Abstract
Background: The existence of specific microbial profiles for different periodontal conditions is still a matter of debate. The aim of this study was to test the hypothesis that 40 bacterial species could be used to classify patients, utilising machine learning, into generalised chronic periodontitis (ChP), generalised aggressive periodontitis (AgP) and periodontal health (PH). Method: Subgingival biofilm samples were collected from patients with AgP, ChP and PH and analysed for their content of 40 bacterial species using checkerboard DNA–DNA hybridisation. Two stages of machine learning were then performed. First of all, we tested whether there was a difference between the composition of bacterial communities in PH and in disease, and then we tested whether a difference existed in the composition of bacterial communities between ChP and AgP. The data were split in each analysis to 70% train and 30% test. A support vector machine (SVM) classifier was used with a linear kernel and a Box constraint of 1. The analysis was divided into two parts. Results: Overall, 435 patients (3,915 samples) were included in the analysis (PH = 53; ChP = 308; AgP = 74). The variance of the healthy samples in all principal component analysis (PCA) directions was smaller than that of the periodontally diseased samples, suggesting that PH is characterised by a uniform bacterial composition and that the bacterial composition of periodontally diseased samples is much more diverse. The relative bacterial load could distinguish between AgP and ChP. Conclusion: An SVC classifier using a panel of 40 bacterial species was able to distinguish between PH, AgP in young individuals and ChP.
Key words: Plaque, oral health, prevention, periodontitis, mathematics
Introduction
Periodontitis is an oral disease driven by deregulated inflammation induced by polymicrobial communities that form on subgingival tooth sites1. The gingival sulcus and periodontal pocket form unique ecological niches for microbial colonisation and the subgingival microbiota drives the inflammatory process that leads to periodontal tissue destruction1., 2..
The existence of different forms of periodontitis is a reality and, over the years, different classification systems have been suggested for these conditions3., 4.. In 1999, the World Workshop of the American Academy of Periodontology (AAP) changed the term ‘adult periodontitis’ to ‘chronic periodontitis’ (ChP) and introduced the term ‘aggressive periodontitis’ (AgP) to define a group of destructive periodontal diseases with a rapid progression3., 5.. This put the emphasis on the rate of disease progression, information rarely available to clinicians6., 7.. Although the classification systems have been continuously under debate, it is generally well accepted that disease in younger patients is different from that in adults, and one possible explanation for this difference is different microbial profiles. If this is the case, a microbiological examination might, in theory, help in the differential diagnosis of AgP in young patients from the more common ChP and would have the potential to help in the diagnosis and treatment of these diseases7., 8..
Since the 1950s, the microbiota of the periodontal diseases has been studied, initially by culture methods and afterwards by molecular techniques. The current knowledge about the microbiota associated with periodontal health or disease has been largely impacted by evaluation of the 40 bacterial species that comprise the microbial complexes described by Socransky et al.9., 10., 11., 12., 13.. Studies using different diagnostic techniques have defined four main periodontal pathogens – the three species from the red complex (Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola) and Aggregatibacter actinomycetemcomitans10., 12., 14., 15., 16.. In addition, several species belonging to different complexes have been associated with periodontal health, in particular those from the genera Actinomyces, Streptococcus and Capnocytophaga16., 17., 18.. It is also important to note that there is moderate evidence in the literature to support the existence of newly identified periodontal pathogens or host-compatible species19., 20., 21., 22. but the role of these species as true pathogens or as markers for periodontal stability are yet to be established, especially by risk assessment and interventional studies. Therefore, the 40 bacterial species defined by Socransky et al.12 are still considered a suitable biological marker for studying the periodontal microbiota associated with periodontal health or disease. In addition, it has been shown that this set of 40 probes would account for 55–60% of the bacteria in subgingival biofilms16.
Machine learning is a discipline of computer science aimed at developing algorithms able to learn from tagged examples instead of performing a predefined explicit routine23., 24., 25., 26.. Specifically, a cost function related to the number of wrongly classified tagged examples is minimised. These approaches are becoming popular in a wide variety of biological applications23., 24., 25., 26.. A common biomedical application of support vector machines (SVM) is the automatic classification of microarray gene expression profiles23., 24.. SVM applied on the gene-expression profile derived from a tumour sample or from peripheral fluid was shown to produce a diagnosis or a prognosis23., 24., 25., 26., 27.. In addition, other biological applications of SVMs involve classifying protein and DNA sequences, microarray expression profiles and mass spectra28. Recently, Nakano et al.25 used SMV to diagnose malodour from oral microbiota and methyl mercaptan levels in saliva. They reported that SVM achieved high accuracy, with sensitivity of 51.1% and specificity of 95.0%. This approach is useful where design of an explicit algorithm is not feasible. Kebschull et al.29 used an SVM approach to explore molecular differences between ChP and AgP, but this method has never been used to assess possible microbiological differences between these two clinical conditions.
The aim of this study was to compare the subgingival microbial profiles of AgP in young patients, in patients with ChP and in periodontally healthy (PH) subjects and to test whether patients’ periodontal status could be classified using the 40 bacterial species of the subgingival microbial complexes12. The hypothesis tested was that this analysis could create a model with the ability to differentiate between these clinical conditions.
Methods
Patient population
Patients who were PH or were diagnosed with generalised ChP or generalised AgP were selected from the database of the Department of Periodontology of Guarulhos University. This study analysed data compiled from large clinical studies that evaluated the subgingival microbiota of PH subjects and patients with periodontitis. Those studies were conducted at Guarulhos University (São Paulo, SP, Brazil) from 2004 to 2015, and followed very similar protocols for selection of participants, sample collection and microbial analysis. The protocols of these studies were approved previously by Guarulhos University’s Ethics Committee in Clinical Research and the research was conducted in full accordance with the World Medical Association Declaration of Helsinki; consent was given by the participants involved in the study.
Clinical examination
The clinical examination was performed by trained and calibrated examiners, as previously described10. The intra-examiner variability in all clinical studies was 0.13–0.21 mm for probing depth (PD) and 0.22–0.31 mm for clinical attachment level (CAL).
Generalised AgP, generalised ChP and PH were diagnosed based on the periodontal classification of the American Academy of Periodontology3 and as described in our previous report10.
Microbiological examination
Subgingival plaque samples were collected from nine non-contiguous interproximal sites per patient, as reported previously by Faveri et al.10 For AgP and ChP groups, three sites at each of the following PD categories were sampled: ≤3 mm; between 4 and 6 mm; and ≥7 mm. Subgingival plaque samples from sites with PD ≤3 mm were collected from subjects in the PH group. Counts of 40 bacterial species were determined in each sample using the checkerboard DNA–DNA hybridisation technique30., 31. at the Laboratory of Microbiology of Guarulhos University. Signals were evaluated visually by a calibrated examiner who compared the signals with standards of 105 and 106 bacterial cells for the test species on the same membrane (k test = 93%). The mean counts of individual bacterial species were averaged within each patient and then across patients in each group. The sum of the individual mean proportions were computed for each microbial complex described by Socransky et al.12
Normalisation and data analysis
The total concentration of each sample was normalised to a value of 1. A principal component analysis (PCA) was performed to visualise the data. Two stages of machine learning were performed. In the first stage, PH patients were compared with all patients with periodontal disease. In the second stage, patients with ChP and AgP were compared. The data were split, in each analysis, to 70% train and 30% test. An SVM classifier was used with a Box constraint of 1 and a linear kernel. The results are presented as a receiver operating characteristics (ROC) curve and their precision is estimated using the area under the curve (AUC). Samples from the same patient were categorically divided to be either in the train or the test set (i.e. no samples from the same patient were used in both the train and the test set).
Note that we did not take into consideration the total bacterial load that might have been affected by the experimental design. Instead, we analysed the proportion of each bacterial species relative to the levels of the 40 species evaluated. The significance of differences among groups was sought using the Kruskal–Wallis test and the Mann–Whitney U-test. The chi-square test was used to compare differences in the distribution of gender.
Results
Table 1 depicts the demographic characteristics and clinical parameters of the study population. A cohort of 435 patients were evaluated: 53 PH, 308 with generalised ChP and 74 with generalised AgP. The mean age of individuals with generalised ChP was significantly higher (45.1 ± 5.9 years) than the mean age of subjects with generalised AgP (27.1 ± 3.1 years) and PH subjects (35.1 ± 9.5 years). No difference was observed in the distribution of gender. The mean PD and CAL and the percentage of sites exhibiting bleeding on probing (BOP), gingival bleeding and suppuration were significantly higher in the ChP and AgP groups than in PH patients. PH individuals and patients with AgP showed less visible plaque (28.1% and 49.5%, respectively) than did patients with ChP (84.6%, P < 0.05).
Table 1.
Demographic characteristics and full-mouth clinical parameters of the patients in experimental groups
| Variables | Experimental groups | P-value* | ||
|---|---|---|---|---|
| Periodontally healthy | Generalised aggressive periodontitis | Generalised chronic periodontitis | ||
| Number of patients | 53 | 74 | 308 | |
| Age (years) | 35.1 ± 9.5A | 27.1 ± 3.1B | 45.1 ± 5.9C | <0.001 |
| Gender (male:female) | 23:30 | 32:42 | 122:186 | NS |
| Probing depth (mm) | 1.9 ± 0.5A | 4.2 ± 1.1B | 4.1 ± 1.3B | <0.001 |
| Clinical attachment level (mm) | 0.7 ± 0.4A | 3.8 ± 1.2B | 3.8 ± 1.1B | <0.001 |
| % Sites with: | ||||
| Plaque | 28.1 ± 7.7A | 49.5 ± 14.2B | 84.6 ± 12.2C | <0.001 |
| Gingival bleeding | 2.0 ± 1.0A | 33.2 ± 12.9B | 34.1 ± 22.1B | <0.001 |
| Bleeding on probing | 2.1 ± 0.8A | 46.1 ± 14.2B | 45.2 ± 27.2B | <0.001 |
| Suppuration | 0 ± 0A | 4.4 ± 3.7B | 3.29 ± 4.1B | <0.001 |
Values are given as number of patients, ratio (male : female) or mean ± standard deviation.
The significance of differences among groups was assessed using the Kruskall–Wallis test (*). The significance of differences between pairs of comparisons was determined using Dunn’s multiple comparison test and the significances are represented by different capital letters.NS, not significant.
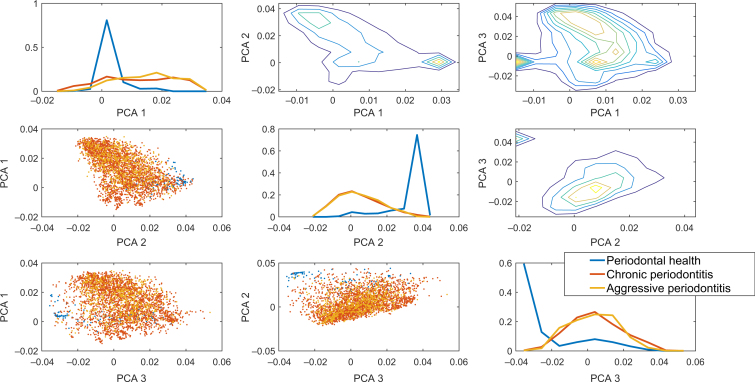
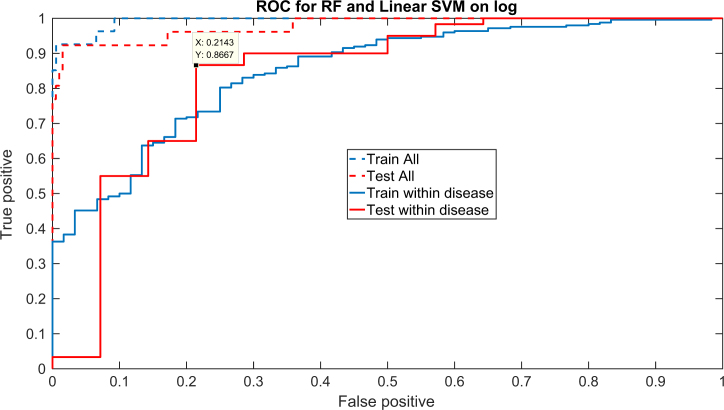
The results for the PCA showed that the variance of the healthy samples in all PCA directions was much smaller than that of the periodontally diseased samples (Figure 1), suggesting that while healthy samples are characterised by a highly uniform bacterial composition, the bacterial composition in periodontally diseased samples is much more diverse. The diversity can be observed in the two conditions studied here. Thus, the variability is within each periodontally involved population. Given the clear difference between the healthy and diseased populations, the classification of samples based on the relative bacterial load was tested. Indeed, a linear SVM supervised classifier produces an AUC of >0.95 on a test set between the diseased and healthy conditions (Figure 2).
Figure 1.
Principal component analysis (PCA) projection of the three conditions (chronic periodontitis, aggressive periodontitis and periodontally healthy). The diagonal plots are the distribution of the principal component weights. One can clearly see that the healthy state is very different from the two other states. Each row is a different principal component. The plots below the diagonal are scatter plots of two principal component vectors. While the healthy condition has a limited variance, the two other conditions have a large variance and mostly overlap. The plots above the diagonal are two-dimensional distributions of the projections on the principal components, as presented by contours. One can clearly see a peak in the distribution representing the healthy state, which is very different from any of the disease states. This is also obvious in the separate blub in the contour plot.
Figure 2.
Receiver operating characteristic curve for chronic periodontitis (ChP) and aggressive periodontitis (AgP) versus healthy (dashed line) and for ChP versus AgP (solid line) using a linear support vector machine. The train and test results are highly similar, showing that there is no over-fitting. The values are above the diagonal showing that a meaningful differentiation can be obtained. The true-positive and false-positive values specified in the text box are just one possible point along the curve.
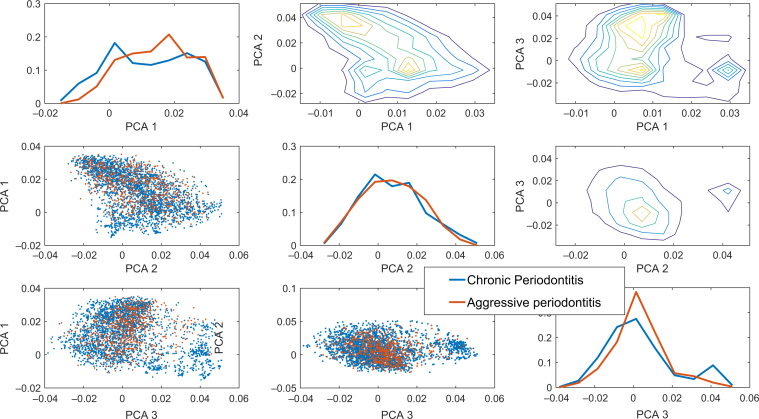
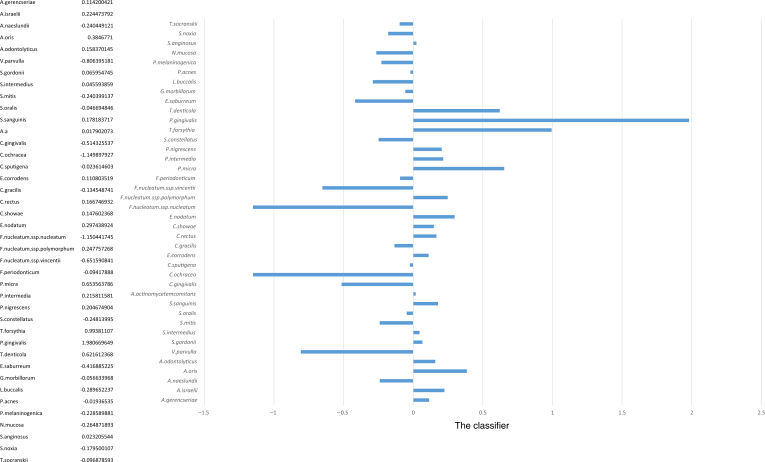
More importantly, the relative bacterial load could distinguish between AgP and ChP with high accuracy. While the difference between these two conditions is smaller than that between health and disease, as can be seen, for example, in the PCA based only on the diseased samples (Figure 3), applying a linear SVM to the two diseased conditions produced a sensitivity of 86%, a specificity of 79% and an AUC of 0.83 on a test set (Figure 2). Thus, not only were the bacterial profiles different between these two diseases, but also this difference was enough to allow a clear distinction between the two conditions. Specifically, a linear SVM was applied to the first 20 PCA vectors (representing over 95% of the variability). The classifier defines a clear direction in the bacterial load concentration space with some bacteria correlated with each of the two diseases (Figure 4). The species P. gingivalis followed by T. forsythia, Fusobacterium ssp. polymorphum, T. denticola and Prevotella intermedia were the five species most representative for ChP. This means that these were the bacteria that could differentiate between chronic and aggressive conditions, favouring ChP. On the other hand, Fusobacterium nucleatum.ssp.nucleatum, Capnocytophaga ochracea, Veillonella parvula, Fusobacterium nucleatum.ssp.vincentii and Capnocytophaga gingivalis were the five species most strongly directed towards AgP. It is noteworthy that the values in Figure 4 do not represent prevalence of the bacterial species but rather their ability to differentiate between ChP (positive values) and AgP (negative values).
Figure 3.
Similarly to Figure 1, when the healthy state is removed. Clear differences can be observed between the two disease states, but the difference is smaller than between healthy and diseased, as observed in Figure 1. Note that here also, there is a clear separate cluster of aggressive periodontitis point (see the third principal component analysis).
Figure 4.
Weights of linear support vector machine classifier for chronic periodontitis (ChP) versus aggressive periodontitis (AgP). Positive weights imply that a high level of the bacterium is correlated with ChP, and the opposite for negative weights, correlated with AgP. The absolute value of the weight represents the contribution of the specific bacterium to the classification. It is noteworthy that the values in Figure 4 do not represent prevalence of the bacterial species but rather their ability to differentiate between ChP (positive values) and AgP (negative values).
Discussion
The hypothesis tested in this study – that an SVM classifier using a panel of 40 bacterial species could differentiate between AgP and ChP – was confirmed.
The SVM analysis, including 435 patients and 3,915 samples, showed that the three red-complex species (P. gingivalis, T. forsythia and T. denticola) had a high weight in the classifying vector to a ChP diagnosis. On the other hand, F. nucleatum ssp. nucleatum, C. ochracea, V. parvulla, F. nucleatum ssp. vincentii and C. gingivalis were the species that allowed classifying or directing the classification towards AgP. These data are in agreement with previous studies showing that pathogens from the red and orange complexes are more associated with the aetiology of ChP9., 12., 32.. Interestingly, other microorganisms not normally associated with AgP were found to allow differentiation towards this condition.
The SVM classifier system used in this study was able to distinguish between generalised AgP and generalised ChP with rather high accuracy. Although the 40 bacterial species evaluated have been used successfully as a biological marker to monitor disease resolution after several periodontal treatments33., 34., 35., when this same microbial panel was analysed using conventional statistical approaches in diagnostic studies, some failed to find major differences between ChP and AgP microbial profiles10., 13., 36.. Faveri et al.10 showed that the composition of the subgingival microbiota did not differ substantially among localised AgP, generalised AgP and ChP. Similar results have been reported by other authors13., 36.. This could be explained by differences in the way in which the data were analysed, in the number of samples evaluated or by the inability to distinguish AgP from ChP clinically using the current classification systems for these conditions7., 37., 38..
To our knowledge, this is the first report in which AgP and ChP could be differentiated according to the subgingival microbial profile. Previous clinical studies and comprehensive reviews have not found major differences between the microbiological and immunological features of these two clinical conditions29., 39., 40.. Nonetheless, it is important to bear in mind that the main inclusion criterion for patients in the two periodontitis groups, in this study, was age, a parameter that is not considered by the current classification of the AAP3. In fact, many clinical researchers in periodontology face the difficulty of selecting volunteers based on characteristics that are not normally available to the clinician. This is even more critical for AgP. The three common features of aggressive periodontal diseases, according to the Consensus Report of the AAP3., 5. are: otherwise clinically healthy patients; familial aggregation; and rapid attachment loss and bone destruction. The first two characteristics are also observed in patients with ChP, and the last one is rarely available to the clinician. Determining the rate of attachment loss while selecting patients for cross-sectional studies is not feasible, leaving researchers with the alternative for using age as a discriminating factor, by estimating ‘rapid periodontal destruction’ if the individual shows advanced disease at an early age. This was the case of the database used in the present study. Therefore, it might be more accurate to state that the statistical model tested in this study is more suitable for differentiating between advanced periodontitis in adults and in young individuals than between ChP and AgP.
Although we understand the infectious nature of periodontitis, the microbial composition and the host response components in relation to the aetiology of the disease have yet to be fully understood41. The efforts to catalogue microbial species associated with disease and exposition of the interspecies interactions in oral biofilm will contribute to our understanding of how these microorganisms may act together and result in either health or disease41. A recent review by Nibali7 stated that as we aim to understand host-associated factors and clinical differences between AgP and ChP, with the hope of designing more effective treatment protocols for these conditions, an interesting insight could be given by studies comparing the microbial composition of these diseases. The use of advanced mathematical approaches evaluating many samples from many patients, similar to the one used in the current report, might shed further light on the differences between those two clinical conditions. Those methods could be applied in the future to differentiate further between subgroups of the diseases (e.g. generalised, localised) and might open new avenues for using population science methods to explore the potential of specific therapeutic interventions.
The results of the present study indicate that an SVM classifier using a panel of 40 bacterial species is able to distinguish between generalised AgP in young patients and generalised ChP.
Acknowledgements
The authors declare no conflict of interest for the above manuscript. This work was supported, in part, by research grants 2007/56413-0, 2007/55291-9, 2009/17677-8, 2010/10384-2 and 2011/23034-2 from São Paulo Research Foundation (FAPESP, Brazil); and research grants 304887/2013-7, 309015/2012-0 and 308124/2013-8 from The National Council for Scientific and Technological Development (CNPq, Brazil).
References
- 1.Hajishengallis G, Hajishengallis E, Kajikawa T, et al. Complement inhibition in pre-clinical models of periodontitis and prospects for clinical application. Semin Immunol. 2016;28:285–291. doi: 10.1016/j.smim.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Tonetti MS, Claffey N, European Workshop in Periodontology group C Advances in the progression of periodontitis and proposal of definitions of a periodontitis case and disease progression for use in risk factor research. Group C consensus report of the 5th European Workshop in Periodontology. J Clin Periodontol. 2005;6:210–213. doi: 10.1111/j.1600-051X.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- 5.Lang NBP, Cullinan M, Jeffcoat M, et al. Consensus report: aggressive periodontitis. Ann Periodontol. 1999;4:53. [Google Scholar]
- 6.Levin L, Baev V, Lev R, et al. Aggressive periodontitis among young Israeli army personnel. J Periodontol. 2006;77:1392–1396. doi: 10.1902/jop.2006.050323. [DOI] [PubMed] [Google Scholar]
- 7.Nibali L. Aggressive Periodontitis: microbes and host response, who to blame? Virulence. 2015;6:223–228. doi: 10.4161/21505594.2014.986407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albandar JM, Tinoco EM. Global epidemiology of periodontal diseases in children and young persons. Periodontology 2000. 2002;29:153–176. doi: 10.1034/j.1600-0757.2002.290108.x. [DOI] [PubMed] [Google Scholar]
- 9.Colombo AP, Teles RP, Torres MC, et al. Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J Periodontol. 2002;73:360–369. doi: 10.1902/jop.2002.73.4.360. [DOI] [PubMed] [Google Scholar]
- 10.Faveri M, Figueiredo LC, Duarte PM, et al. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36:739–749. doi: 10.1111/j.1600-051X.2009.01449.x. [DOI] [PubMed] [Google Scholar]
- 11.Lopez NJ, Socransky SS, Da Silva I, et al. Subgingival microbiota of chilean patients with chronic periodontitis. J Periodontol. 2004;75:717–725. doi: 10.1902/jop.2004.75.5.717. [DOI] [PubMed] [Google Scholar]
- 12.Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 13.Ximenez-Fyvie LA, Almaguer-Flores A, Jacobo-Soto V, et al. Subgingival microbiota of periodontally untreated Mexican subjects with generalized aggressive periodontitis. J Clin Periodontol. 2006;33:869–877. doi: 10.1111/j.1600-051X.2006.01006.x. [DOI] [PubMed] [Google Scholar]
- 14.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feres M, Gursky LC, Faveri M, et al. Clinical and microbiological benefits of strict supragingival plaque control as part of the active phase of periodontal therapy. J Clin Periodontol. 2009;36:857–867. doi: 10.1111/j.1600-051X.2009.01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontology 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 17.Abusleme L, Dupuy AK, Dutzan N, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, et al. Bacterial interactions and successions during plaque development. Periodontology 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 19.Diaz PI, Hoare A, Hong BY. Subgingival microbiome shifts and community dynamics in periodontal diseases. J Calif Dent Assoc. 2016;44:421–435. [PubMed] [Google Scholar]
- 20.Goncalves C, Soares GM, Faveri M, et al. Association of three putative periodontal pathogens with chronic periodontitis in Brazilian subjects. J Appl Oral Sci. 2016;24:181–185. doi: 10.1590/1678-775720150445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveira RR, Fermiano D, Feres M, et al. Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res. 2016;95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Chaparro PJ, Goncalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93:846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan WH, Mohamad MS, Deris S, et al. Identification of informative genes and pathways using an improved penalized support vector machine with a weighting scheme. Comput Biol Med. 2016;77:102–115. doi: 10.1016/j.compbiomed.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Guo J, Hu G, et al. Gene prediction in metagenomic fragments based on the SVM algorithm. BMC Bioinformatics. 2013;14(Suppl 5):S12. doi: 10.1186/1471-2105-14-S5-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano Y, Takeshita T, Kamio N, et al. Supervised machine learning-based classification of oral malodor based on the microbiota in saliva samples. Artif Intell Med. 2014;60:97–101. doi: 10.1016/j.artmed.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 27.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 28.Noble WS. In: Kernel Methods in Computational Biology. Scholkopf B, Tsuda K, Vert J, editors. MIT Press; Cambridge, MA: 2004. Support vector machine applications in computational biology; pp. 71–92. [Google Scholar]
- 29.Kebschull M, Guarnieri P, Demmer RT, et al. Molecular differences between chronic and aggressive periodontitis. J Dent Res. 2013;92:1081–1088. doi: 10.1177/0022034513506011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Socransky SS, Smith C, Martin L, et al. “Checkerboard” DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 31.Mestnik MJ, Feres M, Figueiredo LC, et al. Short-term benefits of the adjunctive use of metronidazole plus amoxicillin in the microbial profile and in the clinical parameters of subjects with generalized aggressive periodontitis. J Clin Periodontol. 2010;37:353–365. doi: 10.1111/j.1600-051X.2010.01538.x. [DOI] [PubMed] [Google Scholar]
- 32.Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontology 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 33.Socransky SS, Haffajee AD, Teles R, et al. Effect of periodontal therapy on the subgingival microbiota over a 2-year monitoring period. I. Overall effect and kinetics of change. J Clin Periodontol. 2013;40:771–780. doi: 10.1111/jcpe.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soares GM, Mendes JA, Silva MP, et al. Metronidazole alone or with amoxicillin as adjuncts to non-surgical treatment of chronic periodontitis: a secondary analysis of microbiological results from a randomized clinical trial. J Clin Periodontol. 2014;41:366–376. doi: 10.1111/jcpe.12217. [DOI] [PubMed] [Google Scholar]
- 35.Tamashiro NS, Duarte PM, Miranda TS, et al. Amoxicillin plus metronidazole therapy for patients with periodontitis and type 2 diabetes: a 2-year randomized controlled trial. J Dent Res. 2016;95:829–836. doi: 10.1177/0022034516639274. [DOI] [PubMed] [Google Scholar]
- 36.Rescala B, Rosalem W, Jr, Teles RP, et al. Immunologic and microbiologic profiles of chronic and aggressive periodontitis subjects. J Periodontol. 2010;81:1308–1316. doi: 10.1902/jop.2010.090643. [DOI] [PubMed] [Google Scholar]
- 37.Gajardo M, Silva N, Gomez L, et al. Prevalence of periodontopathic bacteria in aggressive periodontitis patients in a Chilean population. J Periodontol. 2005;76:289–294. doi: 10.1902/jop.2005.76.2.289. [DOI] [PubMed] [Google Scholar]
- 38.Riep B, Edesi-Neuss L, Claessen F, et al. Are putative periodontal pathogens reliable diagnostic markers? J Clin Microbiol. 2009;47:1705–1711. doi: 10.1128/JCM.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armitage GC. Comparison of the microbiological features of chronic and aggressive periodontitis. Periodontology 2000. 2010;53:70–88. doi: 10.1111/j.1600-0757.2010.00357.x. [DOI] [PubMed] [Google Scholar]
- 40.Duarte PM, Bastos MF, Fermiano D, et al. Do subjects with aggressive and chronic periodontitis exhibit a different cytokine/chemokine profile in the gingival crevicular fluid? A systematic review. J Periodontal Res. 2015;50:18–27. doi: 10.1111/jre.12180. [DOI] [PubMed] [Google Scholar]
- 41.Khan SA, Kong EF, Meiller TF, et al. Periodontal diseases: bug induced, host promoted. PLoS Pathog. 2015;11:e1004952. doi: 10.1371/journal.ppat.1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]