Summary
Humans frequently encounter Staphylococcus aureus (SA) throughout life. Animal studies have yielded SA candidate vaccines; yet all human SA vaccine trials have failed. One notable vaccine “failure” targeted IsdB, critical for host iron acquisition. We explored a fundamental difference between humans and laboratory animals - natural SA exposure. Recapitulating the failed Phase III IsdB vaccine trial, mice previously infected with SA do not mount protective antibody responses to vaccination, unlike naïve animals. Non-protective antibodies exhibit increased α2,3 sialylation that blunts opsonophagocytosis and preferentially target a non-protective IsdB domain. IsdB vaccination of SA-infected mice recalls non-neutralizing humoral responses, further reducing vaccine efficacy through direct antibody competition. IsdB vaccine interference was overcome by immunization against the IsdB heme-binding domain. Purified human IsdB-specific antibodies also blunt IsdB passive immunization and additional SA vaccines are susceptible to SA pre-exposure. Thus, failed anti-SA immunization trials could be explained by non-protective imprint from prior host-SA interaction.
Graphical Abstract
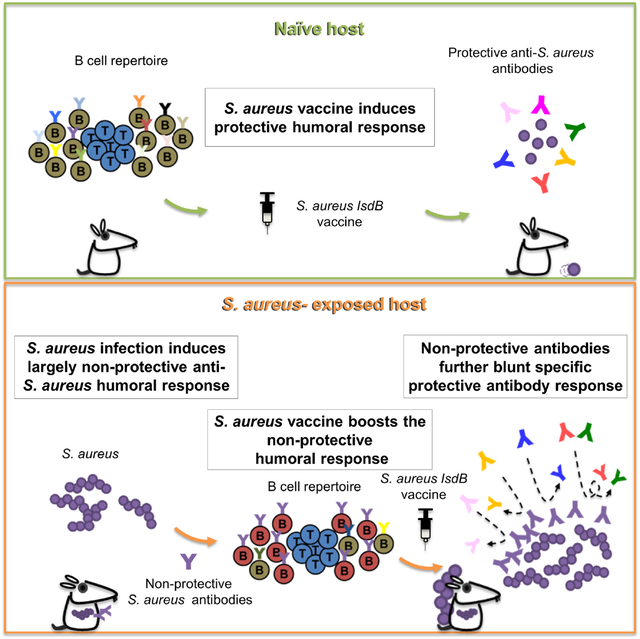
eTOC Blurb
The failure of S. aureus vaccine trials has been a conundrum. Using mice with S. aureus exposure, Tsai et al. find prior infection preprograms the host immune system to respond poorly to vaccines with antibody competition further reducing efficacy. Understanding S. aureus modulation of vaccine responses may facilitate future success.
Introduction
S. aureus (SA) is a major cause of health burden and has been the target of vaccine development for over a century (wright, 1902). Although many successful candidates have emerged from laboratory animal studies, all passive and active immunizations taken to clinical trials have failed in humans without a clear explanation (Miller et al., 2020). We propose that prior exposures to SA in humans account for the ineffectiveness of these vaccines. Whereas mice in a laboratory setting have infrequent exposure to SA, human infants have a colonization rate of over 50% during the first two months of life (Lebon et al., 2008). As a pathobiont, SA elicits abundant antibodies (Ab) against SA in most subjects (Kluytmans et al., 1997). However, these anti-SA Ab likely play a modest role against SA infection as individuals with B cell deficiency are not more susceptible to SA infections than normal subjects (Fowler and Proctor, 2014). SA has developed many mechanisms to evade the host adaptive immune system (Alonzo et al., 2013; Gerlach et al., 2018; Goodyear and Silverman, 2003; Kappler et al., 1989; Sanchez et al., 2017). Thus, we queried if pre-existing immune response to SA can play a role in SA vaccine failures.
To address this question, we turned to one of the most notable SA vaccine “failures” to date that targeted the iron-regulated surface determinant protein B (IsdB), a critical antigen for acquisition of host iron (Fowler et al., 2013). Although the vaccine induced robust titers of IsdB Ab in subjects, the vaccine was ineffective in the Phase 3 clinical trial.
Results
Prior S. aureus infection abrogates protection conferred by IsdB vaccine
To model prior host exposure to SA, we intraperitoneally (i.p,) infected C57BL/6 mice with SA 1–3 times at weekly intervals with an aim to achieve a steady serum IsdB antibody titer (Figure S1A). We then immunized the mice with recombinant IsdB plus alum and evaluated for bacterial burden or mouse mortality (Figure 1A). Corroborating earlier studies (Stranger-Jones et al., 2006), IsdB vaccine was highly efficacious against SA challenge in naïve mice (Figure 1C). However, 2–3 prior SA infections attenuated anti-SA immunity conferred by IsdB vaccine in direct correlation with pre-existing IsdB Ab titers (Figures 1B–E, S1B and S1C). Since 3 SA infections conferred the most robust interference across experiments, we focused on this model for mechanistic studies.
Figure 1. IsdB immunization is not protective in mice previously infected with S. aureus.
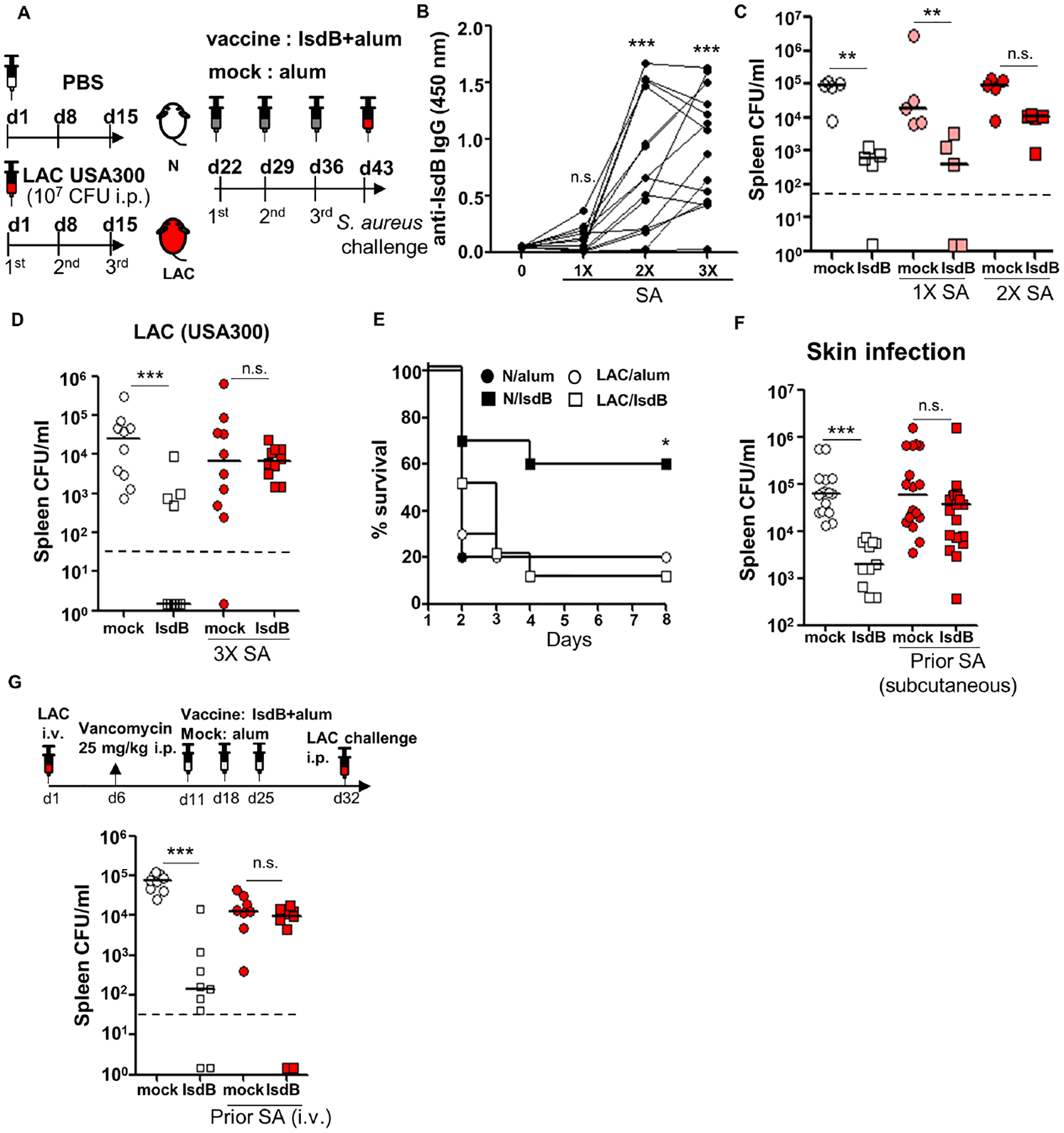
(A) Experimental setting. C57BL/6 mice injected intraperitoneally (i.p.) with 2×107 SA (LAC) or phosphate-buffered saline (PBS) 3 times at 7d intervals, immunized i.p. with IsdB plus alum (IsdB) or alum alone (Mock), then challenged with 2×107 LAC i.p.
(B) Serum anti-IsdB IgG after 1–3 LAC infections (n=15 per condition).
(C and D) Tissue bacterial burden in mice infected 1–3 times with LAC, then immunized and LAC challenged as per Figure 1A. Bacterial burden was measured 24 hrs after the last infection. N/IsdB: Naïve mice vaccinated with IsdB, N/alum: naïve mice given adjuvant alone; LAC/IsdB: LAC-infected mice vaccinated with IsdB; LAC/alum: LAC-infected mice given adjuvant alone (LAC/alum). N=5 for (C) and n=10 for (D) per mouse group.
(E) Kaplan-Meyer plot of mice treated as in Figure 1A with a final LAC challenge dose of 5×107 (LD90) (n=10 per mouse group).
(F) Tissue bacterial burden in mice infected subcutaneously (twice 2 weeks apart) with SA (LAC), IsdB immunized two weeks later and then LAC challenged i.p. per Figure 1A (n=11–19 per mouse group).
(G) Tissue bacterial burden in mice infected once i.v. with SA, treated for 5 days with vancomycin and then immunized and LAC challenged i.p. per Figure 1A (n=9 per mouse group).
Each data point represents an individual mouse; bars denote median and dashed lines indicate the limit of detection (C, D, F and G). n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; one-way ANOVA (B to D, F and G) or long rank test (E). See also Figure S1.
Vaccine efficacy was also blunted by prior SA infections delivered subcutaneously or i.v. (Figures 1F, 1G, S1D and S1E). Unlike human staphylococcal colonization, murine intranasal colonization with SA over 3 weeks failed to induce significant anti IsdB-specific Ab response (Figure S1F). Hence, the effect of intranasal infection on IsdB vaccine was not studied. IsdB vaccine interference was observed in mice inoculated with SA strains LAC, SA113 or Newman when used for both prior infection and final challenge (Figures 1D, S1G and S1H). Interference was dramatic in all mouse genetic backgrounds tested (C57BL/6, CBA, and BALB/c) and equally in both male and female mice (Figures 1D and S1I–S1K).
Vaccine suppression did not depend on innate training mechanisms, as adoptive transfer of B cells from SA-infected mice followed by vaccination of the recipients was sufficient to induce vaccine suppression (Figure 2A and S2A). In addition, vaccine suppression persisted for at least 3 weeks from the time of last SA infection (Figure S2B). Vaccine suppression was antigen-specific based on efficacy of the IsdB vaccine in mice previously infected with the IsdB/HarA mutant that does not induce IsdB cross-reactive Ab (Figures 2B and S2C) (Kuklin et al., 2006). In naïve mice, IsdB vaccination conferred anti-SA protection through B lymphocytes and Ab, but SA-infection prior to vaccination abrogated the ability of the B cells and total IgG to transfer anti-SA immunity (Figures 2C, 2D, S2D and S2E). In comparison, transfer of CD4+ T cells from vaccinated SA-naïve mice was not protective (Figure S2F). Hence, our data demonstrate that prior SA infection by different routes abrogated specific humoral immunity induced by IsdB vaccination.
Figure 2. Parameters of IsdB vaccine and vaccine interference.
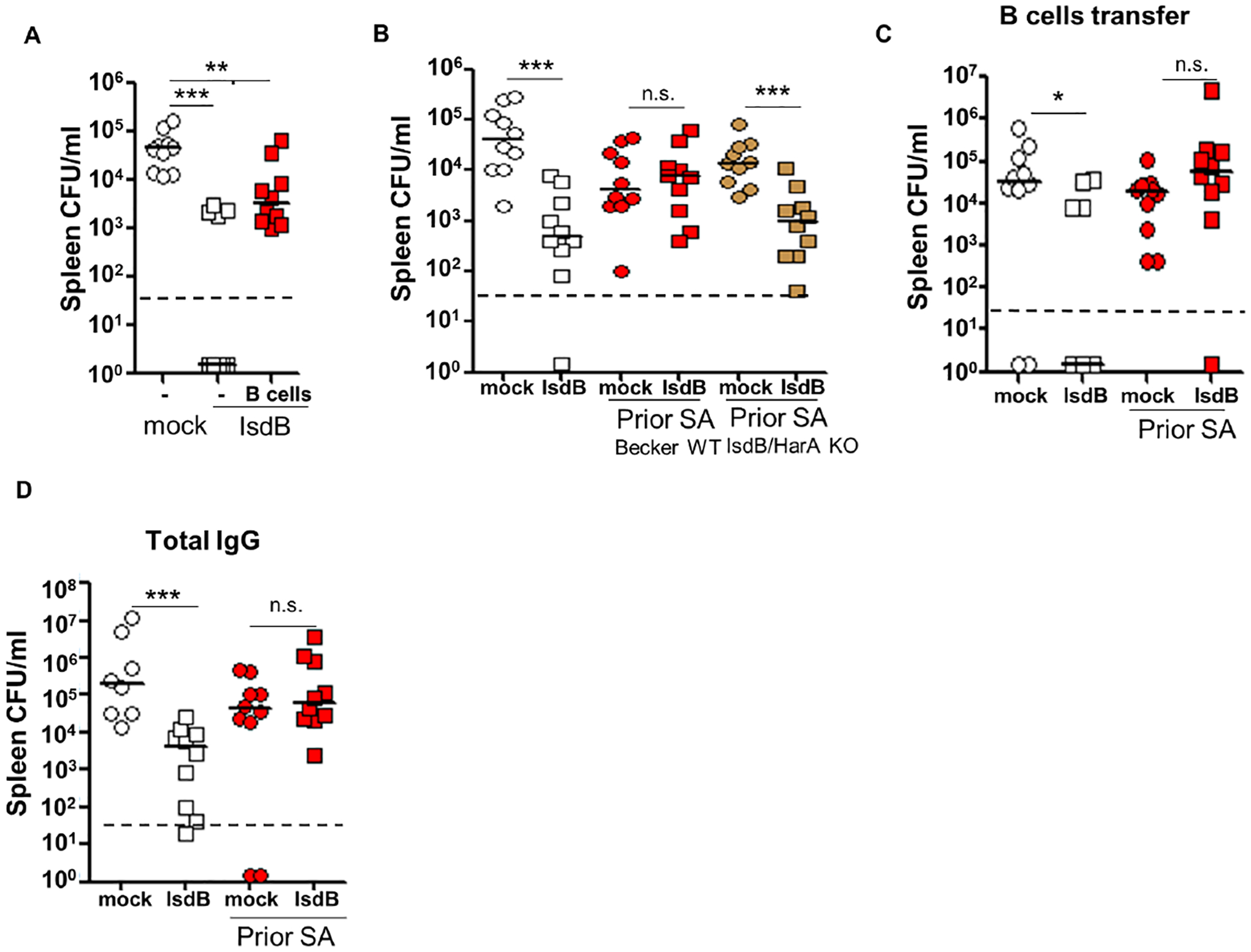
(A) B cells adoptively transferred from SA (LAC) infected mice blunt IsdB vaccine efficacy in the recipient mice. Recipient mice were immunized with IsdB as per Figure 1A, 24h after B cell transfer (n=10 per mouse group).
(B) Lack of vaccine interference when IsdB/HarA Becker strain is used for prior SA infection and WT Becker used in final challenge (n=10 per mouse group).
(C and D) Prior LAC infection abrogates humoral protection conferred by IsdB vaccine. Naïve mice received splenic B cells (n=10 per mouse group) (C) or total IgG (mock, n=8; IsdB, LAC/mock and LAC/IsdB, n=10) (D) i.v. from donor mice infected and immunized as in Figure 1A. The recipient mice were infected 24h later with LAC, and tissue bacterial burden was measured after another 24 hrs. See also Figure S2.
Each data point represents an individual mouse; bars denote median and dashed lines indicate the limit of detection. n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; one-way ANOVA (A to D).
Protective and non-protective IsdB-specific antibodies differ in IsdB target and sialylation
To determine why vaccine-generated Ab were ineffective in mice previously infected with SA, we compared IsdB-specific Ab generated by SA infection, by SA infection followed by IsdB vaccination, and by vaccination alone. We found that Ab titers, isotypes, affinity, and complement binding do not explain the differences in vaccine efficacy between naïve and pre-infected mice (Figures 3A, 3B and S3A–S3F): Specific Ab titers were not different between Naïve-vaccinated mice and SA-infected-vaccinated mice (Figures 3A and S3A–S3D). In fact, IsdB vaccination of SA exposed mice yielded antibodies that bound with higher avidity to IsdB and which bound more complement (Figure 3B and S3F).
Figure 3. Differences in IsdB targets and sialylation contribute to functional differences of IsdB-specific antibodies.
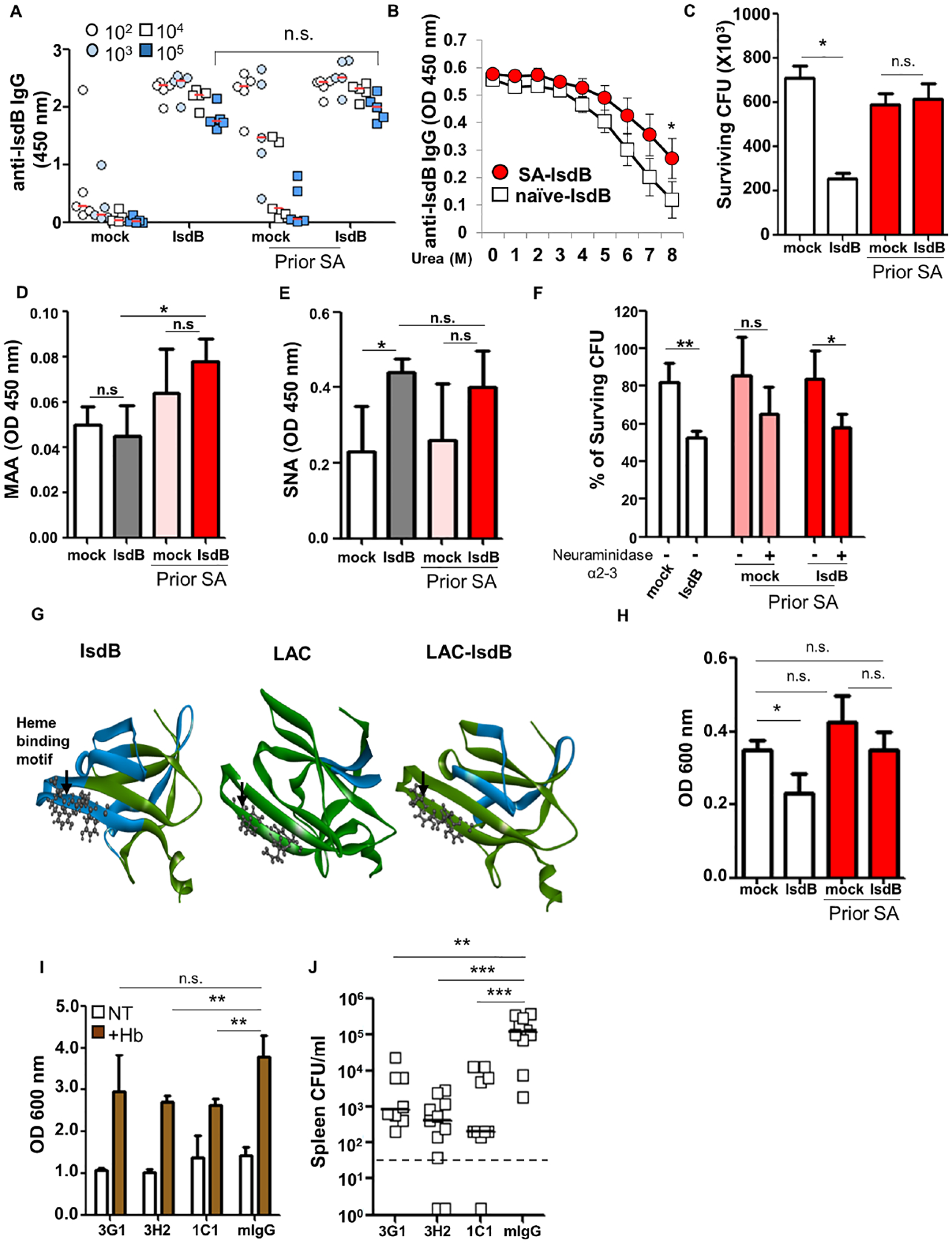
(A) Total serum anti-IsdB IgG on day 8 after IsdB immunization performed as per (Figure 1A) (n=5 per mouse group).
(B) Avidity of IsdB-specific Ab measured in the presence of increasing urea concentrations. (n = 2).
(C) Opsonophagocytic killing (OPK) of SA (LAC) by primary mouse neutrophils in the presence of immunized sera. The graph represents mean values ± SD from three independent experiments.
(D and E) Sialylation of immunized sera IgG was determined by binding to Sambucus Nigra Lectin (SNA, α2–6 sialic acid specific) (H) and Maackia Amurensis Lectin II (MAA, α2–3 sialic acid specific) (I) in a lectin-ELISA.
(F) OPK of SA (LAC) by primary mouse neutrophils in the presence of immunized sera with or without α2–3 neuraminidase for 16 hrs at 4 °C.
(G) Ribbon representation of interaction between immunized mouse sera (from Figure 1A) and overlapping IsdB peptide library. Colors indicate Intensity of Ab binding to specific domains: green (0 or low intensity) to red (over 1000). White arrow points to heme-binding motif.
(H) Heme-dependent growth of SA in the presence of immunized mouse sera. The graph represents mean values ± SD from three independent experiments.
(I) Heme-dependent growth of SA in the presence of mAbs from clones derived from a IsdB-specific B cell hybridoma library, generated by IsdB vaccination of naïve mice. The graph represents mean values ± SD from three independent experiments.
(J) Adoptive transfer of IsdB-specific mAbs from Figure 2E (0.5 mg/kg) prevents SA infection (n=8–11 per mouse group).
Bar represents group median. Each data point represents an individual mouse (A and J); bars denote median and dashed lines indicate the limit of detection (J). Error bars represent means ± SD. n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; One-way ANOVA (A to F, and H to J). See also Figure S3 and S4A.
However, we found that opsonophagocytic killing of SA by neutrophils was more efficient with Ab generated by vaccination of naïve mice (Figure 3C). The finding prompted us to explored Ab glycosylation, particularly sialylation, which has been shown to reduce opsonophagocytosis through blunting of FcgRIII and FcgRIIb binding (Kaneko et al., 2006; Scallon et al., 2007). We showed that Ab generated by IsdB vaccination of SA-infected mice have increased α2,3 sialylation (Figures 3D and 3E), and removal of the sialylation with α2,3 neuraminidase improved opsonophagocytic killing of SA (Figure 3F). Consistent with prior reports (Kaneko et al., 2006; Scallon et al., 2007), binding of the Ab to FcgRIIb also increased with α−2,3 neuraminidase treatment (Figures S3G and S3H). However, there was no difference in FcgRIIb binding between Ab from naïve and SA-infected mice vaccinated with IsdB. Therefore, we conclude that α2,3 sialylation of anti-IsdB Ab likely blunted opsonophagocytosis through interaction with another receptor on myeloid cells.
Analysis of Ab binding to an overlapping IsdB peptide array showed that Ab induced by SA infection or by vaccination in naïve and SA-infected mice mapped to different IsdB subdomains (Figures 3G and S4A). Most notably, naïve mice immunized with IsdB generated abundant Ab that bound to a protective NEAT2 heme-binding domain of IsdB (Bennett et al., 2019). Ab to this region were infrequently induced after SA infection alone and moderately induced after IsdB vaccination of SA-infected mice (Figure S4A). Corroborating these findings, a hemoglobin-dependent SA growth assay revealed that only Ab from naïve mice vaccinated with IsdB restricted growth of SA (Figure 3H). Furthermore, three distinct monoclonal Ab (mAb) from a B cell hybridoma library, generated from naïve mice vaccinated with IsdB and selected based on restriction of growth of SA in the hemoglobin-dependent growth assay, were protective against SA infection in vivo (Figures 3I, 3J and S3I).
IsdB vaccine recalls ineffective antibody memory response in S. aureus-infected mice
The findings of non-protective immunity conferred by both SA infection and SA infection with IsdB vaccination raised the possibility that IsdB vaccine recalls the ineffective IsdB-specific memory response if mice are pre-exposed to SA. To test this hypothesis, we analyzed the complementarity-determining region (CDR3) of IsdB-specific B cells and clonotypes associated with each infection and vaccination regimen. As demonstrated on Alluvial plot and IsdB-specific clonotype network, administration of IsdB vaccine to previously infected mice resulted in the amplification of many IsdB-specific clonotypes found in SA-infected mice. IsdB vaccination of naïve mice generated some of the same clonotypes but produced a significant number of unique clonotypes (Figures 4A, 4B and S4B), consistent with the results of epitope mapping using the IsdB peptide array. Moreover, we found that IsdB-specific IgG response increased sharply within 7 days of vaccinating pre-infected mice compared to vaccination of naïve mice (Figures 4C and S4C–S4E). The increased specific Ab levels are consistent with increased number and intensity of activated Ab secreting cells isolated from mouse splenocytes on day 8 and reflect the recall of the pre-existing humoral response (Figure 4D and S4F). Overall, these findings suggest that, while IsdB vaccine in naïve mice induces a protective neutralizing Ab response to the heme-binding site, the same vaccine in pre-infected hosts recalls a non-protective response predominantly targeting epitopes away from the heme-binding site.
Figure 4. IsdB vaccination recalls the non-protective IsdB-specific response from prior S. aureus infection.
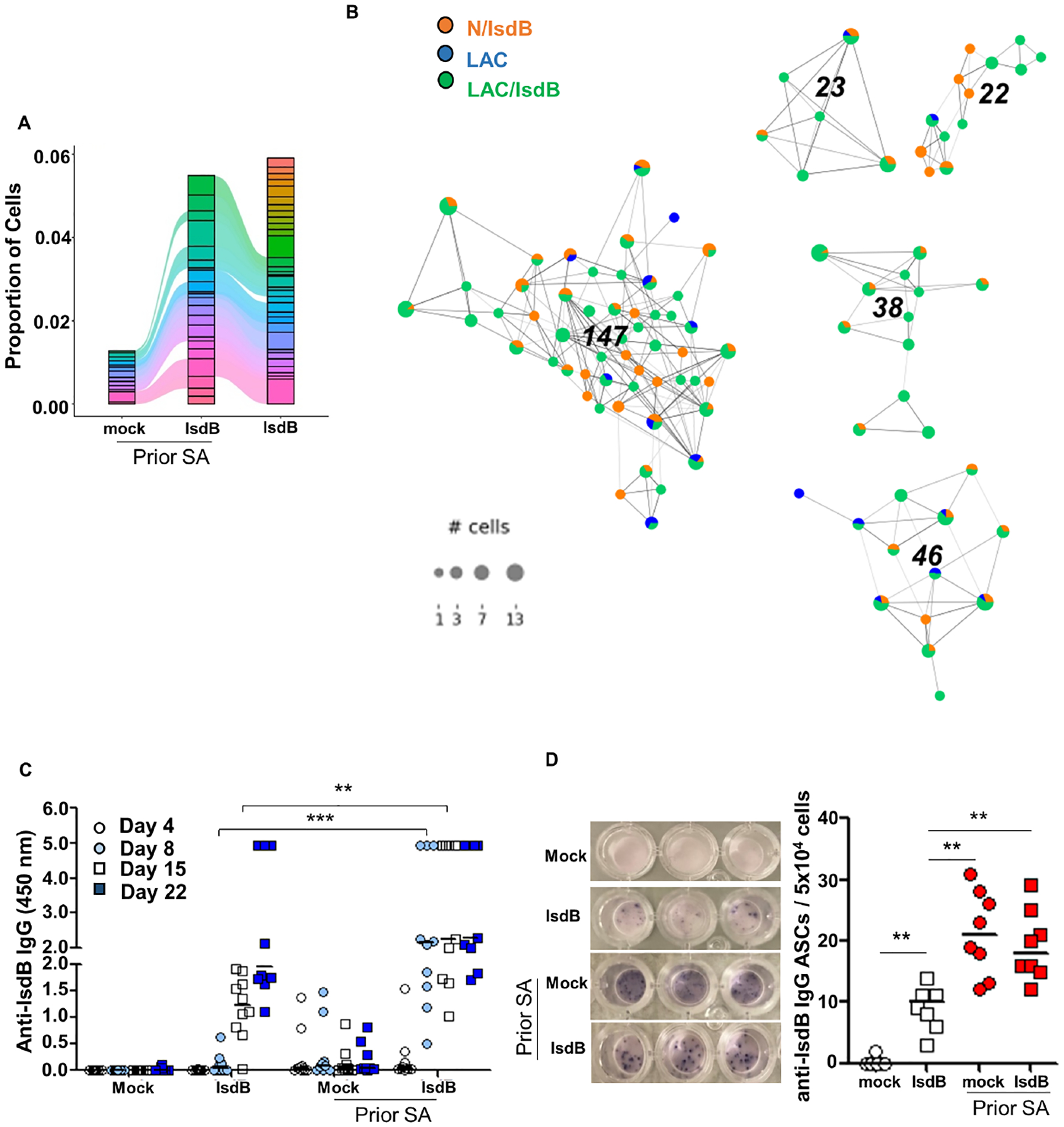
(A) Alluvial plot showing the top 40 of clonotypes from LAC, LAC/IsdB and N/IsdB.
(B) IsdB-specific clonotype network. Each dot represents cells with identical CDR3 sequence. For each clonotype, the numeric clonotype ID is shown in the graph. The size of each dot refers to the number of cells with the same CDR3.
(C) Rapid rise of specific IgG serum titer 7 days after first IsdB vaccination in SA pre-infected mice (n=10 per mouse group).
(D) ELISpot assay showing IsdB-specific IgG-secreting cells (ASCs) per 5×104 spleen B cells isolated 7 Days after the first IsdB vaccination.
Error bars represent means ± SD. n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; one-way ANOVA (C and D). See also Figure S4.
Direct antibody competition reduces vaccine efficacy but can be overcome by immunization against IsdB NEAT2 domain and epitope
IsdB vaccine interference was remarkably effective even though recall of preexisting Ab responses limited but did not prevent de novo priming of IsdB-specific Ab responses to the heme-binding region (Figure 3G). Because both protective and non-protective Ab target the same IsdB protein, we queried if recall of a non-protective Ab response could further blunt IsdB vaccine via antibody competition. In vivo, co-administration of equal volume or quantity of non-protective sera (obtained from SA-infected then vaccinated mice) abolished anti-SA protection conferred by sera from naïve vaccinated mice (Figures 5A, 5B, S5A and S5B).
Figure 5. Competition for IsdB between non-protective and protective antibodies determines outcome of S. aureus infection and overcoming antibody competition.
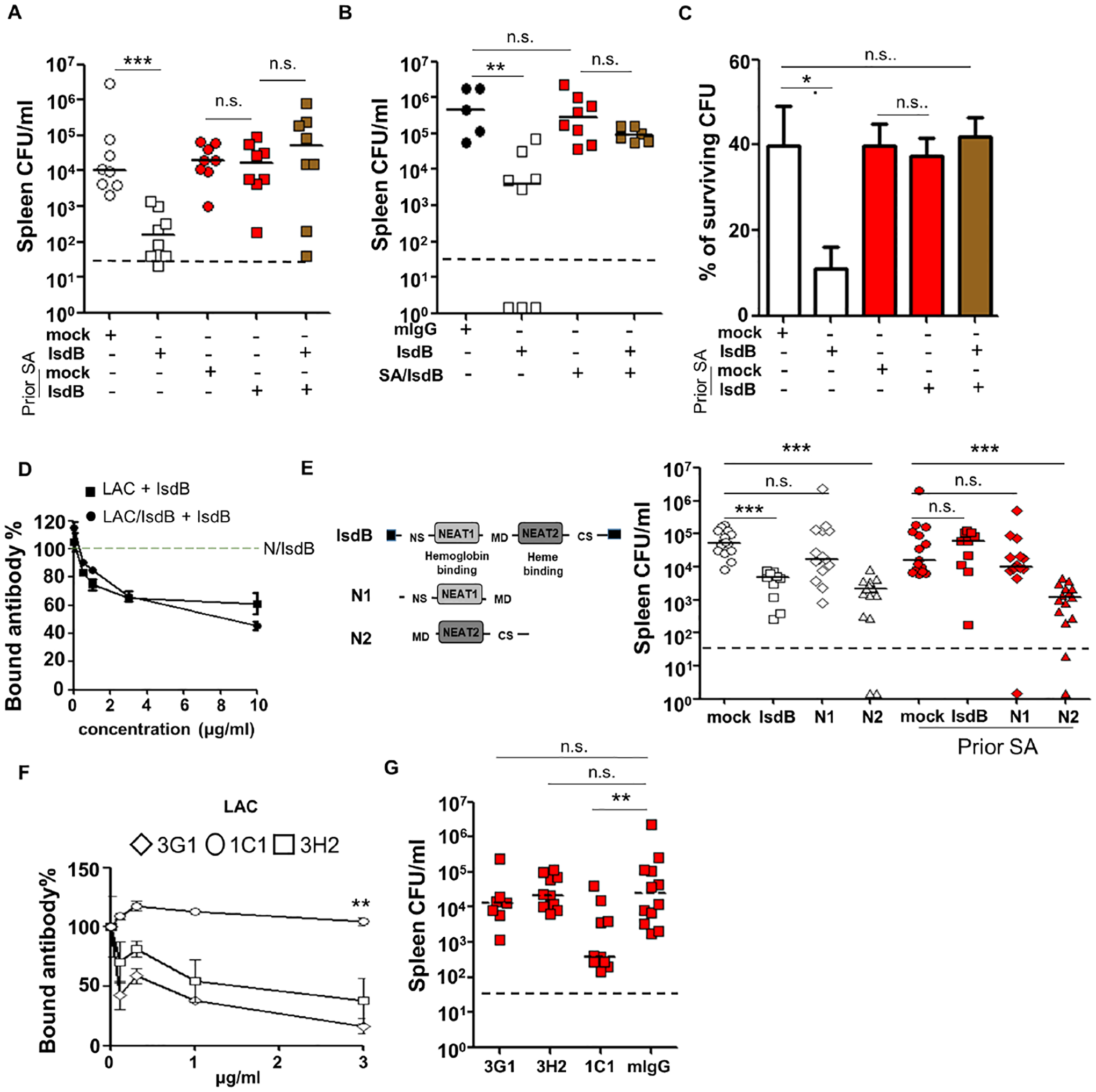
(A) Anti-SA (LAC) immunity of mice co-injected with equal volume (50 μl i.p.) of protective (NT-IsdB) and non-protective (LAC-IsdB) IsdB-specific sera (n=8 per mouse group).
(B) Anti-SA (LAC) immunity of mice co-injected with equal amount (25 μg i.p.) of protective (NT-IsdB) and non-protective (LAC-IsdB) IsdB-specific Ab (n=5–8 per mouse group).
(C) Effect of IsdB-specific Ab competition on mouse neutrophil OPK. Graph represents mean values ± SD from three independent experiments.
(D) IsdB binding to biotinylated IsdB-specific Ab from NT/IsdB (1 μg/ml) in the presence of IsdB-specific Ab from LAC or LAC/IsdB immunized mice in an ELISA plate assay.
(E) Schematic of the IsdB, N1 and N2 proteins used for vaccination. Efficacy of N1 and N2 immunization in naïve and LAC pre-infected mice (n=10–15 per mouse group).
(F) IsdB binding by biotinylated mAbs (0.5 μg/ml) in the absence or presence of IsdB-specific Ab purified from LAC-infected mice, in an ELISA plate assay.
(G) Efficacy of adoptively transferred anti-IsdB mAbs (0.5 mg/kg i.p.) in preventing SA-infection in naïve and SA (LAC) pre-infected mice (n=7–10 per mouse group).
Each data point represents an individual mouse, bars denote median and dashed lines indicate the limit of detection (A, B, E and G). Error bars represent means ± SD. n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; Student’s t test (C and D) or one way ANOVA (A, B and E to G). See also Figure S5.
In vitro, co-administration of the non-protective Ab also inhibited protective Ab functions in opsonophagocytic killing and IsdB antigen binding assays (Figures 5C and 5D), although a 3–10-fold excess of non-protective specific-Ab was required to reduce binding by the protective Ab to the IsdB-coated plate.
To determine if it is possible to overcome Ab suppression mediated by prior SA infection, we evaluated vaccine targeting of the protective heme-binding NEAT2 domain of IsdB alone. We showed that the previously characterized heme-neutralizing mAbs (Figures 3I and 3J) mapped to epitopes on NEAT2 (Figures S5F and S5G), and therefore also tested their ability to overcome Ab competition (Bennett et al., 2019). Immunization of naïve mice with recombinant NEAT2, but not NEAT1, protected against SA challenge in naïve mice (Figures 5E, S5C and S5D), consistent with the prior finding that NEAT2 contains the primary target for neutralization. Importantly, immunization with NEAT2 also overcame suppression by prior SA-infection (Figures 5E and S5D). One of the three NEAT2 epitope-specific mAbs (1C1) also resisted competition by Ab induced by SA infection, both in vitro in a binding assay and in vivo (Figures 5F, 5G, and S5E). We conclude that despite vaccine interference, immunization against the NEAT2 subdomain or a selective epitope of IsdB induces protection in previously infected mice.
The role of human non-protective antibody imprint and susceptibility of other SA vaccines to interference
Next, we queried if pre-existing IsdB-specific Ab from adult humans (Figure 6A) could modify the protective function of IsdB-vaccine generated Ab. IsdB-specific Ab, which are abundant in human sera, decreased binding of vaccine-generated protective specific Ab to 60% at a 10:1 excess molar ratio (Figures 6B). To evaluate the in vivo suppressive functions of human Ab, we developed and validated an adoptive model of human Ab transfer and protection from SA challenge in mice (Tsai et al., 2021). The compatibility of human IgG subclasses for mouse Fc gamma Receptors has been carefully studied and shown to have similar affinity of binding to mouse IgG subclass, which validated the use of the assay (Dekkers et al., 2017). In the model, we showed that both human sera and purified IsdB-specific Ab were non-protective against SA challenge (Figure 6C, 6D, S6A and S6B). When co-administered with vaccine-generated protective mouse sera at near equal volumes, the sera blunted the anti-SA efficacy of the protective Ab (Figures 6C and S6A). To date, many failed staphylococcal vaccine trials have consisted of passive immunizations (Armentrout et al. 2020). Simulating passive immunization in human hosts, we showed that protective purified IsdB-specific Ab lose their protective efficacy when mice are pre-infused with IsdB-specific human Ab at a ratio of close to 1:1 (figure 6D, S6B and S6C). Even when the ratio of protective to non-protective antibodies is raised to 10:1, interference by the human IsdB-specific antibodies remains robust, pointing to the surprisingly effective modulatory role of specific non-protective antibodies (Figure 6E and S6D).
Figure 6. Human serum anti-staphylococcal antibodies blunting of protection conferred by anti-IsdB passive immunization, and interference with FhuD2 and MntC vaccines.
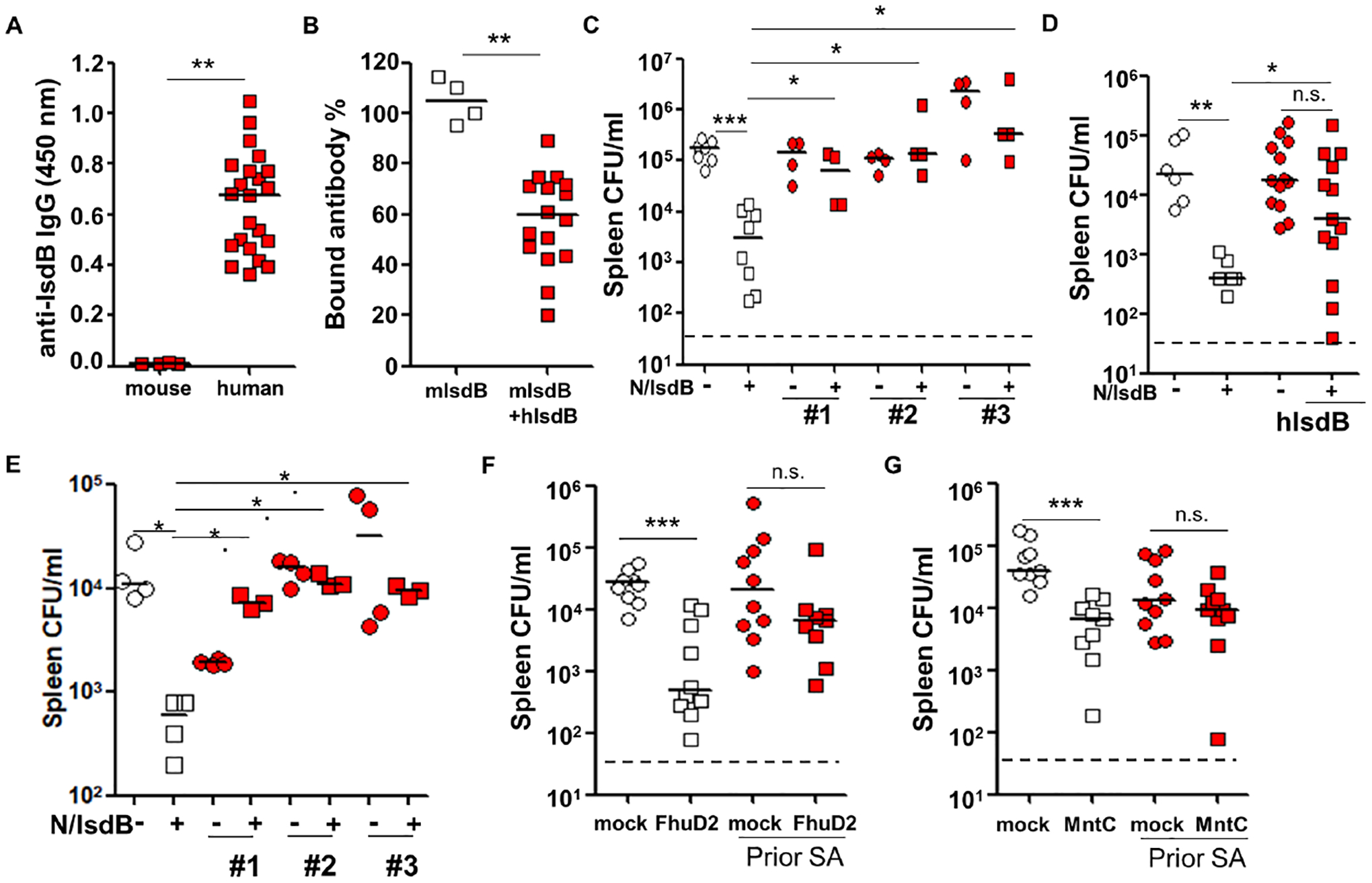
(A) Total anti-IsdB IgG titer in human adult and naïve laboratory mouse sera, diluted to 1:10,000 (mouse sera, n=4; human sera, n=22).
(B) IsdB binding by biotinylated IsdB-specific mouse Ab (1 μg) generated by vaccine in the absence or presence of IsdB-specific human Ab (10 μg) in an ELISA plate assay (human sera, n=16).
(C) Anti-SA protection conferred by co-transfer of IsdB-vaccinated mouse sera (50 μl i.v.) and serum from 3 human donors (100 μl i.p.) (n=4–8 per mouse group).
(C) Anti-SA protection conferred by sequential transfer of purified human IsdB-specific Ab (or control IgG, time = 0 hr) and vaccine-generated IsdB-Ab in mice (time = 16 hr). Ratio of protective to non-protective Ab injected IV: 25 μg to 35 μg. Each red square or circle represents mean SA burden from 3–5 mice (human sera, n=13).
(D) Anti-SA protection conferred by sequential transfer of purified human IsdB-specific Ab (or control IgG, time = 0 hr) and vaccine-generated IsdB-Ab in mice (time = 16 hr). Ratio of protective to non-protective Ab injected IV: 25 μg to 2.5 μg (human Ab, n = 3).
(F and G) Tissue bacterial burden in mice infected 3 times with LAC, then immunized with FhuD2 (F) or MntC (G) and LAC challenged as per Fig. 1A. Bacterial burden is measured 24 hrs after LAC challenge.
Each data point represents an individual mouse serum or human serum (A, B and D). Bars denote median and dashed lines indicate the limit of detection (C, D, F and G). Error bars represent means ± SD. n.s., not significant, *P<0.05, **P<0.01, and ***P< 0.001; Student’s t test (A and B) or one way ANOVA (C to G). See also Figure S6.
Our findings with IsdB vaccine beg the question if prior SA exposure has similar inhibitory effect on other SA vaccines. Therefore, we tested two additional SA vaccines, cell-wall anchored MntC and FhuD2, that conferred good protection against SA challenge when administered to mice (Anderson et al., 2012; Mishra et al., 2012). Notably, MntC, was tested in a multivalent human vaccine trial that was stopped early because of low probability of achieving efficacy objectives (Gurtman et al., 2019), We showed that both vaccines conferred strong protection against SA challenge in naïve mice but were ineffective in mice previously infected with SA (Figure 6F, 6G, S6E and S6F). These findings suggest that the interference principles defined for IsdB might also apply to other SA vaccines.
Discussion
Our study provides a mechanistic framework to explain the failure of the IsdB vaccine trial. SA infection induces preferential generation of Ab to the non-neutralizing NEAT1 domain of IsdB, in contrast to IsdB vaccination in naïve mice which generates robust humoral response to the protective heme-binding NEAT2 domain. In addition, vaccination in SA-infected mice enhances α2,3 sialylation of anti-IsdB antibodies leading to blunted SA killing by opsonophagocytosis.
When administered to mice previously infected with SA, the otherwise protective IsdB vaccine recalls the non-protective antibody response albeit permitting modest priming of Ab response to the heme-binding region. We demonstrated that the recalled non-protective Ab response effectively diminished the protective vaccine response by competitive binding. These findings support recall of non-protective antibody response and antibody competition as two critical mechanisms that could explain the failure of active and passive staphylococcal immunizations. Consistent with these proposed mechanisms, a study of SA-colonized minipigs demonstrated inefficacy of a SA capsular polysaccharide (CPS) vaccine (Fernandez et al., 2022) that has been shown to be protective in mice (Lee et al., 1988; Yoshida et al., 1987).
Induction of the “non-protective imprint” by SA is unsurprising and would serve to promote survival of the pathogen during colonization or infection. We have previously shown that mice could be repeatedly infected with SA without change in SA clearance, a finding we attributed to the modulation of Th17 response (Sanchez et al., 2017). The induction of non-protective antibodies would similarly benefit commensal lifestyle of the pathogen, but importantly would provide a safeguard against protective specific antibody generation by the host.
Vaccine interference by pre-existing anti-SA immunity bears similarity to the concept of original antigenic sin that explains the decreased efficacy of influenza vaccines to influenza strains that have undergone seasonal drift (Francis, 1960). Unlike the original concept where differences in antigenic targets exist between the initial and subsequent antigen exposure, SA IsdB vaccine is generated to a highly conserved antigen and is administered to human subjects with abundant pre-existing specific Ab in the failed trial. Therefore, any protective Ab generated needs to overcome competition from the potentially non-protective Ab. This is arguably a more robust mechanism of interference than that encountered by seasonal influenza vaccines. We showed that targeting of the heme-binding domain and select epitopes of IsdB can overcome this competition.
Unlike the original antigenic sin concept that explains reduced or even deleterious host responses after repeat infections with certain RNA viruses, e.g., influenza, HIV, RSV, and dengue virus, original antigenic sin pertinent to conserved IsdB may find broader application to other SA vaccines and to vaccines against the many pathogens that have adapted to a lifestyle of coexistence with the human host. Taken together, the integration of prior host-pathogen interaction into vaccine studies can help explain the failure of seemingly successful experimental SA vaccines that have been to date studied in silo in naïve animal models.
STAR Methods
Resource Availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, George Liu (gyliu@ucsd.edu).
Materials availability
All unique reagents generated in this study are available upon request made to the Lead Contact.
Data Code and Availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is listed in the key resources table.
All original code has been deposited at https://github.com/LewisLabUCSD/StaphVaccine_BCR and is publicly available as of the date of publication. Please also see the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Key resources table
| REGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models : Organisms/strains | ||
| Mus musculus C57BL/6 | Charles river | Strain # C57BL/6NCrl |
| Mus musculus BALB/c | Charles river | Strain # BALB/cAnNCrl |
| Mus musculus CBA | Charles river | Strain # CBA/CaCrl |
| Primary cells | ||
| Mouse neutrophils | Bone marrow from Mus musculus C57BL/6 | N/A |
| Mouse B cells | Splenocytes from Mus musculus C57BL/6 | N/A |
| Bacterial strains | ||
| Staphylococcus aureus: LAC (USA300) | Dr. Binh Diep | N/A |
| Staphylococcus aureus: Newman | Dr. Ambrose Cheung | N/A |
| S. aureus SA113 | Dr. Fritz Goetz | N/A |
| S. aureus Becker | Dr. Secore (Merck) | N/A |
| S. aureus Becker IsdB/HarA | Dr. Secore (Merck) | N/A |
| E. coli BL21 (DE3) | NEB | Cat # C2527I |
| Oligonucleotides | ||
| 5’IsdB: 5’-GGTCGCGGATCCAACAAACAGCAAAAAGAATTT-3’ | Integrated DNA Technologies | N/A |
| 3’IsdB: 5’-GGTGGTGCTCGAGTTTAGTTTTTACGTTTTCTA-GGTAATAC-3’ | Integrated DNA Technologies | N/A |
| 5’ NEAT1: 5’-CAAGCAGCAGCTGAAGAAACAGG-3’ | Integrated DNA Technologies | N/A |
| 3’ NEAT1: 5’-TTATTTGAATTTATCTGCactgttataaattgg-3’ | Integrated DNA Technologies | N/A |
| 5’ NEAT2: 5’-ACTGAAGAAGATTATAAAGCTGAAA-3’ | Integrated DNA Technologies | N/A |
| 3’ NEAT2: 5’-TTAGTTTTTACGTTTTCTAGGT-3’ | Integrated DNA Technologies | N/A |
| 5’FhuD2: 5’-TAAGGCCTCTGTCGATGAAACTAAATCTTATAAAATGGACT-3’ | Integrated DNA Technologies | N/A |
| 3’ FhuD2: 5’-CAGAATTCGCAAGCTTTATTTTGCAGCTTTAATTAA-3’ | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pET-28a(+) | Novagen | Cat # 69864 |
| pET-6xHN | Takara | Cat # 631433 |
| Antibodies | ||
| horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG | Biolegend | Cat # 405306 |
| alkaline phosphatase (AP)-conjugated anti-mouse IgG | SouthernBiotech | Cat # 1030-04 |
| horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM | SouthernBiotech | Cat # 1021-05 |
| FITC-conjugated anti-C3 polyclonal antibody | MP Biomedicals | Cat # 0855385 |
| FITC-conjugated anti-mouse IgG | Invitrogen | Cat # 31569 |
| 800CW goat anti-mouse IgG | LI-COR | Cat # 926-32210 |
| Chemicals, lectins and recombinant protein | ||
| His60 Ni Superflow Resin | Takara | Cat # 635660 |
| Aluminium hydroxide gel | InvivoGen | Cat # vac-alu-250 |
| Protein A from Staphylococcus aureus | Sigma-Aldrich | Cat # P6031 |
| biotinylated Sambucus Nigra Lectin (SNA) | Vector Laboratories | Cat # B-1305-2 |
| biotinylated Maackia Amurensis Lectin II (MAA) | Vector Laboratories | Cat # B-1265-1 |
| horseradish peroxidase (HRP)-conjugated Avidin | BioLegend | Cat # 405103 |
| recombinant mouse FcgRIIb | Biolegend | Cat # 783304 |
| recombinant mouse FcgRIII | Biolegend | Cat # 790104 |
| complement factors from guinea pig | MP Biomedicals | Cat # 086428 |
| EZ-Link™ Sulfo-NHS-Biotin | ThermoFisher Scientific | Cat # 21217 |
| NHS-activated agarose | ThermoFisher Scientific | Cat # 2697 |
| FluoSpheres | ThermoFisher Scientific | Cat # F8775 |
| Casamino Acids | Sigma-Aldrich | Cat # 2240 |
| 2,2’ Bipyridyl | Sigma-Aldrich | Cat # D216305 |
| hemoglobin | Sigma-Aldrich | Cat # H7379 |
| P3X63Ag8.653 | ATCC | Cat # CRL-1580 |
| polyethylene glycol 1500 | Sigma-Aldrich | Cat # 10783641001 |
| α2-3 Neuraminidase S | NEB | Cat # P0743L |
| Critical Commercial Assays | ||
| MojoSort™ Mouse CD4 T Cell Isolation Kit | Biolegend | Cat # 480033 |
| MojoSort™ Mouse Pan B Cell Isolation Kit II | Biolegend | Cat # 480088 |
| Chromium Single Cell Mouse BCR Amplification Kit | 10xgenomics | Cat # 1000255 |
| Chromium Next GEM Single Cell 5’ Kit v2 | 10xgenomics | Cat # 1000265 |
| Chromium Next GEM Chip K Single Cell Kit | 10xgenomics | Cat # 1000287 |
| RNeasy Mini-kit | QIAGEN | Cat # 74104 |
| High Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat # 4368814 |
| Deposited Data | ||
| BCR-Seq Data | This paper | GEO:GSE193543; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193543 |
| Original Code | This paper | https://github.com/LewisLabUCSD/StaphVaccine_BCR |
| Software and Algorithms | ||
| GraphPad Prism 6.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Microsoft Excel | Microsoft | https://www.microsoft.com/enus/microsoft-365/excel |
| FlowJo (v.10.6.1) | FlowJo | https://www.flowjo.com/ |
| R 4.1.1 | R Core Team | N/A |
| Python 3.7.9 | Python Software Foundation | N/A |
| Cell Ranger 3.1.0 | 10x Genomics | N/A |
| scRepertoire v1.0.2 | https://zenodo.org/record/3856827#.Yoe1qKjMJPY | N/A |
| Seurat V4 | Satija lab | N/A |
| Scripy 0.9.3 | https://github.com/scverse/scirpy | N/A |
| Scanpy 1.9 | https://github.com/theislab/Scanpy | N/A |
Experimental model and subject details
Mouse studies were reviewed and approved by the Institutional Animal Care and Use Committee. Mouse experiments were conducted in accordance with recommendations listed in the Animal Care Program at University of California, San Diego and Cedars-Sinai Medical Center’s regulations and recommendations on animal experiments. Experimentations using human blood were approved by the UCSD Human Research Protection Program. Prior informed consents were obtained from the human subjects. Experimental protocols were approved by the UCSD Biosafety Committee.
C57BL/6, BALB/c, and CBA mice were purchased from Charles River Laboratories. All mice were housed in specific-pathogen free facilities and 6 to 12-week old age-matched female or male mice were used for in vitro and in vivo experiments.
Bacterial strains and culture conditions
S. aureus Becker, S. aureus Becker IsdB/HarA deletion mutant (gifts from Dr. Secore, Merck), S. aureus LAC, Newman, SA113 (courtesy of Dr. Fritz Goetz) were routinely cultured in Todd Hweitt Hweitt broth (THB). Overnight S. aureus cultures were diluted 1:200 in THB and grown to an optical density of 0.8. Bacteria were routinely washed twice in PBS prior to use and inocula were confirmed by determination of CFU on agar plates.
Primary Cells and Cell Line
CD4 T cells or CD19 B cells were isolated from spleen of 10–14 week-old S. aureus-infected female C57BL/6 mice using negative selection kits (Biolegend). Purity of CD4 T cells or CD19 B cells were confirmed by flow cytometry (>95%). P3X cells were maintained in RPMI 1640 media containing Pen/Strep and supplemented with 10% FBS before cell fusion.
Method details
Murine models of S. aureus infection
Model of prior S. aureus infection – For systemic infection, mice were inoculated with S. aureus via intra-peritoneal (i.p.) injection. The mice were reinfected i.p. with the same inoculum on Day 8 and 15 (Figure 1A). For skin infection, mice were injected with S. aureus subcutaneously (s.c.) on Day 1 and 14. For bloodstream infection, mice were inoculated with S. aureus intravenously (i.v.) on Day 1. Unless otherwise stated, 6–8 weeks old female mice were administered 2×107 LAC (USA 300) i.p. for each S. aureus challenge. Spleen and kidneys were harvested 24 hr after the last infection on Day 43, homogenized in phosphate-buffered saline (PBS) and plated on THB agar plates for CFU enumeration. Other S. aureus inoculant were: 5×107 LAC i.p. for the LD90 infection, 2×107 SA113 i.p., 4×107 Newman or Becker (WT or IsdB/HarA mutant) i.p., 4×107 LAC subcutaneous (s.c.), and 1×106 LAC intravenous (i.v.). Staphylococcal antibodies titer was confirmed by ELISA.
Cloning and protein expression
The IsdB gene was amplified from LAC using primers: 5’IsdB (5’-GGTCGCGGATCCAACAAACAGCAAAAAGAATTT-3’) and 3’IsdB (5’-GGTGGTGCTCGAGTTTAGTTTTTACGTTTTCTAGGTAATAC-3’). The PCR product was cloned into pET28 expression vector (Novagen) and expressed as described previously with some modifications (1). Briefly, IsdB-expressing plasmids were used to transform E. coli BL21 (DE3) cells (NEB) to produce a His-tagged protein with 1mM of isopropyl-β-D-thiogalactoside (IPTG) for 2 hours. Recombinant E. coli was centrifuged and suspended in lysis buffer (50 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 2mM MgCl2, 10 mM imidazole, 0.1% Tween 80, 1% Triton ×100, PMSF, lysozyme (2mg/ml). His-tagged IsdB was purified from the clarified lysate by His60 Ni Superflow Resin (Takara, 635660) chromatography. The column was washed with 20 mM Tris-HCl (pH 8.0), 150mM NaCl, 0.1% Tween 80 and His-tagged IsdB was eluted with 300 mM imidazole, 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.1% Tween 80. To generate truncated constructs of IsdB containing single domain (Figure 3E), primers: NEAT1, 5’-CAAGCAGCAGCTGAAGAAACAGG-3’ and 5’-TTA-TTTGAATTTATCTGCactgttataaattgg-3’; NEAT2, 5’-ACTGAAGAAGATTATAAAGCTG-AAA-3’ and 5’-TTAGTTTTTACGTTTTCTAGGT-3’ were applied to PCR reactions using full length IsdB as template. Recombinant IsdB domains were purified as described above. Recombinant MntC was expressed as described previously (Paterson et al., 2020). The fhuD2 gene was amplified from LAC using primers designed by Takara:5’-TAAGGCCTCTGTCGATGAAACTAAATCTTATAAAATGGACT-3’ and 5’-CAGAATTCGCAAGCTTTATTTTGCAGCTTTAATTAA-3’. The PCR product was cloned into pET6xHN vector (Takara, 631433) and expressed.
Immunization with IsdB vaccine
Mice were immunized i.p. three times with IsdB (75μg, 50μg and 50μg) plus aluminum hydroxide (Aluminum hydroxide gel, InvivoGen, vac-alu-250) (450μg per dose) or with aluminum hydroxide alone at 7-day intervals. Mouse sera were screened for reactivity to IsdB by ELISA.
Opsonophagocytic killing assay
Opsonophagocytic killing assay (OPK) was performed as described (Thomer et al., 2016). Mouse neutrophils were isolated from bone marrow by MojoSort™ Mouse Neutrophil Isolation Kit (BioLegend, 480058). Overnight culture of SA LAC was diluted 1:200 in Todd Hewitt broth (THB) and grown to an optical density of 0.6. SA was washed, resuspended in PBS, and incubated with mouse sera at 37°C for 20 min, then added to 105 mouse neutrophils a multiplicity of infection (MOI) of 1:0.5 in the presence of 2% normal mouse serum. Following incubation at 37°C for 1 hr with agitation at 200 rpm, samples were plated on THB agar plates for CFU enumeration.
Antibody avidity assay
Immunized serums were diluted 1:1000 in PBS containing 1% BSA and added to 96-well plates coated with recombinant IsdB (1μg/ml). After incubation for 1 hr, different concentration of urea (0 to 8 M) with 0.05% Tween 20 in PBS were treated for 15 mins. Plates were washed and bound antibodies were detected by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (BioLegend, 405306).
Lectin-ELISA
Lectin-ELISA was described previously (Solkiewicz et al. 2021). Briefly, high binding microtiter plates were coated with 10 μg/ml protein A (Sigma, P6031) in TBS at 4°C overnight and blocked with TBS containing 1%BSA for 1 hr. 5μl of immunized serum were diluted in 100μl of TBS-BSA and incubated for 3 hrs at 37°C. The plates were then incubated with 100μl of biotinylated Sambucus Nigra Lectin (SNA) (1 μg/ml dilution, Vector Laboratories, B-1305–2) or Maackia Amurensis Lectin II (MAA) (4 μg/ml dilution, Vector Laboratories, B-1265–1) in TBS-T for 90 mins at 37°C. Bound lectins were detected by horseradish peroxidase (HRP)-conjugated Avidin (BioLegend, 405103).
FcgRIIb and FcgRIII binding assays
Immunized sera were diluted 1:1000 in PBS containing 1% BSA and added to 96-well plates coated with recombinant mouse FcgRIIb (5 μg/ml dilution, Biolegend, 783304) or mouse FcgRIII (5 μg/ml dilution, Biolegend, 790104) at 25°C for 2 hrs. Plates were washed and bound antibodies were detected by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (BioLegend, 405306).
Antibody-dependent complement deposition assay (ADCD)
Antibody-dependent complement deposition (ADCD) was described previously (Fischinger et al., 2019). Briefly, biotinylated IsdB (EZ-Link™ Sulfo-NHS-Biotin, ThermoFisher Scientific, 21217) was coupled to red fluorescence NeutrAvidin beads (FluoSpheres, ThermoFisher Scientific, F8775). Antigen coupled beads were washed with 0.05% Tween20 in PBS and incubated with 10 μl of immunized serums (1:10 dilution) in 0.1% BSA for 2 hrs at 37°C. Antibodies-beads complex were washed and incubated with 100 μl of complement factors from guinea pig (MP Biomedicals, 086428) for 20 mins at 37°C and washed with 15 mM EDTA/PBS to stop complement reaction. C3 deposition was detected by FITC-conjugated anti-C3 polyclonal antibody (1:100 dilution, MP Biomedicals, 0855385) and subjected to flow analysis.
IsdB ELISA and ELISPOT assay
IsdB-specific antibody levels in human and mouse sera were measured by ELISA as described previously (Kuklin et al., 2006). Briefly, sera were serially diluted in PBS containing 1% BSA and added to 96-well plates coated with recombinant IsdB (1μg/ml). Bound antibodies were detected by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or anti-mouse IgM (SouthernBiotech, 1021–05).
ELISPOT assay was performed on splenocytes isolated from IsdB-immunized mice to quantify IsdB-specific IgG-secreting cells. Splenocytes were serially diluted in RPMI containing 10% FBS from 5×106 cells/well into 96-well PVDF plate (Corning, CLS3508) coated with recombinant IsdB (1μg /well) and incubated for 6 hrs at 37 °C. Afterwards, the cells were detached using PBS with 0.1% Tween20 and the plates were incubated for an hour at 25°C with alkaline phosphatase (AP)-conjugated anti-mouse IgG (SouthernBiotech, 1030–04) and developed with BCIP/NBT solution (MABTECH, 3650–10). Spots were analyzed using ImageJ.
Hemoglobin-dependent S. aureus growth assay
Hemoglobin-dependent growth assay was performed as described previously (Bennett et al., 2019). Briefly, SA LAC was grown in RPMI medium (Gibco, 11835030) containing 0.1 Casamino Acids (Sigma, 2240) and 200μM 2,2’ Bipyridyl (Sigma, D216305). Overnight cultures were washed twice in RPMI containing 500μM 2,2’ Bipyridyl and diluted 1:100 in 200μl of RPMI medium containing 500μM 2,2’ bipyridyl, 25μM ZnCl2, 25μM MnCl2, 1mM MgCl2, 100μM CaCl2, and antibodies or sera with or without 1μM hemoglobin (Sigma, H7379). Bacterial growth at 37°C as measured by OD600 and was recorded using a Perkin Elmer Ensire Alpha plate reader.
Flow cytometric analysis of IsdB antibody binding to S. aureus
For measurement of IsdB antibody binding to SA cell surface, 1×107 LAC were washed and incubated with 10 μl of immunized serum, then FITC-conjugated anti-mouse IgG (Invitrogen, 31569). Fluorescence intensity was analyzed by FACSCanto (BD Biosciences) and FlowJo software.
Epitope mapping using an overlapping IsdB peptide array
The IsdB-binding profile of serum antibodies induced by IsdB immunization in naïve and SA-experienced mice was evaluated using a continuous peptide array composed of 141 15-mer peptides with an 11-residue overlap, covering the full-length IsdB. Peptides were printed on immobilized PVDF membranes. Membranes were blocked with TBST containing 1 % BSA, then pooled serum samples were diluted 1:1000 in blocking solution and added to the peptide array. Bound antibodies were detected by incubation with 800CW goat anti-mouse IgG (LI-COR) and fluorescent intensity was determined by Odyssey Imager (LI-COR). For epitope mapping of mAb 3G1, 3H2 and 1C1, 10 μg/ml of mAb were diluted in blocking buffer and incubated in peptide array of NEAT2 domain (#76-#103) for 1 hr. Bound antibodies were detected by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG.
Adoptive transfer of T lymphocytes, B lymphocytes, sera or purified antibodies
Splenic CD4+ T cells or CD19+ B cells were isolated using kits following instructions provided by the manufacturer (MojoSort, Biolegend, 480033 and 480088). 2×107 T cells or B cells were injected i.v. into recipient mice. IsdB-specific immune sera were generated by SA-infection or immunization as described above. Human sera were obtained from anonymized adult human volunteers. IsdB-specific antibodies were purified from human or mouse sera using immobilized IsdB agarose columns (NHS-activated agarose, ThermoFisher Scientific, 2697).
Generation of anti-IsdB monoclonal antibodies
Monoclonal antibodies were generated as previous described (Bennett et al., 2019). Briefly, mice were immunized i.p. 3 times with 75 μg IsdB formulated with aluminum hydroxide. 20 μg IsdB were injected i.p. 3 days prior to cell fusion. Splenocytes were fused with mouse myeloma partner P3X (ATCC CRL-1580) by using polyethylene glycol 1500 (Sigma, 10783641001) at a ratio of 3:1. The fused cells were plated and screened by ELISA for reactivity to IsdB as previously described. The complementarity-determining region 3 (CDR3) were analyzed as previously described (Tiller et al., 2009).
Preparation of single-cell sequencing
Splenocytes from IsdB immunized or LAC-experienced mice were incubated with phycoerythrin (PE)–labeled IsdB and allophycocyanin-conjugated anti-mouse CD45R/B220. IsdB+B220+ cells were sorted by FACSAria II (BD) and subjected to single-cell preparation by using a Single Cell 5’ Library and Gel Bead kit and Chromium Single Cell A Chip kit, the cell suspension was loaded onto a Chromium single cell controller to generate single-cell gel beads in the emulsion (GEMs) according to the manufacturer’s protocol (10X Genomics). scRNA-seq libraries were constructed using a Chromium Single Cell V(D)J Enrichment Kit, Mouse B Cell following instructions provided by the manufacturer (10X Genomics). The libraries were sequenced using an Illumina Novaseq6000 sequencer with a paired-end 150-bp (PE150) reading strategy (performed by Institute for Genomic Medicine, UCSD).
Generation of anti-IsdB monoclonal antibodies
Monoclonal antibodies were generated as previous described (Bennett et al., 2019). Briefly, mice were immunized i.p. 3 times with 75 μg IsdB formulated with aluminum hydroxide. 20 μg IsdB were injected i.p. 3 days prior to cell fusion. Splenocytes were fused with mouse myeloma partner P3X (ATCC CRL-1580) by using polyethylene glycol 1500 (Sigma) at a ratio of 3:1. The fused cells were plated and screened by ELISA for reactivity to IsdB as previously described. The complementarity-determining region 3 (CDR3) were analyzed as previously described (Tiller et al., 2009).
Preparation of single-cell sequencing
Splenocytes from IsdB immunized or LAC-experienced mice were incubated with phycoerythrin (PE)–labeled IsdB and allophycocyanin-conjugated anti-mouse CD45R/B220. IsdB+ B220+ cells were sorted by FACSAria II (BD) and subjected to single-cell preparation by using a Single Cell 5’ Library and Gel Bead kit and Chromium Single Cell A Chip kit, the cell suspension was loaded onto a Chromium single cell controller to generate single-cell gel beads in the emulsion (GEMs) according to the manufacturer’s protocol (10X Genomics). scRNA-seq libraries were constructed using a Chromium Single Cell V(D)J Enrichment Kit, Mouse B Cell following instructions provided by the manufacturer (10X Genomics). The libraries were sequenced using an Illumina Novaseq6000 sequencer with a paired-end 150-bp (PE150) reading strategy (performed by Institute for Genomic Medicine, UCSD).
Identification of clones and comparisons of clonotypes
The clonal groups were identified by the R package – scRepertoire (Borcherding et al., 2020) based on paired heavy and light chains. To determine clonal groups, we first used the filtered contig annotation obtained from the results performed using the Cell Ranger Single-Cell Software Suites (http://software.10xgenomics.com/single-cell/overview/welcome). Then, for the cells with high quality paired heavy and light chains were sequenced, clones were assigned based on strict definition of clonotype using the CTstrict() function that considers clonally related two sequences with identical V gene usage and > 85% normalized Levenshtein distance of the nucleotide sequence. The clonotype changes between samples were visualized by the compareClonotypes() function with the clones called by amino acid sequence of the CDR3 region.
Clonal lineage analysis (CDR3 neighborhood network)
To indicate convergent BCR evolution, we further used Scirpy (Sturm et al., 2020) to identify clonotype clusters based on CDR3 amino acid sequence similarity. An identified clonotype cluster is a higher-order aggregation of clonotypes that have different CDR3 nucleotide sequences, but might recognize the same antigen because they have the same or similar CDR3 amino acid sequence. Specifically, the clonotype clusters were identified by leveraging the Parasail library (Daily, 2016) to compute pairwise sequence alignments and identify clusters of B cells. The clonotype neighborhood networks was then visualized using Scirpy that makes use of the sparse-matrix implementation from the scipy package (Virtanen et al., 2020).
Quantification and statistical analysis
All of the statistical details of experiments, including the statistical tests used, exact value of n, what n represents, definition of center, and dispersion and precision measures can be found in the figure legends. Two-group analysis used unpaired Student’s t test (two-tailed tests). In vivo experiments were analyzed using non-parametric Mann-Whitney U-test. Comparisons of multiple groups were performed using one-way ANOVA, with Kruskal-Wallis test in the case of missing normality. Log rank test was used for analysis of mouse survival. Data were presented as mean ± standard deviation (SD), unless otherwise indicated. Statistical significance was assigned as ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; p > 0.05: ns (not significant). Analyses were performed using GraphPad Prism 5.
Supplementary Material
Highlights.
Prior S. aureus exposure makes otherwise protective IsdB vaccine non-protective.
IsdB vaccine recalls non-protective humoral memory from prior S. aureus infection.
Specific antibody competition further reduces IsdB vaccine efficacy.
Staphylococcal MntC and FhuD2 vaccines are also blunted by prior S. aureus exposure.
Acknowledgements
We thank S. Secore (Merck) for providing the Becker WT and IsdB/HarA mutant strains, M. Kumaraswamy for suggestions, and W. Parks, M. Bennett, and I. Thomsen for comments on the manuscript. The authors acknowledge funding from the National Institute of Health, grants R01AI127406, R01AI144694, NIGMS (R35 GM119850). Additionally, N.L. received funding from the Novo Nordisk Foundation provided to the Technical University of Denmark (Grant No. NNF20SA0066621).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
CMT and GYL have filed for a patent application for the use of IsdB NEAT2 as a vaccine.
REFERENCES
- Alonzo F 3rd, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, and Torres VJ (2013). CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AS, Scully IL, Timofeyeva Y, Murphy E, McNeil LK, Mininni T, Nunez L, Carriere M, Singer C, Dilts DA, et al. (2012). Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J Infect Dis 205, 1688–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MR, Dong J, Bombardi RG, Soto C, Parrington HM, Nargi RS, Schoeder CT, Nagel MB, Schey KL, Meiler J, et al. (2019). Human VH1–69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherding N, Bormann NL, and Kraus G (2020). scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily J (2016). Parasail: SIMD C library for global, semi-global, and local pairwise sequence alignments. BMC Bioinformatics 17, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers G, Bentlage AEH, Stegmann TC, Howie HL, Lissenberg-Thunnissen S, Zimring J, Rispens T, and Vidarsson G (2017). Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs 9, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Sanders H, Henn J, Wilson JM, Malone D, Buoninfante A, Willms M, Chan R, DuMont AL, McLahan C, et al. (2022). Vaccination With Detoxified Leukocidin AB Reduces Bacterial Load in a Staphylococcus aureus Minipig Deep Surgical Wound Infection Model. J Infect Dis 225, 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger S, Fallon JK, Michell AR, Broge T, Suscovich TJ, Streeck H, and Alter G (2019). A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J Immunol Methods 473, 112630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VG, Allen KB, Moreira ED, Moustafa M, Isgro F, Boucher HW, Corey GR, Carmeli Y, Betts R, Hartzel JS, et al. (2013). Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 309, 1368–1378. [DOI] [PubMed] [Google Scholar]
- Fowler VG Jr., and Proctor RA (2014). Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect 20 Suppl 5, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T (1960). On the doctrine of original antigenic sin. Proc Am Philos Soc 572–578. [Google Scholar]
- Gerlach D, Guo Y, De Castro C, Kim SH, Schlatterer K, Xu FF, Pereira C, Seeberger PH, Ali S, Codee J, et al. (2018). Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 563, 705–709. [DOI] [PubMed] [Google Scholar]
- Goodyear CS, and Silverman GJ (2003). Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal Toxin. J Exp Med 197, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtman A, Begier E, Mohamed N, Baber J, Sabharwal C, Haupt RM, Edwards H, Cooper D, Jansen KU, and Anderson AS (2019). The development of a staphylococcus aureus four antigen vaccine for use prior to elective orthopedic surgery. Hum Vaccin Immunother 15, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Nimmerjahn F, and Ravetch JV (2006). Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 313, 670–673. [DOI] [PubMed] [Google Scholar]
- Kappler J, Kotzin B, Herron L, Gelfand EW, Bigler RD, Boylston A, Carrel S, Posnett DN, Choi Y, and Marrack P (1989). V beta-specific stimulation of human T cells by staphylococcal toxins. Science 244, 811–813. [DOI] [PubMed] [Google Scholar]
- Kluytmans J, van Belkum A, and Verbrugh H (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, Noble L, Brown MJ, Zorman JK, Wang XM, et al. (2006). A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun 74, 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel W, Moll HA, and van Belkum A (2008). Dynamics and determinants of Staphylococcus aureus carriage in infancy: the Generation R Study. J Clin Microbiol 46, 3517–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Perez NE, Hopkins CA, and Pier GB (1988). Purified capsular polysaccharide-induced immunity to Staphylococcus aureus infection. J Infect Dis 157, 723–730. [DOI] [PubMed] [Google Scholar]
- Miller LS, Fowler VG, Shukla SK, Rose WE, and Proctor RA (2020). Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol Rev 44, 123–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RP, Mariotti P, Fiaschi L, Nosari S, Maccari S, Liberatori S, Fontana MR, Pezzicoli A, De Falco MG, Falugi F, et al. (2012). Staphylococcus aureus FhuD2 is involved in the early phase of staphylococcal dissemination and generates protective immunity in mice. J Infect Dis 206, 1041–1049. [DOI] [PubMed] [Google Scholar]
- Paterson MJ, Caldera JR, Nguyen C, Sharma P, Castro AM, Kolar SL, Tsai CM, Limon JJ, Becker CA, Martins GA, et al. (2020). Harnessing antifungal immunity in pursuit of a Staphylococcus aureus vaccine strategy. PLoS Pathog 16, e1008733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Kolar SL, Muller S, Reyes CN, Wolf AJ, Ogawa C, Singhania R, De Carvalho DD, Arditi M, Underhill DM, et al. (2017). O-Acetylation of Peptidoglycan Limits Helper T Cell Priming and Permits Staphylococcus aureus Reinfection. Cell Host Microbe 22, 543–551 e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallon BJ, Tam SH, McCarthy SG, Cai AN, and Raju TS (2007). Higher levels of sialylated Fc glycans in immunoglobulin G molecules can adversely impact functionality. Mol Immunol 44, 1524–1534. [DOI] [PubMed] [Google Scholar]
- Stranger-Jones YK, Bae T, and Schneewind O (2006). Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A 103, 16942–16947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G, Szabo T, Fotakis G, Haider M, Rieder D, Trajanoski Z, and Finotello F (2020). Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics 36, 4817–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomer L, Emolo C, Thammavongsa V, Kim HK, McAdow ME, Yu W, Kieffer M, Schneewind O, and Missiakas D (2016). Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J Exp Med 213, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Busse CE, and Wardemann H (2009). Cloning and expression of murine Ig genes from single B cells. J Immunol Methods 350, 183–193. [DOI] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wright AE (1902). Notes on the Treatment of Furunculosis, Sycosis, and Acne by the Inoculation of a Staphylococcus aureus Vaccine. Lancet, 874–884. [Google Scholar]
- Yoshida K, Umeda A, and Ohshima Y (1987). Induction of resistance in mice by the capsular polysaccharide antigens of Staphylococcus aureus. Microbiol Immunol 31, 649–656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


