; L. Li et al., 2018; Nandakumar et al., 2019; Petrov et al., 2017; Williams et al., 2012). EVs are broadly classified into two categories, exosomes (50–150 nm) and microvesicles (100–1000 nm), distinguished by the cell membrane of origin(Mathieu et al., 2019). Exosomes and microvesicles are also termed small and large EVs, respectively(Crescitelli et al., 2020; Lázaro-Ibáñez et al., 2019; Takov, Yellon, & Davidson, 2019; Temoche-Diaz et al., 2019). After the membrane of the late endosome buds inward, exosomal biogenesis begins with the formation of intraluminal vesicles, which are the intracellular precursors of exosomes(Jones et al., 2018; Mathieu et al., 2019). Before they are released into the extracellular environment as exosomes, the intraluminal vesicles are internalized into multivesicular bodies, transported inward, and fuse with the plasma membrane(Jones et al., 2018; Mathieu et al., 2019; Meldolesi, 2018). Microvesicles are rapidly generated at the plasma membrane by outward budding(Jones et al., 2018; Mathieu et al., 2019; Meldolesi, 2018; Williams et al., 2012). EVs contain many types of biomolecules, such as proteins and nucleic acids. Exosomes can convey signals to a large repertoire of recipient cells either locally or remotely by transferring functional cargos, thus contributing to disease pathogenesis, including infection and inflammation(Bhatnagar et al., 2007; Coelho et al., 2019; Davis et al., 2019; Hui et al., 2018; Jones et al., 2018; L. Li et al., 2018; Nandakumar et al., 2019; Petrov et al., 2017; Williams et al., 2012). EVs inherit cell-type surfaces from their parent cells due to the nature of the endogenous plasma membrane. As a result, different cell-derived exosomes have different cell-type tropisms(Cagno, 2020; Miner & Diamond, 2017). Therefore, direct evidence from a validated single living cell model is crucial for EV biology studies.
Differential ultracentrifugation was historically employed for exosome isolation, but it suffers from aggregation issues and decreased integrity of exosomes after resuspension(Böing et al., 2014; Davis et al., 2019; Lobb et al., 2015; Meldolesi, 2018; Stranska et al., 2018). It is essential to characterize the size and morphology of isolated particles because both quality and quantity are crucial for the outcome of downstream assays involving the functional roles of EVs and their contents. Small EVs have a diameter of less than 200 nm, which prevents the use of optical microscopies for single particle characterization(LeClaire, Gimzewski, & Sharma, 2021). AFM and electron microscopy (EM) are the two nanoscale methods of choice to image and study EVs.
Tapping mode high resolution AFM to image EVs is a label-free and relatively quick technique that does not involve complicated sample preparation. Tapping mode AFM provides a 3-D image of surface structures, including height image or deflection image(Cheng, Nonaka, & Wong, 2019; Sharma, LeClaire, Wohlschlegel, & Gimzewski, 2020; Zhou, Weber, Zhao, Chen, & Sundstrom, 2020), and is commonly used to evaluate the integrity of EVs at the single particle level. Using size-exclusion chromatography (SEC), small EVs (50–150 nm) were purified from Rickettsia-infected mouse plasma and culture media of primary vascular endothelial cells (ECs). Evaluation of single particle morphology using tapping mode AFM images verified the integrity of isolated exosomes from experimental specimens(Liu et al., 2021).
Using AFM, researchers are able to focus on both the biochemical assessment of an EV particle surface(Sharma et al., 2010) and are able to quantify the surface biophysical characteristics of EVs at the single particle level(Bairamukov et al., 2020; M. I. Li et al., 2021; Ridolfi et al., 2020; Sharma et al., 2020), beyond merely assessing the size and counting the particle number. Sharma et al. used force spectroscopy with AFM tips functionalized with anti-CD63 IgGs, and reported evidence of the presence of tetraspanin CD63, an endosomal marker(van Niel et al., 2011), on the exosome surface, directly suggesting an endosomal origin of exosomes instead of a plasma membrane origin(Sharma et al., 2010). By simultaneously acquiring high-resolution tapping mode AFM scanning images combined with force spectroscopy in a liquid environment, the analysis of the mechanical properties of a single EV provides further insights into the biophysical changes between EV subgroups(Bairamukov et al., 2020). AFM offers features that enable the standardization of the functional analysis of EVs, such as label-free quantitative biomechanical profiles that address the regulation of EV uptake in recipient cells(LeClaire et al., 2021; Ma et al., 2016).
3.1. Example of mechanical characterization of EVs
3.1.1. Nanoindentation of EVs using AFM
Nanoindentation is now widely recognized as the method of choice for testing the mechanical properties of thin films and surfaces(van Rosmalen, Roos, & Wuite, 2015). In such experiments, images of the vesicles are first made to characterize the geometry of the vesicles and the location of their centers. The force distance curve on the substrate next to the vesicle is recorded before the vesicle is indented to demonstrate the linear response of the clean tip and cantilever. Nanoindentation proceeds at a slower speed, which mainly results in elastic responses and a better signal-to-noise ratio compared to force distance curves recorded during imaging. Repeated small indents on the same vesicle often produce quantitatively reproducible behavior. Deeper indentation may lead to vesicle damage, such as membrane rupture, which may lead to different responses upon repeated indentation. The indentation of different vesicles is less similar because indentation behavior depends on vesicle size and extent of diffusion, which can vary from vesicle to vesicle(Vorselen, Piontek, Roos, & Wuite, 2020).
3.2. Identification of recipient cell-type tropism of EVs at single living cell level using fluidFM
Transmembrane proteins from parent cells are present on both small and large EVs, while cytosolic proteins and genetic material are contained within the lipid bilayer membrane, which facilitates transportation to remote recipient cells without a loss of bioactivity(Carnino & Jin, 2020; Stahl & Raposo, 2019). There is growing evidence that a variety of cells can serve as parent cells of EVs. EVs have been recognized as cell-type biomarkers because they maintain the same topology of transmembrane proteins as the parent cell plasma membrane(Yáñez-Mó et al., 2015). Once docked on the recipient cell apical surface, the adhesion and internalization are processed in a receptor-dependent manner, in which membrane fusion and endocytosis occur(Mathieu et al., 2019). The level of uptake is proposed to be dependent on the recipient cell type because the nature of the endogenous membrane-derived surface enables exosomes to inherit cell-type surfaces from their parent cells, posing different affinities to different cell types(Chivet et al., 2014; Montecalvo et al., 2012). For example, exosomes from primary neurons are only taken up by other neurons, whereas those from a neuroblastoma cell line bind equally to astrocytes(Chivet et al., 2014). Similarly, bone marrow dendritic cell exosomes were preferentially captured by splenic dendritic cells, rather than by B or T cells(Montecalvo et al., 2012). Exosomes from oligodendroglia precursor cells were taken up by microglia but not by neurons or astrocytes. As a result, different cell-derived exosomes have different cell-type tropisms, potentially like some emerging viruses(Cagno, 2020; Miner & Diamond, 2017). Therefore, direct evidence from a single living cell model is crucial to help identify the cell type of a recipient cell and characterize the mechanism of the cell-type tropisms of EVs. The identification of cell-type tropism of EVs at the level of a single living cell is implementable due to the robustness of fluidic AFM. Since fluidic AFM combines ordinary AFM with microchannel and pressure control into an entire complex, it provides the capability to assess purified cell-type EVs in various cell models at the level of a single cell. Fluidic AFM provides the means to manipulate and dispense dose-dependent EVs, allowing for the assessment of EV uptake by various cell types.
3.3. Single EV analysis using Fluidic AFM
A recent paper describes a single EV analysis technique under light microscopy that enables robust, multiplexed protein biomarker measurements in single EVs(K. Lee et al., 2018). In this method, EVs are immobilized in a microfluidic chamber for immunostaining and imaging. When vesicles are immobilized on the chip surface, the achievable signal-to-noise ratio per vesicle is typically much higher than when the vesicles are free-floating in solution or under flow conditions, indicating better signal quality.
The authors further formalized a graphical cycling program for multiplexed cycling cell and tissue analysis to complement the analysis of nanoscale EVs(Schubert et al., 2006). In detail, the authors experimented with this approach using EVs derived from three isogenic glioblastoma multiforme cell lines. EVs were stained for three protein markers at a time and imaged for four cycles. The results showed a high degree of heterogeneity of biomarkers in the EV population. Data were then visualized using multidimensional data analysis, namely t-distributed stochastic neighborhood embedding (t-SNE); unsupervised clustering revealed the presence of a potential subset of EVs. The results demonstrate the versatility of single EV technology in studying various EV types, and the method generated rich datasets on biomarker expression heterogeneity, marker composition, and the presence of EV subsets(Lin, Fallahi-Sichani, & Sorger, 2015).
4. AFM and fluidFM for single-cell analysis
The dynamics of EV biogenesis and absorption in recipient cells are complex and variable, mostly dominated by cell type and cell state. Conventional methods for EV characterization, which was based on measurements of pooled EV samples, masked the cell-to-cell heterogeneity in EV secretion and absorption(Ji et al., 2019). EV secretion increased proportionally to the number of neighboring cells, suggesting that cell-to-cell interplay may affect EV secretion in paracrine signaling(Ji et al., 2019). Therefore, studying EVs at the single-cell level enables a precise characterization of the heterogeneities of EV biogenesis, secretion, and absorption. AFM and fluidFM provide a single-cell platform that addresses the shortcomings of conventional techniques that rely on pooling samples.
4.1. Exchangeable colloidal fluidFM force spectroscopy
To minimize potential mechanical perturbations on a target cell, AFM cantilevers with micrometer-level size spherical colloids are employed in place of a sharp tip for cell surface nanomechanics. Typical applications of colloidal probes (Fig. 2) include identification of specific biomolecules on targeted individual cell surfaces at the single molecule level(Viljoen et al., 2021), adhesion measurement(Kappl & Butt, 2002; Kuznetsov & Papastavrou, 2012a),(Borkovec et al., 2012), and the study of mechanical properties(Chyasnavichyus, Young, Geryak, & Tsukruk, 2016; McConney, Singamaneni, & Tsukruk, 2010). The procedure to attach the colloidal probe to a microcantilever has not seen significant advancement in the past decades and entails irreversibly immobilizing a spherical particle to the end of a tipless cantilever(Gan, 2007; Kuznetsov & Papastavrou, 2012b; Yuan, Zhang, & Gan, 2017). To obtain a dataset of sufficient statistical rigor requires the attachment of many colloidal probes to different cantilevers. Functionalization of a colloid to a cantilever allows for probing of only one target. Thus, a new conventional AFM cantilever-colloid probe is required to measure different target cells or different receptors, increasing the cost and labor. Also, calibration is required every time a cantilever is exchanged. Furthermore, differences in the mechanical properties between cantilevers that stem from variations in the fabrication process induce potential challenges to comparing results from different cantilevers.
Fig. 2:
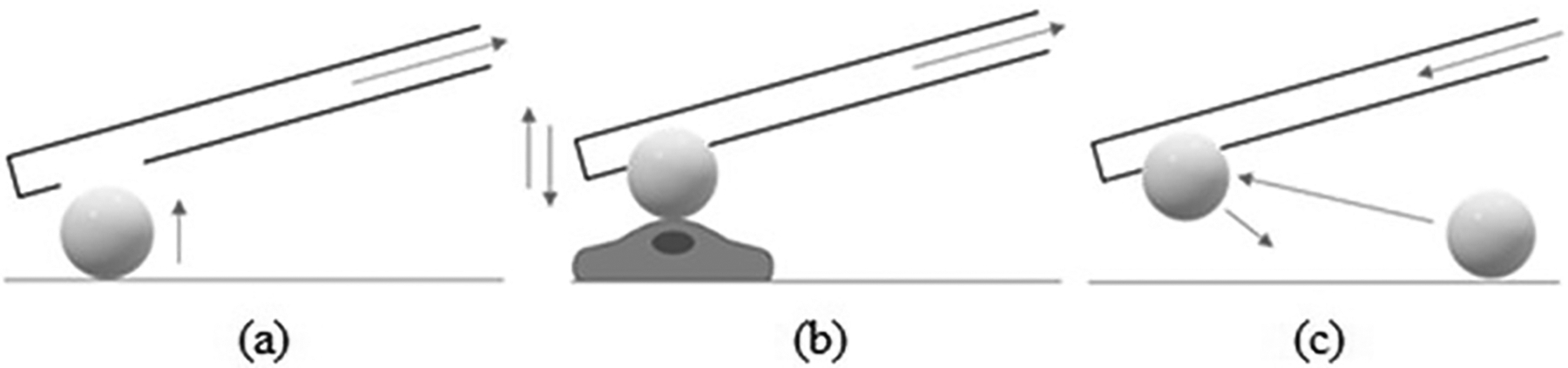
(a) Negative pressure is applied within the microchannel of the cantilever to absorb the bead to the cantilever aperture. (b) The binding force between the bead and the apical surface of the cell is measured by force spectroscopy analysis. (c) Pressure on the microchannel is adjusted to replace the bead.
Unlike conventional AFM cantilevers, fluidFM micropipette cantilevers are reusable. It is possible to probe different targets by functionalizing and replacing microbeads(P. Dörig et al., 2013; Mittelviefhaus, Müller, Zambelli, & Vorholt, 2019). Microbeads are functionalized by mixing with reagents following standard protocols. Microbeads are easily exchanged by applying negative and positive pressure using a fluidic pressure controller (Fig. 2). FluidFM force spectroscopy provides a measure of the binding force between the bead and the apical surface of the cell (i.e., force-distance curve). Using exchangeable colloids and fluidFM force spectroscopy, we recently reported that the intracellular cyclic adenosine monophosphate receptor EPAC1 modulates rickettsial adhesion on host cell surfaces in association with Y23 phosphorylation of the bacterial binding receptor Annexin A2(Su et al., 2021). An example of fast serial adhesion measurements is described in Section 4.1.
4.1.1. Example of colloids for fast cell adhesion measurements
In this section, we further discuss a faster method to sequentially measure adhesion using coated colloid beads(P. Dörig et al., 2013; Angelo Gaitas, 2012; Gerecsei et al., 2019; Potthoff et al., 2012). Aiming to increase adhesion speed without having to exchange beads after each measurement, the functionalized beads are dispersed onto a confluent or near-confluent cell layer in culture and allowed to adhere for a specific time. The fluidic microcantilever is used to apply suction and pull the beads sequentially to measure adhesion. The cells with beads are examined optically to ensure that the beads are adhered to the cells and are not over an area without cells. An example of this methodology is described as follows. Gold nanoshells on 10 μm diameter silica beads were functionalized with a water-soluble cross-linker (DTSSP; 3,3’-dithiobis [sulfosuccinimidyl propionate]). DTSSP adheres to the gold surface by disulfide linkage and covalently to the surface proteins of the cell membrane(A. Gaitas, Malhotra, & Pienta, 2013),(Bennett et al., 2000). DTSSP coated beads showed an ~10-fold increase in the adhesion force to fibroblasts versus uncoated beads. For fluidFM measurements, the protocol depicted in Fig. 3 was used. The spheres were dispersed on a confluent layer of cells growing on a round coverslip and allowed to adhere inside an incubator (steps 1–2 in Fig. 3). The AFM unit is used to guide the cantilever optically near the bead while applying suction (step 3). The AFM unit detects contact, and the bead is attached by the suction applied (step 4, detected by the force-distance curve). The cantilever then pulls the bead upward and away from the cell membrane to measure the force-distance curve. During this pulling phase, the adhesive force strength is measured and is shown in the force-distance curve (step 5). Beads are discharged by applying positive pressure (step 6), and the tip is moved to another bead for another measurement. A cycle of measurements from step 3 to step 6 takes about 3 minutes to complete. Each measurement is an average of 5–10 force-distance curves per pause time (Fig. 4). After employing this approach, the pace of single-cell force spectroscopy was accelerated to up to 200 yeast and 20 mammalian cells per cantilever(Potthoff et al., 2012).
Fig. 3:
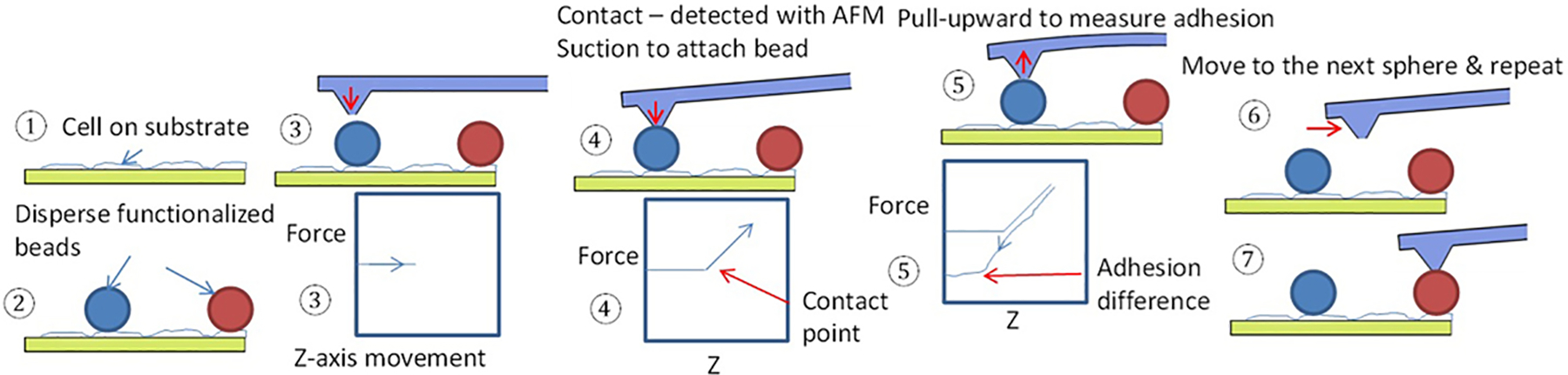
Protocol for high-throughput adhesion measurements using fluidFM.
Fig. 4:
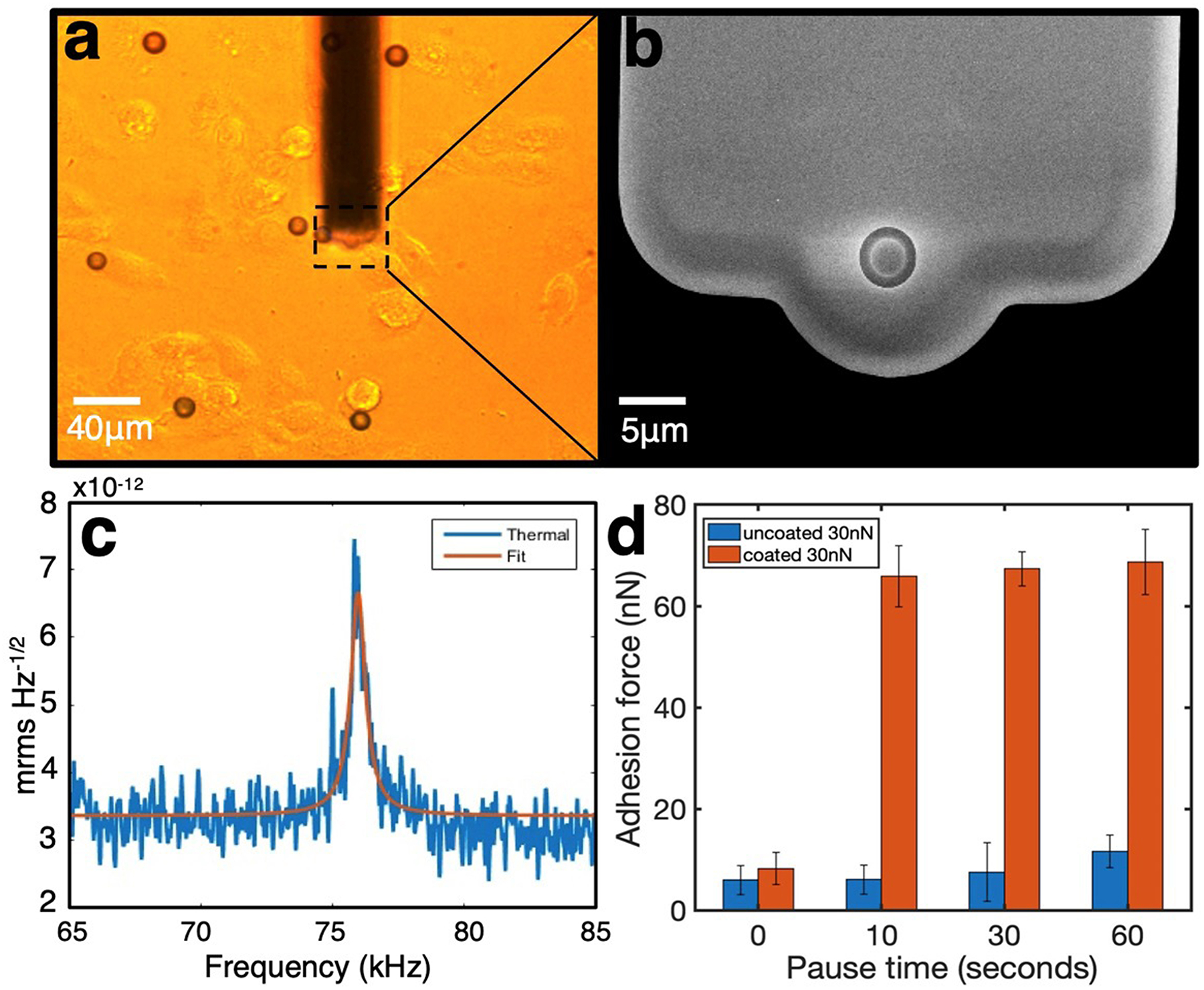
(a) Optical image of fibroblasts with 10 μm beads and a fluidFM cantilever. (b) Scanning electron microscopy image of a fluidFM cantilever (from Cytosurge). (c) Resonance frequency measurement of the cantilever used. (d) Summary of results of the adhesion forces measured in different pause times, conducted with uncoated or DTSSP-coated beads; results are derived from force-distance curves. The fluidic cantilevers (4 μm aperture; spring constant 1.33 N/m) (Cytosurge AG, Switzerland) were mounted on a FlexAFM (Nanosurf).
4.2. Single-cell manipulation
4.2.1. Intracellular injection
Glass micropipettes have been used for intracellular injection(Graessmann & Graessman, 1976; Mueller, Graessmann, & Graessmann, 1980). However, conventional micropipettes lack force feedback for real-time sensing of cell membrane contact and rupture. In addition, delivering small loads in single cells or inside the cell nucleus is particularly challenging(Capecchi, 1980).
FluidFM enables intracellular injection down to the femtoliter level with force feedback, minimizing cell damage(Guillaume-Gentil et al., 2013; Guillaume-Gentil et al., 2014). The load is released by passive diffusion or by applying a positive pressure using a nanopore fluidFM cantilever(Guillaume-Gentil et al., 2013). A recent report describes the use of electrowetting to transfer cytoplasm into nanopipettes on an extremely small scale. The technology can simultaneously analyze mRNA and mitochondrial DNA while maintaining cell viability in a single cell with high throughput sequencing(Actis et al., 2014).
4.2.2. Cell surface dispensing
To precisely observe the initiation of virus particle infection of a single living cell, Stiefel et al. used fluidFM to position individual and multiple virions onto the cell surface(Stiefel et al., 2012). By placing different numbers of virus particles on host cells, they showed that the infection rate grows at a superlinear rate with the number of particles placed on a single cell. This points to a synergy between viral particles, which impacts the early stage of the infection process(Stiefel et al., 2012). Similarly, any small non-biological(Aebersold et al., 2015) or biological(Rodrigues, Fan, Lyon, Wan, & Hu, 2018) particles, including EVs (discussed below), viruses, and bacterial pathogens, can be dispensed onto cellular surfaces. In another study, the fluidFM was combined with a fast-scanning confocal microscope to study host response to viral exposure in real-time(Koehler et al., 2021). In this work, fluidFM was used to attach nanogold particles (400 nm diameter) functionalized with virions(Koehler et al., 2021).
4.2.3. Single-cell sampling
Obtaining single-cell content for downstream analysis without cell lysis, thus enabling for post-extraction monitoring, is challenging. In the past, AFM tips were used to extract mRNA from live cells by chemically modifying the tip surface to immobilize gene-specific primers complementary to the mRNAs of interest (Nawarathna, Turan, & Wickramasinghe, 2009; Osada, Uehara, Kim, & Ikai, 2003). FluidFM enabled single-cell content extraction(Guillaume-Gentil et al., 2014). Guillaume-Gentil et al. inserted a minimally invasive nanopipette cantilever into a single living HeLa cell for cell compartment-selective extraction of the native intracellular fluid(Guillaume-Gentil et al., 2016). The extractions were successfully used for downstream molecular analyses (transmission electron microscopy, enzyme activity assays, and gene expression studies). It is worth noting that cells were viable up to five days post-extraction and that viability was dependent on the volume extracted from cytoplasm and nucleoplasm. In an extension of this work, Chen et al.(Chen et al., 2021) developed a technique called Live-Seq. Standard single-cell RNA sequencing involves lysis, thus providing a snapshot and endpoint measurement. In Live-Seq, repeated cytoplasm extractions from the same cell for downstream RNA sequencing was made possible. Using fluidFM, it is now possible to extract cytoplasm, treat the cells with agents of interest, and repeat the extraction after several hours(Actis et al., 2014).
4.3. Cell-to-cell force analysis
The spatial interaction between hetero- or homogenous cells plays a central role in the pathogenesis of infection and inflammation. Cell-to-cell lateral contacts are critical for tissue homeostasis. The paracellular pathway is an extracellular route across endothelia and epithelia that is generally used for passive transportation of water and small solutes; however, in some cases particles as big as leukocytes may cross it(Cereijido et al., 1993). The epithelial or endothelial barrier is maintained by intercellular multi-protein junctional complexes, either adherens junctions (AJs) and tight junctions (TJs), functionally sealing the lateral space between cells against unbinding forces on the lateral contact sites(Selhuber-Unkel et al., 2010). The interplay between TJs and AJs regulates major rate-limiting paracellular pathways by allowing particles to permeate across the paracellular route(Kawedia et al., 2007) and establishing cell polarity(Hartsock & Nelson, 2008). Dysfunctions, ruptures, and breach of the epithelial or endothelial barriers are major causes of infection and inflammation. Therefore, measuring the lateral binding forces (LBFs) between homogenous cells in response to different stimuli is crucial to understanding the precise biomechanical mechanism underlying intercellular barrier dysfunctions, which is a major outcome of host responses to infections, including inflammation. Traditionally, paracellular permeability can be indirectly evaluated using two methods: measuring trans-endothelial electrical resistance (TEER)(Buchert, Turksen, & Hollande, 2012) and fluorescein tracers after passing through the monolayer(Gong et al., 2013); both methods involve indirect measurements(Srinivasan et al., 2015).
Specific interactions between microbial surface ligands and host receptors account for tissue tropism and influence microbial distribution at the sites of infection. Adherence, however, is also a virulence factor, which requires resistance to the shear stress exerted by flowing blood at the blood-endothelial interface, promoting microbial uptake by ECs to initiate infection(Connell, Hedlund, Agace, & Svanborg, 1997). In bloodstream infections, a major determinant in bacterial disease outcomes is the adherence to ECs by microorganisms that lead to the establishment of metastatic endovascular infections(Claes et al., 2014; Kerdudou et al., 2006; B. C. Lee, Mayer, Leibowitz, Stearns-Kurosawa, & Kurosawa, 2013; McMullen & Freitag, 2015; Mellata, Mitchell, Schödel, Curtiss, & Pier, 2016; Shenoy et al., 2018; Viscoli, 2016). By activating the production of pro-inflammatory cytokines that cause local and systemic inflammation, the attachment of microorganisms to the mucosal surface is a key step in the successful establishment of mucosal infections(He et al., 2019). Thus, the quantification of the adhesion between pathogens and epithelial/endothelial cells is vital, and may lead to notable advances in our understanding of the interplay between microorganisms and the host at the initial stage of infection(Beaussart et al., 2014). In addition, at the site of inflammation, various stimuli induce endothelia on the surface of the luminal blood vessel to become adhesive for leukocytes(He et al., 2019). Following their initial contact with activated endothelia after margination, leukocytes roll along endothelial apical surfaces until they are captured. The adhesion of leukocytes to the vascular endothelium is a hallmark of focal inflammation. A variety of methods have been developed to study pathogen-host and leukocyte adhesions, including in vitro(Bhat et al., 2021), genetic(Joyce, Nelson, & Grinnell, 2004), molecular, and animal methods(Claes et al., 2014; He et al., 2019; Kerdudou et al., 2006). However, direct evidence regarding the biomechanical nature of adhesion is still lacking. Vertical binding force (VBF) measurements between a single bacterium or leukocytes and the target EC reveal the fundamental biomechanical nature of adhesion and its underlying mechanism(He et al., 2019; X. Zhang et al., 2004). In conventional AFM-based, single-cell adhesive force assays, cells are attached to the AFM cantilever to probe adhesive forces with adherent cells or substrates. Various irreversible immobilization strategies have been introduced since the development of this technology(Beaussart et al., 2014). Among these methods, the most straightforward consists of immersing the tip in a cell suspension in order to attach a single cell(Beaussart et al., 2014). However, this might lead to the irreversible attachment of multiple cells. Furthermore, attaching the cells biochemically may result in weak immobilization of the cell to the cantilever.
FluidFM can be used to capture single living cells for adhesion measurements(Su et al., 2021). This approach has been used to study endothelial barrier function by measuring the LBFs between ECs(Sancho, Vandersmissen, Craps, Luttun, & Groll, 2017), and also to dissect the molecular mechanism of Candida albicans adhesion by measuring the LBFs between yeast cells(Dehullu et al., 2019).
4.4. LBFs involving paracellular barrier function
Conventional technologies to directly measure LBFs of living cell-to-cell contacts were not available until a recent report that used fluidFM for the direct measurement of LBFs on a cell monolayer(Sancho et al., 2017). Sancho et al. quantified and compared LBFs between L929 fibroblasts and human endothelial cells from an umbilical artery, and provided evidence that vascular ECs exerted strong intercellular adhesion forces, while fibroblast adhesion forces were not detectable. Furthermore, they reported on the dynamics of the LBFs during endothelial-to-mesenchymal cell transition(Sancho et al., 2017). This study demonstrated the ability to assess EC LBFs in a mature monolayer in physiological settings, providing further evidence that these types of tools can be used to enhance our knowledge of biological processes in developmental biology, tissue regeneration, and disease states like infection and inflammation.
In order to complete measurements of LBFs of ECs, a larger Z-axis travel range piezo is needed, because vascular ECs are exceedingly thin(Alberts et al., 2002) with a relatively larger surface area compared with other epithelial cells(Jaffe, 1987; Wang, Jones, & Clulow, 1994). A large travel range gives the cantilever the ability to move a further distance in the Z-axis direction in a more stabilized form and separate the captured cell from the monolayer, making measuring the LBFs of ECs feasible. A 150 μm Z-axis actuator (Core AFM, NanoSurf, Liestal, Switzerland) is used for the experiments shown in Supplemental Video 2.
Based on the Sancho et al. report(Sancho et al., 2017), the intercellular adhesion forces (i.e., the LBFs) exerted by cells in monolayer after firm adhesion to the substrate and formation of mature intercellular junctions can be measured using fluidFM force spectroscopy. Indeed, the lack of precise quantification of the basal binding force between the cell basal side and the matrix is currently an unavoidable limitation of this approach. Taking advantage of the automation and ease of the measurements, baselines of basal binding forces derived from multiple measurements of the forces required to lift multiple free cells out of the monolayer in the same culture vessel were set, which were then applied in the calculation of the LBF (Fig. 5)(Sancho et al., 2017).
Fig. 5:
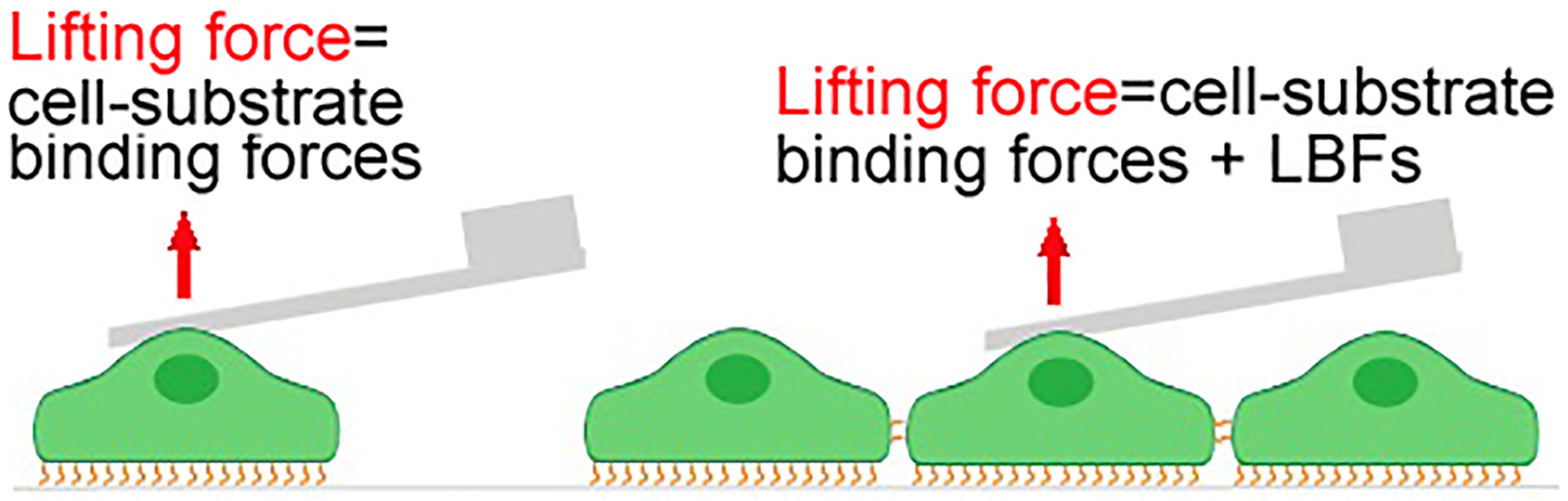
A demonstration of the difference in mechanical interaction between the single-cell model and the monolayer model in the LBF-measuring assay.
4.5. Intercellular adhesion VBFs between heterogeneous cells
The principal of applying fluidFM to study VBFs is that the micropipette acts as a cell probe by capturing a single living cell via negative pressure (Supplemental Video 3), which then interacts with a single living target cell in a liquid environment. This specialized cantilever is connected to the fluidic pressure controller. The opening at the apex of the probe varies from 300 nm to 8 μm. A continuous flow from the fluidic pressure controller creates negative pressure after a cell attaches to the aperture of the micropipette probe, which then becomes a cell probe. After the cell probe is moved onto the apical surface of the target cell, single-point force spectroscopy is performed, and the interacting force between the cell probe (i.e., the micropipette-captured cell) and the target cell is measured in nanonewtons (nN). Thus, VBFs between a single living bacterium or leukocyte and an epithelial cell or EC can be quantified.
4.6. Single-cell mass measurement
Cell mass is a critical parameter impacted by disease dysregulation(Lloyd, 2013) and is indicative of the quantity of fluids, biomolecules, macromolecules, amino acids, lipids, and nucleic acids within a cell(Martínez-Martín et al., 2017). Processes such as cell differentiation, gene expression(Häussinger, 1996), cell shape, metabolism, migration, and proliferation(Lang et al., 1998) can be investigated using measurements of cell mass to study regulation. Rapid cell mass fluctuations may provide insights into basic cellular processes, such as the response to growth stimuli, ATP synthesis, glycolysis, or the transport of water or other substances across the cell membrane(Martínez-Martín et al., 2017). Highly precise mass measurements are required due to the small masses involved, the irregular shapes of cells, and the need to study individual cell behavior. Such higher-resolution measurements are required to advance our understanding of cellular growth, as prior studies into cellular growth patterns have not been conclusive and have shown varying growth curves ranging from linear to exponential(Charvin, 2010; Kubitschek, 1986).
To date, research involving cell growth has mostly relied on volume measurements, as it has been extremely difficult to measure single-cell mass(Bryan, Goranov, Amon, & Manalis, 2010). However, cellular volume and mass may change at different rates, with mass being a better indicator to assay cell growth in single cells(Charvin, 2010). Changes in cell volume have a strong relationship with cell density, which is impacted by osmotic and other processes, whereas cell mass indicates growth as cells acquire new biomass, in particular protein content(Charvin, 2010). Thus, there is a critical need for improved, highly precise methods of dynamic and continual cell mass measurements(Bryan et al., 2010; Charvin, 2010).
Current promising approaches to detecting the mass of single cells that rely on microcantilever resonance frequency changes include microchannel resonators(Burg et al., 2007) and pedestal mass sensors(Park et al., 2010). Techniques employing conventional functionalized AFM cantilevers have been used for mass measurements of single adherent cells(Chien, Jiang, Gong, Li, & Gaitas, 2022; Łabędź, Wańczyk, & Rajfur, 2017).
There are several advantages to using fluidFM in analyzing the mass of single cells. Much like inertial pico-balance resonators(Martínez-Martín et al., 2017), fluidFM achieves high temporal resolution and offers similar mass resolution. Importantly, cell attachment can be achieved without the need for antibodies by physically grabbing the cell in an aqueous environment. This offers a major advantage, as it enables the study of non-adherent cells such as immune cells. Another advantage is that the device can be reused, enabling the measurement of several cells in a short period of time. Potential challenges could be that the behavior of adherent cells (most mammalian cells) is affected if they are suspended, thus pipette attachment may not allow the cells to behave physiologically normal. In addition, the Q factor of the cantilever in liquids drops dramatically compared to air due to high damping in liquids, which decreases sensitivity. However, this is a common problem for all microcantilever techniques performed in a liquid environment. It is worth noting that this is not the case with microchannel resonators(Burg et al., 2007) that operate in air. In an initial proof-of-concept, fluidFM was used to measure the mass of yeast and beads in air, with the ability to catch and release at picogram resolution(Ossola, Dörig, Vörös, Zambelli, & Vassalli, 2016). Nonetheless, techniques that employ fluidFM for single-cell mass measurements require further study.
4.7. Electrophysiology
Patch clamping(Hamill et al., 1981) is used in multiple areas of biology, such as cardiology (cardiomyocytes), neurology/neuroscience (neurons), endocrinology (pancreatic beta cells), and myology (muscle fibers). Ion channels in immune cells play significant roles in directly or indirectly regulating intracellular signaling pathways, cell development, innate and adaptive immune responses, and autoimmunity(Feske, Wulff, & Skolnik, 2015). Ion channels can potentially become pharmacological targets for autoimmune diseases(Feske et al., 2015). Bacteria have many ion channels that respond to chemical and physical alterations(Kralj, Hochbaum, Douglass, & Cohen, 2011; Buechner Martinac, Buechner, Delcour, Adler, & Kung, 1987; Boris Martinac et al., 2014). Classical patch-clamping cannot be performed due to the structure of the bacterial cell wall and the small size of bacterial cells(Kralj et al., 2011; Boris Martinac, Rohde, Cranfield, & Nomura, 2013).
Patch clamping is the gold standard for electrophysiology and offers an accurate and unmatched measurement of ion currents and membrane potentials; however, it is labor intensive and time consuming, requiring an entire day to record two to four cells(Clements & Roquemore, 2017). Furthermore, patch clamping requires lengthy training and expertise due to the difficulty in operating the pipette. Cells and pipettes are prone to damage, requiring frequent replacement. The pipette is guided visually under microscopic observation without force-feedback. Finally, excessive mechanical stress before or during a procedure may affect the results(Hamill & McBride Jr, 1997).
Combining AFM with a fluidic probe for patch clamping has many benefits. The entire measurement can be partially automated. The AFM force-feedback mechanism acts as a feedback touch sensor to detect contact with a cell, significantly reducing the likelihood of cell damage, training time, and time per measurement. This requires compliant cantilevers for nondestructive contact and direct measurements. Ossola et al.(Ossola et al., 2015) reported that a fluidFM cantilever was used for a combination of patch clamping and contraction measurements of cardiomyocytes. The device’s geometry and other factors did not allow for the formation of a GΩ seal (reported in the 10s of MΩ). Furthermore, the high value of the spring constant (1.8 N/m) exerts a force that could be damaging to the cells and could interfere with the measurements. Therefore, while the combination of AFM and fluidic probes is very promising for automation of single-cell patch clamping, several modifications are needed at the device level before patch clamping can be used by a wider user base. The use of MEMS fluidic cantilevers and AFM may result in lower noise compared to conventional patch clamping, less cell content diffusion in the pipette, minimization of mechanical stress, less damage to cells, and device uniformity. In addition, it would enable additional modalities such as cell adhesion, cell contraction, and elasticity to be considered. While fluidFM is low throughput, it can provide high content analysis. For instance, patch clamp AFM could be combined with single-cell content extraction using fluidic probes for single-cell sequencing, thus enabling single-cell physiological and genotypic characterization.
5. Limitations
5.1. AFM limitations
The main limitations of AFM are that it is low throughout (i.e., it can only handle a single cell at a time, at best). Therefore, coupling with other techniques such as light microscopy, scanning fluorescence microscopy, or transmission electron microscopy is required for AFM to be widely applied in the field of cell biology(Friedrichs et al., 2010). However, AFM has nanoscale resolution, ideal for single-cell measurements, and the ability to operate in liquid environments, which are key requirements for biological imaging. The operational range of AFM is suitable for characterizing structures from the molecular to cellular levels. In addition, AFM has the unique ability to measure molecular forces with high sensitivity(Friedrichs et al., 2010; Friedrichs et al., 2013).
Limitations of AFM for the study of biofilms include the inability to obtain a large area survey scan, and the soft and gelatinous nature of the biofilm might be damaged by the imaging of the surface, especially within a liquid environment(Wright, Shah, Powell, & Armstrong, 2010). Another limitation of AFM imaging compared with fluorescence microscopy is its rather poor temporal resolution (typically ~1 minute per image), which is much slower than the time scale at which dynamic processes usually occur in cell biology(Heinisch et al., 2012; Shibata, Yamashita, Uchihashi, Kandori, & Ando, 2010). However, remarkable advances are being made in developing high-speed AFM units that can operate in the millisecond timescale, thus offering new possibilities to explore cellular dynamics(Heinisch et al., 2012; Shibata et al., 2010).
Rateesh et al. further listed limitations from their experience in applying AFM in relevant biomedical fields(Babu & Singh, 2014):
A disadvantage of AFM compared with the scanning electron microscope (SEM) is the single scan image size. This can be improved by using parallel probes.
The relatively slow scanning speed of an AFM is also a limitation. Several fast-acting designs were proposed.
AFM images can also be affected by hysteresis of the piezoelectric material and crosstalk between the x, y, and z axes that may require software enhancement and filtering.
As with any imaging technique, there is the possibility of unavoidable image artifacts, which could be caused by an unsuitable tip, a poor operating environment, or even by the sample itself.
Due to the nature of AFM probes, they cannot normally measure steep walls or overhangs(Babu & Singh, 2014).
5.2. FluidFM limitations during single-cell manipulations
Li et al. discussed the limitations of fluidFM(M. Li, Liu, & Zambelli, 2022). The serial interplay of pressure to hold a cell against the aperture and overpressure to release it is sound and uncomplicated. However, deposition of an anti-fouling coating may be required to minimize cell binding at the aperture. The aspirated cell must be released onto another substrate. This critical action is relatively easy when the new substrate is larger than that of the aperture, but it can cause issues if the new substrate is relatively smaller. There is intrinsic variability in the sharpness of the probe being utilized for each membrane perforation, which may require adjustments to injection and extraction protocols. It is often observed that cell debris remain attached at the aperture edge, despite the use of an anti-fouling coating. Moreover, an anti-fouling coating on the walls of the microchannel is required for extraction to avoid unwanted adsorption of the extracted molecules to the wall surface.
6. Conclusion and future perspectives
In this review, we introduced the use of AFM in EV analysis for host-pathogen studies. We also provided a review of fluidFM development and discussed some new applications, focusing on cell biology applications in cell-pathogen adhesion and cell barrier function as a key host response. FluidFM features a cantilever embedded with micromachined microfluidic channels. Through proper pressure control, targets such as cells or beads can be attached to the tip by suction to conduct desired measurements. Targets are subsequently repelled by overpressure, and the cantilever may be reused for subsequent experiments. The development of fluidFM enables various experimental directions, including fast adhesion measurements, single-cell treatment and sampling, cell-to-cell LBF and VBF analyses, single-cell mass measurements, and patch-clamping measurements. Some of the directions presented in this review are more widely explored, and yet some of the applications are still in their initial stage of development and require further scientific exploration to produce meaningful results. Given that fluidFM was invented relatively recently, it stands as an emerging and prominent candidate for single living cell and EV studies, and there is room for further improvement, optimization, and innovation. One direction for improvement is the development of additionally specialized fluidic cantilevers that serve specific applications, such as patch clamping. Experimentation with new structural materials, such as polymers, may result in more compliant devices. Embedding sensing elements, such as deflection sensors and electrodes, on the cantilevers may enhance their functionality and perhaps even eliminate the need for the AFM optical lever. As AFM innovations are commercialized, they will also contribute to performance enhancement, such as the introduction of photothermal excitation(Marti et al., 1992; Ramos, Mertens, Calleja, & Tamayo, 2008; Umeda, Ishizaki, & Uwai, 1991) in commercial units that significantly improves the AFM performance in liquids.
Supplementary Material
Acknowledgements
We gratefully acknowledge Drs. Zhengchen Su and Edward Nelson for their contributions in helping to establish our fluidFM system capabilities. We also thank Dr. Nelson for assistance with measurements and for insights into the use of AFM and fluidFM. We thank Dr. Kimberly Schuenke for her critical review and editing of the manuscript. This work was supported by NIH grants R01AI121012 (BG), R21AI137785 (BG and AG), R21AI154211(BG), R03AI142406 (BG), R21AI144328 (BG), Sealy Center for Vector Borne and Zoonotic Diseases pilot grant FY21 (BG), and the UTMB Center for Biodefense and Emerging Infectious Diseases pilot grant FY21 (BG). The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Actis P, Maalouf MM, Kim HJ, Lohith A, Vilozny B, Seger RA, & Pourmand N (2014). Compartmental genomics in living cells revealed by single-cell nanobiopsy. ACS nano, 8(1), 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold MJ, Dermutz H, Saenz Cogollo JF, Han H, Demkó L, Zambelli T, & Vörös J (2015). Local chemical stimulation of neurons using fluidfm technology combined with microelectrode arrays. Paper presented at the 19th International Conference on Miniaturized Systems for Chemistry and Life Sciences (μTAS 2015). [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, & Walter P (2002). Molecular biology of the cell (4th ed.). New York, United States: Garland Science. [Google Scholar]
- Alsteens D, Beaussart A, El-Kirat-Chatel S, Sullan RM, & Dufrêne YF (2013). Atomic force microscopy: a new look at pathogens. PLoS Pathog, 9(9), e1003516. doi: 10.1371/journal.ppat.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Asencio R, Thormann E, & Rutland MW (2013). Note: Determination of torsional spring constant of atomic force microscopy cantilevers: combining normal spring constant and classical beam theory. Rev Sci Instrum, 84(9), 096102. doi: 10.1063/1.4820345 [DOI] [PubMed] [Google Scholar]
- Amarouch MY, El Hilaly J, & Mazouzi D (2018). AFM and FluidFM Technologies: Recent Applications in Molecular and Cellular Biology. Scanning, 2018, 7801274. doi: 10.1155/2018/7801274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu R, & Singh E (2014). Atomic force microscopy: A source of investigation in biomedicine. International Journal of Electronic and Electrical Engineering, 7(1), 59–66. [Google Scholar]
- Bairamukov V, Bukatin A, Landa S, Burdakov V, Shtam T, Chelnokova I, … Starodubtseva M (2020). Biomechanical Properties of Blood Plasma Extracellular Vesicles Revealed by Atomic Force Microscopy. Biology (Basel), 10(1). doi: 10.3390/biology10010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A, El-Kirat-Chatel S, Sullan RM, Alsteens D, Herman P, Derclaye S, & Dufrêne YF (2014). Quantifying the forces guiding microbial cell adhesion using single-cell force spectroscopy. Nat Protoc, 9(5), 1049–1055. doi: 10.1038/nprot.2014.066 [DOI] [PubMed] [Google Scholar]
- Bennett KL, Kussmann M, Björk P, Godzwon M, Mikkelsen M, Sørensen P, & Roepstorff P (2000). Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping--a novel approach to assess intermolecular protein contacts. Protein Sci, 9(8), 1503–1518. doi: 10.1110/ps.9.8.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Lewis A, Harootunian A, Isaacson M, & Kratschmer E (1986). Near field scanning optical microscopy (NSOM): development and biophysical applications. Biophysical journal, 49(1), 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SV, Price JDW, & Dahms TES (2021). AFM-Based Correlative Microscopy Illuminates Human Pathogens. Front Cell Infect Microbiol, 11, 655501. doi: 10.3389/fcimb.2021.655501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S, Shinagawa K, Castellino FJ, & Schorey JS (2007). Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood, 110(9), 3234–3244. doi: 10.1182/blood-2007-03-079152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, & Gerber C (1986). Atomic force microscope. Physical review letters, 56(9), 930. [DOI] [PubMed] [Google Scholar]
- Böing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, & Nieuwland R (2014). Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles, 3. doi: 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovec M, Szilagyi I, Popa I, Finessi M, Sinha P, Maroni P, & Papastavrou G (2012). Investigating forces between charged particles in the presence of oppositely charged polyelectrolytes with the multi-particle colloidal probe technique. Adv Colloid Interface Sci, 179–182, 85–98. doi: 10.1016/j.cis.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Bowers V, Fisher L, Francis G, & Williams K (1989). A micromechanical technique for monitoring cell—substrate adhesiveness: measurements of the strength of red blood cell adhesion to glass and polymer test surfaces. Journal of biomedical materials research, 23(12), 1453–1473. [DOI] [PubMed] [Google Scholar]
- Bryan AK, Goranov A, Amon A, & Manalis SR (2010). Measurement of mass, density, and volume during the cell cycle of yeast. Proceedings of the National Academy of Sciences, 107(3), 999. doi: 10.1073/pnas.0901851107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert M, Turksen K, & Hollande F (2012). Methods to examine tight junction physiology in cancer stem cells: TEER, paracellular permeability, and dilution potential measurements. Stem cell reviews and reports, 8(3), 1030–1034. [DOI] [PubMed] [Google Scholar]
- Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, … Manalis SR (2007). Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature, 446(7139), 1066–1069. doi: 10.1038/nature05741 [DOI] [PubMed] [Google Scholar]
- Cagno V (2020). SARS-CoV-2 cellular tropism. Lancet Microbe, 1(1), e2–e3. doi: 10.1016/S2666-5247(20)30008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR (1980). High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell, 22(2), 479–488. [DOI] [PubMed] [Google Scholar]
- Carnino JM, & Jin Y (2020). Intercellular Communication via Extracellular Vesicle Cargo MicroRNAs: Challenges for Experimental Design. Crit Care Med, 48(12), e1364–e1365. doi: 10.1097/CCM.0000000000004564 [DOI] [PubMed] [Google Scholar]
- Cereijido M, Ruiz O, González-Mariscal L, Contreras RG, Susana Balda M, & García-Villegas MR (1993). The Paracellular Pathway. In Audus KL & Raub TJ (Eds.), Biological Barriers to Protein Delivery (pp. 3–21). Boston, MA: Springer US. [Google Scholar]
- Charvin G (2010). Measuring the growth rate of cells, one at a time. Nature Methods, 7(5), 363–363. doi: 10.1038/nmeth0510-363 [DOI] [PubMed] [Google Scholar]
- Chen W, Guillaume-Gentil O, Dainese R, Rainer PY, Zachara M, Gabelein CG, … Deplancke B (2021). Genome-wide molecular recording using Live-seq. bioRxiv. [Google Scholar]
- Cheng J, Nonaka T, & Wong DTW (2019). Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials, 12(4). doi: 10.3390/ma12040654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesla SE, Selvaraj P, & Zhu C (1998). Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophysical journal, 75(3), 1553–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C-C, Jiang J, Gong B, Li T, & Gaitas A (2022). AFM Microfluidic Cantilevers as Weight Sensors for Single Cell Mass Measurements. bioRxiv, 2022.2002.2021.481347. doi: 10.1101/2022.02.21.481347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivet M, Javalet C, Laulagnier K, Blot B, Hemming FJ, & Sadoul R (2014). Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J Extracell Vesicles, 3, 24722. doi: 10.3402/jev.v3.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyasnavichyus M, Young SL, Geryak R, & Tsukruk VV (2016). Probing elastic properties of soft materials with AFM: Data analysis for different tip geometries. Polymer, 102, 317–325. [Google Scholar]
- Claes J, Vanassche T, Peetermans M, Liesenborghs L, Vandenbriele C, Vanhoorelbeke K, … Verhamme P (2014). Adhesion of Staphylococcus aureus to the vessel wall under flow is mediated by von Willebrand factor-binding protein. Blood, 124(10), 1669–1676. doi: 10.1182/blood-2014-02-558890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M, & Roquemore L (2017). Stem Cell-Derived Models in Toxicology: Springer. [Google Scholar]
- Coelho C, Brown L, Maryam M, Vij R, Smith DFQ, Burnet MC, … Casadevall A (2019). virulence factors, including listeriolysin O, are secreted in biologically active extracellular vesicles. J Biol Chem, 294(4), 1202–1217. doi: 10.1074/jbc.RA118.006472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N, Sarkar S, Hondroulis E, Sabhachandani P, & Konry T (2017). Quantification of intercellular adhesion forces measured by fluid force microscopy. Talanta, 174, 409–413. doi: 10.1016/j.talanta.2017.06.038 [DOI] [PubMed] [Google Scholar]
- Connell H, Hedlund M, Agace W, & Svanborg C (1997). Bacterial Attachment To Uro-Epithelial Cells: Mechanisms and Consequences. Advances in Dental Research, 11(1), 50–58. doi: 10.1177/08959374970110011701 [DOI] [PubMed] [Google Scholar]
- Crescitelli R, Lässer C, Jang SC, Cvjetkovic A, Malmhäll C, Karimi N, … Lötvall J (2020). Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. J Extracell Vesicles, 9(1), 1722433. doi: 10.1080/20013078.2020.1722433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CN, Phillips H, Tomes JJ, Swain MT, Wilkinson TJ, Brophy PM, & Morphew RM (2019). The importance of extracellular vesicle purification for downstream analysis: A comparison of differential centrifugation and size exclusion chromatography for helminth pathogens. PLoS Negl Trop Dis, 13(2), e0007191. doi: 10.1371/journal.pntd.0007191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehullu J, Valotteau C, Herman-Bausier P, Garcia-Sherman M, Mittelviefhaus M, Vorholt JA, … Dufrêne YF (2019). Fluidic Force Microscopy Demonstrates That Homophilic Adhesion by Candida albicans Als Proteins Is Mediated by Amyloid Bonds between Cells. Nano Lett, 19(6), 3846–3853. doi: 10.1021/acs.nanolett.9b01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deladi S, Tas N, Berenschot J, De Boer J, De Boer M, Krijnen G, & Elwenspoek M (2005). Micromachined fountain pen as a tool for atomic force microscope-based nanoelectrochemical metal deposition. Paper presented at the 18th IEEE International Conference on Micro Electro Mechanical Systems, 2005. MEMS 2005. [Google Scholar]
- Deladi S, Tas NR, Berenschot JW, Krijnen GJ, de Boer MJ, De Boer J, … Elwenspoek MC (2004). Micromachined fountain pen for atomic force microscope-based nanopatterning. Applied Physics Letters, 85(22), 5361–5363. [Google Scholar]
- Dörig P, Ossola D, Truong AM, Graf M, Stauffer F, Vörös J, & Zambelli T (2013). Exchangeable colloidal AFM probes for the quantification of irreversible and long-term interactions. Biophys J, 105(2), 463–472. doi: 10.1016/j.bpj.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörig P, Stiefel P, Behr P, Sarajlic E, Bijl D, Gabi M, … Zambelli T (2010). Force-controlled spatial manipulation of viable mammalian cells and micro-organisms by means of FluidFM technology. Applied Physics Letters, 97(2), 023701. [Google Scholar]
- Dufrêne YF (2008). Atomic force microscopy and chemical force microscopy of microbial cells. Nature Protocols, 3(7), 1132–1138. doi: 10.1038/nprot.2008.101 [DOI] [PubMed] [Google Scholar]
- Feske S, Wulff H, & Skolnik EY (2015). Ion channels in innate and adaptive immunity. Annual review of immunology, 33, 291–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish G, Bouevitch O, Kokotov S, Lieberman K, Palanker D, Turovets I, & Lewis A (1995). Ultrafast response micropipette-based submicrometer thermocouple. Review of Scientific Instruments, 66(5), 3300–3306. [Google Scholar]
- Francis G, Fisher L, Gamble R, & Gingell D (1987). Direct measurement of cell detachment force on single cells using a new electromechanical method. Journal of cell science, 87(4), 519–523. [DOI] [PubMed] [Google Scholar]
- Friedrichs J, Helenius J, & Muller DJ (2010). Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nature Protocols, 5(7), 1353–1361. doi: 10.1038/nprot.2010.89 [DOI] [PubMed] [Google Scholar]
- Friedrichs J, Legate KR, Schubert R, Bharadwaj M, Werner C, Müller DJ, & Benoit M (2013). A practical guide to quantify cell adhesion using single-cell force spectroscopy. Methods, 60(2), 169–178. doi: 10.1016/j.ymeth.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Gaitas A (2012). USA Patent No.
- Gaitas A, & Hower RW (2014). SU-8 microcantilever with an aperture, fluidic channel, and sensing mechanisms for biological and other applications. Journal of Micro/Nanolithography, MEMS, and MOEMS, 13(3), 030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitas A, Malhotra R, & Pienta K (2013). A method to measure cellular adhesion utilizing a polymer micro-cantilever. Appl Phys Lett, 103(12), 123702. doi: 10.1063/1.4821946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y (2007). Invited review article: a review of techniques for attaching micro- and nanoparticles to a probe’s tip for surface force and near-field optical measurements. Rev Sci Instrum, 78(8), 081101. doi: 10.1063/1.2754076 [DOI] [PubMed] [Google Scholar]
- Garcia-Martin R, Wang G, Brandão BB, Zanotto TM, Shah S, Kumar Patel S, … Kahn CR (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature, 601(7893), 446–451. doi: 10.1038/s41586-021-04234-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecsei T, Erdődi I, Peter B, Hős C, Kurunczi S, Derényi I, … Horvath R (2019). Adhesion force measurements on functionalized microbeads: An in-depth comparison of computer controlled micropipette and fluidic force microscopy. J Colloid Interface Sci, 555, 245–253. doi: 10.1016/j.jcis.2019.07.102 [DOI] [PubMed] [Google Scholar]
- Gong B, Lee YS, Lee I, Shelite TR, Kunkeaw N, Xu G, … Walker DH (2013). Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect Dis, 13, 285. doi: 10.1186/1471-2334-13-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessmann M, & Graessman A (1976). “ Early” simian-virus-40-specific RNA contains information for tumor antigen formation and chromatin replication. Proceedings of the National Academy of Sciences, 73(2), 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume-Gentil O, Grindberg RV, Kooger R, Dorwling-Carter L, Martinez V, Ossola D, … Vorholt JA (2016). Tunable Single-Cell Extraction for Molecular Analyses. Cell, 166(2), 506–516. doi: 10.1016/j.cell.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Guillaume-Gentil O, Potthoff E, Ossola D, Dörig P, Zambelli T, & Vorholt JA (2013). Force-controlled fluidic injection into single cell nuclei. Small, 9(11), 1904–1907. doi: 10.1002/smll.201202276 [DOI] [PubMed] [Google Scholar]
- Guillaume-Gentil O, Potthoff E, Ossola D, Franz CM, Zambelli T, & Vorholt JA (2014). Force-controlled manipulation of single cells: from AFM to FluidFM. Trends Biotechnol, 32(7), 381–388. doi: 10.1016/j.tibtech.2014.04.008 [DOI] [PubMed] [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, & Sigworth FJ (1981). Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Archiv, 391(2), 85–100. [DOI] [PubMed] [Google Scholar]
- Hamill OP, & McBride DW Jr (1997). Induced membrane hypo/hyper-mechanosensitivity: a limitation of patch-clamp recording. Annual Review of Physiology, 59(1), 621–631. [DOI] [PubMed] [Google Scholar]
- Han H, Martinez V, Aebersold MJ, Lüchtefeld I, Polesel-Maris J, Vörös J, & Zambelli T (2018). Force controlled SU-8 micropipettes fabricated with a sideways process. Journal of Micromechanics and Microengineering, 28(9), 095015. [Google Scholar]
- Han H, Martinez V, Forro C, Polesel-Maris J, Voeroes J, & Zambelli T (2020). Integration of silver nanowires into SU-8 hollow cantilevers for piezoresistive-based sensing. Sensors and Actuators A: Physical, 301, 111748. [Google Scholar]
- Hartsock A, & Nelson WJ (2008). Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochimica et biophysica acta, 1778(3), 660–669. doi: 10.1016/j.bbamem.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häussinger D (1996). The role of cellular hydration in the regulation of cell function. Biochem J, 313 (Pt 3), 697–710. doi: 10.1042/bj3130697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang W, Chang Q, Su Z, Gong D, Zhou Y, … Gong B (2019). A new role for host annexin A2 in establishing bacterial adhesion to vascular endothelial cells: lines of evidence from atomic force microscopy and an in vivo study. Lab Invest. doi: 10.1038/s41374-019-0284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinisch JJ, Lipke PN, Beaussart A, El Kirat Chatel S, Dupres V, Alsteens D, & Dufrêne YF (2012). Atomic force microscopy - looking at mechanosensors on the cell surface. J Cell Sci, 125(Pt 18), 4189–4195. doi: 10.1242/jcs.106005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug T, Biss T, De Rooij N, & Staufer U (2005). Generic fabrication technology for transparent and suspended microfluidic and nanofluidic channels. Paper presented at the The 13th International Conference on Solid-State Sensors, Actuators and Microsystems, 2005. Digest of Technical Papers. TRANSDUCERS’05. [Google Scholar]
- Hui WW, Hercik K, Belsare S, Alugubelly N, Clapp B, Rinaldi C, & Edelmann MJ (2018). Salmonella enterica Serovar Typhimurium Alters the Extracellular Proteome of Macrophages and Leads to the Production of Proinflammatory Exosomes. Infect Immun, 86(2). doi: 10.1128/IAI.00386-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes WF, Doty NJ, Zarembinski TI, Schwartz MP, Toepke MW, Murphy WL, … Cady NC (2014). Micropatterning of 3D Microenvironments for Living Biosensor Applications. Biosensors, 4(1). doi: 10.3390/bios4010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe EA (1987). Cell biology of endothelial cells. Hum Pathol, 18(3), 234–239. doi: 10.1016/s0046-8177(87)80005-9 [DOI] [PubMed] [Google Scholar]
- Jani PK, Schwaner E, Kajdácsi E, Debreczeni ML, Ungai-Salánki R, Dobó J, … Cervenak L (2016). Complement MASP-1 enhances adhesion between endothelial cells and neutrophils by up-regulating E-selectin expression. Mol Immunol, 75, 38–47. doi: 10.1016/j.molimm.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Ji Y, Qi D, Li L, Su H, Li X, Luo Y, … Lu Y (2019). Multiplexed profiling of single-cell extracellular vesicles secretion. Proc Natl Acad Sci U S A, 116(13), 5979–5984. doi: 10.1073/pnas.1814348116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LB, Bell CR, Bibb KE, Gu L, Coats MT, & Matthews QL (2018). Pathogens and Their Effect on Exosome Biogenesis and Composition. Biomedicines, 6(3). doi: 10.3390/biomedicines6030079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce DE, Nelson DR, & Grinnell BW (2004). Leukocyte and endothelial cell interactions in sepsis: relevance of the protein C pathway. Crit Care Med, 32(5 Suppl), S280–286. doi: 10.1097/01.ccm.0000128037.72072.22 [DOI] [PubMed] [Google Scholar]
- Kappl M, & Butt HJ (2002). The colloidal probe technique and its application to adhesion force measurements. Particle & Particle Systems Characterization: Measurement and Description of Particle Properties and Behavior in Powders and Other Disperse Systems, 19(3), 129–143. [Google Scholar]
- Kawedia JD, Nieman ML, Boivin GP, Melvin JE, Kikuchi K-I, Hand AR, … Menon AG (2007). Interaction between transcellular and paracellular water transport pathways through Aquaporin 5 and the tight junction complex. Proceedings of the National Academy of Sciences, 104(9), 3621. doi: 10.1073/pnas.0608384104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdudou S, Laschke MW, Sinha B, Preissner KT, Menger MD, & Herrmann M (2006). Fibronectin binding proteins contribute to the adherence of Staphylococcus aureus to intact endothelium in vivo. Thromb Haemost, 96(2), 183–189. [PubMed] [Google Scholar]
- Kim K, Ke C, Moldovan N, & Espinosa H (2003). Massively parallel multi-tip nanoscale writer with fluidic capabilities-fountain pen nanolithography (FPN). Paper presented at the Proc. 4th Int. Symp. on MEMS and Nanotechnology. [Google Scholar]
- Koehler M, Petitjean SJ, Yang J, Aravamudhan P, Somoulay X, Giudice CL, … Alsteens D (2021). Reovirus directly engages integrin to recruit clathrin for entry into host cells. Nature communications, 12(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Környei Z, Beke S, Mihálffy T, Jelitai M, Kovács KJ, Szabó Z, & Szabó B (2013). Cell sorting in a Petri dish controlled by computer vision. Sci Rep, 3, 1088. doi: 10.1038/srep01088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Hochbaum DR, Douglass AD, & Cohen AE (2011). Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science, 333(6040), 345–348. [DOI] [PubMed] [Google Scholar]
- Kramer RC, Verlinden EJ, Angeloni L, Van Den Heuvel A, Fratila-Apachitei LE, Van Der Maarel SM, & Ghatkesar MK (2020). Multiscale 3D-printing of microfluidic AFM cantilevers. Lab on a chip, 20(2), 311–319. [DOI] [PubMed] [Google Scholar]
- Kubitschek HE (1986). Increase in cell mass during the division cycle of Escherichia coli B/rA. J Bacteriol, 168(2), 613–618. doi: 10.1128/jb.168.2.613-618.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov V, & Papastavrou G (2012a). Adhesion of colloidal particles on modified electrodes. Langmuir, 28(48), 16567–16579. doi: 10.1021/la3029726 [DOI] [PubMed] [Google Scholar]
- Kuznetsov V, & Papastavrou G (2012b). Note: mechanically and chemically stable colloidal probes from silica particles for atomic force microscopy. Rev Sci Instrum, 83(11), 116103. doi: 10.1063/1.4765299 [DOI] [PubMed] [Google Scholar]
- Łabędź B, Wańczyk A, & Rajfur Z (2017). Precise mass determination of single cell with cantilever-based microbiosensor system. PLoS One, 12(11), e0188388. doi: 10.1371/journal.pone.0188388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Busch GL, Ritter M, Völkl H, Waldegger S, Gulbins E, & Häussinger D (1998). Functional significance of cell volume regulatory mechanisms. Physiol Rev, 78(1), 247–306. doi: 10.1152/physrev.1998.78.1.247 [DOI] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E, Lässer C, Shelke GV, Crescitelli R, Jang SC, Cvjetkovic A, … Lötvall J (2019). DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles, 8(1), 1656993. doi: 10.1080/20013078.2019.1656993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClaire M, Gimzewski J, & Sharma S (2021). A review of the biomechanical properties of single extracellular vesicles. Nano Select, 2(1), 1–15. [Google Scholar]
- Lee BC, Mayer CL, Leibowitz CS, Stearns-Kurosawa DJ, & Kurosawa S (2013). Quiescent complement in nonhuman primates during E coli Shiga toxin-induced hemolytic uremic syndrome and thrombotic microangiopathy. Blood, 122(5), 803–806. doi: 10.1182/blood-2013-03-490060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, … Weissleder R (2018). Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano, 12(1), 494–503. doi: 10.1021/acsnano.7b07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Isaacson M, Muray A, & Harootunian A (1983). Scanning Optical Spectral Microscopy with 500A Spatial-Resolution. Paper presented at the Biophysical journal. [Google Scholar]
- Lewis A, Kheifetz Y, Shambrodt E, Radko A, Khatchatryan E, & Sukenik C (1999). Fountain pen nanochemistry: Atomic force control of chrome etching. Applied Physics Letters, 75(17), 2689–2691. [Google Scholar]
- Li L, Cheng Y, Emrich S, & Schorey J (2018). Activation of endothelial cells by extracellular vesicles derived from Mycobacterium tuberculosis infected macrophages or mice. PLoS One, 13(5), e0198337. doi: 10.1371/journal.pone.0198337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu L, & Zambelli T (2022). FluidFM for single-cell biophysics. Nano Research, 15(2), 773–786. doi: 10.1007/s12274-021-3573-y [DOI] [Google Scholar]
- Li MI, Xu X, Xi N, Wang W, Xing X, & Liu L (2021). Multiparametric atomic force microscopy imaging of single native exosomes. Acta Biochim Biophys Sin (Shanghai), 53(3), 385–388. doi: 10.1093/abbs/gmaa172 [DOI] [PubMed] [Google Scholar]
- Lieberman K, & Lewis A (1993). Simultaneous scanning tunneling and optical near-field imaging with a micropipette. Applied Physics Letters, 62(12), 1335–1337. [Google Scholar]
- Lieberman K, Lewis A, Fish G, Shalom S, Jovin TM, Schaper A, & Cohen SR (1994). Multifunctional, micropipette based force cantilevers for scanned probe microscopy. Applied Physics Letters, 65(5), 648–650. [Google Scholar]
- Lin JR, Fallahi-Sichani M, & Sorger PK (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun, 6, 8390. doi: 10.1038/ncomms9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou C, Su Z, Chang Q, Qiu Y, Bei J, … Gong B (2021). Endothelial Exosome Plays a Functional Role during Rickettsial Infection. mBio, 12(3). doi: 10.1128/mBio.00769-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC (2013). The regulation of cell size. Cell, 154(6), 1194–1205. doi: 10.1016/j.cell.2013.08.053 [DOI] [PubMed] [Google Scholar]
- Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, & Möller A (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles, 4, 27031. doi: 10.3402/jev.v4.27031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakina EB, & Waugh RE (2004). Micromechanical tests of adhesion dynamics between neutrophils and immobilized ICAM-1. Biophys J, 86(2), 1223–1233. doi: 10.1016/S0006-3495(04)74196-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhu Y, Kim S, Liu Q, Byrley P, Wei Y, … Liu M (2016). Sharp-Tip Silver Nanowires Mounted on Cantilevers for High-Aspect-Ratio High-Resolution Imaging. Nano letters, 16(11), 6896–6902. doi: 10.1021/acs.nanolett.6b02802 [DOI] [PubMed] [Google Scholar]
- Mager MD, LaPointe V, & Stevens MM (2011). Exploring and exploiting chemistry at the cell surface. Nature chemistry, 3(8), 582–589. doi: 10.1038/nchem.1090 [DOI] [PubMed] [Google Scholar]
- Marti O, Ruf A, Hipp M, Bielefeldt H, Colchero J, & Mlynek J (1992). Mechanical and thermal effects of laser irradiation on force microscope cantilevers. Ultramicroscopy, 42, 345–350. [Google Scholar]
- Martinac B, Buechner M, Delcour AH, Adler J, & Kung C (1987). Pressure-sensitive ion channel in Escherichia coli. Proceedings of the National Academy of Sciences, 84(8), 2297–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B, Nomura T, Chi G, Petrov E, Rohde PR, Battle AR, … Carne S (2014). Bacterial mechanosensitive channels: models for studying mechanosensory transduction. Antioxidants & redox signaling, 20(6), 952–969. [DOI] [PubMed] [Google Scholar]
- Martinac B, Rohde PR, Cranfield CG, & Nomura T (2013). Patch Clamp Electrophysiology for the Study of Bacterial Ion Channels in Giant Spheroplasts of E. coli. In Delcour AH (Ed.), Bacterial Cell Surfaces: Methods and Protocols (pp. 367–380). Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Martínez-Martín D, Fläschner G, Gaub B, Martin S, Newton R, Beerli C, … Müller DJ (2017). Inertial picobalance reveals fast mass fluctuations in mammalian cells. Nature, 550(7677), 500–505. doi: 10.1038/nature24288 [DOI] [PubMed] [Google Scholar]
- Martinez V, Behr P, Drechsler U, Polesel-Maris J, Potthoff E, Vörös J, & Zambelli T (2016). SU-8 hollow cantilevers for AFM cell adhesion studies. Journal of Micromechanics and Microengineering, 26(5), 055006. [Google Scholar]
- Martinez V, Forró C, Weydert S, Aebersold MJ, Dermutz H, Guillaume-Gentil O, … Demkó L (2016). Controlled single-cell deposition and patterning by highly flexible hollow cantilevers. Lab Chip, 16(9), 1663–1674. doi: 10.1039/c5lc01466b [DOI] [PubMed] [Google Scholar]
- Martinez V, Forró C, Weydert S, Aebersold MJ, Dermutz H, Guillaume-Gentil O, … Demkó L (2016). Controlled single-cell deposition and patterning by highly flexible hollow cantilevers. Lab on a chip, 16(9), 1663–1674. [DOI] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, & Théry C (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol, 21(1), 9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- McConney ME, Singamaneni S, & Tsukruk VV (2010). Probing soft matter with the atomic force microscopies: imaging and force spectroscopy. Polymer Reviews, 50(3), 235–286. [Google Scholar]
- McMullen PD, & Freitag NE (2015). Assessing bacterial invasion of cardiac cells in culture and heart colonization in infected mice using Listeria monocytogenes. J Vis Exp(99), e52497. doi: 10.3791/52497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Gabi M, Behr P, Studer P, Vörös J, Niedermann P, … Zambelli T (2009). FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett, 9(6), 2501–2507. doi: 10.1021/nl901384x [DOI] [PubMed] [Google Scholar]
- Meister A, Gabi M, Behr P, Studer P, Vörös J. n., Niedermann P, … Heinzelmann H (2009). FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano letters, 9(6), 2501–2507. [DOI] [PubMed] [Google Scholar]
- Meister A, Jeney S, Liley M, Akiyama T, Staufer U, De Rooij N, & Heinzelmann H (2003). Nanoscale dispensing of liquids through cantilevered probes. Microelectronic Engineering, 67, 644–650. [Google Scholar]
- Meldolesi J (2018). Exosomes and Ectosomes in Intercellular Communication. Curr Biol, 28(8), R435–R444. doi: 10.1016/j.cub.2018.01.059 [DOI] [PubMed] [Google Scholar]
- Mellata M, Mitchell NM, Schödel F, Curtiss R, & Pier GB (2016). Novel vaccine antigen combinations elicit protective immune responses against Escherichia coli sepsis. Vaccine, 34(5), 656–662. doi: 10.1016/j.vaccine.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, & Diamond MS (2017). Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe, 21(2), 134–142. doi: 10.1016/j.chom.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelviefhaus M, Müller DB, Zambelli T, & Vorholt JA (2019). A modular atomic force microscopy approach reveals a large range of hydrophobic adhesion forces among bacterial members of the leaf microbiota. ISME J, 13(7), 1878–1882. doi: 10.1038/s41396-019-0404-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, … Morelli AE (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood, 119(3), 756–766. doi: 10.1182/blood-2011-02-338004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussy F, Neumann A, & Zingg W (1990). The force of detachment of endothelial cells from different solid surfaces. ASAIO transactions, 36(3), M568–572. [PubMed] [Google Scholar]
- Mueller C, Graessmann A, & Graessmann M (1980). Microinjection: turning living cells into test tubes. Trends in Biochemical Sciences, 5(3), 60–62. doi: 10.1016/0968-0004(80)90068-7 [DOI] [Google Scholar]
- Nagy Á, Kámán J, Horváth R, & Bonyár A (2019). Spring constant and sensitivity calibration of FluidFM micropipette cantilevers for force spectroscopy measurements. Sci Rep, 9(1), 10287. doi: 10.1038/s41598-019-46691-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar R, Tschismarov R, Meissner F, Prabakaran T, Krissanaprasit A, Farahani E, … Paludan SR (2019). Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling. Nat Microbiol, 4(4), 701–713. doi: 10.1038/s41564-019-0367-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawarathna D, Turan T, & Wickramasinghe HK (2009). Selective probing of mRNA expression levels within a living cell. Applied Physics Letters, 95(8), 083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgovan N, Ungai-Salánki R, Lukácsi S, Sándor N, Bajtay Z, Erdei A, … Horvath R (2016). Adhesion kinetics of human primary monocytes, dendritic cells, and macrophages: Dynamic cell adhesion measurements with a label-free optical biosensor and their comparison with end-point assays. Biointerphases, 11(3), 031001. doi: 10.1116/1.4954789 [DOI] [PubMed] [Google Scholar]
- Osada T, Uehara H, Kim H, & Ikai A (2003). mRNA analysis of single living cells. Journal of nanobiotechnology, 1(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossola D, Amarouch M-Y, Behr P, Vörös J. n., Abriel H, & Zambelli T (2015). Force-controlled patch clamp of beating cardiac cells. Nano letters, 15(3), 1743–1750. [DOI] [PubMed] [Google Scholar]
- Ossola D, Dörig P, Vörös J, Zambelli T, & Vassalli M (2016). Serial weighting of micro-objects with resonant microchanneled cantilevers. Nanotechnology, 27(41), 415502. [DOI] [PubMed] [Google Scholar]
- Palmer CP, Mycielska ME, Burcu H, Osman K, Collins T, Beckerman R, … Djamgoz MB (2008). Single cell adhesion measuring apparatus (SCAMA): application to cancer cell lines of different metastatic potential and voltage-gated Na+ channel expression. European Biophysics Journal, 37(4), 359–368. [DOI] [PubMed] [Google Scholar]
- Park K, Millet LJ, Kim N, Li H, Jin X, Popescu G, … Bashir R (2010). Measurement of adherent cell mass and growth. Proc Natl Acad Sci U S A, 107(48), 20691–20696. doi: 10.1073/pnas.1011365107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AI, Kay SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, … The RNAcentral Consortium. (2017). RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res, 45(D1), D128–D134. doi: 10.1093/nar/gkw1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff E, Guillaume-Gentil O, Ossola D, Polesel-Maris J, LeibundGut-Landmann S, Zambelli T, & Vorholt JA (2012). Rapid and serial quantification of adhesion forces of yeast and Mammalian cells. PLoS One, 7(12), e52712. doi: 10.1371/journal.pone.0052712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos D, Mertens J, Calleja M, & Tamayo J (2008). Phototermal self-excitation of nanomechanical resonators in liquids. Applied Physics Letters, 92(17), 173108. [Google Scholar]
- Ridolfi A, Brucale M, Montis C, Caselli L, Paolini L, Borup A, … Valle F (2020). AFM-Based High-Throughput Nanomechanical Screening of Single Extracellular Vesicles. Anal Chem, 92(15), 10274–10282. doi: 10.1021/acs.analchem.9b05716 [DOI] [PubMed] [Google Scholar]
- Rodrigues M, Fan J, Lyon C, Wan M, & Hu Y (2018). Role of Extracellular Vesicles in Viral and Bacterial Infections: Pathogenesis, Diagnostics, and Therapeutics. Theranostics, 8(10), 2709–2721. doi: 10.7150/thno.20576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftics A, Türk B, Sulyok A, Nagy N, Gerecsei T, Szekacs I, … Horvath R (2019). Biomimetic Dextran-Based Hydrogel Layers for Cell Micropatterning over Large Areas Using the FluidFM BOT Technology. Langmuir, 35(6), 2412–2421. doi: 10.1021/acs.langmuir.8b03249 [DOI] [PubMed] [Google Scholar]
- Salánki R, Gerecsei T, Orgovan N, Sándor N, Péter B, Bajtay Z, … Szabó B (2014). Automated single cell sorting and deposition in submicroliter drops. Applied Physics Letters, 105(8), 083703. doi: 10.1063/1.4893922 [DOI] [Google Scholar]
- Sancho A, Vandersmissen I, Craps S, Luttun A, & Groll J (2017). A new strategy to measure intercellular adhesion forces in mature cell-cell contacts. Scientific reports, 7(1), 46152. doi: 10.1038/srep46152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön P, Geerlings J. l., Tas N, & Sarajlic E (2013). AFM Cantilever with in situ renewable mercury microelectrode. Analytical chemistry, 85(19), 8937–8942. [DOI] [PubMed] [Google Scholar]
- Schorey JS, & Harding CV (2016). Extracellular vesicles and infectious diseases: new complexity to an old story. J Clin Invest, 126(4), 1181–1189. doi: 10.1172/JCI81132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Böckelmann R, Malykh Y, … Dress AW (2006). Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol, 24(10), 1270–1278. doi: 10.1038/nbt1250 [DOI] [PubMed] [Google Scholar]
- Selhuber-Unkel C, Erdmann T, López-García M, Kessler H, Schwarz US, & Spatz JP (2010). Cell adhesion strength is controlled by intermolecular spacing of adhesion receptors. Biophysical journal, 98(4), 543–551. doi: 10.1016/j.bpj.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, LeClaire M, Wohlschlegel J, & Gimzewski J (2020). Impact of isolation methods on the biophysical heterogeneity of single extracellular vesicles. Scientific reports, 10(1), 13327. doi: 10.1038/s41598-020-70245-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, & Gimzewski JK (2010). Structural-Mechanical Characterization of Nanoparticle Exosomes in Human Saliva, Using Correlative AFM, FESEM, and Force Spectroscopy. ACS nano, 4(4), 1921–1926. doi: 10.1021/nn901824n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy AT, Beno SM, Brissac T, Bell JW, Novak L, & Orihuela CJ (2018). Severity and properties of cardiac damage caused by Streptococcus pneumoniae are strain dependent. PLoS One, 13(9), e0204032. doi: 10.1371/journal.pone.0204032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yamashita H, Uchihashi T, Kandori H, & Ando T (2010). High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat Nanotechnol, 5(3), 208–212. doi: 10.1038/nnano.2010.7 [DOI] [PubMed] [Google Scholar]
- Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, & Hickman JJ (2015). TEER measurement techniques for in vitro barrier model systems. J Lab Autom, 20(2), 107–126. doi: 10.1177/2211068214561025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl PD, & Raposo G (2019). Extracellular Vesicles: Exosomes and Microvesicles, Integrators of Homeostasis. Physiology (Bethesda), 34(3), 169–177. doi: 10.1152/physiol.00045.2018 [DOI] [PubMed] [Google Scholar]
- Stiefel P, Schmidt FI, Dörig P, Behr P, Zambelli T, Vorholt JA, & Mercer J (2012). Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett, 12(8), 4219–4227. doi: 10.1021/nl3018109 [DOI] [PubMed] [Google Scholar]
- Stranska R, Gysbrechts L, Wouters J, Vermeersch P, Bloch K, Dierickx D, … Snoeck R (2018). Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med, 16(1), 1. doi: 10.1186/s12967-017-1374-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Shelite TR, Qiu Y, Chang Q, Wakamiya M, Bei J, … Gong B (2021). Host EPAC1 Modulates Rickettsial Adhesion to Vascular Endothelial Cells via Regulation of ANXA2 Y23 Phosphorylation. Pathogens, 10(10). doi: 10.3390/pathogens10101307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztilkovics M, Gerecsei T, Peter B, Saftics A, Kurunczi S, Szekacs I, … Horvath R (2020). Single-cell adhesion force kinetics of cell populations from combined label-free optical biosensor and robotic fluidic force microscopy. Sci Rep, 10(1), 61. doi: 10.1038/s41598-019-56898-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha H, Marks RS, Gheber LA, Rousso I, Newman J, Sukenik C, & Lewis A (2003). Protein printing with an atomic force sensing nanofountainpen. Applied Physics Letters, 83(5), 1041–1043. [Google Scholar]
- Takov K, Yellon DM, & Davidson SM (2019). Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J Extracell Vesicles, 8(1), 1560809. doi: 10.1080/20013078.2018.1560809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, & Schekman R (2019). Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife, 8. doi: 10.7554/eLife.47544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N, Ishizaki S, & Uwai H (1991). Scanning attractive force microscope using photothermal vibration. Journal of Vacuum Science & Technology B: Microelectronics and Nanometer Structures Processing, Measurement, and Phenomena, 9(2), 1318–1322. [Google Scholar]
- Ungai-Salánki R, Haty E, Gerecsei T, Francz B, Béres B, Sztilkovics M, … Horvath R (2021). Single-cell adhesion strength and contact density drops in the M phase of cancer cells. Sci Rep, 11(1), 18500. doi: 10.1038/s41598-021-97734-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungai-Salánki R, Peter B, Gerecsei T, Orgovan N, Horvath R, & Szabó B (2019). A practical review on the measurement tools for cellular adhesion force. Advances in Colloid and Interface Science, 269, 309–333. doi: 10.1016/j.cis.2019.05.005 [DOI] [PubMed] [Google Scholar]
- van Oorschot R, Garza HHP, Derks RJ, Staufer U, & Ghatkesar MK (2015). A microfluidic AFM cantilever based dispensing and aspiration platform. EPJ Techniques and Instrumentation, 2(1), 1–11.26146600 [Google Scholar]
- van Rosmalen MG, Roos WH, & Wuite GJ (2015). Material properties of viral nanocages explored by atomic force microscopy. Methods Mol Biol, 1252, 115–137. doi: 10.1007/978-1-4939-2131-7_11 [DOI] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, … Raposo G (2011). The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Developmental Cell, 21(4), 708–721. doi: 10.1016/j.devcel.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden E, Madadelahi M, Sarajlic E, Shamloo A, Engel A, Staufer U, & Ghatkesar M (2020). Volume and concentration dosing in picolitres using a two-channel microfluidic AFM cantilever. Nanoscale, 12(18), 10292–10305. [DOI] [PubMed] [Google Scholar]
- Viljoen A, Mathelié-Guinlet M, Ray A, Strohmeyer N, Oh YJ, Hinterdorfer P, … Dufrêne YF (2021). Force spectroscopy of single cells using atomic force microscopy. Nature Reviews Methods Primers, 1(1), 63. doi: 10.1038/s43586-021-00062-x [DOI] [Google Scholar]
- Viscoli C (2016). Bloodstream Infections: The peak of the iceberg. Virulence, 7(3), 248–251. doi: 10.1080/21505594.2016.1152440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorselen D, Piontek MC, Roos WH, & Wuite GJL (2020). Mechanical Characterization of Liposomes and Extracellular Vesicles, a Protocol. Front Mol Biosci, 7, 139. doi: 10.3389/fmolb.2020.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Jones RC, & Clulow J (1994). Surface area of apical and basolateral plasmalemma of epithelial cells of the ductuli efferentes testis of the rat. Cell Tissue Res, 276(3), 581–586. doi: 10.1007/BF00343956 [DOI] [PubMed] [Google Scholar]
- Williams MA, Schmidt RL, & Lenz LL (2012). Early events regulating immunity and pathogenesis during Listeria monocytogenes infection. Trends Immunol, 33(10), 488–495. doi: 10.1016/j.it.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Shah MK, Powell LC, & Armstrong I (2010). Application of AFM from microbial cell to biofilm. Scanning, 32(3), 134–149. doi: 10.1002/sca.20193 [DOI] [PubMed] [Google Scholar]
- Wysotzki P, Sancho A, Gimsa J, & Groll J (2020). A comparative analysis of detachment forces and energies in initial and mature cell-material interaction. Colloids Surf B Biointerfaces, 190, 110894. doi: 10.1016/j.colsurfb.2020.110894 [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, … De Wever O (2015). Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles, 4, 27066. doi: 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CC, Zhang D, & Gan Y (2017). Invited Review Article: Tip modification methods for tip-enhanced Raman spectroscopy (TERS) and colloidal probe technique: A 10 year update (2006–2016) review. Rev Sci Instrum, 88(3), 031101. doi: 10.1063/1.4978929 [DOI] [PubMed] [Google Scholar]
- Zhang D, Lee H, & Jin Y (2020). Delivery of Functional Small RNAs via Extracellular Vesicles In Vitro and In Vivo. Methods Mol Biol, 2115, 107–117. doi: 10.1007/978-1-0716-0290-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen A, De Leon D, Li H, Noiri E, Moy VT, & Goligorsky MS (2004). Atomic force microscopy measurement of leukocyte-endothelial interaction. Am J Physiol Heart Circ Physiol, 286(1), H359–367. doi: 10.1152/ajpheart.00491.2003 [DOI] [PubMed] [Google Scholar]
- Zhou M, Weber SR, Zhao Y, Chen H, & Sundstrom JM (2020). Chapter 2 - Methods for exosome isolation and characterization. In Edelstein L, Smythies J, Quesenberry P, & Noble D (Eds.), Exosomes (pp. 23–38): Academic Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


