Abstract
Accurate identification of the presence, absence or possibility of relevant entities in clinical notes is important for healthcare professionals to quickly understand crucial clinical information. This introduces the task of assertion classification - to correctly identify the assertion status of an entity in the unstructured clinical notes. Recent rule-based and machine-learning approaches suffer from labor-intensive pattern engineering and severe class bias toward majority classes. To solve this problem, in this study, we propose a prompt-based learning approach, which treats the assertion classification task as a masked language auto-completion problem. We evaluated the model on six datasets. Our prompt-based method achieved a micro-averaged F-1 of 0.954 on the i2b2 2010 assertion dataset, with ~1.8% improvements over previous works. In particular, our model showed excellence in detecting classes with few instances (few-shot). Evaluations on five external datasets showcase the outstanding generalizability of the prompt-based method to unseen data. To examine the rationality of our model, we further introduced two rationale faithfulness metrics: comprehensiveness and sufficiency. The results reveal that compared to the “pre-train, fine-tune” procedure, our prompt-based model has a stronger capability of identifying the comprehensive (~63.93%) and sufficient (~11.75%) linguistic features from free text. We further evaluated the model-agnostic explanations using LIME. The results imply a better rationale agreement between our model and human beings (~71.93% in average F-1), which demonstrates the superior trustworthiness of our model.
Keywords: Prompt-based learning, Concept assertion, Deep learning, NLP
1. Introduction
Assertion classification is the task of classifying the assertion status of clinical concepts expressed in natural languages, such as a diagnosis or condition being present, absent, or possible [1]. It is of substantial importance to the understanding of Electronic Health Records (EHRs) and has shown the great potential to benefit various clinical applications since the assertion status is a critical contextual property to automated clinical reasoning [2]. However, assertion classification has long been a challenging task due to the imbalance in the class distribution and the unstructured nature of clinical notes [3]. For example, classifying Possible assertions is particularly difficult because they have a much smaller occurring frequency than the Present and Absent assertions, and they are often expressed vaguely [2,4].
Various approaches have been explored for assertion classification. Some earliest attempts handled this task via hand-crafted rules and carefully designed heuristics [5,6]. For example, Chapman et al. [5] posited that medical language is lexically less ambiguous, and hence their model used a simple regular expression algorithm to detect negation cues (NegEx). Peng et al. [6] enhanced NegEx and utilized Universal Dependency patterns to design the rules. Rule-based approaches usually achieve a high precision but are often cited for a low recall due to the rigid hand-crafted patterns. While it is feasible to manually identify and implement high-quality patterns to achieve good precision, it is often impractical to exhaustively design all patterns necessary for a high recall. To overcome this limitation, machine learning approaches were explored, such as Conditional Random Fields [7] and Support Vector Machines [8–11].
More recently, several deep learning methods were introduced for assertion classification in the biomedical domain. Qian et al. [12] considered bringing the advantage of Convolutional Neural Networks to identifying the scopes of negations in clinical texts. Many others explored bidirectional Long-Short Term Memory for negation recognition [3,13–15]. Nowadays, transformer-based methods have become dominant [1,4]. While conventional deep learning methods demonstrated excellent performances, they typically rely on large amounts of labeled data to learn the distinguishing class features and are often hampered when the dataset is small or imbalanced.
To relieve these limitations, we introduce a powerful prompt-based learning approach for its proven capability of performing few-shot learning and rapid adaptation to new tasks with only a limited number of labeled examples. Prompting methods have shown success in various natural language tasks [16,17], such as knowledge probing [18,19], question answering [20], and textual entailment [21]. However, to the best of our knowledge, no previous work introduces prompt-based approaches to assertion classification. Our prompt-based learning method treats the assertion classification task as a masked language auto-completion problem. The model probabilistically generates a textual response to a given prompt defined by a task-specific template [22]. In this way, we can manipulate the model behaviors so that the pre-trained language model (LM) can learn to classify the assertion types. Prompting framework allows us to utilize the LMs pre-trained on massive amounts of raw text, and to perform few-shot or even zero-shot learning by defining a new prompting function, which enables us to adapt to new tasks with few or no supervised data [22,23], reducing or obviating the need for large, supervised datasets. We trained a prompt-based model on the i2b2 2010 assertion dataset [24], and evaluated its performances on six datasets, including the i2b2 2010 assertion dataset, i2b2 2012 assertion dataset [25], MIMIC-III assertion dataset [4,26], BioScope [27], NegEx [28] and Chia [29]. The observed results demonstrated our prompt model’s superior classification capability and generalizability over the state-of-the-art approaches.
Beyond evaluating the performances of the NLP models, research interest has recently grown in revealing why models make specific predictions [30]. Model’s rationality measures how well the rationales (i.e., a snippet that supports outputs) provided by models align with human rationales, and the degree to which the provided rationales influence the corresponding predictions [30] Metrics such as precision, recall, and F-1 score can only measure partial quality and quantity aspects of model predictions, but cannot evaluate properties of the model’s rationality. Hence, the effectiveness of these NLP systems is limited by their current inability to explain their decisions to human beings, especially in clinical practices. To quantify the model’s rationality for model comparisons and progress tracking, we introduced two rationale faithfulness evaluation metrics, comprehensiveness and sufficiency, which measure to what extent the model adheres to human rationales. We further evaluated the alignments between the model explanations and the human rationales, and the results show the superior trustworthiness of our prompt-based method in terms of its better alignment with human rationales, compared to the state-of-the-art models. We believe that our prompt-based method provides a reasonable start featuring human rationales for assertion classification.
We will make our code and model publicly available to facilitate future research.1
2. Material and methods
2.1. Task of assertion classification
Assertion classification is the task of classifying if the patient has or had a given condition. Following the definition in the work of Uzuner et al. [24], the outcomes are Present, Absent, Possible, Conditional, Hypothetical, and Not Associated (Table 1).
Table 1.
Examples of assertion types. Concepts are italicized.
| Assertion type | Example |
|---|---|
| Present | Severe systolic HTN is noted. |
| Absent | There is no pericardial effusion. |
| Possible | High CO and low SVR suggestive of sepsis. |
| Conditional | Narcotics can cause constipation. |
| Hypothetical | Return to the emergency room if he experiences any chest pain. |
| Not Associated | Father had MI at 42. |
In this work, we take an input sentence x with a given concept mention e and predict a label l from a fixed label set , based on a model P(l|x, e;θ). For example, given an input x = “This is very likely to be an asthma exacerbation” and e = “an asthma exacerbation”, we aim to predict a label l = “Possible” out of a 6-class label set.
2.2. Datasets
In this study, we included six independent datasets (Table 2).
Table 2.
Statistics of the datasets. Size - the number of notes..
| Dataset | Size | Present | Absent | Possible | Hypothetical | Conditional | Not Associated | Total |
|---|---|---|---|---|---|---|---|---|
| i2b2 2010 Train | 170 | 4,624 | 1,596 | 309 | 382 | 73 | 89 | 7,073 |
| i2b2 2010 Test | 256 | 8,604 | 2,592 | 646 | 442 | 148 | 131 | 12,563 |
| i2b2 2012 Test | 119 | 3,360 | 640 | 245 | – | 64 | – | 4,309 |
| BioScope | 1,954 | 5,338 | 899 | 1,368 | – | – | – | 7,605 |
| MIMIC-III | 239 | 3,392 | 1,243 | 365 | – | – | – | 5,000 |
| NegEx | 116 | 1,885 | 491 | – | – | – | – | 2,376 |
| Chia | 1,000 | 1,057 | 1,057 | – | – | – | – | 2,114 |
i2b2 2010 assertion dataset. annotates a corpus of assertions in discharge summaries and progress reports from three institutions [24]. Six assertion types of medical concepts in clinical notes were manually annotated, including Present, Absent, Possible, Hypothetical, Conditional and Not Associated with the Patient. In the released version, there are 170 annotated clinical notes in the training set and 256 notes in the test set. Table 2 reveals that the class distribution is highly imbalanced. For example, the number of training instances for Present is about 50 times more than the number of instances for Conditional and Not Associated with the Patient.
i2b2 2012 assertion dataset. contains 189 annotated notes in the training set and 119 notes in the test set from de-identified discharge summaries [25]. In the i2b2 2012 assertion dataset, clinical concepts were annotated with polarity attributes (whether an event was positive or negative) and modality attributes (whether an event occurred or not). We defined three assertion types. A concept is Present if its polarity is “positive” and its modality is “factual”, Absent if its polarity is “negative”, and Possible if its polarity is “positive” and its modality is “possible”. In this study, we only used the test set to assess the generalizability of the proposed model.
BioScope. provides a corpus of 3 assertion types (i.e., Present, Absent, Possible) annotated by two independent linguist annotators following the guidelines set up by a chief linguist [27]. The corpus consists of medical free texts, biological full papers and biological scientific abstracts, resulting in 1,954 notes.
MIMIC-III assertion dataset. annotates 3 assertion types (i.e., Present, Absent, Possible) in 239 clinical notes, including 92 discharge summaries, 49 nursing notes, 23 physician notes, and 75 radiology reports [4]. The dataset follows the same annotation guidelines as the i2b2 2010/VA challenge [24]. The detailed statistics of MIMIC-III subsets can be found in Table B.1 in Appendix A.
NegEx. annotates the Present and Absent assertion types in 116 de-identified discharge summaries dictated at two medical ICU’s at the University of Pittsburgh Medical Center. Assertions of medical concepts were first identified by a regular expression algorithm and then verified by three physicians.
Chia. is a large-scale corpus of patient eligibility criteria extracted from 1,000 interventional, Phase IV clinical trials registered on ClinicalTrials.gov [29]. From this dataset, a concept is Absent if there is a “has_negation” relation between this concept and a trigger word (e.g., “cannot”). In this study, we obtained 1,057 Absent concepts, and sampled the same number of Present concepts.
2.3. Prompt-based assertion classification
Given an input sentence x = {w0, …, wn} with a given concept mention e = {wi, …, wj}, the prompt function fprompt(x, e) will convert the input to a prompt xprompt, which is a textual string that includes a one-token answer slot [MASK]. The LM takes xprompt as the input, maps it to a sequence of token embeddings, and learns to select one answer z for the [MASK] token that can be mapped to the label space (Fig. 1).
Fig. 1.
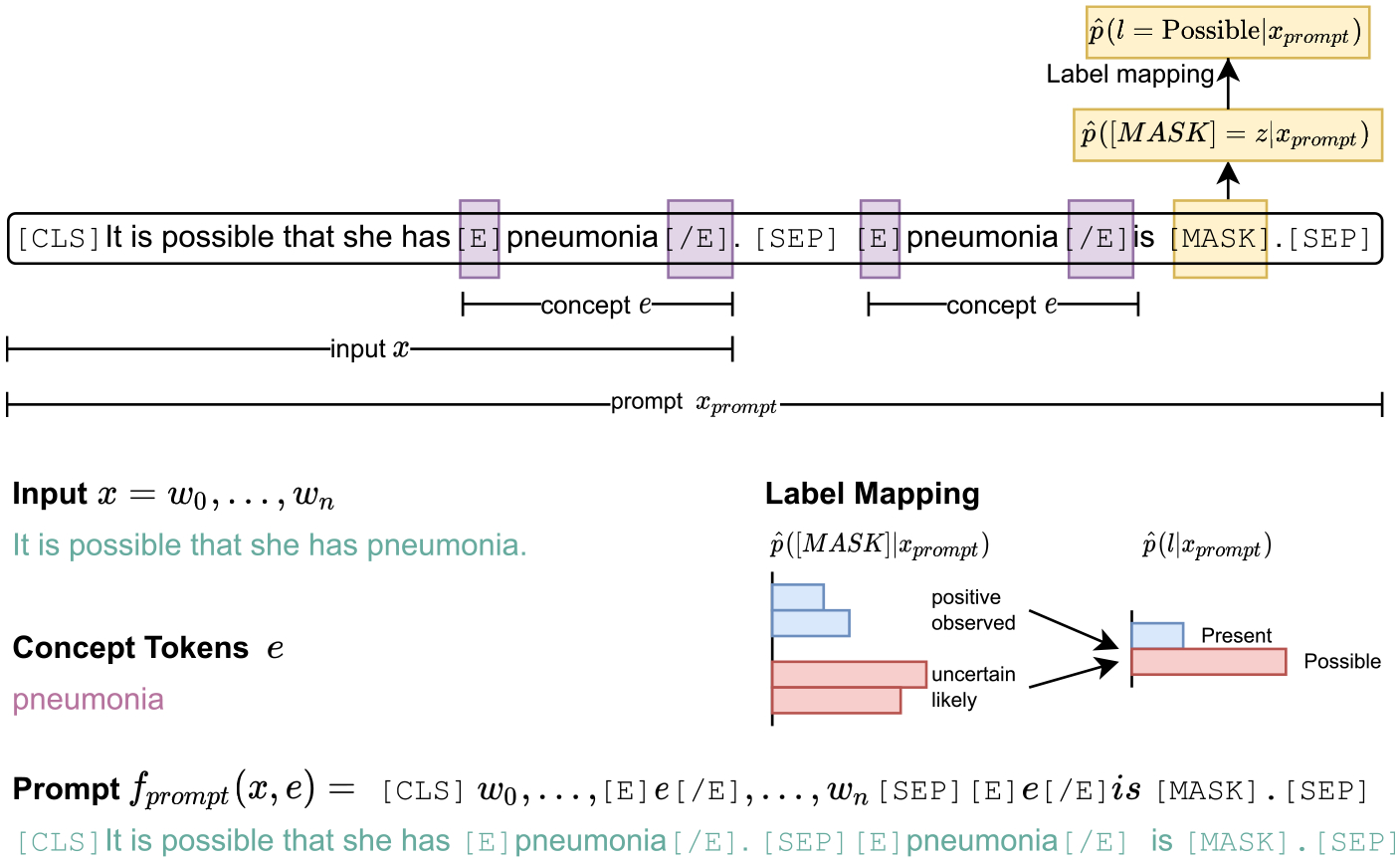
Prompt-based assertion classification.
Prompt function.
The most natural way to create prompts is to manually create intuitive templates based on human introspection. In this work, we designed the prompt function fprompt(x, e) = “[CLS] w0… [E] e [/E]… wn [SEP] [E] e [/E] is [MASK] [SEP]” to generate a prompt xprompt for a sentence x. Specifically, we surrounded the concept-of-interest tokens e in the input sentence with special indicator tokens [E] and [/E], whose embeddings were randomly initialized. We then concatenated the sentence with a prompting snippet “[E] e [/E] is [MASK]”, where the concept tokens were also surrounded by [E] and [/E]. The [SEP] token in the middle helps the model understand which part of xprompt belongs to the input sentence and which part belongs to the prompting question.
One sample input sequence looks like this: “[CLS] It is possible that she has [E] pneumonia [/E]. [SEP] [E] pneumonia [/E] is [MASK]. [SEP]”, where “pneumonia” is the concept we focus on in this case.
Answer search and label mapping.
LMs learned to search for the highest-scored word to fill in the answer slot [MASK] in xprompt. For each specific prompt function, we defined as a set of permissible values for z, such as “positive” and “negative”. The highest-scored answer z can be further mapped to in the label space.
Training details.
We trained a task-specific head by maximizing the log-probability of the correct label at the masked token, given the hidden vector of [MASK]. Taking xprompt as the input, LM’s probability of predicting assertion label l is:
where is the correct label, maps the label l to the word z in the answer vocabulary, h[MASK] is the hidden vector of the [MASK] token, and W represents the trainable weights. We fine-tuned the LM to minimize the cross-entropy loss.
2.4. Measuring rationality
Inspired by DeYoung et al. [30], we introduced two rationale faithfulness evaluation metrics comprehensiveness (do we need every sentence token to make a correct prediction?), and sufficiency (do the linguistic scopes contain enough information to make a correct prediction?) to provide reasonable comparisons of specific aspects of model’s rationality (Fig. 2). To better understand on what grounds our model took the decision, we adopted Local Interpretable Model-agnostic Explanations (LIME) [31] to explain the model predictions and further compared how the identified model explanations aligned with the human rationales.
Fig. 2.
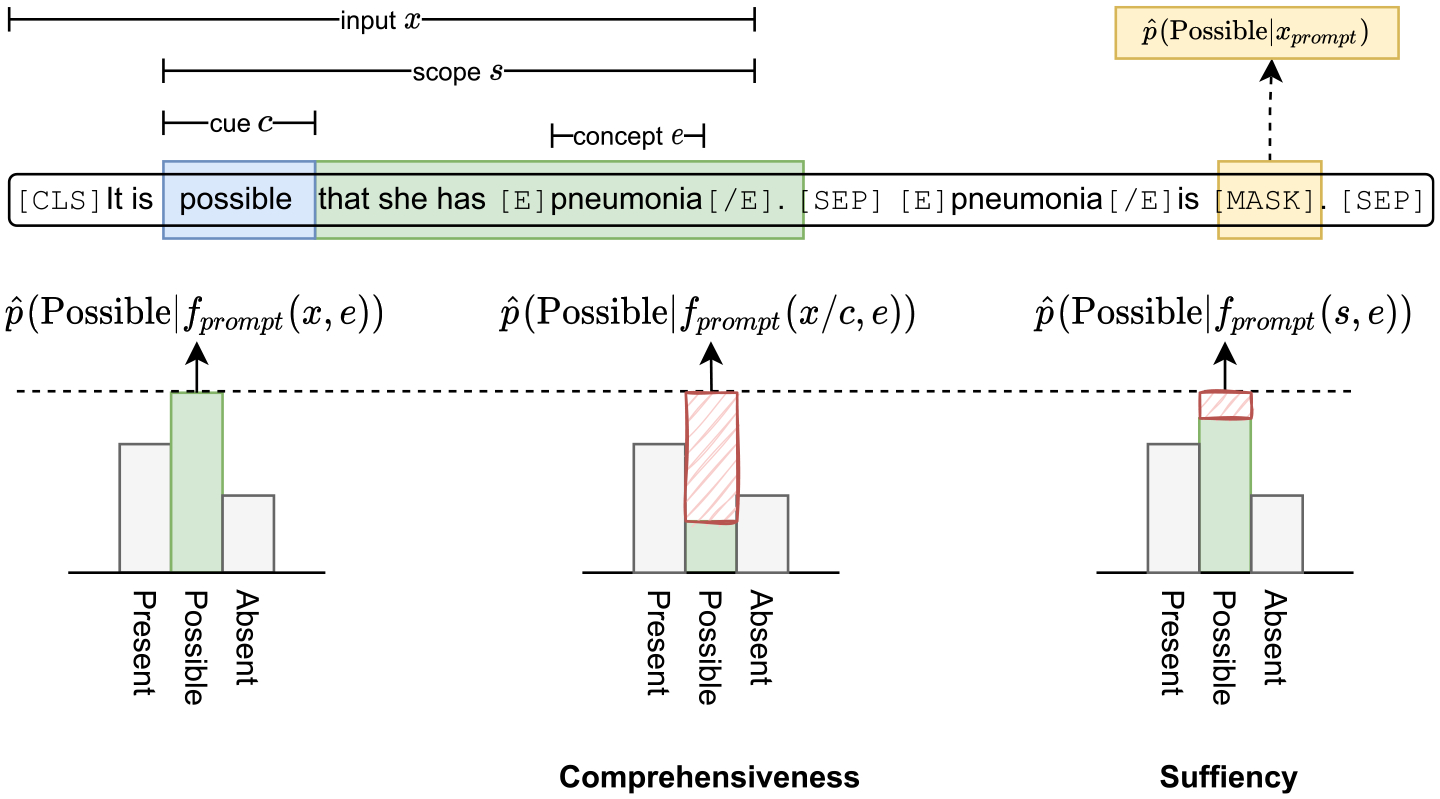
Comprehensiveness and sufficiency demonstrations. x is the input sentence, e is the concept of interest, c is the assertion semantic cue, s is the assertion scope containing the cue phrase and the objects it applies to.
Comprehensiveness.
Cue phrases are often used in natural language to provide key semantic information about a target [32]. For example, in the sentence “This is very likely to be an asthma exacerbation”, the phrase “very likely” can be semantically perceived as cue words of a Possible assertion to “an asthma exacerbation” by human beings. To measure the rationale’s comprehensiveness, we constructed a counterexample for the input sentence x with a concept mention e, by removing the assertion cues c from the text, resulting in x/c. In our setting, let be the original prediction probability of our model for the class l. comprehensiveness is defined as the changes in the model’s predicted probabilities for the same class:
| (1) |
A higher comprehensiveness score reflects a more severe confidence drop in the model when removing the linguistic cues, which implies that the removed rationales are more influential in making the prediction. Therefore, the model with a higher comprehensiveness score tends to focus on the linguistic cues more heavily when making the prediction, indicating better adherence to human rationales.
Sufficiency.
A linguistic scope contains the semantic operator (i.e., cue phrase) and the objects it applies to [27]. For example, in the sentence “Right middle lobe abnormalities suggest airways disease rather than bacterial pneumonia”, the preposition “rather than” affects the interpretation of “bacterial pneumonia” instead of “airways disease”, since “airways disease” is not within its semantic scope. sufficiency is defined to evaluate to what degree the linguistic scopes are sufficient for our model to make a correct prediction. Denote s as the linguistic scope of the assertion in sentence x, and sufficiency can be formulated as:
| (2) |
The sufficiency score can be used to imply a model’s capability of grasping the sufficient rationales from the text: a lower sufficiency score suggests that the model is more capable of making correct predictions by solely using the assertion scopes, hence a better capability of capturing sufficient features from the unstructured text.
LIME-based explanations.
LIME is a local interpretability model that can explain the predictions of any classifier. To obtain the explanations of a black-box model which has a complex decision function f, LIME samples instances, acquires predictions using f, and assigns continuous importance scores to tokens by the proximity to the instance being explained [31]. For example, in the sentence “Findings suggesting viral or reactive airway disease”, LIME assigns an importance score 0.52 to the token “suggesting” and 0.24 to the token “or” to explain the Possible assertion of “airway disease”. In this study, we converted the soft importance scores into discrete rationales by taking the top–n values. We set n to 1 and 5, given the fact that the ground truth human rationales are short in length. We counted a token as a true positive if it overlaps with any ground truth cue words; otherwise, a false positive. We used these definitions to measure the token-level precision, recall, and F-1 score. A higher F-1 score implies a better agreement of the model rationales with the human rationales, hence a more trustworthy model.
2.5. Experimental settings
There are various pre-trained LMs available for prompt-based learning, and we selected the BioBERT [33] which was additionally pre-trained on discharge summaries and further fine-tuned on the i2b2 2010 training data [4]. AdamW optimizer [34] and weighted Cross-Entropy loss were adopted. We used a learning rate of 10−6, batch size of 8, and 10 epochs of training with Early-Stopping enabled to prevent overfitting. Intel Core i9-9960X 16 cores processor, NVIDIA Quadro RTX 5000 GPU and a memory size of 128G were used in this work. Following the i2b2 2020 challenge task, we used the precision, recall, F-1, and micro F-1 (for multi-class) to evaluate the model performance.
3. Results and discussions
3.1. Assertion classification
3.1.1. Evaluation on the i2b2 2010 dataset
We selected the best-performed models in the i2b2 2010/VA challenge [24] as our baseline models, including Roberts et al. [8], Jiang et al. [9], Demner-Fushman et al. [10], Clark et al. [7], de Bruijn et al. [11].
We also compared our model with a feature-based Logistic Regression model and a fine-tuned ClinicalBERT model [33]. Table 3 shows that our proposed prompt model outperformed baseline methods by a large margin in terms of micro F-1 (> 1.8%). The BERT model achieved the second-best micro F-1 of 0.936. However, it failed to classify the few-shot classes, such as Conditional and Not Associated. In contrast, our prompt model boosted the classification performances of almost all classes, especially those few-shot ones, presenting a notable 1.85% improvement in the Hypothetical class and a 1.4% improvement in the Conditional class, demonstrating its superior capability of few-shot learning. We also noticed that compared to Demner-Fushman et al. [10], our prompt model reported a 9.6% lower F-1 in the Possible class.
Table 3.
Results on the i2b2 2010 dataset. The best scores are bolded.
| Model | Present | Absent | Hypothetical | Possible | Conditional | Not Associated | micro F-1 |
|---|---|---|---|---|---|---|---|
| Logistic Regression | 0.900 | 0.842 | 0.833 | 0.464 | 0.471 | 0.596 | 0.850 |
| Roberts et al. [8]* | 0.962 | 0.947 | 0.895 | 0.684 | 0.423 | 0.861 | 0.928 |
| Jiang et al. [9]* | 0.960 | 0.954 | 0.904 | 0.666 | 0.391 | 0.863 | 0.931 |
| Demner et al. [10]⋄ | 0.957 | 0.940 | 0.626 | 0.859 | 0.384 | 0.835 | 0.933 |
| Clark et al.[7]* | 0.958 | 0.937 | 0.890 | 0.630 | 0.422 | 0.869 | 0.934 |
| de Bruijin et al. [11]⋄ | 0.959 | 0.942 | 0.884 | 0.643 | 0.263 | 0.824 | 0.936 |
| BERT model | 0.959 | 0.955 | 0.902 | 0.760 | 0.000 | 0.000 | 0.936 |
| Prompt model | 0.971 | 0.968 | 0.921 | 0.763 | 0.485 | 0.875 | 0.954 |
- the numbers were reported in the i2b2 2010/VA challenge [24], and were not directly comparable with our model.
- the numbers are computed based on the reported confusion matrices from the original paper, and were not directly comparable with our model.
The detailed class-wise precision and recall scores can be found in Table B.2 in Appendix A. We observed drastic improvements in the recall scores in most classes, except for a 2.5% drop in the Present class. The tremendous recall boosts in the few-shot classes were particularly notable, 42% for Conditional and 11.7% for Not Associated. It is noticeable that there was a 4% increase on the precision score of Present, but a 0.2%–63.2% precision drop was also observed in other classes. The improved recall scores showed our model’s superior capability of identifying false negatives over state-of-the-art methods, which is critical in clinical practices.
We looked at some instances in the Possible class where our model failed to make correct predictions (Table 4, cases 1–3). In cases 1 and 2, our prompt model was not able to identify the possibleness reflected by the mentions of “consistent with”, mistakenly classifying the concepts to be Present. In case 3, our model sensed the hypothesis from the mention of “to reassess for”, hence classifying the “recurrent pleural effusion” to be Hypothetical.
Table 4.
Error cases. Concepts are italicized.
| 1. | This was consistent with scar. |
| 2. | Examination revealed an apicovaginal lesion consistent with recurrent tumor. |
| 3. | Over the next several days the patient remained in the hospital to reassess for recurrent pleural effusion. |
| 4. | When her pacemaker was in a sinus rhythm without a beta blocker, she had significant angina. |
| 5. | The patient will have these symptoms when the eyes are closed. |
| 6. | She could not walk a few yards without developing symptoms. |
| 7. | A question of a SULFA allergy. |
We also looked at some error cases in the few-shot Conditional class (Table 4, cases 4–7). In cases 4 and 5, there are certain conditions described in the clause following “when”, only under which the concepts-of-interest hold, but our model failed to identify the conditional prerequisites and mistakenly classified them into Present. In case 6, “symptoms” is conditional on “walk a few yards”, but our prompt method classified it as Absent. The double negative “could not walk a few yards” and “without developing symptoms” makes it more difficult to classify the assertion of “symptoms”. In case 7, “allergy” is considered to be Conditional itself according to the annotation guideline, but our model classified it to be Possible.
3.1.2. Evaluations on external datasets
We further evaluated our model on five external datasets. We selected several baseline models for comparison, including one feature-based Logistic Regression model, two rule-based systems (NegEx [5] and RadText [35]), and a BERT-based model [4]. The class-wise performance comparisons in Tables 5 and 6 show that our model demonstrated the best performances in almost all classes on all test sets. In Table 5, compared to the rule-based system RadText, drastic improvements can be observed on nearly all datasets. Compared to van Aken et al. [4], our prompt-based method achieved a noticeable 2% micro F-1 improvement on the BioScope dataset and reported comparable performances on other datasets. In Table 6, Chapman et al. [5] outperformed other methods on the NegEx dataset, but our prompt method showed superior or comparable results to other baselines, presenting a 0.3%–1.6% improvement in micro F-1 on the remaining test sets.
Table 5.
Results of three-class assertion classification. The best scores are bolded.
| Dataset | Model | Present | Absent | Possible | micro F-1 |
|---|---|---|---|---|---|
| i2b2 2010 | Logistic Regression | 0.926 | 0.866 | 0.468 | 0.888 |
| RadText [35] | 0.897 | 0.706 | 0.420 | 0.839 | |
| BERT model [4] | 0.977 | 0.967 | 0.756 | 0.964 | |
| Prompt model | 0.980 | 0.975 | 0.769 | 0.966 | |
| i2b2 2012 | Logistic Regression | 0.921 | 0.782 | 0.548 | 0.874 |
| RadText [35] | 0.898 | 0.607 | 0.348 | 0.829 | |
| BERT model [4] | 0.955 | 0.866 | 0.652 | 0.924 | |
| Prompt model | 0.956 | 0.875 | 0.656 | 0.927 | |
| BioScope | Logistic Regression | 0.945 | 0.780 | 0.720 | 0.877 |
| RadText [35] | 0.836 | 0.631 | 0.432 | 0.735 | |
| BERT model [4] | 0.951 | 0.835 | 0.732 | 0.892 | |
| Prompt model | 0.966 | 0.823 | 0.811 | 0.912 | |
| MIMIC-III | Logistic Regression | 0.899 | 0.846 | 0.454 | 0.855 |
| RadText [35] | 0.880 | 0.700 | 0.420 | 0.816 | |
| BERT model [4] | 0.951 | 0.937 | 0.621 | 0.927 | |
| Prompt model | 0.950 | 0.933 | 0.662 | 0.927 |
Table 6.
Results of two-class assertion classification. The best scores are bolded.
| Dataset | Model | Present | Absent | micro F-1 |
|---|---|---|---|---|
| i2b2 2010 | Logistic Regression | 0.926 | 0.866 | 0.911 |
| NegEx [5] | 0.925 | 0.836 | 0.906 | |
| RadText [35] | 0.897 | 0.706 | 0.858 | |
| BERT model [4] | 0.977 | 0.967 | 0.975 | |
| Prompt model | 0.980 | 0.975 | 0.978 | |
| i2b2 2012 | Logistic Regression | 0.921 | 0.782 | 0.893 |
| NegEx [5] | 0.937 | 0.815 | 0.917 | |
| RadText [35] | 0.898 | 0.607 | 0.853 | |
| BERT model [4] | 0.955 | 0.866 | 0.940 | |
| Prompt model | 0.956 | 0.875 | 0.943 | |
| BioScope | Logistic Regression | 0.945 | 0.780 | 0.914 |
| NegEx [5] | 0.879 | 0.621 | 0.847 | |
| RadText [35] | 0.836 | 0.631 | 0.789 | |
| BERT model [4] | 0.951 | 0.835 | 0.928 | |
| Prompt model | 0.966 | 0.823 | 0.938 | |
| MIMIC-III | Logistic Regression | 0.899 | 0.846 | 0.883 |
| NegEx [5] | 0.908 | 0.863 | 0.896 | |
| RadText [35] | 0.880 | 0.700 | 0.890 | |
| BERT model [4] | 0.951 | 0.937 | 0.947 | |
| Prompt model | 0.950 | 0.933 | 0.950 | |
| NegEx | Logistic Regression | 0.926 | 0.821 | 0.889 |
| NegEx [5] | 0.983 | 0.931 | 0.972 | |
| RadText [35] | 0.817 | 0.530 | 0.734 | |
| BERT model [4] | 0.926 | 0.815 | 0.890 | |
| Prompt model | 0.940 | 0.821 | 0.938 | |
| Chia | Logistic Regression | 0.693 | 0.540 | 0.609 |
| NegEx [5] | 0.763 | 0.612 | 0.705 | |
| RadText [35] | 0.703 | 0.430 | 0.609 | |
| BERT model [4] | 0.763 | 0.619 | 0.708 | |
| Prompt model | 0.772 | 0.652 | 0.724 |
The comparisons of micro F-1 scores and the detailed class-wise performance comparisons of MIMIC-III subsets can be found in Tables B.4 and B.5 in Appendix, respectively. Our proposed model had a slightly lower micro F-1 score than van Aken et al. [4] on the Physician Letters subset, but performed better on the other three subsets. The external evaluations showcased the prompt method’s outstanding generalizability to unseen data.
Comparing Tables 3, 5, and 6, we also observed that the performance improvement of our prompt model on the 3-type or 2-type assertion classification was not as substantial as that on the 6-type classification. One potential reason is that the class distribution of Present, Absent, and Possible is more balanced than other assertion types. For example, there are only 73 and 89 training instances for Conditional and Not Associated assertion types in the i2b2 2010 training set. The comparison demonstrates that prompt-based learning can achieve better and more robust performances than the standard fine-tuning learning, especially for the few-shot learning task. This capability might be useful in the clinical domain, where we often have a few training examples for a new task. In such a situation, prompting may offer a feasible alternative methodology.
3.2. Measuring rationality
We then evaluated the models’ rationality. Both the annotated linguistic scope and cue information of the concepts are required to compute the comprehensiveness and sufficiency. We utilized two datasets for measuring rationality, BioScope and an annotated corpus from i2b2 2010 dataset (the annotation details can be found in Appendix A). Note that both datasets only annotated the scopes and cues for the Possible and Absent concepts.
Fig. 3(a) compares the comprehensiveness scores of the Logistic Regression model, fine-tuned BERT model and our prompt-based model on the BioScope dataset. Here, we hypothesize that removing the linguistic cues ought to decrease the model’s confidence in classifying assertions. The results show that the confidence in predicting Absent of the prompt model dropped by 79.03% (79.03% comprehensiveness), while the confidence of the BERT model dropped by 69.09% (69.09% comprehensiveness) and the confidence of the Logistic Regression model only dropped by 14.99% (14.99% comprehensiveness). Similarly, the confidence in predicting Possible of the prompt model dropped by 48.84% (48.84% comprehensiveness), while the confidences of the BERT model and the Logistic Regression model dropped by 42.52% (42.52% comprehensiveness) and 30% (30% comprehensiveness) respectively. Here is one example, “Increase in markings centrally with streaky disease in lingula that has the appearance most suggestive of atelectasis, less likely early infiltrate”. After removing the linguistic cue “suggestive” from the input, the Logistic Regression model’s confidence of classifying “atelectasis” as Possible only dropped by 28.6%, the fine-tuned BERT model’s confidence dropped by 62.3%, while our prompt model’s confidence dropped by 98.9%. Fig. 3(c) compares the comprehensiveness scores on the annotated i2b2 2010 corpus. The prompt model is observed to yield a higher comprehensiveness than other models. The results prove that the prompt model is better at capturing comprehensive features that are aligned with human rationales to make predictions.
Fig. 3.
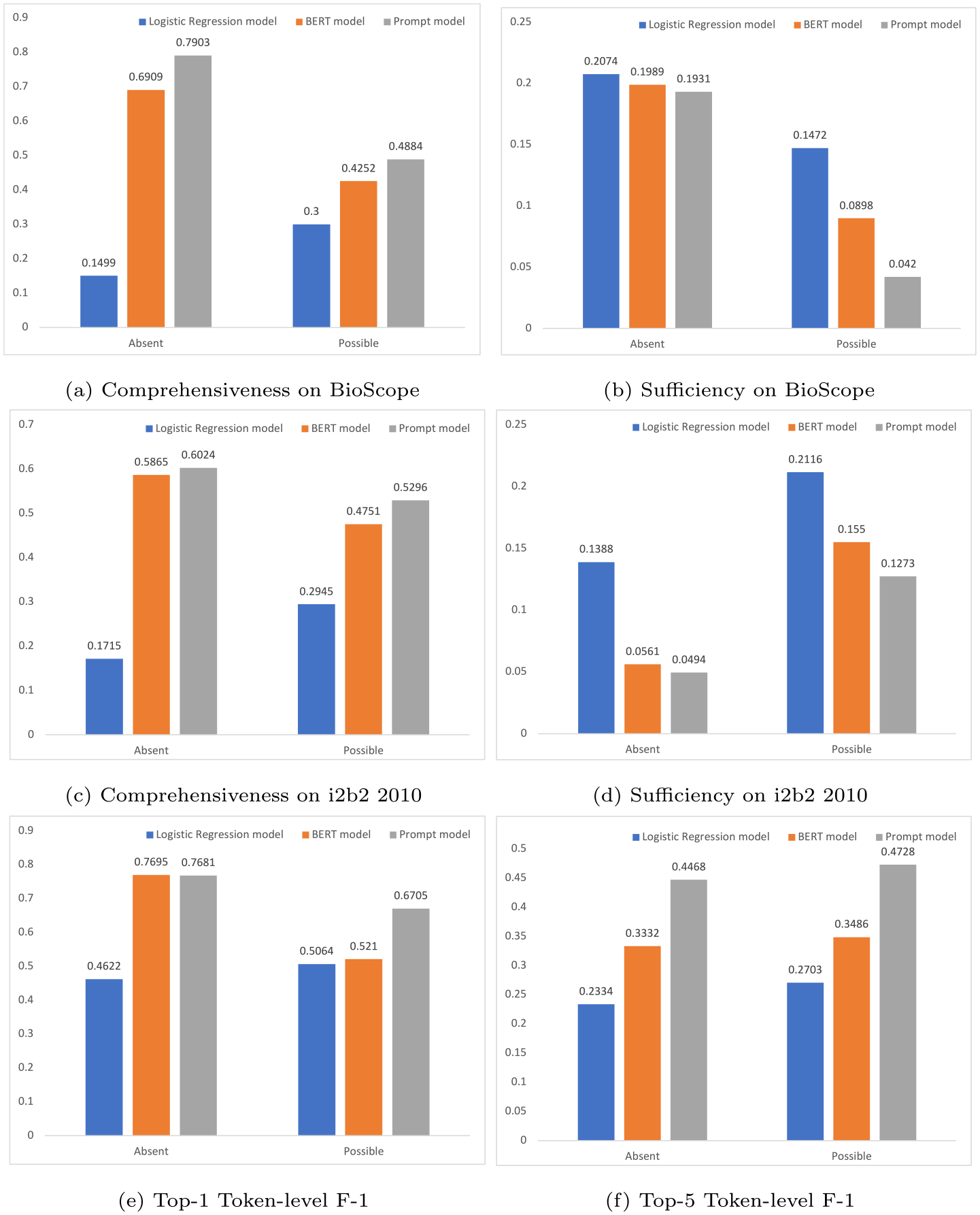
Comparisons of model rationality. (a) comprehensiveness comparisons on BioScope. A higher comprehensiveness score implies a more important role the linguistic cues play in the model’s prediction. (b) sufficiency comparisons on BioScope. A lower sufficiency score implies the model’s better capability of capturing sufficient features. (c) comprehensiveness comparisons on i2b2 2010. (d) sufficiency comparisons on i2b2 2010. (e) Top-1 token-level F-1 comparisons on BioScope. A higher score implies the highest-scored model rationale token has a better agreement with the ground truth rationales. (f) Top-5 token-level F-1 comparisons on BioScope. A higher score implies that the top-5 model rationale tokens are better aligned with the ground truth rationales.
Fig. 3(b) compares the sufficiency scores of the Logistic Regression model, the fine-tuned BERT model and our prompt-based model on the BioScope dataset. Here, we hypothesize that the model should be able to come to a similar prediction (i.e., a smaller confidence drop) using only the linguistic scopes. The results show that the prompt model’s confidence in predicting Absent dropped by 19.31% (19.31% sufficiency), while the confidences of the BERT model and the Logistic Regression model dropped by 19.89% (19.89% sufficiency) and 20.74% (20.74% sufficiency) respectively. Similarly, the confidence in predicting Possible of the prompt model dropped by 4.20% (4.20% sufficiency), while the confidences of the BERT model and the Logistic Regression model dropped by 8.98% (8.98% sufficiency) and 14.72% (14.72% sufficiency) respectively. Here we also look at one example, “Scattered perihilar air space opacity with questionable left lower lobe opacity”. When only using the linguistic scopes “questionable left lower lobe opacity” as the input sentence, the fine-tuned BERT model’s confidence of classifying “lower lobe opacity” as Possible dropped drastically by 8.2%, the Logistic Regression model’s confidence dropped by 6.7%, while our prompt model’s confidence dropped by 0.4%, almost unchanged. Fig. 3(d) compares the sufficiency scores on the i2b2 2010 corpus. We can observe that the prompt model reports a smaller confidence drop than the other two models when using only the linguistic scope information. The results suggest that the linguistic scopes are more adequate for a prompt model to make a prediction than for the BERT model and the feature-based machine learning model.
Fig. 3(e) compares the top-1 token-level F-1 scores of the Logistic Regression model, the fine-tuned BERT model and our prompt-based model on the BioScope dataset. The results show that the BERT model and the prompt model were comparable in terms of the Absent class F-1 scores, while the Logistic Regression model reported a much lower Absent class F-1 score. Our prompt-based model reported an F-1 score of 0.6705 in the Possible class, which was 14.95% higher than that of the fine-tuned BERT model. Fig. 3(f) compares the top-5 token-level F-1 scores the Logistic Regression model, the fine-tuned BERT model and our prompt-based model on the BioScope dataset. Our prompt-based model reported F-1 scores of 0.4468 and 0.4728, respectively, in the Absent class and Possible class, which were respectively 11.36% and 12.42% better than that of the fine-tuned BERT model, 21.34% and 20.25% better than that of the Logistic Regression model. The results imply a better rationale agreement between the prompt-based model and human beings, demonstrating superior model trustworthiness when compared to the BERT model and the feature-based machine learning model.
In summary, the evaluations show that our prompt method has better rationality for its faithfulness to the human rationales, and it is more trustworthy in terms of its rationale agreement with human beings.
3.3. Ablation study
We conducted several ablation studies to understand the effects of prompt engineering, label mapping and LM backbones in prompt-based learning.
3.3.1. Prompt engineering
We explored three types of prompt templates to evaluate the impact of prompt engineering (Table 7). Note that we kept all other model elements identical while the prompt template was the only variable here. P1 is to ask LMs to fill the assertion words in the [MASK] token based on their impressions of the whole sentence. P2 provides the concept-of-interest together with a list of potential assertion types. It then asks LMs to choose one from the list. P3 provides LMs with the concept and asks LMs to fill in the assertion words. The evaluation was conducted on the i2b2 2010 test set. According to the results, P3 performed the best (a micro F-1 of 0.954), 0.5% higher than P1, and 0.4% higher than P2.
Table 7.
Micro F-1 comparisons of different prompt templates.
| Prompt Template | micro F-1 |
|---|---|
| P1: [MASK]. | 0.949 |
| P2: Is [E] concept [/E] present, absent, possible, hypothetical, conditional or N/A? [MASK]. | 0.950 |
| P3: [E] concept [/E] is [MASK]. | 0.954 |
3.3.2. Label mapping
In this study, several label mapping approaches were explored (Table 8). M1 maps a single-letter answer to a classification label in a one-to-one manner. For example, the single-letter answer “P” maps to the label Present. M2 also does the mapping in a one-to-one fashion, but instead of using a single letter as the answer, it uses a single word. For example, the answer “positive” maps to the Present label. M3 further extends M2 to a single-word many-to-one mapping. For example, both the answers “positive” and “present” can be mapped to the Present label. Among the three mapping approaches, M1 and M2 gave comparable performances, but M3 showed a 0.5% relative performance drop. The detailed mappings can be found in Table B.6 in Appendix A.
Table 8.
Micro F-1 comparisons of different answer mappings.
| Label Mapping | micro F-1 |
|---|---|
| M1: Single-letter one-to-one mapping | 0.954 |
| M2: Single-word one-to-one mapping | 0.954 |
| M3: Single-word many-to-one mapping | 0.949 |
3.3.3. Backbone models
Our prompt-based method employs a LM as the backbone. Hence the model performance can vary when using different pre-trained LMs. To explore the impact of backbone models, we selected four pre-trained LMs, including BERT [36], BlueBERT [37], ClinicalBERT [33], and BioBERT+Discharge Summaries model [4]. We trained four prompt-based models on the assertion classification task, and compared their micro F-1 scores (Table 9). BioBERT+Discharge Summaries model performed 0.6% higher than the BERT model, 0.1% higher than the BlueBERT model and ClinicalBERT model.
Table 9.
Micro F-1 comparisons of the prompt model using different backbone models.
| Backbone Model | micro F-1 |
|---|---|
| BERT | 0.948 |
| BlueBERT | 0.953 |
| ClinicalBERT | 0.953 |
| BioBERT+Discharge summaries | 0.954 |
In the ablation studies, though only three prompt templates and three label mapping approaches were evaluated, the reported 0.5% micro F-1 differences were not trivial. This implied that the importance of appropriate prompt engineering and answer designing should not be neglected. A more sophisticated prompt engineering (e.g., soft prompt templates [38]) and label mapping design (e.g., soft answer tokens [39]) in building the clinical NLP application could potentially further improve the outcome. In the ablation study of backbone models, performance differences were identified among different LMs, and it is noticeable that BERT models fine-tuned on clinical notes or medical corpus demonstrated a better performance than the base BERT model.
4. Conclusions
In this work, we introduced the prompt-based method to the assertion classification task. Noticeable improvements were observed in the evaluations of six datasets, proving the effectiveness of prompting methods, especially in few-shot learning, compared to conventional supervised or fine-tuning methods. By introducing two rationale faithfulness metrics to measure our model’s rationality, we showed that our model demonstrated better adherence and faithfulness to human rationales. The evaluations of LIME-based explanations implied a better rationale alignment between our prompt-based model and human beings, which further proved better trustworthiness of our model. Through ablation studies, we showed the importance of prompt engineering and label mapping but found no significant performance differences using variant backbones. Compared to conventional machine learning-based systems, our method requires less exhausting feature engineering; compared to BERT-based systems, our method features better classification performances and explainability; compared to traditional rule-based systems, our method is way less labor-intensive while possessing a more efficient inference capability. This enables our methods to better assist healthcare professionals to quickly understand crucial clinical information from clinical notes in several applications. For example, our prompt-based method can be incorporated with radiology report analysis for efficient assertion classification. Negative and uncertain assertions of medical findings are frequent in radiology reports [40]. Since they may indicate the absence or uncertainty of findings mentioned in the radiology report, identifying them is as important as identifying those positive ones. In our previous work, we developed NegBio [6,41]. It conducts pattern definition utilizing universal dependencies and graph traversal search using subgraph matching, so that the scope for negation/uncertainty is no longer restricted to the fixed word distance [42]. While NegBio has been widely used to harvest labels from radiology reports and construct chest X-ray databases such as NIH Chest X-ray and MIMIC, it is often impractical to exhaustively design high-quality patterns necessary for a new dataset, let alone to accommodate a new note type. Our prompt-based method provides an opportunity for high-performance assertion classification since it was trained on a diverse set of note types that covers various writing styles. In the future, we plan to integrate our model into clinical NLP pipelines, such as RadText [35], cTAKES [43], and medspaCy [44].
One limitation of our work is that the manual design of prompts and answers could inject bias into evaluations. Also, manually defining prompt templates may fail to discover optimal prompts. Automatic prompt generation methods and automated answer space searches can be further explored. Though our model demonstrated noticeable improvements in recall scores, we cannot neglect the performance drops in the precision scores. When evaluating the rationality of our model, we masked out some parts of the input sentence, which produced incomplete sentences. Such a perturbation could lead to several issues. For example, the corrupted sentence could fall off the distribution of the training data [45]. Furthermore, there are ongoing discussions about LIME’s stability and robustness issues, that LIME can be stable when explaining linear models, but this may not be the case for non-linear models [46]. More sophisticated post-hoc explanation methods can be explored. We hope our results could encourage future work to address these limitations to further explore the potential of prompt-based learning.
Acknowledgments
This work is supported by the National Library of Medicine, USA under Award No. 4R00LM013001, Amazon Diagnostic Development Initiative 2022, Cornell Multi-investigator Seed Grant 2022, and the NSF AI Institute for Foundations of Machine Learning (IFML).
Appendix A. Annotating rationales of the i2b2 2010 dataset
We randomly sampled 50 instances from the i2b2 2010 dataset, 34 of which were Absent assertions and 16 were Possible assertions. Two independent annotators annotated the cues and scopes of these 50 instances following the annotation guidelines of BioScope. Cases of the agreement were accepted without further checking, while differences between the two were resolved by a third expert, yielding the gold standard labeling of the corpus. After removing the ambiguous instances, there were 31 Absent assertions and 15 Possible assertions left. We measured the consistency level of the annotations using inter-annotator agreement analysis. We defined the inter-annotator agreement rate as the overall F-measures of one annotation, treating the second one as the gold standard [47]. Precision is the number of correct answers divided by the total number of answers a system has predicted. Recall is the number of correct answers divided by the total number of answers in the gold standard. We report high inter-annotator agreements (0.9787 for cue annotations and 0.9375 for scope annotations).
Appendix B
See Tables B.1–B.6.
Table B.1.
Statistics of the MIMIC-III assertion dataset.
| Note type | Present | Absent | Possible | Total |
|---|---|---|---|---|
| Discharge summaries | 2,610 | 980 | 250 | 3,840 |
| Nursing letters | 293 | 59 | 14 | 366 |
| Physician letters | 204 | 66 | 34 | 304 |
| Radiology reports | 285 | 138 | 67 | 490 |
Table B.2.
Results of (P)recision, (R)ecall, and F-1 for each assertion type on the i2b2 2010 dataset. The best scores are bolded.
| Model | Present | Absent | Hypothetical | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F-1 | P | R | F-1 | P | R | F-1 | |
| Logistic Regression | 0.921 | 0.883 | 0.900 | 0.809 | 0.882 | 0.842 | 0.844 | 0.810 | 0.833 |
| Roberts et al. [8]* | 0.944 | 0.980 | 0.962 | 0.959 | 0.934 | 0.947 | 0.921 | 0.870 | 0.895 |
| Jiang et al. [9]* | 0.943 | 0.977 | 0.960 | 0.962 | 0.946 | 0.954 | 0.939 | 0.872 | 0.904 |
| Demner et al. [10]⋄ | 0.932 | 0.983 | 0.957 | 0.958 | 0.923 | 0.940 | 0.815 | 0.509 | 0.626 |
| Clark et al. [7]* | 0.937 | 0.980 | 0.958 | 0.955 | 0.920 | 0.937 | 0.924 | 0.859 | 0.890 |
| de Bruijin et al. [11]⋄ | 0.938 | 0.981 | 0.959 | 0.951 | 0.934 | 0.942 | 0.909 | 0.861 | 0.884 |
| BERT model | 0.936 | 0.983 | 0.959 | 0.967 | 0.943 | 0.955 | 0.906 | 0.898 | 0.902 |
| Prompt-based | 0.984 | 0.958 | 0.971 | 0.965 | 0.971 | 0.968 | 0.907 | 0.935 | 0.921 |
| Possible | Conditional | Not Associated | |||||||
| Logistic Regression | 0.441 | 0.482 | 0.464 | 0.462 | 0.487 | 0.471 | 0.501 | 0.735 | 0.596 |
| Roberts et al. [8]* | 0.816 | 0.589 | 0.684 | 0.729 | 0.298 | 0.423 | 0.915 | 0.814 | 0.861 |
| Jiang et al. [9]* | 0.761 | 0.593 | 0.666 | 0.714 | 0.270 | 0.391 | 0.962 | 0.782 | 0.863 |
| Demner et al. [10]⋄ | 0.937 | 0.792 | 0.859 | 0.759 | 0.257 | 0.384 | 0.917 | 0.766 | 0.835 |
| Clark et al. [7]* | 0.772 | 0.532 | 0.630 | 0.803 | 0.287 | 0.422 | 0.983 | 0.780 | 0.869 |
| de Bruijin et al. [11]⋄ | 0.818 | 0.530 | 0.643 | 0.963 | 0.152 | 0.263 | 0.955 | 0.724 | 0.824 |
| BERT model | 0.818 | 0.709 | 0.760 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Prompt-based | 0.709 | 0.825 | 0.763 | 0.331 | 0.907 | 0.485 | 0.824 | 0.931 | 0.875 |
- the numbers are from original paper, and were not directly comparable with our model.
- the numbers are computed based on the reported confusion matrices from the original paper, and were not directly comparable with our model.
Table B.3.
Results of (P)recision, (R)ecall, and F-1 on the external evaluation datasets.
| Dataset | Model | Present | Absent | Possible | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F-1 | P | R | F-1 | P | R | F-1 | ||
| i2b2 2010 | Logistic Regression | 0.934 | 0.918 | 0.926 | 0.835 | 0.900 | 0.866 | 0.490 | 0.447 | 0.468 |
| NegEx [5] | 0.881 | 0.975 | 0.925 | 0.885 | 0.792 | 0.836 | – | – | – | |
| RadText [35] | 0.859 | 0.939 | 0.897 | 0.792 | 0.637 | 0.706 | 0.599 | 0.323 | 0.420 | |
| BERT model [4] | 0.968 | 0.986 | 0.977 | 0.969 | 0.966 | 0.967 | 0.874 | 0.666 | 0.756 | |
| Prompt-based | 0.975 | 0.985 | 0.980 | 0.973 | 0.976 | 0.975 | 0.835 | 0.712 | 0.769 | |
| i2b2 2012 | Logistic Regression | 0.944 | 0.899 | 0.921 | 0.725 | 0.847 | 0.782 | 0.508 | 0.595 | 0.548 |
| NegEx [5] | 0.913 | 0.962 | 0.937 | 0.779 | 0.855 | 0.815 | – | – | – | |
| RadText [35] | 0.881 | 0.916 | 0.898 | 0.627 | 0.588 | 0.607 | 0.454 | 0.282 | 0.348 | |
| BERT model [4] | 0.959 | 0.951 | 0.955 | 0.831 | 0.905 | 0.866 | 0.693 | 0.616 | 0.652 | |
| Prompt-based | 0.961 | 0.951 | 0.956 | 0.846 | 0.906 | 0.875 | 0.671 | 0.641 | 0.656 | |
| BioScope | Logistic Regression | 0.904 | 0.989 | 0.945 | 0.724 | 0.847 | 0.780 | 0.919 | 0.592 | 0.720 |
| NegEx [5] | 0.784 | 0.999 | 0.879 | 0.658 | 0.587 | 0.621 | – | – | – | |
| RadText [35] | 0.804 | 0.871 | 0.836 | 0.495 | 0.870 | 0.631 | 0.912 | 0.283 | 0.432 | |
| BERT model [4] | 0.911 | 0.994 | 0.951 | 0.766 | 0.947 | 0.835 | 0.985 | 0.583 | 0.732 | |
| Prompt-based | 0.941 | 0.991 | 0.966 | 0.752 | 0.908 | 0.823 | 0.961 | 0.702 | 0.811 | |
| MIMIC-III | Logistic Regression | 0.920 | 0.879 | 0.899 | 0.782 | 0.921 | 0.846 | 0.507 | 0.411 | 0.454 |
| NegEx [5] | 0.867 | 0.954 | 0.908 | 0.855 | 0.871 | 0.863 | – | – | – | |
| RadText | 0.819 | 0.950 | 0.880 | 0.847 | 0.597 | 0.700 | 0.609 | 0.321 | 0.420 | |
| BERT model [4] | 0.937 | 0.965 | 0.951 | 0.929 | 0.945 | 0.937 | 0.775 | 0.518 | 0.621 | |
| Prompt-based | 0.946 | 0.953 | 0.950 | 0.922 | 0.945 | 0.933 | 0.722 | 0.611 | 0.662 | |
| NegEx | Logistic Regression | 0.985 | 0.874 | 0.926 | 0.725 | 0.945 | 0.821 | – | – | – |
| NegEx [5] | 0.977 | 0.988 | 0.983 | 0.951 | 0.912 | 0.931 | – | – | – | |
| RadText [35] | 0.901 | 0.748 | 0.817 | 0.434 | 0.680 | 0.530 | – | – | – | |
| BERT model [4] | 0.993 | 0.867 | 0.926 | 0.700 | 0.976 | 0.815 | – | – | – | |
| Prompt-based | 0.975 | 0.907 | 0.940 | 0.747 | 0.912 | 0.821 | – | – | – | |
| Chia | Logistic Regression | 0.606 | 0.810 | 0.693 | 0.798 | 0.408 | 0.540 | – | – | – |
| NegEx [5] | 0.639 | 0.946 | 0.763 | 0.896 | 0.465 | 0.612 | – | – | – | |
| RadText [35] | 0.570 | 0.916 | 0.703 | 0.803 | 0.293 | 0.430 | – | – | – | |
| BERT model [4] | 0.640 | 0.944 | 0.763 | 0.915 | 0.467 | 0.619 | – | – | – | |
| Prompt-based | 0.669 | 0.913 | 0.772 | 0.894 | 0.513 | 0.652 | – | – | – | |
Table B.4.
Micro F-1s on the MIMIC-III assertion dataset.
Table B.5.
Results of (P)recision, (R)ecall, and F-1 on the MIMIC-III assertion dataset. The best scores are bolded.
| Note type | Model | Present | Absent | Possible | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F-1 | P | R | F-1 | P | R | F-1 | ||
| Discharge summaries | Logistic Regression | 0.923 | 0.890 | 0.906 | 0.792 | 0.917 | 0.850 | 0.521 | 0.396 | 0.450 |
| NegEx [5] | 0.876 | 0.961 | 0.917 | 0.881 | 0.878 | 0.879 | – | – | – | |
| RadText [35] | 0.817 | 0.947 | 0.877 | 0.836 | 0.584 | 0.688 | 0.608 | 0.316 | 0.416 | |
| BERT model [4] | 0.941 | 0.961 | 0.951 | 0.920 | 0.948 | 0.934 | 0.727 | 0.480 | 0.578 | |
| Prompt-based | 0.949 | 0.948 | 0.949 | 0.916 | 0.951 | 0.933 | 0.678 | 0.580 | 0.625 | |
| Nursing letters | Logistic Regression | 0.916 | 0.860 | 0.887 | 0.639 | 0.780 | 0.702 | 0.105 | 0.143 | 0.121 |
| NegEx [5] | 0.931 | 0.966 | 0.948 | 0.839 | 0.881 | 0.860 | – | – | – | |
| RadText [35] | 0.875 | 0.956 | 0.914 | 0.719 | 0.390 | 0.506 | 0.429 | 0.429 | 0.429 | |
| BERT model [4] | 0.980 | 0.983 | 0.981 | 0.966 | 0.949 | 0.957 | 0.786 | 0.786 | 0.786 | |
| Prompt-based | 0.983 | 0.980 | 0.981 | 0.950 | 0.966 | 0.958 | 0.714 | 0.714 | 0.714 | |
| Physician letters | Logistic Regression | 0.912 | 0.814 | 0.860 | 0.628 | 0.952 | 0.756 | 0.682 | 0.469 | 0.556 |
| NegEx [5] | 0.809 | 0.912 | 0.857 | 0.703 | 0.788 | 0.743 | – | – | – | |
| RadText [35] | 0.781 | 0.980 | 0.870 | 0.897 | 0.530 | 0.667 | 0.889 | 0.235 | 0.372 | |
| BERT model [4] | 0.908 | 0.971 | 0.938 | 0.934 | 0.864 | 0.898 | 0.880 | 0.647 | 0.746 | |
| Prompt-based | 0.895 | 0.956 | 0.924 | 0.887 | 0.833 | 0.859 | 0.875 | 0.618 | 0.724 | |
| Radiology reports | Logistic Regression | 0.887 | 0.856 | 0.871 | 0.876 | 0.971 | 0.921 | 0.516 | 0.478 | 0.496 |
| NegEx [5] | 0.767 | 0.902 | 0.829 | 0.768 | 0.862 | 0.812 | – | – | – | |
| RadText [35] | 0.821 | 0.951 | 0.881 | 0.923 | 0.783 | 0.847 | 0.558 | 0.358 | 0.436 | |
| BERT model [4] | 0.886 | 0.979 | 0.930 | 0.978 | 0.957 | 0.967 | 0.900 | 0.537 | 0.673 | |
| Prompt-based | 0.923 | 0.968 | 0.945 | 0.970 | 0.942 | 0.956 | 0.825 | 0.702 | 0.758 | |
Table B.6.
Label mappings.
| Present | Absent | Hypothetical | |
|---|---|---|---|
| M1 | P | N | H |
| M2 | Present | Absent | Hypothetical |
| M3 | P, Positive, Present | N, Negative, Absent | H, Hypothetical, Imaginary |
| Possible | Conditional | Not Associated | |
| M1 | U | C | O |
| M2 | Possible | Conditional | Not-Associated |
| M3 | U, Possible, Uncertain | C, Conditional, Consequent | O, Not-Associated, Irrelevant |
Footnotes
CRediT authorship contribution statement
Song Wang: Conceptualization, Methodology, Software, Validation, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Liyan Tang: Methodology, Writing – review & editing. Akash Majety: Investigation, Writing – review & editing. Justin F. Rousseau: Writing – review & editing. George Shih: Writing – review & editing. Ying Ding: Funding acquisition, Writing – review & editing. Yifan Peng: Conceptualization, Resources, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Khandelwal A, Sawant ST, NegBERT: A transfer learning approach for negation detection and scope resolution, in: Proceedings of the 12th Language Resources and Evaluation Conference, 2020, pp. 5739–5748. [Google Scholar]
- [2].Narayanan S, Achan P, Rangan PV, Rajan SP, Unified concept and assertion detection using contextual multi-task learning in a clinical decision support system, J. Biomed. Inform 122 (2021) 103898. [DOI] [PubMed] [Google Scholar]
- [3].Chen L, Attention-based deep learning system for negation and assertion detection in clinical notes, Int. J. Artif. Intell. Appl 10 (2019) 1–9. [Google Scholar]
- [4].van Aken B, Trajanovska I, Siu A, Mayrdorfer M, Budde K, Loeser A, Assertion detection in clinical notes: medical language models to the rescue?, in: Proceedings of the Second Workshop on Natural Language Processing for Medical Conversations, Association for Computational Linguistics, Online, 2021, pp. 35–40, 10.18653/v1/2021.nlpmc-1.5, https://aclanthology.org/2021.nlpmc-1.5. [DOI] [Google Scholar]
- [5].Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B, A simple algorithm for identifying negated findings and diseases in discharge summaries, J. Biomed. Inform 34 (2001) 301–310. [DOI] [PubMed] [Google Scholar]
- [6].Peng Y, Wang X, Lu L, Bagheri M, Summers RM, Lu Z, NegBio: a high-performance tool for negation and uncertainty detection in radiology reports, AMIA Summits Transl. Sci. Proc 2018 (2018) 188–196. [PMC free article] [PubMed] [Google Scholar]
- [7].Clark C, Aberdeen J, Coarr M, Tresner-Kirsch D, Wellner B, Yeh A, Hirschman L, Determining assertion status for medical problems in clinical records, in: Proceedings of the 2010 I2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data, 2010. [Google Scholar]
- [8].Roberts K, Harabagiu SM, A flexible framework for deriving assertions from electronic medical records, J. Am. Med. Inform. Assoc. : JAMIA 18 5 (2011) 568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiang M, Chen Y, Liu M, Rosenbloom S, Mani S, Denny J, Qi W, A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries, J. Am. Med. Inform. Assoc. : JAMIA 18 (2011) 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Demner-Fushman D, Apostolova E, Doğan RI, cois Michel Lang F, Mork JG, Névéol A, Shooshan SE, Simpson MS, Aronson AR, NLM’s system description for the fourth i2b2/VA challenge, in: Proceedings of the 2010 I2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data, 2010. [Google Scholar]
- [11].de Bruijn B, Cherry C, Kiritchenko S, Martin JD, Zhu X-D, Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010, J. Am. Med. Inform. Assoc. : JAMIA 18 (2011) 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qian Z, Li P, Zhu Q, Zhou G, Luo Z, Luo W, Speculation and negation scope detection via convolutional neural networks, in: Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Association for Computational Linguistics, Austin, Texas, 2016, pp. 815–825, 10.18653/v1/D16-1078, URL: https://aclanthology.org/D16-1078. [DOI] [Google Scholar]
- [13].Fancellu F, Lopez A, Webber B, Neural networks for negation scope detection, in: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Association for Computational Linguistics, Berlin, Germany, 2016, pp. 495–504, 10.18653/v1/P16-1047, URL: https://aclanthology.org/P16-1047. [DOI] [Google Scholar]
- [14].Taylor S, Harabagiu S, The role of a deep-learning method for negation detection in patient cohort identification from electroencephalography reports, AMIA … Annu. Symp. Proc. AMIA Symp 2018 (2018) 1018–1027. [PMC free article] [PubMed] [Google Scholar]
- [15].Bhatia P, Celikkaya B, Khalilia M, Joint entity extraction and assertion detection for clinical text, in: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Association for Computational Linguistics, Florence, Italy, 2019, pp. 954–959, 10.18653/v1/P19-1091, https://aclanthology.org/P19-1091. [DOI] [Google Scholar]
- [16].Radford A, Narasimhan K, Improving language understanding by generative pre-training, 2018.
- [17].Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I, et al. , Language models are unsupervised multitask learners, OpenAI blog 1 (8) (2019) 9. [Google Scholar]
- [18].Zhong Z, Friedman D, Chen D, Factual probing is [MASK]: Learning vs. Learning to recall, in: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Online, 2021, pp. 5017–5033, 10.18653/v1/2021.naacl-main.398, https://aclanthology.org/2021.naacl-main.398. [DOI] [Google Scholar]
- [19].Qin G, Eisner J, Learning how to ask: Querying LMs with mixtures of soft prompts, in: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Online, 2021, pp. 5203–5212, 10.18653/v1/2021.naacl-main.410, https://aclanthology.org/2021.naacl-main.410. [DOI] [Google Scholar]
- [20].Zhong R, Lee K, Zhang Z, Klein D, Adapting language models for zero-shot learning by meta-tuning on dataset and prompt collections, in: Findings of the Association for Computational Linguistics: EMNLP 2021, Association for Computational Linguistics, Punta Cana, Dominican Republic, 2021, pp. 2856–2878, 10.18653/v1/2021.findings-emnlp.244, https://aclanthology.org/2021.findings-emnlp.244. [DOI] [Google Scholar]
- [21].Wang S, Fang H, Khabsa M, Mao H, Ma H, Entailment as few-shot learner, 2021, arXiv, arXiv:2104.14690. [Google Scholar]
- [22].Liu P, Yuan W, Fu J, Jiang Z, Hayashi H, Neubig G, Pre-train, prompt, and predict: A systematic survey of prompting methods in natural language processing, 2021, arXiv, arXiv:2107.13586. [Google Scholar]
- [23].Gao T, Fisch A, Chen D, Making pre-trained language models better few-shot learners, in: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Association for Computational Linguistics, Online, 2021, pp. 3816–3830, 10.18653/v1/2021.acl-long.295, https://aclanthology.org/2021.acl-long.295. [DOI] [Google Scholar]
- [24].Uzuner O, South B, Shen S, DuVall S, 2010 I2b2/VA challenge on concepts, assertions, and relations in clinical text, J. Am. Med. Inform. Assoc. : JAMIA 18 (2011) 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun W, Rumshisky A, Uzuner O, Evaluating temporal relations in clinical text: 2012 i2b2 challenge, J. Am. Med. Inform. Assoc. : JAMIA 20 5 (2013) 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson A, Pollard T, Shen L, Lehman L.-w., Feng M, Ghassemi M, Moody B, Szolovits P, Celi L, Mark R, MIMIC-III, a freely accessible critical care database, Sci. Data 3 (2016) 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Szarvas G, Vincze V, Farkas R, Csirik J, The BioScope corpus: Annotation for negation, uncertainty and their scope in biomedical texts, in: Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing, Association for Computational Linguistics, Columbus, Ohio, 2008, pp. 38–45, URL: https://aclanthology.org/W08-0606. [Google Scholar]
- [28].Chapman WW, Dowling JN, Chu D, Context: An algorithm for identifying contextual features from clinical text, in: Biological, translational, and clinical language processing, Association for Computational Linguistics, Prague, Czech Republic, 2007, pp. 81–88, https://aclanthology.org/W07-1011. [Google Scholar]
- [29].Kury FSP, Butler AM, Yuan C, heng Fu L, Sun Y, Liu H, Sim I, Carini S, Weng C, Chia, a large annotated corpus of clinical trial eligibility criteria, Sci. Data 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].DeYoung J, Jain S, Rajani NF, Lehman E, Xiong C, Socher R, Wallace BC, Eraser: a benchmark to evaluate rationalized nlp models, in: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Association for Computational Linguistics, Online, 2020, pp. 4443–4458, 10.18653/v1/2020.acl-main.408, https://aclanthology.org/2020.acl-main.408. [DOI] [Google Scholar]
- [31].Ribeiro MT, Singh S, Guestrin C, “Why should I trust you?”: Explaining the predictions of any classifier, in: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations, Association for Computational Linguistics, San Diego, California, 2016, pp. 97–101, 10.18653/v1/N16-3020, https://aclanthology.org/N16-3020. [DOI] [Google Scholar]
- [32].Boyle M, Semantic cue, in: Kreutzer JS, DeLuca J, Caplan B (Eds.), Encyclopedia of Clinical Neuropsychology, Springer International Publishing, Cham, 2018, pp. 3119–3120, 10.1007/978-3-319-57111-9_921. [DOI] [Google Scholar]
- [33].Alsentzer E, Murphy J, Boag W, Weng W-H, Jindi D, Naumann T, McDermott M, Publicly available clinical BERT embeddings, in: Proceedings of the 2nd Clinical Natural Language Processing Workshop, Association for Computational Linguistics, Minneapolis, Minnesota, USA, 2019, pp. 72–78, 10.18653/v1/W19-1909, https://aclanthology.org/W19-1909. [DOI] [Google Scholar]
- [34].Loshchilov I, Hutter F, Decoupled weight decay regularization, in: ICLR, 2019. [Google Scholar]
- [35].Wang S, Lin M, Ding Y, Shih G, Lu Z, Peng Y, Radiology text analysis system (RadText): Architecture and evaluation, in: IEEE International Conference on Healthcare Informatics, 2022, URL: https://github.com/bionlplab/radtext. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Devlin J, Chang M-W, Lee K, Toutanova K, BERT: Pre-training of deep bidirectional transformers for language understanding, in: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Association for Computational Linguistics, Minneapolis, Minnesota, 2019, pp. 4171–4186, 10.18653/v1/N19-1423. [DOI] [Google Scholar]
- [37].Peng Y, Yan S, Lu Z, Transfer learning in biomedical natural language processing: An evaluation of BERT and ELMo on ten benchmarking datasets, in: Proceedings of the 18th BioNLP Workshop and Shared Task, Association for Computational Linguistics, Florence, Italy, 2019, pp. 58–65, 10.18653/v1/W19-5006, https://aclanthology.org/W19-5006. [DOI] [Google Scholar]
- [38].Qin G, Eisner J, Learning how to ask: Querying LMs with mixtures of soft prompts, in: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Online, 2021, pp. 5203–5212, 10.18653/v1/2021.naacl-main.410, URL: https://aclanthology.org/2021.naacl-main.410. [DOI] [Google Scholar]
- [39].Hambardzumyan K, Khachatrian H, May J, WARP: Word-level adversarial reprogramming, in: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Association for Computational Linguistics, Online, 2021, pp. 4921–4933, 10.18653/v1/2021.acl-long.381, URL: https://aclanthology.org/2021.acl-long.381. [DOI] [Google Scholar]
- [40].Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG, Evaluation of negation phrases in narrative clinical reports, Proc. AMIA Symp (2001) 105–109. [PMC free article] [PubMed] [Google Scholar]
- [41].Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM, ChestX-Ray8: Hospital-scale chest X-Ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR, 2017, pp. 3462–3471. [Google Scholar]
- [42].de Marneffe M-C, Dozat T, Silveira N, Haverinen K, Ginter F, Nivre J, Manning CD, Universal stanford dependencies: A cross-linguistic typology, in: Proceedings of the Ninth International Conference on Language Resources and Evaluation, LREC’14, European Language Resources Association (ELRA), Reykjavik, Iceland, 2014, pp. 4585–4592, URL: http://www.lrec-conf.org/proceedings/lrec2014/pdf/1062_Paper.pdf. [Google Scholar]
- [43].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Schuler KK, Chute CG, Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications, J. Ame. Med. Inform. Assoc. : JAMIA 17 5 (2010) 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Eyre H, Chapman AB, Peterson KS, Shi J, Alba PR, Jones MM, Box TL, DuVall SL, Patterson OV, AMIA Annual Symposium Proceedings, 2021, American Medical Informatics Association, 2021, p. 438. [PMC free article] [PubMed] [Google Scholar]
- [45].Bastings J, Filippova K, The elephant in the interpretability room: Why use attention as explanation when we have saliency methods? in: Proceedings of the Third BlackboxNLP Workshop on Analyzing and Interpreting Neural Networks for NLP, Association for Computational Linguistics, Online, 2020, pp. 149–155, 10.18653/v1/2020.blackboxnlp-1.14, URL: https://aclanthology.org/2020.blackboxnlp-1.14. [DOI] [Google Scholar]
- [46].Alvarez-Melis D, Jaakkola T, On the robustness of interpretability methods, 2018, arXiv, arXiv:1806.08049. [Google Scholar]
- [47].Deléger L, Li Q, Lingren T, Kaiser M, Molnár K, Stoutenborough L, Kouril M, Marsolo KA, Solti I, Building gold standard corpora for medical natural language processing tasks, AMIA … Annu. Symp. Proc. AMIA Symp 2012 (2012) 144–153. [PMC free article] [PubMed] [Google Scholar]


