Abstract
The deduced product of the Bacillus subtilis ytvP gene is similar to that of ORF13, a gene of unknown function in the Lactococcus lactis histidine biosynthesis operon. A B. subtilis ytvP mutant was auxotrophic for histidine. The only enzyme of the histidine biosynthesis pathway that remained uncharacterized in B. subtilis was histidinol phosphate phosphatase (HolPase), catalyzing the penultimate step of this pathway. HolPase activity could not be detected in crude extracts of the ytvP mutant, while purified glutathione S-transferase-YtvP fusion protein exhibited strong HolPase activity. These observations demonstrated that HolPase is encoded by ytvP in B. subtilis and led us to rename this gene hisJ. Together with the HolPase of Saccharomyces cerevisiae and the presumed HolPases of L. lactis and Schizosaccharomyces pombe, HisJ constitutes a family of related enzymes that are not homologous to the HolPases of Escherichia coli, Salmonella typhimurium, and Haemophilus influenzae.
In Bacillus subtilis, all enzymes involved in the histidine biosynthesis pathway have been characterized at the genetic level, except histidinol phosphate phosphatase (HolPase) (10, 18). This enzyme (l-histidinol-phosphate phosphohydrolase; EC 3.1.3.15) catalyzes dephosphorylation of histidinol phosphate to histidinol, the direct precursor of histidine. In the well-studied prokaryotes Escherichia coli, Salmonella typhimurium, and Haemophilus influenzae, HolPase activity is associated with the N-terminal domain of the HisB bifunctional enzyme. The C-terminal domain of HisB exhibits imidazoleglycerol phosphate dehydratase activity (4, 5, 22). However, this does not appear to be a common feature of these enzymes, as searches for similarities with the E. coli HisB bifunctional enzyme revealed the existence, in many organisms across all three kingdoms (eubacteria, archaea, and eukaryotes), of imidazoleglycerol phosphate dehydratases not fused to any other protein domain. For most of these organisms, nothing is known about the HolPase. A search for homologues to the N-terminal domain of the E. coli enzyme does not reveal any well-conserved domain, except in the S. typhimurium and H. influenzae bifunctional enzymes, and only the more distantly related Streptomyces lincolnensis LmbK protein has been associated with this family as a putative HolPase (19). A HolPase has been characterized at the genetic, physiological, and biochemical levels in the yeast Saccharomyces cerevisiae (8, 14), but its sequence has no similarity with the corresponding E. coli enzyme (12).
The complete genome of B. subtilis has been sequenced; among the 4,100 open reading frames identified, about 70% (termed y genes) correspond to putative genes for which the function has not been ascertained (10). The goal of the B. subtilis systematic functional analysis program is to assign a function to these genes. The applied strategy consists of constructing a collection of mutants that correspond to each of these genes and looking for a phenotype. We are currently testing whether the utilization of various carbon sources is affected in these mutants.
In the work presented here, we demonstrate that the ytvP gene encodes the B. subtilis HolPase, thus completing our knowledge of the histidine biosynthesis pathway in this organism.
The B. subtilis ytvP mutant BFA1037 is devoid of HolPase activity.
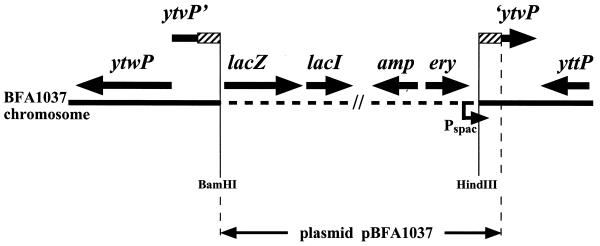
B. subtilis BFA1037 [trpC2 ytvP′::pBFA1037 (Eryr)] is a ytvP mutant, obtained by single crossing-over integration of plasmid pBFA1037 into the chromosome of the reference strain 168 Marburg (trpC2). The integrative plasmid pBFA1037 was derived from pMUTIN2mcs (21) by insertion, between its unique HindIII and BamHI sites, of a 218-bp HindIII/BamHI ytvP internal fragment amplified by PCR with B. subtilis 168 chromosomal DNA as a template and primers ytvP-H (5′-GCCGAAGCTTGGACAGCTTAGCTTAGCATACGG-3′) and ytvP-B (5′-CGCGGATCCACCGCTTCAATGCTTCC-3′). Correct integration of a single copy of plasmid pBFA1037 into the ytvP gene, confirmed by Southern blot analysis (17), led to disruption of ytvP and created a ytvp′-lacZ transcriptional fusion (Fig. 1).
FIG. 1.
Structure of the ytvP chromosomal region of mutant BFA1037. Integration of plasmid pBFA1037 led to disruption of ytvP and created a ytvP′-lacZ transcriptional fusion. The 218-bp ytvP internal fragment (symbolized by hatching) used as integration platform is duplicated. In addition to genes conferring resistance to ampicillin (amp) and erythromycin (ery), plasmid pMUTIN2mcs and its pBFA derivatives also carry a lacI gene and a Pspac isopropyl-β-d-thiogalactopyranoside-inducible promoter (broken arrow), which minimizes potential polar effects on expression of genes downstream of the integration site (21). Genes are symbolized by arrows indicating their orientation (not drawn to scale).
During growth tests performed in chemically defined media, we observed that strain BFA1037 was unable to grow in liquid MM medium (1) containing succinate (15 mM) and glutamate (30 mM) as carbon sources and supplemented with tryptophan (25 mg/liter), whereas the wild-type strain 168 could grow under these conditions (Table 1). The same observation was made with either glucose, glucitol, glycerol, gluconate, asparagine, proline, or arginine as a carbon source, and in all cases, growth of the ytvP mutant could be restored to the wild-type level by adding Casamino Acids (data not shown). This suggested that the mutant was auxotrophic for one or several amino acids.
TABLE 1.
Growth of the B. subtilis wild-type strain and the ytvP mutant
| Growth medium supplementa | Doubling time (min)b
|
||
|---|---|---|---|
| 168 (wild type) | BFA1037 (ytvP mutant) | Complemented BFA1037 (ytvP mutant with amyE′::ytvP) | |
| None | 136 | >750 | 138 |
| Histidine (50 mg/liter) | 132 | 135 | 127 |
| Histidinol (50 mg/liter) | 128 | 408 | 126 |
| Histidinol (250 mg/liter) | 124 | 301 | 110 |
The growth medium was MM containing succinate and glutamate plus tryptophan.
Values correspond to a typical experiment. Similar results were reproducibly obtained in several experiments.
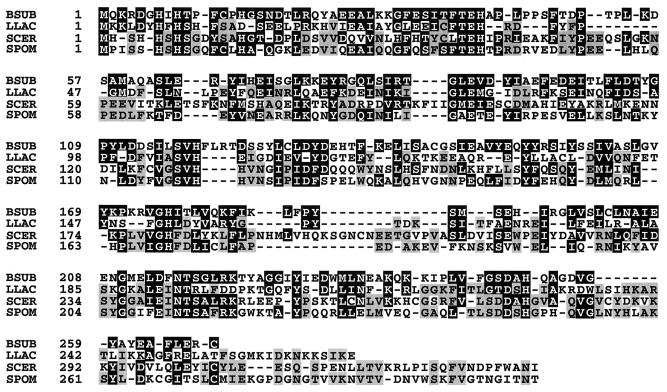
The ytvP gene (position 3030690 to 3031496 of the B. subtilis genome [10]) seems to be monocistronic. It encodes a 268-amino-acid protein, which exhibits weak but significant similarity (28% identity) to the product of ORF13, the last gene of the Lactococcus lactis histidine biosynthesis operon (7) (Fig. 2). Although the function of this L. lactis protein remains unknown, location of its structural gene suggested that in B. subtilis YtvP could be implicated in histidine biosynthesis. We observed that addition of histidine could indeed restore the growth of strain BFA1037 to wild-type level (Table 1). Growth could also be significantly improved by the addition of histidinol, although not as efficiently as with histidine (Table 1). These results showed that strain BFA1037 is auxotrophic for histidine and suggested that it is affected in HolPase activity.
FIG. 2.
Alignment of the B. subtilis YtvP (HisJ) protein (BSUB; accession no. PIR:D70002), the S. cerevisiae His9p HolPase (SCER; accession no. PIR:S56280), and the presumed HolPases L. lactis ORF13 gene product (LLAC; accession no. PIR:D47754) and S. pombe His9p homologue (SPOM; accession no. SPTREMBLNEW:E1316727). Residues conserved (identity and conservative change) in at least two of these proteins are against a black background (residues conserved in YtvP) or a grey background (residues not conserved in YtvP); groups of conservative change considered are G and A; D and E; F and Y; I, L, V, and M; K and R; N and Q; and S and T. Dashes indicate gaps introduced to maximize alignment.
A 1,054-bp EcoRI/BamHI fragment carrying the entire wild-type ytvP coding sequence and promoter region was amplified by PCR with primers ptvpE (5′-CCTGAATTCCAATGACAAACTCCATAATG-3′) and ftvpB (5′-CGCGGATCCGGACAGGAGACGTGG-3′) from B. subtilis strain 168 chromosomal DNA. This fragment was cloned into plasmid pDG1662 and integrated into the amyE locus of strain MO1099, following previously described methods (9). The amyE′::ytvP chromosomal region of the resulting strain was then transferred into BFA1037, which led to full complementation of the growth defect of this strain (Table 1). This showed that the phenotype of BFA1037 was due to the ytvP mutation.
We compared HolPase activity in extracts of the wild-type strain 168 and of the mutant strain BFA1037. For this purpose, B. subtilis cells were grown at 37°C with shaking in liquid medium (MM containing succinate and glutamate plus tryptophan, supplemented with histidine [50 mg/liter] when required), harvested by centrifugation when the culture reached an A600 of 0.5, and concentrated 10-fold in triethanolamine-HCl buffer (0.1 M triethanolamine-HCl, 10 mM Na2EDTA [pH 7.5]). Glass beads (diameter, 0.10 to 0.11 mm; Braun Sciencetec) were added to the suspension (about 0.5 g/ml), and the cells were broken by vigorous vortexing followed by sonication. After centrifugation, the supernatant was applied to a prepacked Sephadex G-50 column (Pharmacia, Uppsala, Sweden) pre-equilibrated with triethanolamine-HCl buffer. The phosphate concentration of each fraction was estimated with an ascorbate/ammonium molybdate color-developing reagent (13). Fractions containing phosphate-free proteins were collected, and protein concentration was determined by the Bradford method (3). HolPase activity was assayed as previously described (13). No significant HolPase activity (<0.4 U/mg of protein) could be detected in extracts of the ytvP mutant strain BFA1037 grown in the presence of histidine, whereas extracts of the wild-type strain grown with and without histidine exhibited in both cases significant and roughly equal levels of HolPase activity (6.4 and 6.9 U/mg of protein, respectively).
From these results, we concluded that the ytvP gene encodes either the B. subtilis HolPase or a positive regulator of it.
ytvP encodes HolPase activity.
An 812-bp BamHI/EcoRI fragment carrying the entire ytvP sequence was amplified by PCR with primers 5′ytvP-Bam (5′-CGCGGATCCATGCAAAAGCGAGACGGAC-3′) and 3′ytvP-Eco (5′-CCGGAATTCTAGCACCGTTCCAAAAACG-3′) from B. subtilis 168 chromosomal DNA. This fragment was cloned between the BamHI and EcoRI sites of plasmid pGEX-2T (Pharmacia), generating pGST::YtvP, a plasmid allowing overproduction of a glutathione S-transferase (GST)-YtvP fusion protein. Purification of GST and GST-YtvP proteins from E. coli TG1 [supE thi hsdD5 Δ(lac-proAB) (F′ traD36 proAB lacIq ZΔM15)] transformed respectively with pGEX-2T and pGST::YtvP was performed, following previously described methods (20). As estimated by polyacrylamide gel electrophoresis (11), the purity of both proteins was over 95%, and the apparent molecular mass of each purified product corresponded to that expected (26 and 57 kDa for GST and GST-YtvP, respectively) (results not shown). In vitro assays established tha the GST-YtvP fusion protein possesses strong HolPase activity (840.0 U/mg of protein versus <0.1 U/mg of protein for GST), thus demonstrating that in B. subtilis this activity is encoded by ytvP. We therefore propose to rename this gene hisJ.
As mentioned above, the protein most closely related to HisJ is the product of ORF13 of the L. lactis his operon. The structural gene encoding HolPase has not been identified in this bacterium (18). It has been proposed that this activity would be performed by the product of ORF8, based on its homology with enzymes catalyzing phosphorylation of a hydroxyl group, which is the opposite reaction to the dephosphorylation of such a group as carried out by HolPase (7). From the work presented here, it seems more likely that L. lactis HolPase is encoded by ORF13. The B. subtilis HisJ protein is also related to the S. cerevisiae HolPase and its Schizosaccharomyces pombe homologue, although more distantly (24 and 23% identity, respectively) (Fig. 2). These four proteins seem therefore to constitute a family of distantly related HolPases, distinct from the E. coli-type HolPases. As recently reported, this HisJ family belongs to a novel enzymatic superfamily designated PHP, which also comprises DNA polymerase domains (2). It has been suggested that these domains possess intrinsic phosphatase activity that hydrolyzes the pyrophosphate released during nucleotide polymerization. Among the PHP HolPases, HolPase activity had been demonstrated only for His9p of S. cerevisiae. The PHP superfamily also comprises numerous proteins of unknown function. While several of these seem to be clearly related to DNA polymerases (e.g., the 570-amino-acid B. subtilis YshC protein), some, such as the 211-amino-acid Archaeoglobus fulgidus AF1233 protein or the 246-amino-acid Anaerocellum thermophilum YOR4 protein, might belong to the His9p/HisJ HolPase family or even to other families of phosphatases. Interestingly, the E. coli-type HolPases belong to another superfamily of phosphohydrolases. This “DDDD” family also includes several classes of nonspecific acid phosphatases, phosphoglycolate phosphatases, phosphoserine phosphatases, and trehalose-6-phosphatases, but no DNA polymerases (19). In evolutionary terms, the existence of these two superfamilies could reflect the existence of two different ancestral nonspecific phosphatases that have evolved through fusion to other domains having specific properties, such as polymerase or binding of a particular substrate.
Identification of the hisJ HolPase structural gene in B. subtilis completes our knowledge of the histidine biosynthesis pathway in this bacterium. This also provides strong evidence for the role of the L. lactis ORF13 gene and will help with the identification of the functions of related gene products in several other organisms.
Transcription of hisJ is repressed neither by histidine nor by histidinol.
In L. lactis, the ORF13 gene belongs to the his operon, the transcription of which is enhanced 15-fold in cultures grown in the absence of histidine (6). The hisJ gene characterized in this work does not belong to the B. subtilis his operon and seems to be monocistronic. The two neighboring genes (yttP and ytwP) are oriented in the opposite direction (Fig. 1), and their inactivation does not confer auxotrophy to histidine (data not shown). We did not observe any significant difference in the level of HolPase activity in B. subtilis wild type grown in the absence or in the presence of 50 mg of histidine per liter (see above) or even 250 mg of histidine per liter (data not shown). This suggested that neither HisJ enzymatic activity nor hisJ transcription is regulated by this amino acid.
We also investigated potential regulation of hisJ transcription in the mutant strain BFA1037, in which a hisJ (ytvP)′-lacZ transcriptional fusion is under the control of the hisJ promoter (Fig. 1). For these β-galactosidase assays, extracts were prepared and enzymatic activity was determined as previously described (15, 16). The same level of β-galactosidase activity (about 20 to 25 Miller units/mg of protein) was measured in extracts of the mutant grown in liquid MM containing succinate and glutamate plus tryptophan, supplemented with various concentrations of histidine or histidinol (β-galactosidase activity in extracts of the wild-type strain 168 grown in the same conditions was <0.5 Miller units/mg of protein). In order to exclude potential effects due to hyperaccumulation of histidinol phosphate (the substrate for HolPase) in the mutant strain BFA1037, this experiment was also performed with the complemented strain. As mentioned above, this strain carries a wild-type hisJ gene in the amyE ectopic locus and is prototrophic for histidine (Table 1). The same level of β-galactosidase activity (about 15 to 20 Miller units/mg of protein) was measured in extracts of this strain grown in liquid MM containing succinate and glutamate plus tryptophan which was either not supplemented or supplemented with various concentrations of histidine or histidinol (up to 1,250 mg/liter), or even in rich medium (Luria broth). Thus, it appeared that transcription of hisJ is not repressed by histidine or by histidinol.
Acknowledgments
We thank Christine Delorme for helpful advice concerning the HolPase assay, Matthieu Simon for technical assistance, and Josef Deutscher for comments on the manuscript.
This work was supported by funds from the Centre National de la Recherche Scientifique and the Institut National de la Recherche Agronomique and was performed within the “Systematic Function Analysis of the Bacillus subtilis Genes” European Program BIO4-CT95-0278.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin E V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Carlomagno M S, Chiariotti L, Alifano P, Nappo A G, Bruni C B. Structure and function of the Salmonella typhimurium and Escherichia coli K-12 histidine operon. J Mol Biol. 1988;203:585–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- 5.Chiariotti L, Nappo A G, Carlomagno M S, Bruni C B. Gene structure in the histidine operon of Escherichia coli. Identification and nucleotide sequence of the hisB gene. Mol Gen Genet. 1986;202:42–47. doi: 10.1007/BF00330514. [DOI] [PubMed] [Google Scholar]
- 6.Delorme C. Polymorphisme de l’opéron histidine chez les lactocoques. Thèse de doctorat. Paris, France: Université Paris VII; 1992. [Google Scholar]
- 7.Delorme C, Ehrlich S D, Renault P. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6571–6579. doi: 10.1128/jb.174.20.6571-6579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Robichon-Szulmajster H, Surdin-Kerjan Y. Nucleic acid and protein synthesis in yeasts: regulation of synthesis and activity. In: Rose A H, Harrison J S, editors. The yeasts, vol. 2: physiology and biochemistry of yeasts. London, United Kingdom: Academic Press; 1971. pp. 335–418. [Google Scholar]
- 9.Guérout-Fleury A-M, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 10.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Malone R E, Kim S, Bullard S A, Lundquist S, Hutchings-Crow L, Cramton S, Lutfiyya L, Lee J. Analysis of a recombination hotspot for gene conversion occurring at the HIS2 gene of Saccharomyces cerevisiae. Genetics. 1994;137:5–18. doi: 10.1093/genetics/137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin R G, Berberich M A, Ames B N, Davis W W, Goldberger R F, Yourno J D. Enzymes and intermediates of histidine biosynthesis in Salmonella typhimurium. Methods Enzymol. 1971;17B:3–44. [Google Scholar]
- 14.Millay R H, Houston L L. Purification and properties of yeast histidinol phosphate phosphatase. Biochemistry. 1973;12:2591–2596. doi: 10.1021/bi00738a007. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Piggot P J, Curtis C A M. Analysis of the regulation of gene expression during Bacillus subtilis sporulation by manipulation of the copy number of spo-lacZ fusions. J Bacteriol. 1987;169:1260–1266. doi: 10.1128/jb.169.3.1260-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Sonenshein A L. Introduction to metabolic pathways. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 127–132. [Google Scholar]
- 19.Thaller M C, Schippa S, Rossolini G M. Conserved sequence motifs among bacterial, eukaryotic and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 1998;7:1647–1652. doi: 10.1002/pro.5560070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tortosa P, Aymerich S, Lindner C, Saier M H, Jr, Reizer J, Le Coq D. Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J Biol Chem. 1997;272:17230–17237. doi: 10.1074/jbc.272.27.17230. [DOI] [PubMed] [Google Scholar]
- 21.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 22.Winkler M E. Biosynthesis of histidine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 485–505. [Google Scholar]