Abstract
Engineered microbes are rapidly being developed for the delivery of therapeutic modalities to sites of disease. Escherichia coli Nissle 1917 (EcN), a genetically tractable probiotic with a well-established human safety record, is emerging as a favored chassis. Here we summarize the latest progress in rationally engineered variants of EcN for the treatment of infectious diseases, metabolic disorders, and inflammatory bowel diseases when administered orally, as well as cancers when injected directly into tumors or the systemic circulation. We also discuss emerging studies that raise potential safety concerns regarding these EcN-based strains as therapeutics due to their secretion of a genotoxic colibactin that can promote the formation of DNA double-stranded breaks in mammalian DNA.
E. coli Nissle 1917 is a chassis of choice for developing smart microbes
Over a century ago, during the pre-antibiotic era, in his quest to identify E. coli from healthy individuals that inhibit the in vitro growth of pathogenic E. coli and related species, Dr. Alfred Nissle isolated one such strain from a World War I soldier1, 2. This strain, E. coli Nissle 1917 (EcN), was subsequently found to have both intrinsic anti-bacterial and anti-inflammatory properties (for review, see3). This led to its long history of use in Europe and Canada, under the trade name of Mutaflor®, for the treatment of inflammatory bowel diseases (IBD) (see Glossary)3. Given its proven human safety record, GRAS (generally regarded as safe) status, genetic tractability and the growing tools of synthetic biology (Box 1), EcN is emerging as a common chassis for the development of smart microbes. While the focus of this review is on EcN-based therapeutics, efforts are also underway to develop variants of other gastrointestinal commensal organisms, including Lactococcus, Lactobacillus, and Bacteroides species (see Charbonneau et al4 and Kelly et al5 for recent reviews). At this point, it is unclear which platforms and chassis will prove to be most efficacious. Here we provide a summary of recently developed EcN variants engineered for the treatment of inflammatory, infectious, and metabolic diseases and cancer (Table 1). In addition, we highlight avenues for further development of EcN-based therapeutics and review recent safety concerns regarding its use.
Box 1: Synthetic biology.
Synthetic biology involves redesigning organisms or cells with new or enhanced abilities as well as the generation of new biological parts, devices or systems. In the context of engineering smart microbes, synthetic circuits are used to develop variants with specific therapeutic or diagnostic properties. These circuits can be expressed via natural or synthetic promoters that control their expression in space and/or time, including ones that respond to environmental cues, e.g., oxygen tension and/or pH, as well as markers of disease, e.g., inflammation. These gene circuits can be further fine-tuned by using Boolean logic gates to process multiple inputs (for review, see66). Engineered circuits are typically initially encoded on plasmids maintained via antibiotic resistance during their initial characterization in the lab. However, as exposing mice and humans to antibiotics is not suitable in most disease settings, they are often transferred to plasmids that can be maintained by other means before advancing to efficacy studies in preclinical models. One strategy is to introduce the gene circuits onto the bacterial chromosome, a process that for which an efficient bacterial conjugation-based method for the markerless integration of genes into the EcN chromosome has recently been described77. Nevertheless, a limitation of chromosomal integration is that the copy number of the insert genes is much lower than when present on most plasmids. When thought to be a potential issue, EcN has been engineered to maintain plasmids via auxotrophic selection. In addition, plasmids can be maintained via toxin/antitoxin systems and recent evidence suggests that endogenous EcN plasmids that have been genetically manipulated are stably maintained78. Lastly, synthetic based approaches are being used to generate variants of genetically modified EcN, and other smart microbes, with biocontainment strategies. A common approach has been to engineer EcN strains that are auxotrophic for metabolites that can be added to media in the laboratory but are not available in the environment. More sophisticated forms of biocontainment have or are currently being developed, including circuits that act as switches to prevent activity of smart microbes by killing them or halting their engineered function. In addition, to ensure lack of genetic transfer of materials form engineered strains, variants with altered genetic codes are being explored (for review see76)
Table 1.
Summary of recently developed EcN therapeutic variants
| EcN Variant | Target | Genetic properties | Mechanism | Site of genetic modifications | Preclinical and clinical findings | Clinical trial number | Ref. |
|---|---|---|---|---|---|---|---|
| Bacterial infections | |||||||
| EcN J25 | Non-typhoidal Salmonella (NTS) | Constitutively secretes MccJ25 | Targets killing of NTS | Plasmid (antibiotic selection) | Limits NTS colonization in a turkey model | 12 | |
| EcN-pttrMcH47 | Non-typhoidal Salmonella | Secretes Mcc47 upon sensing tetrathionate, a reporter of gut inflammation | Targets killing of NTS | Plasmid (antibiotic selection) | Limits S. typhimurium growth in an in vitro killing assay | 16 | |
| EcN BHA | Enterococci gut colonization | Constitutively secretes enterocin A, enterocin B and hiracin JM79, each fused to an N-terminal MccV secretion sequence, via the MccV ABC transporter | Targets killing of Enterococci | Plasmid (antibiotic selection) | Limits Enterococci gastrointestinal colonization in a mouse model | 17 | |
| Sense-Kill-EcN SED | Pseudomonas aeruginosa gut colonization | LasR, is activated upon binding QS after which promotes secretion of Lysis E7, Pyocin S5 and DsbB Lysis E7 promotes EcN lysis Pyocin S5 kills P. aeruginosa DspB degrades biofilms |
Disrupts biofilms and promotes killing of P. aeruginosa | Plasmid (auxotrophic selection) | Limits P. aeruginosa colonization in the gastrointestinal tracts of C. elegans and mice | 18 | |
| Inflammatory bowel disease | |||||||
| PBP8 CsgA-TFF3 | Ulcerative colitis | Displays CsgA-TFF3 fibers on surface of EcN | Displayed TFFs promote epithelial restitution and mucosal healing | Plasmid (antibiotic selection) | Efficacy in mice in a DSS model of colitis | 26 | |
| EGF-EcN | Ulcerative colitis NSAID-induced ulcerative injuries |
Constitutively expresses a lipase ABC transporter from Erwinia chrysanthemi Secretes EGF fused to a C-terminal LARD (lipase ABC transporter recognition domain) |
Secreted EGF stimulates cell migration and extracellular matrix formation to promote mucosal healing | Chromosomal integration | Efficacy in mice in a DSS model of colitis and NSAID-mediated small intestines injury model | 27 | |
| Lresb pDGAT | Ulcerative colitis | UCMs convert NIR to blue light Expresses Ag43, a biofilm promoting protein, when exposed to blue light NIR exposure and timing of administration of bacteria can be used to promote colonization to specific sites in GI tract TGF-ß1 fused to OmpA secretion signal sequence.. |
Spatiotemporal colonization controlled by optogenetic regulation Mucosal healing mediated via secretion of TGF-ß1 |
Plasmid (antibiotic selection) | Efficacy in mice in a chronic DSS colitis model | 28 | |
| Metabolic diseases | |||||||
| SYNB1020 | Hyperammonemia | Lacks ArgR, protein involved in feedback inhibition of arginine biosynthesis Encodes feedback resistant mutant of ArgA (N-acetylglutamate synthase), first enzyme in the arginine biosynthesis pathway ArgA regulated via Pfnrs Thymine auxotroph |
Enhanced conversion of ammonia to L-arginine Unable to replicate in gut or environment (in absence of thymine) | Chromosomal integration | Leads to decreased systemic ammonia levels in the spfash mouse model of ornithine transcarbamylase deficiency, a server urea cycle disorder as well as in a mouse thioacetamide-induced liver injury model. Phase 1 study; healthy volunteers administered the strain; increases in urinary nitrate, a reporter of ammonia metabolism Phase 2a study in cirrhosis patients; terminated; lack of efficacy |
NCT03447730 | 33 |
| SYNB1618 | PKU | Encodes 3 proteins involved in phenylalanine metabolism: (1) Phe: promotes import of Phe into EcN, (2) PAL: converts Phe to trans-cinnamate (3) LAAD, converts Phe to phenylpyruvate Phe and PAL controlled via Pfnrs PAL controlled via PBAD Diaminopinelic acid auxotroph |
Promotes phenylalanine metabolism Unable to replicate (in absence of DAP supplementation) |
Chromosomal integration | Efficacy in the Pahenu2/enu2 PKU mouse model and in dietary challenge of monkeys Phase 2 study in patients with PKU; safe and well tolerated; increases in Phe metabolites in plasma and urine providing proof of mechanism |
NCT04534842 | 34, 36 |
| Cancer | |||||||
| SYNB1891 | Cancer treatment | Expresses dacA from Listeria monocytogenes under the control of Pfnrs promoter Thymine and diaminopinelic acid auxotroph |
Produces cAMP, a STING agonist, that induces antigen presenting cells (APCs) Unable to replicate in gut or environment (in absence of thymine) |
Chromosomal integration | Efficacy in mouse tumor models (CT26, A20, B16.F10) Activates human APCs Ongoing phase I trial in patients with advanced/metastatic solid tumors and lymphoma |
NCT04167137 | 43 |
| SLIC | Cancer treatment | Encodes a quorum sensing lysis system Bacterial lysis results in release of nanobodies and cytokines | Synchronized delivery into tumor microenvironment of nanobodies that block immune checkpoint inhibitors, and GM-CSF, a cytokine that stimulates the production and differentiate of myeloid cells | Chromosomal integration Plasmid (toxin/antitoxin selection) |
Efficacy in A20 and CT26 mouse tumor models | 45 | |
| EdaI1-HlpA | Cancer prevention | Constitutively surface displays S. gallolyticus HlpA fused to ice nuclease protein membrane localization domain Constitutively secretes myrosinase fused to YebF secretion signal |
HlpA promotes binding to heparin sulfate proteoglycan on the surface of colorectal tumors. Myrosinase converts glucosinolate to sulforaphane, an inhibitor of cancer cell growth |
Plasmid (auxotrophic selection) | Efficacy in AOM/DSS mouse CRC model | 46 | |
| L-Arg EcN | Cancer treatment | Lacks ArgR, protein involved in feedback inhibition of arginine biosynthesis Encodes feedback resistant mutant of ArgA (N-acetylglutamate synthase), first enzyme in the arginine biosynthesis pathway ArgA regulated via Pfnrs |
Elevated levels of production of L-arginine promote T cell infiltration of tumors | Chromosomal integration | Efficacy in mouse MC38 and B16-OVA (+ T cells) tumor models. | 48 | |
Abbreviations: AOM, azoxymethane; AHL, acyl homoserine lactone; NSAID, nonsteroidal anti-inflammatory drug.
Enhancing the antimicrobial properties of EcN
A recent area of development of EcN-based therapies is the generation of variants engineered to secrete antimicrobial proteins into the intestinal lumen that target the killing of gastrointestinal bacterial pathogens, i.e., Salmonella, as well as commensal strains that can lead to the development of opportunistic infections, i.e., Enterococci6 and Pseudomonas aeruginosa. The overall strategy of each of these groups has been to outfit EcN to release bacteriocins, ribosomally-synthesized secreted antimicrobial proteins that inhibit the growth of related bacterial species7, 8.
EcN encodes two small bacteriocins, i.e., microcins, only one of which has been established to be expressed and secreted, in this case, within the inflamed gut or when grown under iron-limiting conditions in the laboratory9. Each of the EcN strains discussed below has been engineered to enhance the repertoire of EcN bacteriocins and to expand the conditions under which they are released. This is not as straight forward as heterologously expressing the gene encoding a bacteriocin in EcN. Rather, it is necessary to additionally outfit EcN such that it can process and secrete the bacteriocins.
The secretion of microcins across the inner membrane of EcN and other Gram-negative bacteria is mediated by dedicated ABC transporters, complex transmembrane proteins, after which they are secreted via porins present in the outer membrane into the surrounding extracellular space (for review, see10, 11). Two groups independently engineered EcN to secrete either MccJ2512 or MccH4713, each of which target the killing of E. coli and Salmonella. In the first case, Forkus and colleagues developed EcN MccJ25, EcN that secretes MccJ25. To enable high level expression of Mcc25, the gene encoding its precursor, mcjA, was placed with that encoding ProTeOn, a synthetic transcription factor, in an operon that is controlled by ProTeOn, thus generating a positive feedback loop (Fig. 1A)14. The enzymes needed for its processing and the ABC transporter were positioned downstream of this operon under the control of a constitutive promoter. In the second case, Palmer and colleagues developed EcN-pttrMcH47, EcN that secretes MccH47, a microcin that also targets E coli and Salmonella. In this case, they engineered EcN to express MccH47 upon sensing tetrathionate, a reporter of gut inflammation15. This was accomplished by engineering a hybrid operon, in which genes encoding proteins needed for sensing tetrathionate are positioned upstream of those involved in MccH47 biosynthesis and secretion (Fig. 1B)16.
Figure 1. Variants of EcN engineered with enhanced antimicrobial properties.
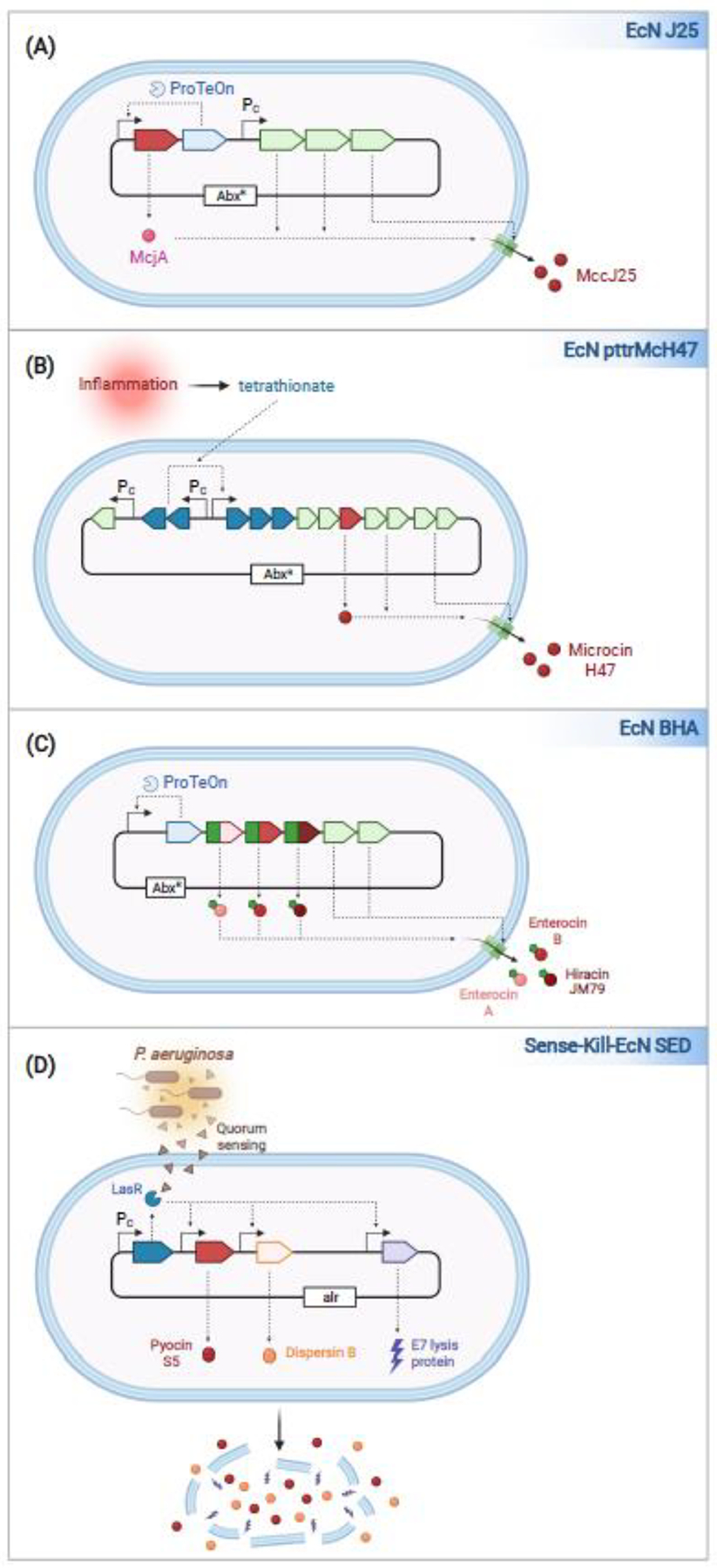
(a) EcN J25 is engineered to constitutively secrete the microcin MccJ25. The expression of McjA, the MccJ25 precursor, and ProTeOn, are controlled via a ProTeOn-dependent promoter, thus generating a positive feedback loop. Genes encoding the proteins needed for J25 biosynthesis and transport are in an operon present on the same plasmid under the control of a constitutive promoter maintained via antibiotic selection. (b) EcN pttrMcH47 is engineered to secrete the microcin Mcc47 upon sensing tetrathionate, a reporter of inflammation. Genes encoding the proteins needed for tetrathionate sensing are positioned upstream of those involved in MccH47 biosynthesis and export. (c) EcN BHA is engineered to constitutively secrete Enterocins A and B, and Hiracin, each fused to a MccV N-terminal secretion tag. The expression of all three bacteriocins, the proteins that mediate their secretion, and ProTeOn are controlled via the expression of a ProTeOn-dependent promoter on a plasmid maintained via antibiotic selection. (d) Sense-Kill-EcN SED is engineered to produce molecules that eradicate Pseudomonas aeruginosa. Sense-Kill-EcN SED constitutively expresses LasR, a transcription factor activated upon binding a quorum-sensing (QS) molecule produced by P. aeruginosa. Activated LasR promotes the transcription of pyocin S5, E7 lysis protein, and Dispersin B. E7 proceeds to lyse the cells, releasing pyocin S5 and Dispersin B. The genes encoding the ‘sense-kill’ circuit are encoded on a plasmid that is maintained via auxotrophic selection.
In the next two instances, the investigators developed platforms that enable EcN to secrete bacteriocins from more distantly related species, i.e., Gram-positive Enterococci and non-enteric Gram-negative Pseudomonas aeruginosa, both of which can cause difficult to treat antibiotic-resistant opportunistic infections. In the case of the Gram-positive enterocins, Geldart and colleagues fused three Enterococci specific bacteriocins, enterocin A, enterocin B and hiracin JM79, to an N-terminal E. coli microcin V secretion tag. These amino acids are cleaved off as the hybrid microcins are secreted across the MccV autotransporter13. They then introduced the genes encoding these fusion proteins into the MPES (Modular Peptide Expression System), whereby they are encoded in a positive feedback circuit with those encoding ProTeOn and the MccV ABC transporter (Fig. 1C)17.
In the case of the pyocins, Hwang and colleagues developed Sense-Kill-EcN SED, EcN engineered to specifically sense and target Pseudomonas aeruginosa (Fig. 1D)18, 19. While P. aeruginosa is not typically a component of the gut microbiota, it can be found in the intestines of patients treated with broad-spectrum antibiotics. Sense-Kill-EcN is designed to expresses four heterologous proteins: LasR, a transcription factor activated upon binding to a small quorum-sensing (QS) molecule secreted by P. aeruginosa; Lysis E7, a phage bacterial lysis protein, Dispersin B (DspB), a biofilm disruptor, and Pyocin S, a P. aeruginosa-bacteriocin. E7-mediated lysis of Sense-Kill-EcN SED releases DspB and Pyocin S, which work synergistically to eradicate P. aeruginosa.
When engineering EcN to secrete bacteriocins, high-level expression/secretion of these antimicrobial proteins is likely desired. Thus, motivating the placement of these gene circuits on multi-copy plasmids. Using a variety of experimental approaches, each of the five bacteriocin-secreting strain was demonstrated to secrete functional bacteriocins and four of the strains were studied in preclinical models where they have shown variable levels of efficacy in eradicating the intestinal colonization of the targeted bacterium. Mixed results were observed when investigating the ability of EcN MccJ25 to limit the colonization of Salmonella enteritidis in the gastrointestinal tract of turkeys and of EcN BHA to limit the growth of E. faecium or E. faecalis, two common etiologies of vancomycin resistant enterococci (VRE), in the intestines of mice. The bacteriocins in both these strains are encoded on a plasmid that is maintained in EcN via antibiotic selection. It seems likely that the mixed results reflect plasmid loss, a possibility that was not investigated.
In preclinical Caenorhabditis elegans and mouse models of gastrointestinal Pseudomonas infections, Sense-Kill-EcN SED reduced intestinal colonization when administered as a prophylactic or therapeutic intervention. In this case, to ensure maintenance of the plasmid-encoded ‘sense-kill’ circuit, the investigators used an auxotrophic selection scheme. This was achieved by developing a variant of EcN incapable of synthesizing D-alanine, an essential bacterial cell wall component, such that growth of the strain is dependent on the presence of D-alanine supplementation or the introduction of an alanine racemase-encoding plasmid.
These recently developed strains demonstrate how outfitting EcN to secrete bacteriocins can serve as an intervention for the prevention and treatment of intestinal infections. They build upon earlier work whereby a variant of EcN was engineered to secrete CAI-1, a quorum sensing factor which limits the expression of Vibrio cholerae virulence factors, was demonstrated to limit V. cholerae virulence in a mouse model20. In the future it is likely that EcN will be engineered to secrete factors that synergize to inhibit growth of GI pathogens and commensals, including bacteriocins, quorum sensing molecule and proteins that inhibit specific virulence determinants, i.e., nanobodies that inhibit the activity of bacterial toxins21 and essential secreted virulence determinants22.
Engineering EcN for the treatment of inflammatory bowel disease
Inflammatory bowel diseases (IBD) are commonly treated with systemically administered agents that suppress inflammation. Some of the more immunosuppressive and effective agents, including monoclonal antibodies that neutralize TNFα, are associated with an increased risk of developing life-threatening infections as well as lymphoma. Due to its inherent anti-inflammatory properties, EcN is used for the treatment of patients with IBD, where it has been demonstrated to be as effective as an oral agent in limiting IBD flares23. However, EcN is not always effective24, 25. To enhance its therapeutic efficacy, several groups recently sought to enhance its ability to promote intestinal mucosal healing by engineering EcN to produce molecules naturally involved in this process. These engineered variants have the potential of acting directly at the main site of disease, the intestines, thus limiting off-target effects. Each group has developed an innovative means to enable EcN to secrete or display a therapeutic payload, as EcN, like most Gram-negative bacteria, does not secrete substantial levels of proteins into its surroundings.
In the first study, Praveschotinunt and colleagues developed the PATCH (Probiotic Associated Therapeutic Curli Hybrid) system26, a platform for the display of high concentrations of proteins on the surface of EcN. This is accomplished by fusing self-assembling protein (CsgA) subunits to a protein of interest. Once secreted, the CsgA fusion proteins form Curli, nanofibers that extend from the surface of the bacteria. PBP8 CsgA-TFF3 is a variant of EcN that express CsgA fused to TFF3, a trefoil factor that promotes mucosal healing, along with the proteins needed to promote export of CsgA-TFF3. This gene circuit is carried on a plasmid maintained via antibiotic selection and expressed via an arabinose-dependent (PBAD) promoter (Fig 2A)26. In the second study, Yu and colleagues introduced the genes encoding a lipase ABC transporter from Erwinia chrysanthemi into the chromosome of EcN27, which they engineered to recognize and secrete transforming growth factor beta (TGF-ß1), another protein that promotes mucosal healing. In this way they generated EGF-EcN, a variant that constitutively secretes TGF-ß1 into the intestinal lumen (Fig. 2B).
Figure 2. Engineered EcN variants for the treatment of inflammatory bowel disease.
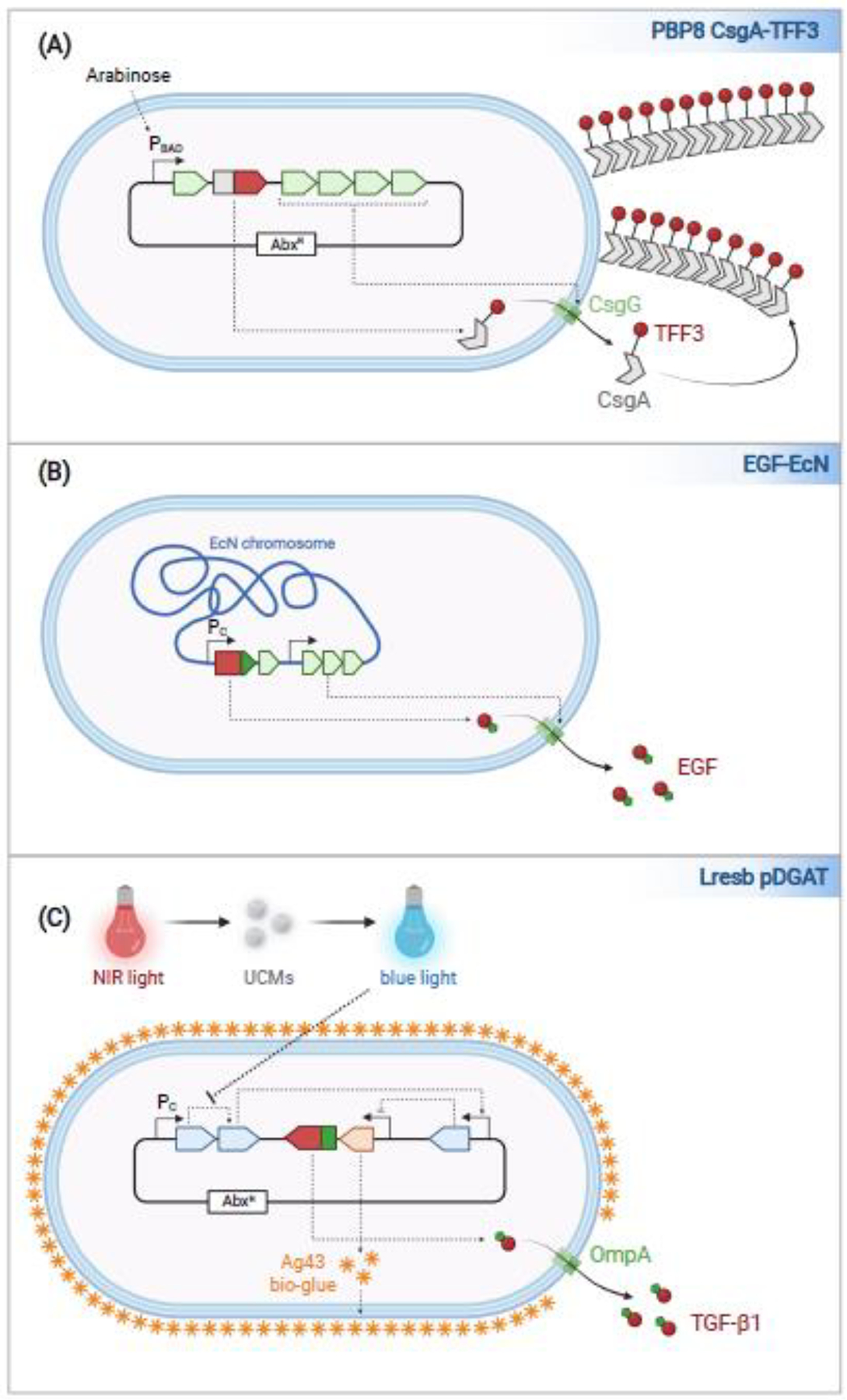
(a) PBP8 CsgA-TFF3 is engineered to display TFF3 on its surface. TFF3 is fused to CsgA such that it is assembled and exposed on curli fibers attached to EcN. The expression of the plasmid-encoded CsgA-TFF3, and other components of the curli operon, including the assembly/transport component CsgG, is expressed in an arabinose-dependent manner from a plasmid. (b) EGF-EcN is engineered to constitutively express a lipase ABC transporter. EGF is fused to a C-terminal LARD (Lipase ABC transporter Recognition Domain). The genes encoding the transporter and EGF are integrated into the EcN chromosome. (c) Lresb pDGAT is engineered with a blue-light responsive circuit that controls expression of Ag43, an adhesin (‘bio-glue’) that promotes EcN biofilm formation. Since blue-light does not penetrate the intestines, upconversion molecules (UCMs) that convert the NIR light to blue light are co-administered with the bacteria. Lresb pDGAT secrete TGF-ß1 fused to an OmpA secretion signal peptide. The genes encoding this system are encoded on a plasmid maintained via antibiotic selection.
The therapeutic efficacy of PBP8 CsgA-TFF3 and EGF-EcN were each investigated in a mouse dextran sodium sulphate (DSS)-induced colitis model of IBD. PBP8 CsgA-TFF3 demonstrated some improvement in markers of colitis and a modest, but statistically non-significant, reduction in histopathology scores. While treatment with EGF-EcN was efficacious in both preventing and treating DSS-induced colitis in mice, the magnitude of protection was more effective when administered as a prophylactic intervention. While these platforms have demonstrated proof of concept, there is room for improvement for each, which potentially could be achieved by increasing the levels of TFF3 and TGF-ß1 that are in proximity to the intestinal mucosa.
In the third study, Cui and colleagues focused on targeting EcN colonization to specific loci in the gastrointestinal tract28. Thus, they engineered EcN to express, in a blue-light responsive manner, Ag43, an adhesin that promotes biofilm formation, that they propose enables the bacteria to form “bio-glues” that enhance bacteria survival in the gastrointestinal tract (Fig. 2C). By co-administering Lresb EcN with particles that contain upconversion microgels (UCMs), which convert near-infrared light (NIR) to blue light, they found that it is possible to promote the expression of Ag43 within the gastrointestinal tract by exposing the mice to a NIR light. By controlling the timing of exposure of mice to NIR post-administration of EcN and UCMs, colonization of the small intestines versus the colon can be favored, although the magnitude of increased localized colonization to each region was low. Nevertheless, a variant engineered to constitutively secrete TGF-ß1 by fusion to an OmpA signal sequence was effective in ameliorating disease in a DSS-induced chronic colitis model
EcN-, Lactococcus lactis- and Saccharomyces cerevisiae-based platforms targeting the modulation of cytokines29, 30, purines (e.g., ATP)31, and the microbiota32 have also demonstrated success in preclinical models of colitis. In the future, it is likely that each of these platforms, and others, could be expanded for the site-specific delivery of multiple therapeutic payloads, including those that promote mucosal healing as well as block the activity of pro-inflammatory cytokines.
Rewiring EcN for the processing of toxic metabolites.
EcN-based therapeutics are emerging as a platform for the treatment of diseases associated with the accumulation of toxic metabolites. The strategy in this situation is to enhance the ability of EcN within the gut to convert the toxic metabolites, including ammonia and phenylalanine into non-toxic forms, thereby limiting their absorption and distribution via systemic circulation.
Patients with extensive liver disease, i.e., cirrhosis, and those with inborn mutations in ammonia-metabolizing enzymes (urea cycle disorders) develop hyperammonemia. If not lowered, circulating ammonia can lead to neurologic sequelae, including the development of hepatic encephalopathy. To enhance the consumption of ammonia in the intestines, Kurtz and colleagues rewired EcN to efficiently channel gut ammonia into the L-arginine (L-arg) biosynthesis pathway33. The resulting strain, SYNB1020, lacks ArgR, a negative regulator of this biosynthetic pathway, and expresses a feedback-resistant variant of ArgA, a L-arg biosynthesis enzyme. The modified ArgA is expressed via the hypoxic induced promoter PfnrS, which promotes high-level expression but only when the bacteria are grown in anaerobic conditions, including the gut. To ensure that this strain does not replicate within the gut or once shed in stool, it was also engineered as a thymine auxotroph.
Patients with Phenylketonuria (PKU), an inborn error of phenylalanine metabolism, also have issues with toxic metabolites. In this case, they have elevated levels of circulating phenylamine which like ammonia can lead to the development of neurologic damage. These patients are deficient in the production of phenylalanine hydroxylase (PAH), a key enzyme involved in phenylalanine degradation. The current mainstay of therapy is strict limitation of dietary protein. To enhance the consumption of phenylalanine in the intestines, Isabella and colleagues have developed SYNB1618 and SYNB1934, variants of EcN engineered with three chromosomally inserted gene circuits that promote phenylalanine metabolism34, 35. Within the hypoxic gut environment, both expresses a phenylalanine transporter (PheP), which promotes uptake of the amino acid into bacteria, and phenylalanine ammonia lyase (PAL), an enzyme that converts phenylalanine to a non-toxic metabolite, trans-cinnamate, each under the control of the hypoxic induced promoter PfnrS. The genetically engineered PAL variant present in SYNB1934 is approximately two-fold more active than that found in SYNB161835. Both strains also express L-amino acid deaminase (LAAD), a protein that metabolizes phenylalanine to non-toxic phenylpyruvate under aerobic conditions, via an arabinose inducible promoter. Arabinose is added to the fermentation process such that LAAD is expressed when SYNB1618 initially enters the gastrointestinal tract.
SYNB1020, SYNB1618, and SYNB1934 have been engineered to be auxotrophic for thymine and/or diaminopimelic acid, which ensures that they cannot replicate outside of the gut, unless grown in supplemented media. This is a biocontainment strategy to alleviate concerns regarding their introduction into the environment. In terms of therapeutic efficacy, oral administration of each of the three strains demonstrated efficacy in preclinical mouse models of their respective target diseases. SYNB1618 also exhibited efficacy in a non-human primate model34.
All three strains have advanced to human trials. SYNB1020 was well tolerated in a phase 1 trial and was associated with increased levels of circulating nitrate, a biomarker of strain activity. However, a phase 2a trial with the strain was terminated due to lack of improvement in circulating ammonia levels in cirrhotic patients, although the strain did lower levels of circulating nitrate, suggesting that the strain was active, but likely not as metabolically active as needed. Results from a SYNB1618 phase 1/2a indicated that it is well tolerated and increases levels of predicted metabolites in plasma and urine36. A phase 2 clinical trial is currently underway to test efficacy of SYNB1618 and the second-generation, more potent SYNB1934 in patients with PKU. Whether or not these strains have metabolic capacity to achieve therapeutic efficacy in the target patient population, remains to be seen. However, if needed, Adolfsen and colleagues have developed an innovative platform to screen their greater than 1-million member mutant PAL library, which they can use to screen for variants with increased activity within the hypoxic acidic environment of the gut35.
Outfitting EcN for the treatment of cancer
The pioneering studies of William Coley in the late 19th century revealed the potential of pathogenic bacteria as anticancer agents and engineering bacteria as anticancer agents has become a burgeoning area of research, for review see37, 38. EcN shows a propensity to home to and proliferate to titers on the order of 108–1010 CFU/g in tumors. However, its presence does not induce a marked immune response or tumor eradication39–42. These observations have led to a surge in efforts to engineer EcN to promote anti-tumor immunity (Box 2), which we review below.
Box 2: Anti-tumor immunity.
The term anti-tumor immunity refers to innate and adaptive immune responses, which lead to tumor eradication. This can be achieved therapeutically using non-specific immunotherapies, vaccines, adoptive-cell therapy and more recently through the blockade of immune checkpoint molecules. Immune checkpoint molecules act to prevent over-activation of the immune system during the host immune response to an invading pathogen and to promote tolerance to self-antigens. However, many malignant tumors alter the expression of these molecules to prevent their recognition by the host immune system. Three of the best-studied checkpoint molecules in this scenario are PD-1 (programmed death protein 1), PD-L1 (programmed death ligand 1) and CTLA-4 (cytotoxic T-lymphocyte-associated antigen 4)79–81. Monoclonal antibodies that bind to and inhibit PD-1, PD-L1 and CTLA-4 promote tumor clearance primarily through the activation of T cells and are showing extraordinary promise in clinical trials, particularly in the treatment of melanoma, renal cell, and lung cancer. However, immune checkpoint inhibitor drugs are administered intravenously which typically results in overactivation of the immune system in areas of the body outside the tumor resulting in immune adverse events (e.g., colitis, hepatitis)82, 83, an observation that has prompted investigators to develop targeted approaches for their delivery, such as the EcN-based platforms discussed herein. An additional limitation of checkpoint inhibitors is that they are less effective in cold tumors i.e., tumors that are poorly infiltrated by T cells. In this regard, new approaches that mimic or activate the innate immune system are being developed though with same goal of ultimately inducing anti-tumor immunity. For example, type I interferons (IFN) can promote antitumor immunity through a variety of pathways84 and increasing their production within the tumor microenvironment can be achieved using agonists of STING (Stimulator of interferon genes). The pharmaceutical industry is developing STING agonists with the rationale of activating tumor cells or tumor-infiltrating immune cells (including dendritic cells)85, and approaches such as the EcN-based strategies discussed herein offer a targeted mechanism of intratumoral STING activation.
STING (STimulator of INterferon Genes) agonists are emerging as a new means for the treatment of tumors, as they act to boost intratumoral immune responses. As a potential means to deliver STING agonists directly to tumors, Leventhal and colleagues developed SYNB1891, an EcN variant that expresses the Listeria monocytogenes Diadenylate cyclase (DacA) enzyme via the Pfnrs hypoxia-induced promoter and thus produces high levels of cyclic AMP, a STING agonist (Fig. 3A)43. SYNB1891, like SYNB1618 is auxotrophic for thymine and diaminopimelic acid. In vitro studies showed that the uptake of SYNB1891 into phagocytic cells triggered the secretion of interferons, while in mouse studies, the direct injection of the strain into a cold mouse tumor promoted its clearance and induced the development of immunological memory that prevented tumor relapse. SYNB1891 is currently being tested in a phase I clinical trial.
Figure 3. EcN variants developed for the treatment of cancer.
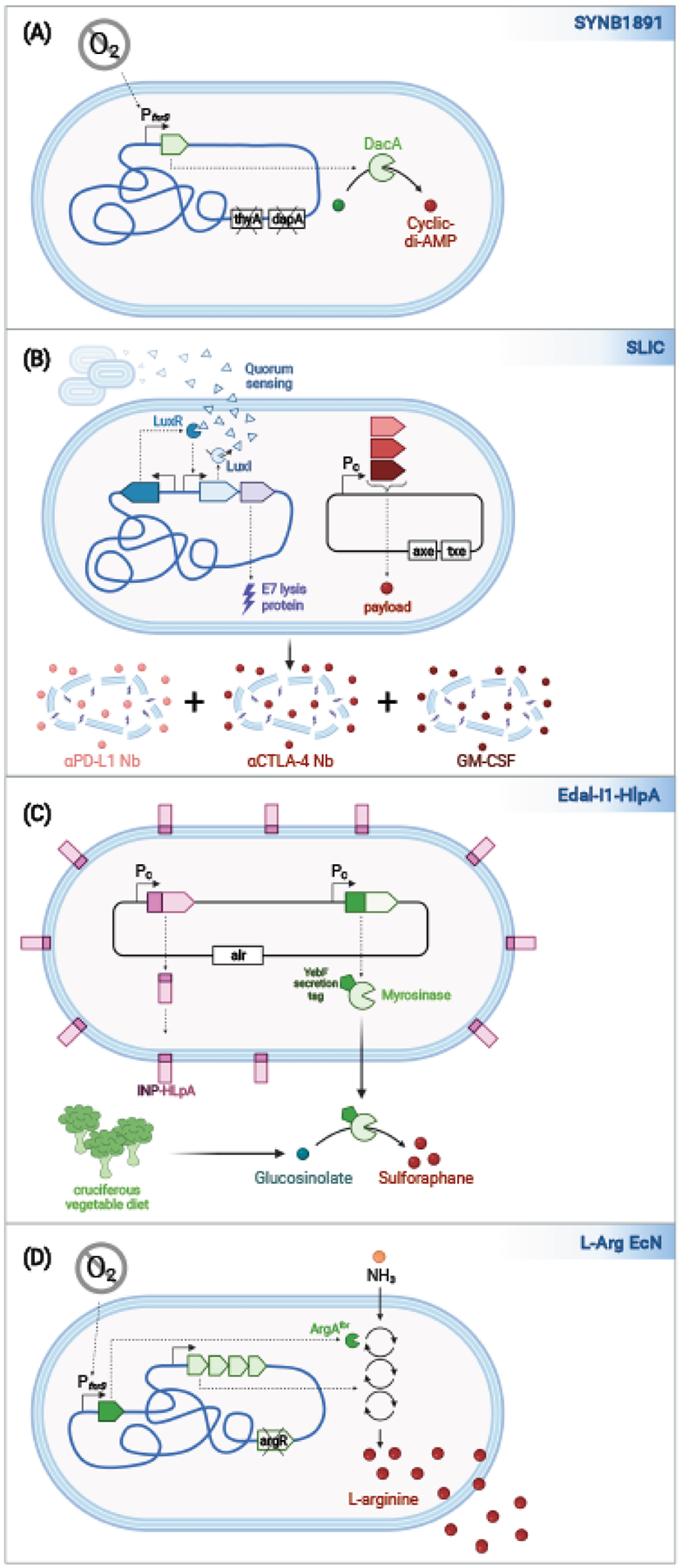
(a) SYNB1891 engineered to express Listeria monocytogenes DacA, which promotes the biosynthesis of cyclic di-AMP. dacA is encoded in the chromosome under the control PfnrS, a hypoxia-induced promoter. For biocontainment, SYNB1891 lacks thyA and dapA. (b) SLIC variants of EcN each contain a chromosomally encoded lysis circuit, whereby the expression of the QS molecule by LuxI promotes activation of LuxR, which promotes the expression of additional LuxI and the E7 lysis protein. When the bacteria reach a certain density, they coordinately lyse releasing therapeutic payloads. The payload proteins are constitutively expressed and encoded on a plasmid maintained via a toxin-antitoxin system. Strains expressing different payloads can be used in combination, as exemplified by the coordinated release of anti-PD-L1 and anti-CTLA-4 nanobodies and recombinant GM-CSF for the treatment of cancer. (c) Eda-l1-HlpA secretes myrosinase, an enzyme that catalyzes the conversion of glucosinolate to sulphoraphane, and HlpA, a protein that promotes binding to cancer cells. Myrosinase is targeted for secretion via the Sec pathway and OMP porins via fusion to a YebF secretion tag. HlpA is exposed on the other surface of the E. coli via fusion to INP (ice nuclease protein). Both proteins are constitutively expressed from a plasmid maintained via auxotrophic selection. (d) L-Arg EcN is engineered to convert ammonia to L-arginine when grown under hypoxic conditions. L-Arg no longer encodes argR, a repressor of the L-argnine biosynthesis, and carries a feedback-resistant mutant of ArgA, the first enzyme in this biosynthesis pathway, under the control of the PfnrS promoter.
Therapies that block immune checkpoint molecules (e.g., PD-L1 and CTLA-4) have revolutionized the treatment of hot immunogenic tumors; however, their systemic administration is associated with off-target effects, which lead to immune-related adverse events (irAEs). To enable the direct delivery of therapeutic payloads to tumors, Gurbatri and colleagues developed EcN with a SLIC (synchronized lysing integrated circuit) platform (Fig. 3B)44, 45. This circuit, like that present in Sense-Kill EcN SED, uses quorum-sensing to trigger EcN lysis via expression of the E7 lysis protein. However, in this case, EcN itself are the source of the QS-molecule. SLIC contain a synthetic operon whereby the gene encoding LuxI, the protein that produces the QS molecule, is located upstream of that encoding the E7 lysis protein. LuxR, the transcription factor activated upon by the QS-molecule, is constitutively expressed. QS molecules produced by SLIC diffuse from one bacterium to another such that once a critical threshold concentration is reached the bacteria undergo synchronized lysis. SLIC strains that constitutively express anti-PD-L1 Nb (nanobody), anti-CTLA-4 Nb and granulocyte-macrophage colony-stimulating factor (GM-CSF) were tested individually and in combination for their ability to efficiently promote anti-tumor immunity and tumor growth in several mouse cancer models. When used in combination the Nb-producing strains were efficacious in the treatment of hot tumors, while all three strains were needed in the case of cold tumors.
EcN has also been engineered to enhance the availability of the phytochemical sulphoraphanes, an anti-tumor compound derived from glucosinolates found in cruciferous vegetables. Ho and colleagues engineered Eda-l1-HlpA to constitutively secrete horseradish myrosinase fused to a YebF secretion sequence46. This strain also is designed to express Streptococcus gallolyticus HlpA on its outer surface. HlpA promotes interactions with heparin sulfate proteoglycan on the surface of colorectal tumors46. To display HlpA on the surface of EcN it is fused to an ice nuclease protein (Fig. 3C). When administered with a cruciferous vegetable diet, mice receiving the resulting strain Eda-l1-HlpA exhibited smaller and fewer colorectal tumors in a chemically induced model of colorectal cancer (CRC).
Given evidence that the limited availability of L-arginine in tumors restricts T cell responses47, Canale and colleagues investigated whether L-Arg EcN, an EcN variant optimized for the conversion of ammonia to L-Arginine, promotes T cell activation and inhibits tumor growth48. This strain contains the same modifications in this biosynthetic pathway as described above for SYNB1020. When directly injected into tumors, L-Arg EcN reached similar titers as unmodified EcN, but, in contrast, was associated with elevated levels of intratumor arginine and increased tumor-infiltrating T cells. While the presence of the strain alone did not affect tumor growth, co-administration of anti-PD-L1 blocking antibodies led to a significant reduction in the growth of immunogenic hot tumors. When T cells engineered to target the tumor were added, this intervention also inhibited growth of poorly immunogenic cold tumors.
Here we have chosen primarily to focus on engineered EcN variants that promote the development of anti-tumor immune responses. In addition, other engineered EcN strategies have recently been developed for the targeted delivery of chemotherapeutic agents including doxorubicin49 and P53 and Tum-550 to tumors. Collectively, these promising findings pave the way for EcN-based cancer therapeutics and raise interesting questions. For example, what EcN determinants promote its homing to and replication in tumors? Does the presence of the engineered EcN lead to immune responses that result in its clearance from tumors? Is there a risk of infection associated with their administration, particularly when given to those who are immunosuppressed due to their malignancies or related treatments?
Revisiting the safety of EcN
EcN has been administered to patients for over a century and there have been few reports of adverse events in the literature. However, like other members of the B2 phylogenetic group of E. coli, EcN contains a pks genomic island. This island encodes enzymes that mediate the synthesis of colibactin, a genotoxin capable of alkylating DNA and inducing double-strand breaks51. This is of concern, as in several mouse models, other colibactin-producing E. coli have been observed to promote the development of colorectal cancer (CRC)52–57, and human organoids exposed to certain pks+ E. coli exhibit colibactin-dependent mutational signatures also found in human CRCs58.
Whether EcN is pro-tumorigenic is not yet known. It was originally thought that EcN does not produce colibactin59. However, colibactin was recently detected in EcN supernatants and cell lines exposed to EcN have been found to exhibit evidence of colibactin-dependent DNA cross-linking and mutations60. In animal models where EcN has been orally administered to germ-free anexic mice and 8-day old SPF (specific pathogen free) mice, their intestinal epithelial cells exhibited evidence of DNA damage60, which was not observed when mice were inoculated with variants that carry mutations in clbA or clbP, genes encoded in the pks island that are directly involved in colibactin synthesis61, 62. In contrast, no evidence of DNA damage was found in gastrointestinal cells of conventionally-raised adult rats colonized with EcN59.
Why is it that EcN is associated with DNA breaks in the intestinal epithelial cells of mice but not rats, and which is most representative of the situation in humans? DNA damage was monitored via different assays in the two models, perhaps explaining their disparate findings60. An alternative possibility is that the high titers of EcN present in the colon of anexic adult mice [~109 colony forming units (CFU)/g feces] and presumably 8-day old SPF mice [titers not reported], 6 hours post oral inoculation with ~108 CFU, result in the exposure of intestinal epithelial cells to non-physiological high levels of colibactin. This effect could be further magnified in the germ-free mice and pups, as the colonic mucous layer of these mice is less developed as compared to that of conventionally raised mice63, and direct contact between colibactin and host cells, which even a thin lay mucus can inhibit, has been reported to be required for the formation of DNA breaks64.
To the best of our knowledge, EcN has never been demonstrated to promote tumorigenesis in a mouse model, and there is no evidence, correlative or otherwise, linking EcN with colorectal tumors in humans. An easy “fix” might seem to remove the pks island from EcN; however, there is evidence that this region is also involved in mediating its anti-inflammatory65 and its anti-bacterial properties. Interestingly, a point mutation in clbP a pks island gene, has been identified that impairs the production of colibactin, but not its microcins60. Encouragingly, this variant of EcN is not impaired in colonization of the gastrointestinal tracts of mice, suggesting that, if needed, this modification could be introduced into engineered EcN to alleviate concerns regarding their production of colibactin.
Concluding remarks and future perspectives
It is early and exciting times in the development of smart microbes as therapeutics. In this review, we provide a deep-dive into recently developed EcN-based smart microbes engineered with enhanced intrinsic anti-bacterial and anti-inflammatory properties as well as new therapeutic modalities. While a few have advanced to clinical trials, essentially nothing is known regarding how these strains will work in humans and, in most cases, before proceeding in this direction, more proof-of-principle experiments are needed in animal models (see Outstanding Questions).
Outstanding Questions.
Which types of preclinical animal and in vitro cell culture-based models will prove to be most reflective of EcN-based therapeutics in human disease?
How will EcN interactions with the host microbiota influence their effectiveness?
When colonization in humans is a desired modality, will the engineered gene circuits in EcN remain stable?
Will the presence of the genomic pks island in E. coli prove to be a counter-indication for the use of these smart microbes as a therapeutic platform?
Will engineered EcN variants trigger immune responses that limit their repeated use over time?
How many modifications can be introduced into EcN to improve upon its therapeutic efficacy before imparting a fitness cost?
For what additional targets and disease indications can engineered EcN-based platforms be developed?
As these strains are further developed, it will be important to consider ways in which their therapeutic efficacy can be improved. For example, approaches to optimally regulate therapeutic delivery by smart microbes in both space and time is an area of active investigation (for reviews see66–68). In this context, an active area of research is the refinement of ‘sense and respond systems’ activated in response to environmental cues, e.g., by pH or oxygen levels38, the presence of an invading pathogen69, or an inflammatory marker15, 70. Another area of active investigation is the development of ‘user activated’ biologic circuits triggered by exposure to light28, ultrasound71 or Wi-Fi72). In parallel, to promote binding to specific regions of the intestines and potentially types of tumors, EcN could be engineered to express designer adhesins that promote their binding to specific cell types73.
In parallel with these efforts, it is essential to ensure biosafety and biocontainment. The current variants in clinical trials are auxotrophic strains that do not replicate in patients or the environment. However, as it is envisioned that the efficacy of some variants will be dependent on their prolonged colonization and ability to replicate, before proceeding to clinical trials, there need to be ways to ensure clearance of the bacteria from patients if they become associated with adverse reactions, as well as means to ensure that shed bacteria are not viable in the environment. While the administration of antibiotics to patients could address the first issue, it is not ideal given that these agents will perturb the gut microbiota. To address the latter there are extensive efforts underway to generate strains with temperature-sensitive kill switches that are engaged upon sensing a change in the environment, i.e., ambient temperatures encountered after fecal elimination74, 75, as well as additional approaches as recently reviewed in76.
In summary, as exemplified by the progress summarized here, engineered EcN variants hold great promise as an affordable means to provide high specificity therapeutic interventions to sites of disease. While there are outstanding questions regarding the long-term utility of engineered EcN variants as therapeutics, given EcN’s genetic tractability it will likely be possible to modify the strains as needed to uncouple their therapeutic and potentially adverse effects. Overall, recent progress in this field paves the way for an innovative platform for the treatment of a variety of diseases that performs with increased efficacy and is associated with decreased off-target effects.
Highlights.
Due to its genetic tractability, track record of safety in humans, ability to home to, survive and replicate in the gastrointestinal tract and tumors, E. coli Nissle 1917 has become a favored chassis for engineering ‘smart microbes’ that deliver therapeutic modalities to sites of disease.
EcN variants engineered with new therapeutic properties via alterations in its metabolic pathways or the introduction of in situ drug delivery systems are showing promise for the treatment of bacterial infections, inflammatory bowel diseases, metabolic disorders and cancer.
Clinical trials are underway to test the safety and efficacy of EcN-based smart microbes for inborn diseases of metabolism and cancer.
EcN synthesizes a genotoxin called colibactin capable of causing DNA double-strand breaks in mammalian cells, raising safety concerns about its use clinically.
Acknowledgments
Supported by NIH grants (AI064285, AI144716, DK113599) and the Brit d’Arbeloff Research Scholar award to C.F.L and a Crohn’s & Colitis Foundation Research Fellow Award, award number 654758, to J.P.L. All figures created with BioRender.com.
Glossary
- Auxotrophic
organism that requires supplemental nutrients to support its growth
- B2 phylogenetic group of E. coli
one of four main genetically related lineages of E. coli
- Bacteriocins
bacterial secreted peptides that inhibit growth of related bacteria
- Biofilm
a mono- or poly-microbial aggregation of bacteria that adhere to a surface
- Carbapenem
broad-spectrum antibiotic reserved for the treatment of multi-drug resistant bacterial pathogens
- DSS-induced colitis
when administered via the drinking water to mice or rats, dextran sodium sulfate (DSS) induces inflammation and injury localized to the distal colon resembling human ulcerative colitis
- Enterobacteriaceae
large family of Gram-negative bacteria that includes pathogens and symbionts
- Epidermal growth factor
a 6 kDa protein that stimulates cell growth and differentiation by binding to its receptor, EGFR
- Granulocyte-macrophage colony-stimulating factor
a growth factor that stimulates stem cells to produce granulocytes and monocytes
- Hyperammonemia
occurs in patients with inborn urea cycle disorders, and those with cirrhosis, late-stage liver disease in which healthy liver tissue is irreversible replaced with scar tissue
- Ice nucleation localization tag
N-terminal domain of Pseudomonas syringae that enables display of heterologous fused proteins on the outer cell envelope of E. coli
- Immune privileged
tissues able to tolerate the introduction of antigens without eliciting an inflammatory immune response
- Immunological memory
the ability of the immune system to respond more rapidly and effectively to antigen(s) to which the system has previously been exposed
- Inflammatory bowel diseases (IBD)
disorders that involve chronic inflammation of the gastrointestinal tract
- Microcins
small <10 kDa bacteriocins
- Mucosal healing
in IBD, refers to a state of remission when active disease is not directly visualizing in the lining of the digestive tract
- OmpA
a porin located in the outer membrane of E. coli and other Gram-negative bacteria
- Opportunistic infections
Infections caused by microbes normally held in check by the immune system, occur in people with weakened immune systems
- Phenylketonuria
a genetic disorder associated with a deficiency in phenylalanine hyrodroxylase, an enzyme that promotes the metabolism of phenylalanine
- pks genomic island: harbor
segment of the genome that encodes proteins that produce colibactin, a polyketide-peptide genotoxin
- Positive feedback loop
when the product of a reaction (e.g., a biological pathway) leads to an increase in that reaction
- Quorum sensing
cell-to-cell communication through small molecules
- Transforming growth factor beta
a multifunctional cytokine known to regulate normal cell growth, development, and tissue remodeling following injury
- Trefoil factors (TFFs)
7–12 kDa proteins secreted by mucus-producing cells that promote epithelial restitution
- Smart microbes
microbes engineered to enhance a particular property or endow it with a new property
- STING
an endoplasmic reticulum protein that serves as a signaling hub, receiving input from pattern recognition receptors, the majority of which sense ectopic DNA in the cytosol. STING signaling leads to type I interferon production
- VRE (vancomycin-resistant enterococci)
species commensal bacteria that can cause antibiotic resistant infections
- YebF
a 13 kDa protein of unknown function secreted across the inner membrane of E. coli via the Sec pathway and the outer membrane via outer membrane proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations
C.F.L. is a member of the Scientific Advisory Board of Synlogic, Inc.
References
- 1.Nißle A Weiteres über Grundlagen und Praxis der Mutaflorbehandlung. Dtsch Med Wochenschr 51, 1809–1813 (1925). [Google Scholar]
- 2.Nissle A Die antagonistische behandlung chronischer darmstörungen mit colibakterien. Med Klin 2, 29–33 (1918). [Google Scholar]
- 3.Sonnenborn U & Schulze J The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microbial Ecology in Health and Disease 21, 122–158 (2009). [Google Scholar]
- 4.Charbonneau MR, Isabella VM, Li N & Kurtz CB Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun 11, 1738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly VW, Liang BK & Sirk SJ Living Therapeutics: The Next Frontier of Precision Medicine. ACS synthetic biology 9, 3184–3201 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Cattoir V The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr Opin Microbiol 65, 73–80 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Heilbronner S, Krismer B, Brötz-Oesterhelt H & Peschel A The microbiome-shaping roles of bacteriocins. Nature reviews. Microbiology 19, 726–739 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Baquero F, Lanza VF, Baquero MR, Del Campo R & Bravo-Vázquez DA Microcins in Enterobacteriaceae: Peptide Antimicrobials in the Eco-Active Intestinal Chemosphere. Front Microbiol 10, 2261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sassone-Corsi M et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits SHJ, Schmitt L & Beis K Self-immunity to antibacterial peptides by ABC transporters. FEBS Lett 594, 3920–3942 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Beis K & Rebuffat S Multifaceted ABC transporters associated to microcin and bacteriocin export. Res Microbiol 170, 399–406 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Forkus B, Ritter S, Vlysidis M, Geldart K & Kaznessis YN Antimicrobial Probiotics Reduce Salmonella enterica in Turkey Gastrointestinal Tracts. Sci Rep 7, 40695 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geldart K, Forkus B, McChesney E, McCue M & Kaznessis YN pMPES: A Modular Peptide Expression System for the Delivery of Antimicrobial Peptides to the Site of Gastrointestinal Infections Using Probiotics. Pharmaceuticals (Basel) 9, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volzing K, Biliouris K & Kaznessis YN proTeOn and proTeOff, new protein devices that inducibly activate bacterial gene expression. ACS Chem Biol 6, 1107–1116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daeffler KN et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer JD et al. Engineered Probiotic for the Inhibition of Salmonella via Tetrathionate-Induced Production of Microcin H47. ACS Infect Dis 4, 39–45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geldart KG et al. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract. Bioeng Transl Med 3, 197–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang IY et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun 8, 15028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang IY et al. Reprogramming microbes to be pathogen-seeking killers. ACS synthetic biology 3, 228–237 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Duan F & March JC Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci U S A 107, 11260–11264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K et al. A probiotic yeast-based immunotherapy against Clostridioides difficile infection. Sci Transl Med 12, eaax4905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruano-Gallego D et al. A nanobody targeting the translocated intimin receptor inhibits the attachment of enterohemorrhagic E. coli to human colonic mucosa. PLoS Pathog 15, e1008031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruis W et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53, 1617–1623 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derwa Y, Gracie DJ, Hamlin PJ & Ford AC Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther 46, 389–400 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Jia K, Tong X, Wang R & Song X The clinical effects of probiotics for inflammatory bowel disease: A meta-analysis. Medicine (Baltimore) 97, e13792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Praveschotinunt P et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat Commun 10, 5580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M, Kim J, Ahn JH & Moon Y Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF. JCI Insight 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui M et al. NIR light-responsive bacteria with live bio-glue coatings for precise colonization in the gut. Cell Rep 36, 109690 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Vandenbroucke K et al. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal immunology 3, 49–56 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Steidler L et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science (New York, N.Y.) 289, 1352–1355 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Scott BM et al. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nature Medicine 27, 1212–1222 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Yan X et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cellular & Molecular Immunology 18, 2344–2357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurtz CB et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med 11, eaau7975 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Isabella VM et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol 36, 857–864 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Adolfsen KJ et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat Commun 12, 6215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puurunen MK et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nature Metabolism 3, 1125–1132 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Forbes NS Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 10, 785–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chien T et al. Enhancing the tropism of bacteria via genetically programmed biosensors. Nature Biomedical Engineering (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill PJ et al. Magnetic resonance imaging of tumors colonized with bacterial ferritin-expressing Escherichia coli. PloS one 6, e25409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stritzker J, Hill PJ, Gentschev I & Szalay AA Myristoylation negative msbB-mutants of probiotic E. coli Nissle 1917 retain tumor specific colonization properties but show less side effects in immunocompetent mice. Bioengineered bugs 1, 139–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stritzker J et al. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol 297, 151–162 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y et al. Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4T1 breast tumors through expression of azurin protein. Appl Environ Microbiol 78, 7603–7610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leventhal DS et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat Commun 11, 2739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Din MO et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurbatri CR et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho CL et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nature Biomedical Engineering 2, 27–37 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Geiger R et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canale FP et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 598, 662–666 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y et al. E. coli Nissle 1917-Derived Minicells for Targeted Delivery of Chemotherapeutic Drug to Hypoxic Regions for Cancer Therapy. Theranostics 8, 1690–1705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L et al. Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum-5 to solid tumors for cancer therapy. J Biol Eng 13, 58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nougayrède J-P et al. A Toxic Friend: Genotoxic and Mutagenic Activity of the Probiotic Strain Escherichia coli Nissle 1917. mSphere 6, e00624–00621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arthur JC et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (New York, N.Y.) 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arthur JC et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 5, 4724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomkovich S et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer research 77, 2620–2632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cougnoux A et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63, 1932–1942 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A & Bonnet R The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut microbes 5, 675–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonnet M et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clinical cancer research : an official journal of the American Association for Cancer Research 20, 859–867 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Pleguezuelos-Manzano C et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubbert S, Klinkert B, Schimiczek M, Wassenaar TM & Bunau RV No Genotoxicity Is Detectable for Escherichia coli Strain Nissle 1917 by Standard In Vitro and In Vivo Tests. Eur J Microbiol Immunol (Bp) 10, 11–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nougayrède JP et al. A Toxic Friend: Genotoxic and Mutagenic Activity of the Probiotic Strain Escherichia coli Nissle 1917. mSphere 6, e0062421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin P et al. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog 9, e1003437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massip C et al. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLOS Pathogens 15, e1008029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson MEV et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe 18, 582–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reuter C, Alzheimer M, Walles H & Oelschlaeger TA An adherent mucus layer attenuates the genotoxic effect of colibactin. Cell Microbiol 20 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Olier M et al. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut microbes 3, 501–509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cubillos-Ruiz A et al. Engineering living therapeutics with synthetic biology. Nat Rev Drug Discov 20, 941–960 (2021). [DOI] [PubMed] [Google Scholar]
- 67.McNerney MP, Doiron KE, Ng TL, Chang TZ & Silver PA Theranostic cells: emerging clinical applications of synthetic biology. Nature Reviews Genetics 22, 730–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riglar DT & Silver PA Engineering bacteria for diagnostic and therapeutic applications. Nature reviews. Microbiology 16, 214–225 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Saeidi N et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol 7, 521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woo SG et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens Bioelectron 168, 112523 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Bourdeau RW et al. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 553, 86–90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mimee M et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science (New York, N.Y.) 360, 915–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piñero-Lambea C et al. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS synthetic biology 4, 463–473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan CTY, Lee JW, Cameron DE, Bashor CJ & Collins JJ ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nature Chemical Biology 12, 82–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stirling F et al. Rational Design of Evolutionarily Stable Microbial Kill Switches. Mol Cell 68, 686–697.e683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JW, Chan CTY, Slomovic S & Collins JJ Next-generation biocontainment systems for engineered organisms. Nat Chem Biol 14, 530–537 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Seco EM & Fernández L Efficient markerless integration of genes in the chromosome of probiotic E. coli Nissle 1917 by bacterial conjugation. Microb Biotechnol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kan A, Gelfat I, Emani S, Praveschotinunt P & Joshi NS Plasmid Vectors for in Vivo Selection-Free Use with the Probiotic E. coli Nissle 1917. ACS synthetic biology 10, 94–106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Naidoo J, Page DB & Wolchok JD Immune modulation for cancer therapy. British journal of cancer 111, 2214–2219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Page DB, Postow MA, Callahan MK, Allison JP & Wolchok JD Immune modulation in cancer with antibodies. Annual review of medicine 65, 185–202 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Shin DS & Ribas A The evolution of checkpoint blockade as a cancer therapy: what’s here, what’s next? Current opinion in immunology 33C, 23–35 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Postow MA, Callahan MK & Wolchok JD Immune Checkpoint Blockade in Cancer Therapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vudattu NK et al. Humanized Mice as a Model for Aberrant Responses in Human T Cell Immunotherapy. The Journal of Immunology (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Musella M, Manic G, De Maria R, Vitale I & Sistigu A Type-I-interferons in infection and cancer: Unanticipated dynamics with therapeutic implications. Oncoimmunology 6, e1314424–e1314424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amouzegar A, Chelvanambi M, Filderman JN, Storkus WJ & Luke JJ STING Agonists as Cancer Therapeutics. Cancers 13, 2695 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


