Abstract
The circadian clock is an intracellular timekeeping device that drives daily rhythms in diverse and extensive processes throughout the body. The mechanism of this clock is comprised of a core transcription/translation negative feedback loop, that is modulated by a complex set of additional interlocking feedback loops. Pharmacological manipulation of the clock may be valuable for many maladies, including treating jet lag, shift work and related sleep disorders, but also various metabolic diseases and cancer. Here we review recent identification of small molecule clock modulators and discuss the biochemical features of the core clock that may be amenable for future drug discovery.
Keywords: CLOCK, BMAL1, PER, CRY, rhythms
The Circadian Clock Broadly Controls Physiology
Life on earth evolved to adapt to the earth’s rotation around the sun, taking advantage of the predictability of day and night. The result is an endogenous cyclic mechanism with a period of approximately 24 hours known as the circadian clock (see Glossary). Rhythms in the environment, such as light/dark cycles, entrain these clocks and therefore determine the correct phase. In animals, a “central” clock in the brain drives rhythms in behavior and coordinates the “peripheral” clocks found in cells throughout the body. The circadian clock mechanism is composed of several interlocking transcription/translation feedback loops (TTFLs) that act in concert with the core negative feedback loop (hereafter referred to as core loop) to generate the daily rhythms [1].
At the heart of this core loop in mammals, the activator proteins, CLOCK and BMAL1, form a heterodimer and drive the expression of multiple genes, including the repressors CRYPTOCHROME 1 and 2 (CRY1 and CRY2) and PERIOD 1, 2, and 3 (PER1, PER2, and PER3). The CRYs and PERs (in complexes with other proteins) then translocate to the nucleus and interact with CLOCK:BMAL1 to inhibit their own transcription (see FIG. 1), which takes approximately 24 hours to complete the cycle. This core loop is modulated by other interlocking loops, including an auxiliary loop consisting of REV-ERB and ROR that control the expression of CLOCK and BMAL1. In addition, these proteins are post translationally regulated by proteins such as Casein kinase 1ε/δ and Casein kinase 2, which are relevant to this review, in addition to a number of other kinases and ubiquitin ligases that contribute to stability, activity, and localization [2]. The full circadian machinery is much more complex than this, involving many other proteins and loops that will not be discussed here but are reviewed in [1].
Figure 1. Graphical depiction of the core transcription translation feedback loop.
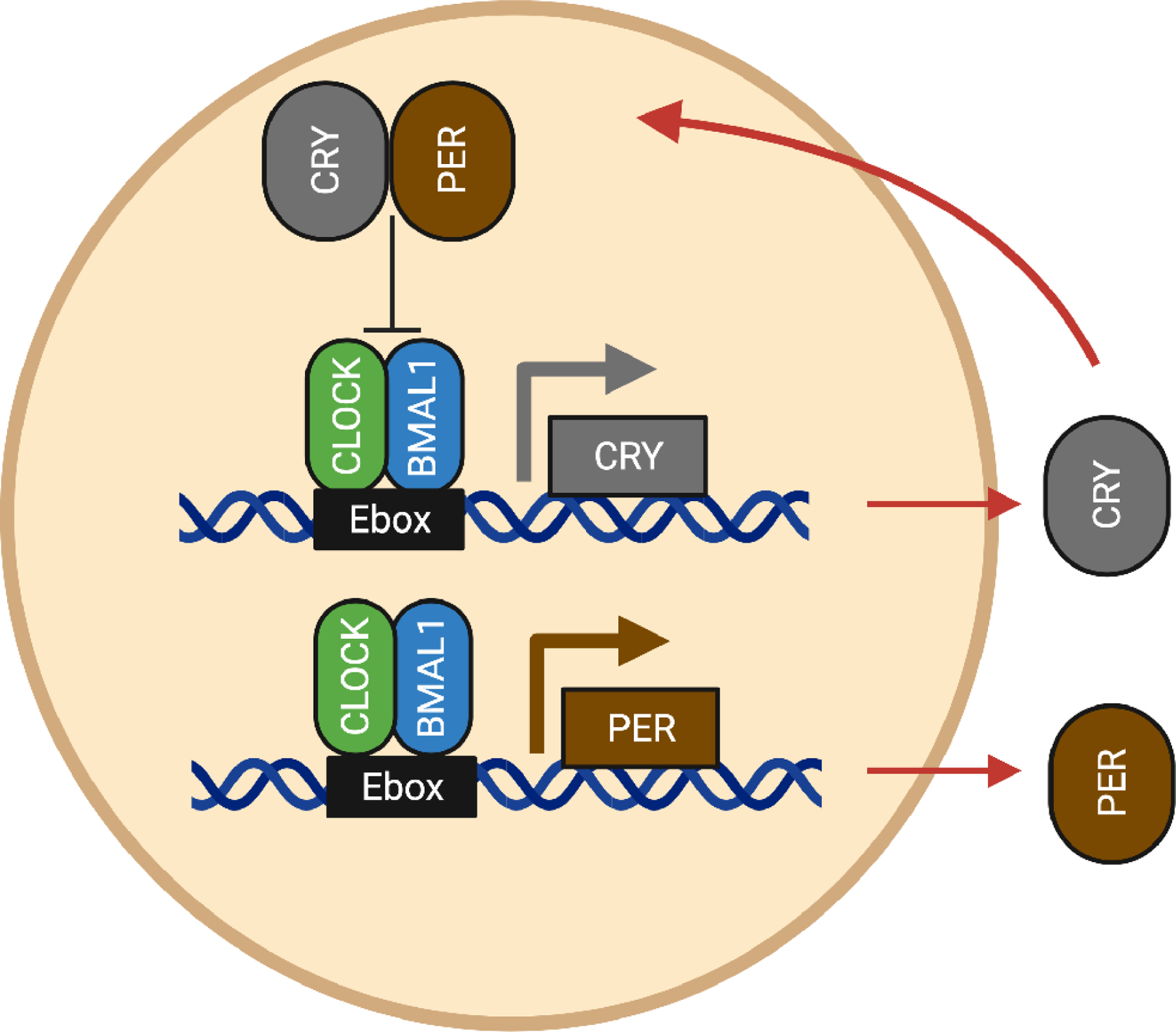
CLOCK and BMAL drive the expression of CRY and PER which then relocate to the nucleus to bind CLOCK and BMAL and stop their own expression. This cycle takes roughly 24 hours.
The circadian clock drives daily oscillations of thousands of genes, a large percentage of the total transcriptome [3, 4], and therefore deviations in the circadian clock mechanism can have consequences for physiology and behavior. As a consequence, many diseases have been linked to alterations or disruptions in circadian rhythms, such as cancers, metabolic disorders, heart disease, sleep disorders, and several types of mental illness [5, 6]. Furthermore, aging has also been related to the decline in the amplitude of the circadian rhythm [7, 8]. In addition, as was first shown in the 70s [9], many drugs targeting serious diseases have different efficacy depending on the time of day at which they are given. Using RNA-sequencing (RNA-seq) and DNA arrays to study the circadian oscillations in transcript expression in 12 different mouse organs, it was shown that more than half of the top-selling drugs act on proteins from rhythmic genes [4]. The importance of circadian timing in medicine has been increasingly demonstrated, and grown into a field of research known as circadian medicine [6, 10, 11].
Because circadian rhythms impact physiology so broadly, it is of interest to develop drugs that target the circadian clock. However, the choice of targets and the types of small molecule effectors can result in altering the clock in several different ways, and therefore strategies will need to depend on the desired outcome. For example, for improved health in aging, boosting the amplitude of the circadian rhythm could be beneficial, while for sleep disorders resulting from a lengthened circadian rhythm, drugs that shift the clock ahead would be of interest, and in some types of cancers (e.g., glioblastoma), inhibition of the clock may stop cancer stem cell proliferation [12]. Devising these strategies is aided by the increased understanding of the circadian clock mechanism, which has been greatly enhanced in recent years by the identification of atomic level structures of many of the key components [13–24] and by increased interrogation of the components and stoichiometry of the activator and repressor complexes and how they are regulated [22, 25–29].
In this review we will examine the biochemical and structural features of the core clock proteins to reveal where compounds could interact and discuss the predicted effects. In addition, we will discuss the efforts that have resulted in identifying small molecules that alter clock function through these core clock proteins (see Box 1 for common assays used in these screens). Successful identification of potent and bioavailable compounds that target the core clock will be of great use in dissecting the intricacies of this mechanism as well as aid future endeavors in pharmacological interventions for the circadian clock.
The simplified circadian landscape
Taking a closer look at the molecular components in the TTFL can help us understand how the desired effect of a therapeutic agent might be obtained. FIG. 2 shows an overview of the core circadian proteins in the different states of the TTFL, adapted from [1]. The repression phase consists of an initial repressive phase at about Circadian Time 16 (CT16) in which CLOCK:BMAL1:CRY1/2:PER1/2 (and associated proteins) are bound to DNA, followed by the poised phase at CT24, where the complex consists of CLOCK:BMAL1:CRY1. Following the degradation of CRY1, the repression is relieved and allowing the onset of the transcription phase [3].
Figure 2. Molecular assemblies at the E box at three different times of day [1].
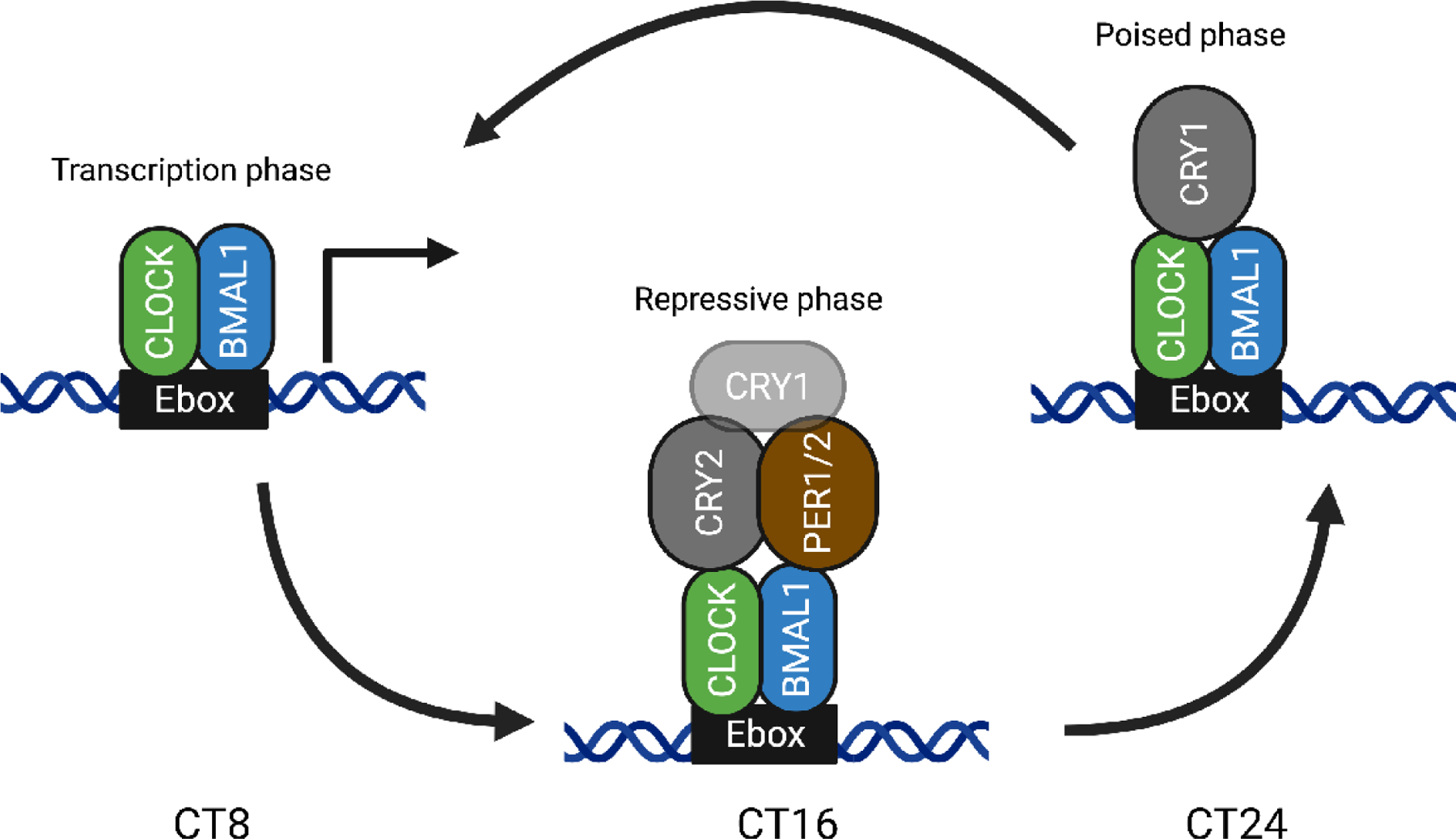
At CT8, the CLOCK and BMAL1 are bound to the E box driving gene expression, known as the transcription phase. At CT16, PER1/2 and CRY2 (along with some CRY1) have bound CLOCK and BMAL1 to stop the expression, known as the repressive phase, and at CT24, CRY1 is the predominate CRY and has replaced CRY2 and PER1/2, maintaining the repressed state (poised state) until transactivation begins again.
Inhibiting the proteins in each of the three states will have different outcomes [28]. For instance, inhibiting CRY2 will extend the time spent in the transcriptional phase, with extended transcription of the circadian controlled genes in this simplified picture. Similarly, inhibiting CRY1 will also extend the transcriptional phase, but it will do so primarily by decreasing the poised phase and allowing the transcriptional phase to begin prematurely, making it a distinctively different outcome. This is consistent with the free-running period of CRY knockout mice, where CRY1 knockout mice show a 1.2 hour shortening of period; whereas, CRY2 knockout mice show a 0.8 hour longer period [30, 31]. Exemplifying that even though both proteins are in the negative arm of the feedback loop, inhibiting them does not result in the same outcome. This highlights the importance of understanding which state of the molecular clock is being repressed or enhanced when considering which of the core clock proteins to target for therapeutical intervention, which we shall discuss below.
The activators of the TTFL
Perhaps the most obvious targets for regulation of the circadian clock are the activator proteins of the TTFL, CLOCK and BMAL1. These proteins directly drive gene expression, and compounds that would inhibit their activity (through inhibition of DNA binding, prevention of CLOCK/BMAL1 dimerization or inhibition of their interaction with other positive transcription factors) would be predicted to decrease the amplitude of circadian rhythmicity and reduce the gene expression levels of their many targets. Conversely, compounds that prevent their interaction with the repressor proteins would increase expression of their target genes and might extend the time of the active phase. CLOCK and BMAL1 belong to a family of proteins called the bHLH-PAS proteins, many of which have been strongly implicated in human disease.
The bHLH-PAS transcription factor family is named after the distinct structural domains of which they consist. An N-terminal basic Helix-Loop-Helix (bHLH) domain is followed by two tandem PER-ARNT-SIM (PAS) domains, while the remaining portions of the proteins are largely unstructured [32]. In general, the bHLH-PAS transcription factor family can be divided into two classes based on their heterodimerization patterns. Class I includes three Hypoxia-inducible factor (HIF)a proteins, HIF-1a, HIF-2a, and HIF-3a, neuronal PAS domain proteins (NPAS1-4), aryl hydrocarbon receptor (AHR), AHR repressor (AHRR) single-minded proteins (SIM1-2), and CLOCK. Class II includes aryl hydrocarbon receptor nuclear translocator (ARNT, ARNT2), BMAL1 (ARNTL), and BMAL2 (ARNTL2). Heterodimerization between a protein in class I and class II produce a transcription factor capable of gene regulation, as shown in FIG. 3A [32].
Figure 3. Classification of bHLH-PAS transcription factors.
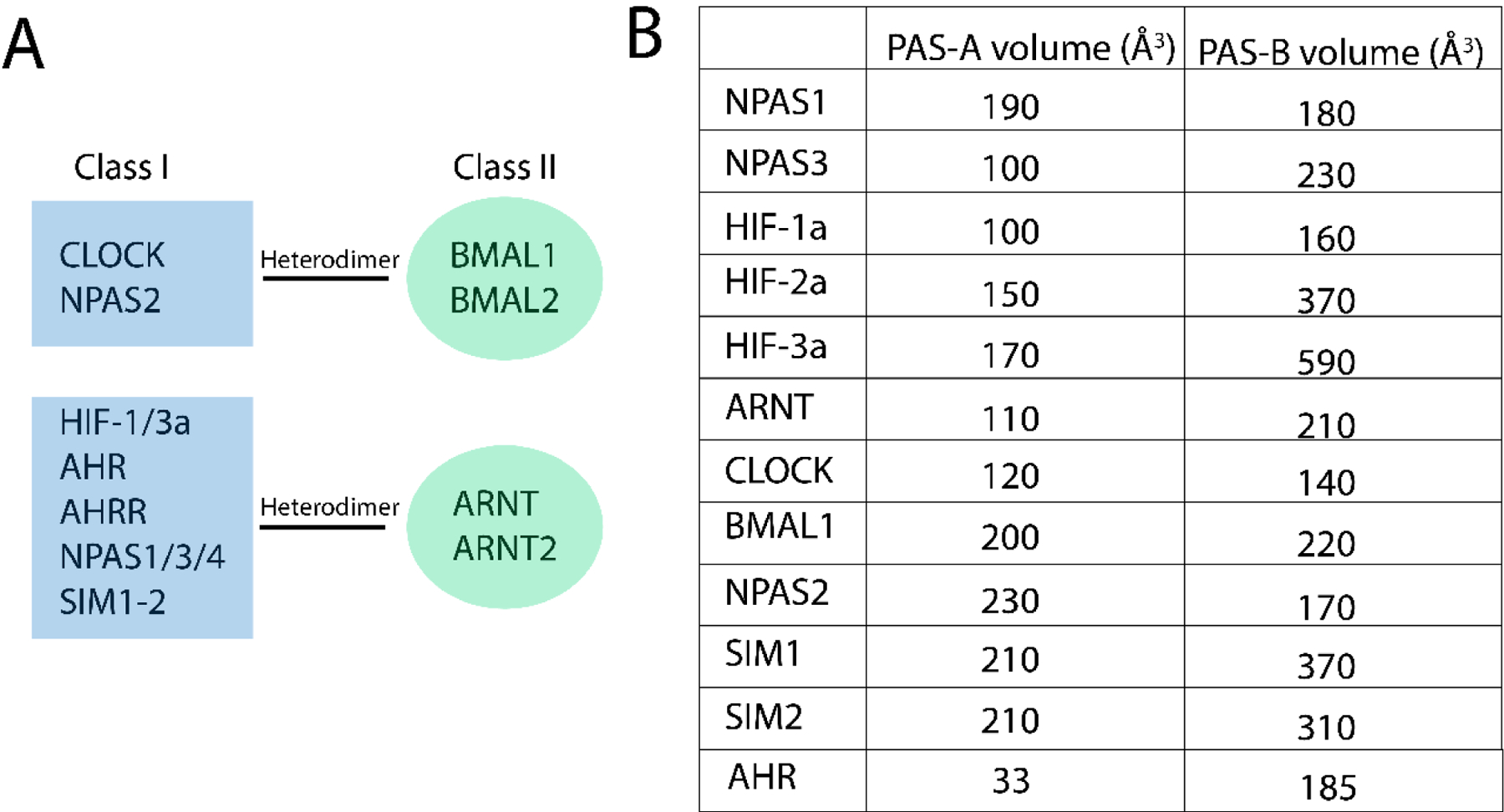
A: Heterodimerization patterns of class I and class II transcription factors. B: Volumes of the PAS-A and PAS-B pockets, all adapted from [39] except for AHR. The AHR PAS-A and PAS-B volumes were calculated using the Epock VMD plugin [96], with standard parameters; Grid resolution 1.0, Contig. Cutoff 4.0, and Grid sphere size 1.4. The PAS-A structure taken from PDB ID: 4M4X [97]. The inclusion selection was set to “chain A and not resid 107 to 120” using the sphere method with a radius if 8 Å. The PAS-B structure was obtained with AlphaFold2.1 [98], using resid 270 to 396 and the reduced database. The model has a pLDDT of 92.1 which is described as “Very high confidence”. The inclusion selection in epock was set to “all” using the sphere method with a radius of 10 Å. The inclusion parameters were set based on visual inspection to ensure that volumes outside of the PAS-B pocket were not included.
As mentioned before, many of these proteins have been implicated in human diseases. Loss-of-function mutations in SIM1 have been linked to obesity, SIM2 is associated with cancers, HIF proteins regulate key genetic programs required for tumor control, and disruption of CLOCK and BMAL1 can lead to various metabolic diseases as well as sleep disorders [1, 33].
PAS B pockets as ligand-binding domains
Because of their involvement in disease, some of these proteins have already been targeted for drug screens. For example, the HIF-2α transcription factor has been the target of a successful drug screen [34, 35] and has been shown to bind small molecules in the PAS-B pocket that act as inhibitors of the HIF2α:ARNT transcription factor. AHR has also been investigated, and multiple activators of the protein have been found [36, 37]. AHR is slightly different since it has been found to be ligand-activated as opposed to the other bHLH-PAS transcription factors [38]. These compounds are believed to bind the PAS-B pocket, similar to HIF-2α.
Despite the success in finding compounds that modulate these other family members, only one screen has found a potential compound for CLOCK or BMAL1 to date. A reason for this could be that the volumes of their PAS domain pockets are small compared to HIF-2α and AHR. The size of the binding pocket does, to some extent, predict the ability to bind small molecules; if the pocket is very small, it will be difficult to accommodate compounds that are protein specific. FIG 3B shows a comparison of the volume of the PAS-A/B pockets [39] for the bHLH-PAS proteins. Looking at HIF-2a that is known to bind small compounds in the PAS-B pocket, it features a PAS-B binding pockets with a volume of 370 Å3, while the circadian analogue, CLOCK, has a volume of 140 Å3. BMAL1 has a bigger PAS-B pocket with a volume of 220 Å3 making it closer to HIF-2a, but still 150 Å3 smaller. The smaller PAS-B binding pockets could make CLOCK and BMAL1 more problematic targets than some of the other bHLH-PAS proteins. Despite the small PAS pockets, both NPAS2 and CLOCK have been reported to bind the large molecule HEME, although this binding is suggested to be axial, which may be why these pockets can accommodate a ligand of that size [40–42]. Further, the AHR protein is activated by ligands, such as dioxins, which are smaller in size compared to HEME [43] and the ligand binding domain of AHR is found to encompass the PAS-B pocket [44].
Additional targets in CLOCK and BMAL1
In addition to the PAS-B pockets, there are other regions of the CLOCK/BMAL1 structures that may be amenable to perturbation by small molecules. For example, multiple mutations have been reported that inhibit the dimerization of CLOCK:BMAL1, most of which are along the interface of the two proteins, as shown in FIG. 4. Interestingly, the most effective single-point mutations are in the bHLH region: L57E and L74E in mouse CLOCK and L95E and L115E in mouse BMAL1 [14]. Targeting the interface in the PAS domains seems to require multiple mutations to achieve the same effect, likely due to the extensive interface between them. One such pair is the double mutation CLOCK:L113E+BMAL1:I137D in the PAS-A, while in the PAS-B CLOCK:W284A+BMAL:W427A also appears to lower the dimerization ability. While these mutations in the interface do seem to alter the interaction of CLOCK and BMAL1, targeting them for drug screens could be problematic given the flexibility of the proteins and the need for them to be in monomeric form to be targeted.
Figure 4. Secondary structure of mouse CLOCK:BMAL1 with bHLH and PAS domains indicated.
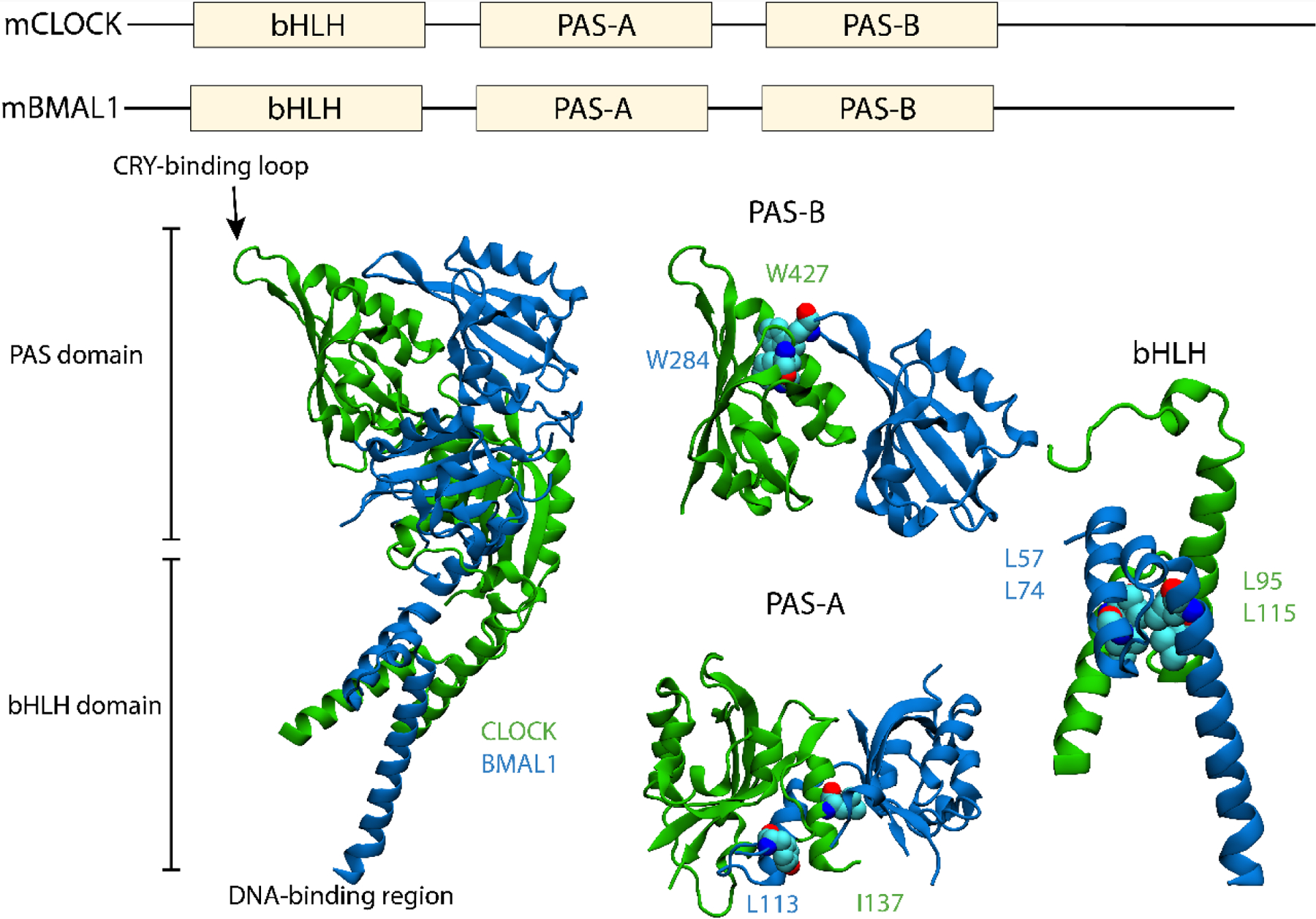
Residues that are known to disrupt binding have been highlighted in PAS-B, PAS-A and the bHLH regions separately. PDB ID: 4F3L, adapted from [14].
Despite this challenge, a recent paper reports success in targeting the interface between CLOCK and BMAL1 [45]. Using an in silico screen searching the entire structure of CLOCK, Doruk et al. docked 2 million compounds and identified 100 compounds with predicted binding affinities in the −7 to −10 kcal/mol range. One of these compounds, CLK8, is predicted to bind the bHLH region of CLOCK, not the PAS-B domain. Testing these compounds in U2OS cells, they find CLK8 disrupts CLOCK:BMAL1 interaction and enhances the circadian amplitude but did not alter the period length [45].
The repressors of the TTFL
A different approach to modulating the circadian clock is to target the core repressive components of the TTFL, which consists of the proteins CRY and PER (Figure 2). Inhibition of these proteins are predicted to prolong the transcriptional phase of the clock, while agonists would increase the length of the repressive phase.
CRYPTOCHROME
CRYPTOCHROMES are flavoproteins structurally related to PHOTOLYASE, which functions as a light-dependent DNA repair protein, although CRYPTOCHROME has no photolyase activity. There are four types of CRYs, CRY1, CRY2, CRY4, and DASH-CRY, the latter of which will not be discussed in the scope of this review. Unlike the activators of the TTFL, CRYs have many different functions; in plants, CRYPTOCHROMES function as photosensitive regulators of growth [46], in Drosophila melanogaster, CRY is a photosensitive circadian regulator [47], and in mammals, CRY1 and CRY2 are non-photosensitive repressors of the TTFL [48]. In recent years, it has become more apparent that CRY4 likely functions as a magnetoreceptive protein in migratory birds, responsible for sensing magnetic fields [49].
Structurally CRY consist of two domains, the Photolyase Homology Region (PHR) domain and the C-terminal tail. The PHR domain features two binding pockets, the primary and secondary binding pocket, shown in FIG. 5 top, while the C-terminal domain is unstructured. The primary binding pocket is known to bind an FAD co-factor [50] and also facilitate interactions with FBXL3/21. The secondary binding pocket in photolyases bind a small molecule cofactor that acts as a light antennae (such as methenyltetrahydrofolate) [51], but in mammalian CRYs, this pocket mediates interactions with CLOCK via the CLOCK:TRP362 loop [14, 22, 29]. Although the PHR domains of CRY1 and CRY2 are well conserved, the disorderd C-terminal tails are completely distinct and the function of these tails are not well understood. The fact that CRYs have two well defined binding pockets makes them very attractive as targets for drug screens.
Figure 5. Sequence of mouse CRY2 and PER2 along with the resolved secondary structures.
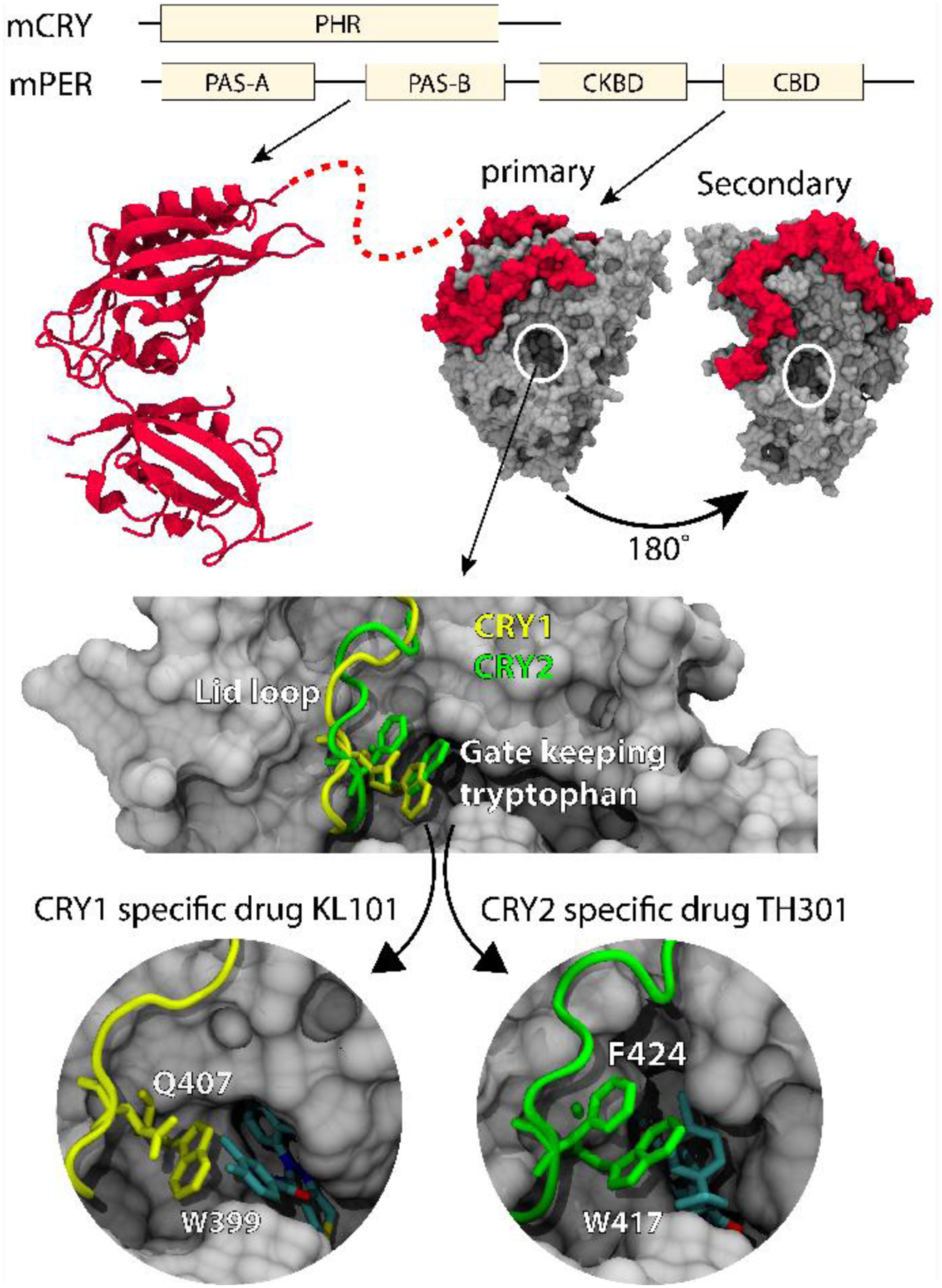
(TOP) The left-hand side shows the two PAS domains in PER2 (PDB ID: 3GDI from [13]), while the right-hand side shows the CRY-binding domain bound to the structure of CRY2 (PDB ID: 4U8H from [20]). The structure of the casein kinase binding domain is not known, as indicated by the dotted line. (BOTTOM) Binding of isoform specific compounds for CRY1 and CRY2. Top. Primary binding pocket of CRY1 with the lid-loop shown as a tube and the gate keeping tryptophans and stabilizing amino acids shown in the molecular representation for CRY1 (yellow) and CRY2 (green). Bottom left. Interaction of the gatekeeping tryptophan W399 with the CRY1 specific drug KL101 and the stabilizing amino acid Q407, shown in the binding pocket of CRY1. Bottom right. Interaction of the gatekeeping tryptophan W417 with the CRY2 specific drug TH301 and the stabilizing amino acid F424, shown in the binding pocket of CRY2. Adapted from [57], PDB IDs: 7D0M, 7D0N, 6KX6, 6KX8.
The initial CRY agonist (KL001) was identified in a cell-based phenotypic screen designed to find compounds that altered circadian rhythmicity. KL001 works by inhibiting the interaction of the ubiquitin ligases FBXL3 and FBXL21 with CRY [16, 52], which also bind the primary binding pocket of CRY and tag them for degradation. Binding small compounds in the binding pocket lengthens the lifetime of CRY, leading to a longer repressive phase of the clock [53]. Systematic structure activity relationship (SAR) analysis using KL001 as a starting point produced additional compounds with higher potency and improved in vivo exposure and have been shown to be efficacious in vivo in improving glucose tolerance in the db/db mouse model of diabetes [54]. Another group has used in silico docking screens and molecular dynamics simulations to identify additional compounds that are predicted to bind the primary pocket and also cause period lengthening [55].
The secondary pocket in CRY1 and CRY2 is highly homologous and differs only by 5 amino acids, all of which are conservative differences. Even with this high similarity, it has been shown that CRY1 is able to bind and repress CLOCK on its own, while CRY2 requires PER2 to bind CLOCK effectively [22, 29]. Binding small molecules to the secondary pocket could help extend the transcriptional phase of the clock by preventing repression of CLOCK/BMAL1 activity either at the beginning (CRY2) or ending (CRY1) of the repressive phase.
The high sequence similarity between CRY1 and CRY2 made it difficult to design isoform-specific inhibitors, but several compounds have been developed that bind selectively to CRY1 (KL101 and KL201) or CRY2 (TH301) and showed isoform-specific regulation [23, 56]. Surprisingly, crystal structures of CRY1 and CRY2 in complex with the compounds showed that they interact with the same residues in the primary pocket of each protein and suggested that the specificity comes from the disordered and unique C-terminal tails. However, swapping of the C-terminal tails did not reverse the specificity, but rather caused loss of binding, indicating that some combination of the PHR and tail must be important. Indeed, using improved crystal structures with higher electron density in the primary pocket, coupled with molecular dynamics simulations, it was revealed that although the compounds interact with the same residues in the pockets, the orientation of these residues is distinct between CRY1 and CRY2. This work identified a critical “gatekeeper” tryptophan in both proteins that interacts with a residue in the neighboring loop structure (the “lid loop”) in distinct ways in the two proteins (FIG. 5, bottom; [57]). This paper proposes that these conformational differences may be facilitated by the C-terminal tails. Although the disordered tails are not represented in these crystal structures of mammalian CRYs, in Drosophila the CRY tail interacts with the analogous tryptophan and lid loop structure [58], supporting this idea. Additional phenotypic screens have now identified CRY1 specific agonists that do not require the C-terminal tail for specificity (TH303 and TH129) [24, 59]. These compounds also rearranged the lid loop structure through a unique interaction of the benzophenone moiety in the compounds with F409 in the lid loop of CRY1 [24].
These isoform-specific compounds are of great value for use as tools to dissect the similar and distinct functions of these two CRY protein isoforms. Additional chemical modification of TH129, in which a photo-switchable derivative (azobenzene) replaced the benzophenone moiety, resulted in a novel switchable compound called GO1423 [24]. This allows reversible control of circadian period length by light and further expands the toolbox, making spatial and temporal manipulation of CRY1 function now possible.
In addition to their great value as tool compounds and in potential metabolic regulation, modulators of CRY are currently being considered for targeting the circadian clock in glioblastoma tumors which are known to hijack the circadian clock to promote growth in the cancer stem cells [12]. CRY stabilizers that cause inhibition of CLOCK:BMAL1 are expected to help suppress tumor growth.
Finally, other groups have identified additional CRY modulators that act to weakly inhibit CRY1 and CRY2 interactions with BMAL1. Using a CLOCK/BMAL1 transcriptional activity screen with ~1000 drug-like compounds, Chun et al [60] identified KS15, which produced about 2-fold enhancement of CLOCK/BMAL1 transcriptional activation. Counterintuitively, this inhibitor weakly dampens the BMAL1-luciferase rhythms in cells and has no effect on period length [60, 61].
PERIOD
mPER1 and mPER2 are 1290 and 1255 amino acid proteins, respectively, with two N-terminal PAS domains that can mediate protein-protein interactions such as homodimerization and heterodimerization [15, 62] (FIG 5). At the C-terminal end of the protein, the CRY binding domain (CBD) is present, which facilitates the interaction with CRY to form the repressive complex with CLOCK:BMAL1. The 128 amino acids of the CBD of PER2 have been solved in complex with CRY1 or CRY2 [19, 20]; PER2 CBD binds to CRY by wrapping around the protein, close to the secondary binding pocket as seen in FIG. 5 top. Lastly, there is also the Casein Kinase-Binding domain (CKBD) which is located between the PAS domains and the CBD. This domain facilitates interactions with Casein kinaseδ/ε (CKδ/ε) [25, 63], but the structure of this domain is currently unknown.
Mutations in the PAS domain of PER2 proteins have long been known to alter circadian rhythms in mice [64] and recently it has been shown that mutating even single amino acids in the PAS domain of PER2 can alter the circadian rhythmicity [65], showing the importance of the PAS domains in PER. This makes it tempting to speculate that the PAS domains in PER can be targeted in the same way as the bHLH-PAS proteins to disrupt the protein-protein interaction. Unlike the PAS domains, the CBD is an unlikely target as it is short and without any motifs that could bind potential compounds. The CKBD could be of potential interest, however the structure of the domain is unknown making it difficult to predict compound binding.
Other potential targets
Outside of the four core circadian proteins discussed above, many additional components of the clock could, of course, also be potential drug targets for modulation of circadian rhythms, affecting period length, robustness of rhythmicity, or resetting properties. These possibilities are too numerous to describe in detail here, but we will briefly discuss the four additional circadian proteins for which small molecule effectors have already been identified: REV-ERBα/β, RORα/β/γ, and Casein kinase 1 (CK1) and CK2.
REV-ERBs and RORs
REV-ERBα/β and RORα/β/γ are part of the nuclear receptor superfamily of transcription factors and form part of an auxiliary circadian loop that provides added robustness to the clock by controlling the expression of CLOCK and BMAL1 [66, 67]. Both act through the same enhancer element, but ROR activates transcription and REV-ERBs repress transcription. Antagonists of REV-ERB are expected to boost circadian transcription, as are agonists of ROR.
Like AHR, REV-ERBs and RORs are known to be ligand-regulated, with heme being an endogenous ligand for REV-ERB [68, 69]. The physiological ligands for RORs are less well understood, but sterols such as cholesterol have been implicated [70–72]. Binding of heme to REV-ERBs enhances recruitment of its corepressor complex containing nuclear corepressor (NCoR) and histone deacetylase 3 (HDAC3). Following this discovery, a biochemical screen for synthetic ligands was done, using a fluorescence resonance energy transfer (FRET) assay to measure the recruitment of NCoR identifying the first synthetic agonist, known as GSK4112 [73], which was further investigated using cellular assays and mouse models [74]. However, unfavorable pharmacokinetic (PK) properties limited the use of this compound in vivo. Since that time, a number of agonists as well as a few antagonists have been developed and tested by several different groups (reviewed in [75]). Although a number of these have shown promising results in cellular and in vivo applications, there have been many difficulties achieving good bioavailability and other optimal PK profiles and the effects of some vary between cell lines and tissues. In addition, off-target effects of some of these compounds have been reported [76, 77]. Finally, apart from work using in silico docking and computational predictions [78], detailed work on binding site or mechanism of binding is lacking for nearly all these compounds.
The first synthetic agonist for ROR was found to be the diabetic drug thiazolidinedione [79], and later an inverse agonist was identified for RORs [80] which was used as the scaffold for developing further agonists and inverse agonists [81, 82]. Another compound was found independently from an unbiased screen for circadian effectors – a high throughput screen of 200,000 compounds using PER2-luciferase reporter cells [83]; this compound is called Nobiletin and is a natural compound found in citrus peels. Deconvolution of its target revealed that it is an agonist of ROR and binds competitively to the ligand binding domains of RORα and RORγ with higher affinity for RORγ [84]. Nobiletin acts as a potent clock amplitude enhancer, and produces many beneficial effects on metabolism and increased longevity in mice (for example, see [84, 85]).
Casein kinases
Casein kinases are a family of serine/threonine kinases involved in regulating a set of diverse physiological activities including circadian rhythms, DNA repair, apoptosis, and cell differentiation and have been implicated in diseases including many types of cancer, Alzheimer’s Disease, and depression [86]. The Casein kinase family consists of CK1 which has many isoforms CK1α/β/γ1/γ2/γ3/δ/ϵ, as well as splice variants of these, and CK2. Implication of the CK proteins in circadian rhythms has been well characterized for CK1ε/δ and CK2 which interact with PER1/2 and phosphorylate multiple sites of the protein controlling the stability [87]. Furthermore, CK1δ is a stable component of both the cytosolic CRY/PER and the nuclear CLOCK/BMAL1/CRY/PER megacomplexes [25].
A high-throughput phenotypic screen followed by chemical optimization of hit compounds led to the discovery a period-lengthening compound longdaysin [88]. Target deconvolution revealed that longdaysin bound and inhibited both CK1α and CK1δ (as well as ERK2). Building upon the purine scaffold of longdaysin, a more potent compound NCC007 was discovered and shown to cause period lengthening of behavioral rhythms in mice [89]. Recently, azobenzene derivatives of longdaysin have yielded photosensitive switchable CK1 inhibitor that allows spatiotemporal regulation of period length through reversible inhibition of this kinase [90].
In addition, due to CK1 proteins’ broad role in disease, many CK1 inhibitors have been developed for reasons outside of the circadian clock field, with varying specificity and off-target effects [91]. Circadian biologists have made use of these compounds to interrogate the roles of the CK1 enzymes in the circadian clock. The discovery of PF670462, an inhibitor of CKIε/δ, by Pfizer was initially shown to have phase delaying effects in mice [92]. Further efforts revealed an isoform selective compound, PF4800567, that exhibits a 22-fold selectivity for CKIε [93]. The use of these two compounds revealed that inhibition of CK1ε alone produced only minimal effects on the circadian period length, while inhibition of CK1δ produced significantly increased period lengths in both cultured cells and in vivo in mice [93, 94]. These inhibitors, combined with specific genetic knockouts revealed that it is the CK1δ isoform that plays a dominant role in period determination. The use of isoform-specific inhibitors is a great example of how specific modulators can be tools for dissecting molecular mechanisms of the clock.
Finally, a cell-based phenotypic screen has also recently identified CK2 inhibitors CX-4945 and GO289, that both lengthen circadian period in U2OS cells and also inhibit the growth of human renal cell carcinoma and mouse leukemia cells [95]. These new tools will undoubtedly reveal new information about the role of CK2 in the circadian clock.
Concluding remarks
In this review, we have discussed the current state of drug development that targets the core circadian clock. Although the majority of compounds identified to date target REV-ERB/ROR and CK1 and CK2, new exciting efforts are focusing on the core transcriptional activators and repressors. Here, we have discussed newly understood structural aspects of the core circadian proteins CLOCK, BMAL1, CRY, and PER and how these may be exploited to develop molecules that target the circadian clock in different ways. Such molecules will be invaluable tools for dissecting the roles of the individual components of the clock mechanism. In addition, since circadian clocks drive extensive aspects of physiology and behavior and alterations in these clocks can have many different deleterious effects, therapeutic uses of these molecules are an exciting possibility. Indeed, the development of pharmacological modulators are of great interest for many different maladies, including those arising from jet lag and shift work, as well as metabolic diseases and many types of cancer. As our knowledge of the mechanism of the circadian clock becomes more sophisticated, more opportunities for successful drug development strategies continue to emerge.
Outstanding Questions.
The complete makeup and stoichiometry of the activation and repression complexes of the core clock mechanism are not completely known and will vary over the course of the circadian cycle. Will there be additional components of these complex that will be amenable to pharmacological manipulation?
Intracellular processes, such as timing of degradation or nuclear import have major effects on the circadian clock. Can these processes be modulated effectively by small molecules?
Highlights.
The circadian rhythm governs many physiological processes and has been linked to diseases such as cancers and metabolic disorders.
Pharmacological manipulation of circadian rhythms can have profoundly different effects depending on whether the positive or negative arm of the loop are targeted and in which direction.
The biochemical features of the core circadian clock components reveal many possible avenues for pharmacological modulation.
Identification of compounds that target the core circadian proteins will provide tools for further dissection of the clock mechanism and will have broad therapeutic potential.
Text box. Common assays that have been used to screen for circadian modulators.
Luciferase reporter gene driven by a circadian promoter (ie. Per1/2 or BMAL1 promoters) containing an E-box element have been used for both acute measurements of CLOCK/BMAL1 transcriptional activity from cell lysates (left) and for cell-based circadian screens right [53, 59, 83, 99] (Figure IA). The acute approach can be used to measure effects of potential compounds on the transcriptional activity of CLOCK/BMAL1 [60] or a specific activity or function, such as effects of compounds on interactions (e.g., ability of REV-EB to bind NCoR; [73]). In the circadian screening approach, the effects of compounds on the period, amplitude or phase of circadian rhythms are examined by measuring rhythmicity over several days in cell culture. In contrast, in silico screens can be used to find compounds that are predicted computationally to interact with circadian proteins such as recently reported for CLOCK [45] (Figure IB).
Box 1, Figure I.
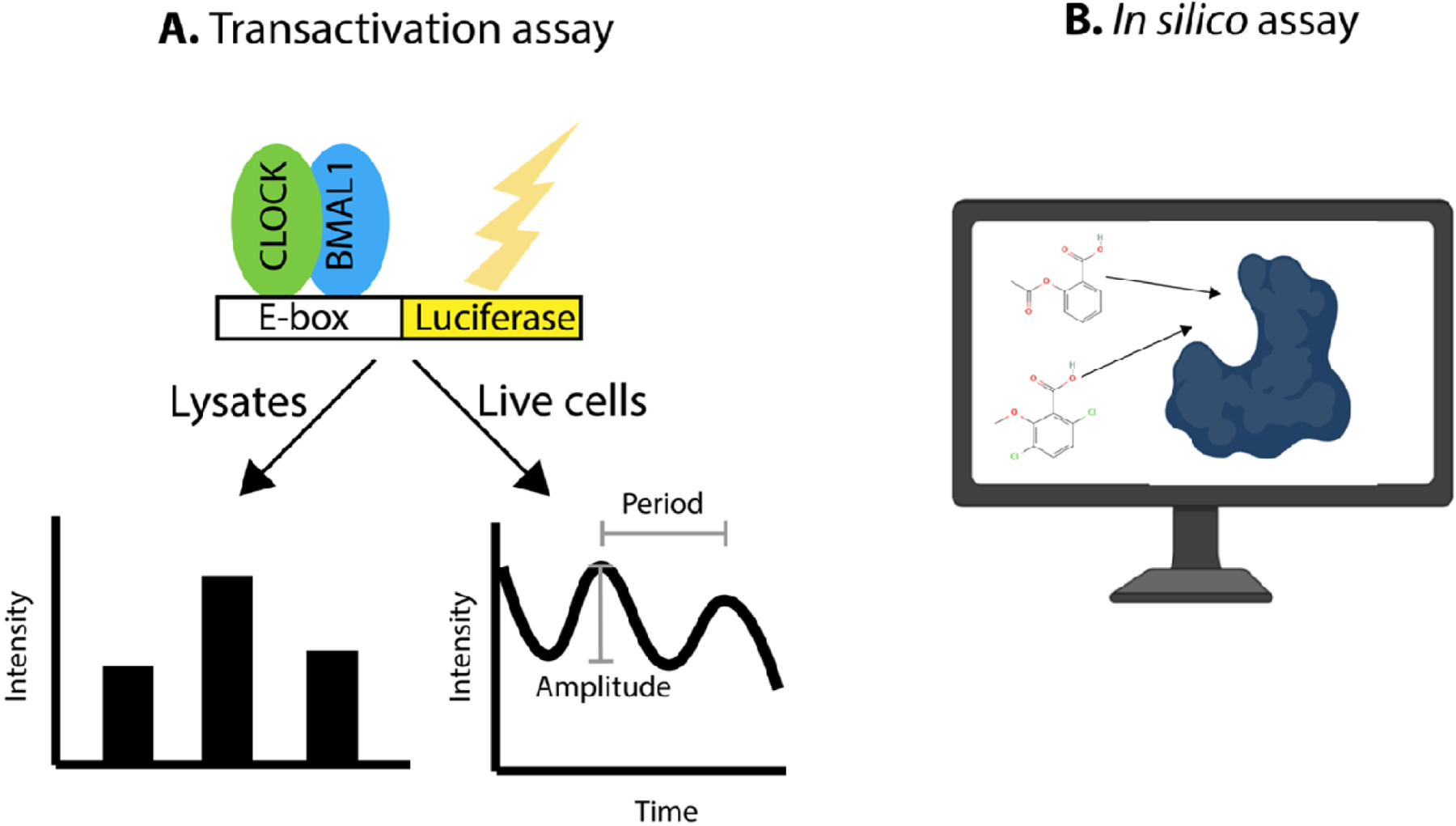
Common assays that have been used to screen for circadian modulators.
Glossary
- Amplitude
the difference between the trough and the peak of the rhythm
- Circadian clock
an internal intracellular mechanism that oscillates with an approximately 24-hour period and drives 24-hour rhythms
- Circadian medicine
alignment of treatments with the patient’s circadian rhythms
- Circadian oscillations
rhythms of approximately 24-hour periods that are driven by the circadian clock
- Circadian Time (CT)
standardized time that is based on the free-running (internal) circadian rhythm of the organism (for example if the organism is in constant conditions). CT0 is defined as the beginning of the activity in a diurnal animal and CT12 is defined as beginning of the rest phase
- Entrainment
the alignment or synchronization of internal rhythms to environmental cycles such as light:dark cycles
- Period
the length of one complete cycle
- Phase
the timing of a particular point in the rhythm (for example the “peak” or “trough”) with regard to a fixed point (for example beginning of the light phase).
- Zeitgeber Time (ZT)
time that is based on the light:dark cycles (or other environmental input) to which the organism is exposed. ZT0 is defined as the beginning of the light phase (dawn) and ZT12 is defined as beginning of the dark phase (dusk).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosby P and Partch CL (2020) New insights into non-transcriptional regulation of mammalian core clock proteins. J Cell Sci 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koike N, et al. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, et al. (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111, 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jagannath A, et al. (2017) The genetics of circadian rhythms, sleep and health. Hum Mol Genet 26, R128–R138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allada R and Bass J (2021) Circadian Mechanisms in Medicine. N Engl J Med 384, 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood S and Amir S (2017) The aging clock: circadian rhythms and later life. J Clin Invest 127, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta-Rodriguez VA, et al. (2021) Importance of circadian timing for aging and longevity. Nat Commun 12, 2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haus E, et al. (1972) Increased tolerance of leukemic mice to arabinosyl cytosine with schedule adjusted to circadian system. Science 177, 80–82 [DOI] [PubMed] [Google Scholar]
- 10.Cederroth CR, et al. (2019) Medicine in the Fourth Dimension. Cell Metab 30, 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancar A and Van Gelder RN (2021) Clocks, cancer, and chronochemotherapy. Science 371 [DOI] [PubMed] [Google Scholar]
- 12.Dong Z, et al. (2019) Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov 9, 1556–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennig S, et al. (2009) Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol 7, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang N, et al. (2012) Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucera N, et al. (2012) Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci U S A 109, 3311–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing W, et al. (2013) SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czarna A, et al. (2013) Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 [DOI] [PubMed] [Google Scholar]
- 18.Nangle S, et al. (2013) Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res 23, 1417–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmalen I, et al. (2014) Interaction of circadian clock proteins CRY1 and PER2 is modulated by zinc binding and disulfide bond formation. Cell 157, 1203–1215 [DOI] [PubMed] [Google Scholar]
- 20.Nangle SN, et al. (2014) Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife 3, e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael AK, et al. (2017) Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci U S A 114, 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fribourgh JL, et al. (2020) Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller S, et al. (2020) Isoform-selective regulation of mammalian cryptochromes. Nat Chem Biol 16, 676–685 [DOI] [PubMed] [Google Scholar]
- 24.Kolarski D, et al. (2021) Photopharmacological Manipulation of Mammalian CRY1 for Regulation of the Circadian Clock. J Am Chem Soc 143, 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryal RP, et al. (2017) Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell 67, 770–782 e776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parico GCG, et al. (2020) The human CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1. Proc Natl Acad Sci U S A 117, 27971–27979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, et al. (2021) Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc Natl Acad Sci U S A 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosensweig C and Green CB (2020) Periodicity, repression, and the molecular architecture of the mammalian circadian clock. Eur J Neurosci 51, 139–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosensweig C, et al. (2018) An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat Commun 9, 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Horst GT, et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 [DOI] [PubMed] [Google Scholar]
- 31.Vitaterna MH, et al. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 96, 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D and Rastinejad F (2017) Structural characterization of mammalian bHLH-PAS transcription factors. Curr Opin Struct Biol 43, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bersten DC, et al. (2013) bHLH-PAS proteins in cancer. Nat Rev Cancer 13, 827–841 [DOI] [PubMed] [Google Scholar]
- 34.Scheuermann TH, et al. (2013) Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol 9, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace EM, et al. (2016) A Small-Molecule Antagonist of HIF2alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer Res 76, 5491–5500 [DOI] [PubMed] [Google Scholar]
- 36.Baker JR, et al. (2020) The aryl hydrocarbon receptor (AhR) as a breast cancer drug target. Med Res Rev 40, 972–1001 [DOI] [PubMed] [Google Scholar]
- 37.Puyskens A, et al. (2020) Aryl Hydrocarbon Receptor Modulation by Tuberculosis Drugs Impairs Host Defense and Treatment Outcomes. Cell Host Microbe 27, 238–248 e237 [DOI] [PubMed] [Google Scholar]
- 38.Kawajiri K and Fujii-Kuriyama Y (2017) The aryl hydrocarbon receptor: a multifunctional chemical sensor for host defense and homeostatic maintenance. Exp Anim 66, 75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu D, et al. (2016) NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dioum EM, et al. (2002) NPAS2: a gas-responsive transcription factor. Science 298, 2385–2387 [DOI] [PubMed] [Google Scholar]
- 41.Lukat-Rodgers GS, et al. (2010) Heme-based sensing by the mammalian circadian protein CLOCK. Inorg Chem 49, 6349–6365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchida T, et al. (2012) Effects of the bHLH domain on axial coordination of heme in the PAS-A domain of neuronal PAS domain protein 2 (NPAS2): conversion from His119/Cys170 coordination to His119/His171 coordination. J Inorg Biochem 108, 188–195 [DOI] [PubMed] [Google Scholar]
- 43.Sorg O (2014) AhR signalling and dioxin toxicity. Toxicol Lett 230, 225–233 [DOI] [PubMed] [Google Scholar]
- 44.Fukunaga BN, et al. (1995) Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem 270, 29270–29278 [DOI] [PubMed] [Google Scholar]
- 45.Doruk YU, et al. (2020) A CLOCK-binding small molecule disrupts the interaction between CLOCK and BMAL1 and enhances circadian rhythm amplitude. J Biol Chem 295, 3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad M, et al. (1998) Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emery P, et al. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 [DOI] [PubMed] [Google Scholar]
- 48.Griffin EA Jr., et al. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 [DOI] [PubMed] [Google Scholar]
- 49.Xu J, et al. (2021) Magnetic sensitivity of cryptochrome 4 from a migratory songbird. Nature 594, 535–540 [DOI] [PubMed] [Google Scholar]
- 50.Cashmore AR, et al. (1999) Cryptochromes: blue light receptors for plants and animals. Science 284, 760–765 [DOI] [PubMed] [Google Scholar]
- 51.Kavakli IH, et al. (2019) DNA repair by photolyases. Adv Protein Chem Struct Biol 115, 1–19 [DOI] [PubMed] [Google Scholar]
- 52.Yoo SH, et al. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirota T, et al. (2012) Identification of small molecule activators of cryptochrome. Science 337, 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Humphries PS, et al. (2018) Carbazole-containing amides and ureas: Discovery of cryptochrome modulators as antihyperglycemic agents. Bioorg Med Chem Lett 28, 293–297 [DOI] [PubMed] [Google Scholar]
- 55.Gul S, et al. (2021) Structure-based design and classifications of small molecules regulating the circadian rhythm period. Sci Rep 11, 18510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller S, et al. (2020) An Isoform-Selective Modulator of Cryptochrome 1 Regulates Circadian Rhythms in Mammals. Cell Chem Biol 27, 1192–1198 e1195 [DOI] [PubMed] [Google Scholar]
- 57.Miller S, et al. (2021) Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation. Proc Natl Acad Sci U S A 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levy C, et al. (2013) Updated structure of Drosophila cryptochrome. Nature 495, E3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirota T and Kay SA (2015) Identification of small-molecule modulators of the circadian clock. Methods Enzymol 551, 267–282 [DOI] [PubMed] [Google Scholar]
- 60.Chun SK, et al. (2014) Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem Biol 9, 703–710 [DOI] [PubMed] [Google Scholar]
- 61.Jang J, et al. (2018) The cryptochrome inhibitor KS15 enhances E-box-mediated transcription by disrupting the feedback action of a circadian transcription-repressor complex. Life Sci 200, 49–55 [DOI] [PubMed] [Google Scholar]
- 62.Yagita K, et al. (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev 14, 1353–1363 [PMC free article] [PubMed] [Google Scholar]
- 63.Lee C, et al. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 64.Zheng B, et al. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173 [DOI] [PubMed] [Google Scholar]
- 65.Militi S, et al. (2016) Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking. Proc Natl Acad Sci U S A 113, 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preitner N, et al. (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 67.Sato TK, et al. (2004) A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 [DOI] [PubMed] [Google Scholar]
- 68.Reinking J, et al. (2005) The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 122, 195–207 [DOI] [PubMed] [Google Scholar]
- 69.Raghuram S, et al. (2007) Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin L, et al. (2010) Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol 24, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kallen J, et al. (2004) Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem 279, 14033–14038 [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, et al. (2010) A second class of nuclear receptors for oxysterols: Regulation of RORalpha and RORgamma activity by 24S-hydroxycholesterol (cerebrosterol). Biochim Biophys Acta 1801, 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng QJ, et al. (2008) Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci 121, 3629–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grant D, et al. (2010) GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbalpha. ACS Chem Biol 5, 925–932 [DOI] [PubMed] [Google Scholar]
- 75.Uriz-Huarte A, et al. (2020) The transcriptional repressor REV-ERB as a novel target for disease. Bioorg Med Chem Lett 30, 127395. [DOI] [PubMed] [Google Scholar]
- 76.Dierickx P, et al. (2019) SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci U S A 116, 12147–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trump RP, et al. (2013) Optimized chemical probes for REV-ERBalpha. J Med Chem 56, 4729–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westermaier Y, et al. (2017) Binding mode prediction and MD/MMPBSA-based free energy ranking for agonists of REV-ERBalpha/NCoR. J Comput Aided Mol Des 31, 755–775 [DOI] [PubMed] [Google Scholar]
- 79.Missbach M, et al. (1996) Thiazolidine diones, specific ligands of the nuclear receptor retinoid Z receptor/retinoid acid receptor-related orphan receptor alpha with potent antiarthritic activity. J Biol Chem 271, 13515–13522 [DOI] [PubMed] [Google Scholar]
- 80.Kumar N, et al. (2010) The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethy l]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol 77, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, et al. (2010) Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS Chem Biol 5, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar N, et al. (2011) Identification of SR3335 (ML-176): a synthetic RORalpha selective inverse agonist. ACS Chem Biol 6, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Z, et al. (2012) Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A 109, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He B, et al. (2016) The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab 23, 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nohara K, et al. (2019) Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat Commun 10, 3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schittek B and Sinnberg T (2014) Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis. Mol Cancer 13, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Philpott JM, et al. (2021) Biochemical mechanisms of period control within the mammalian circadian clock. Semin Cell Dev Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirota T, et al. (2010) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol 8, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JW, et al. (2019) Chemical Control of Mammalian Circadian Behavior through Dual Inhibition of Casein Kinase Ialpha and delta. J Med Chem 62, 1989–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kolarski D, et al. (2021) Reversible modulation of circadian time with chronophotopharmacology. Nat Commun 12, 3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li SS, et al. (2021) Recent Advances in the Development of Casein Kinase 1 Inhibitors. Curr Med Chem 28, 1585–1604 [DOI] [PubMed] [Google Scholar]
- 92.Badura L, et al. (2007) An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther 322, 730–738 [DOI] [PubMed] [Google Scholar]
- 93.Walton KM, et al. (2009) Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther 330, 430–439 [DOI] [PubMed] [Google Scholar]
- 94.Meng QJ, et al. (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A 107, 15240–15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oshima T, et al. (2019) Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci Adv 5, eaau9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laurent B, et al. (2015) Epock: rapid analysis of protein pocket dynamics. Bioinformatics 31, 1478–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu D, et al. (2013) Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol Cell Biol 33, 4346–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jumper J, et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]


