Abstract
BarA of Streptomyces virginiae is a specific receptor protein for virginiae butanolide (VB), one of the γ-butyrolactone autoregulators of the Streptomyces species, and acts as a transcriptional regulator controlling both virginiamycin production and VB biosynthesis. The downstream gene barB, the transcription of which is under the tight control of the VB-BarA system, was found to be transcribed as a polycistronic mRNA with its downstream region, and DNA sequencing revealed a 1,554-bp open reading frame (ORF) beginning at 161 bp downstream of the barB termination codon. The ORF product showed high homology (68 to 73%) to drug efflux proteins having 14 transmembrane segments and was named varS (for S. virginiae antibiotic resistance). Heterologous expression of varS with S. lividans as a host resulted in virginiamycin S-specific resistance, suggesting that varS encoded a virginiamycin S-specific transport protein. Northern blot analysis indicated that the bicistronic transcript of barB-varS appeared 1 to 2 h before the onset of virginiamycin M1 and S production, at which time VB was produced, while exogenously added virginiamycin S apparently induced the monocistronic varS transcript.
Streptomycetes are gram-positive filamentous bacteria that are well known for producing a vast array of bioactive compounds, including more than 70% of commercially important antibiotics. The production of antibiotics by these organisms is regulated by a variety of physiological and nutritional conditions and is coordinated with processes of morphological differentiation, such as the formation of aerial mycelia and spores. Despite the long years of research on antibiotics driven by their commercial importance, the overall regulatory pathway governing antibiotic production is still poorly understood. A detailed knowledge of the signal cascade and the genetic components involved in antibiotic production should permit the construction of strains that can overproduce these commercially important compounds.
Antibiotic production and/or morphological differentiation are controlled in some Streptomyces species by low-molecular-weight compounds called butyrolactone autoregulators (21). Their effectiveness at extremely low concentrations, as well as the presence in these species of specific receptor proteins, implies that they should be regarded as Streptomyces hormones. To date, 10 butyrolactone autoregulators have been isolated and their structures have been elucidated chemically (25). Virginiae butanolide (VB) (11, 19, 24) and the corresponding receptor protein (BarA) (14) of Streptomyces virginiae have been among the most frequently studied. In S. virginiae, the VB-BarA system regulates the coordinate production of two structurally different compounds (15), virginiamycin M1 (VM1) and virginiamycin S (VS), a pair of antibiotics showing strong synergistic bactericidal activity.
In our previous in vitro (9) and in vivo (9, 13) analyses to clarify how the VB signal is transmitted into the cell to result, ultimately, in virginiamycin production, we demonstrated that the VB-specific receptor BarA is a DNA-binding protein acting as a transcriptional repressor; the binding of VB to DNA-bound BarA caused the dissociation of BarA from the promoter region of a target gene(s), enabling the transcription of the target gene(s) to occur. One of the target genes, designated barB, was located immediately downstream of the barA gene. However, transcriptional analysis suggested that barB and its downstream region were of a polycistronic nature, indicating that the barB downstream region also contains the target gene of BarA. To obtain clues to the overall signal-transmitting pathway governing virginiamycin production, the barB downstream region containing the plausible target gene of BarA was analyzed in detail in this study.
Strains, growth conditions, and plasmids.
S. virginiae (strain MAFF 10-06014; National Food Research Institute, Ministry of Agriculture, Forestry, and Fisheries, Tsukuba, Japan) was grown at 28°C as described previously (8, 24). Streptomyces strains were grown at 28°C in yeast extract-malt extract liquid medium for preparation of protoplasts (7), in tryptic soy broth (Oxoid, Hampshire, United Kingdom) for preparation of plasmid DNA, on agar medium R5 (7) for spore formation, and on agar medium NE (12) for determination of sensitivity to several antibiotics. S. lividans TK21 (7) was used as a host for cloning with Streptomyces plasmid pIJ486 (23) or pIJ4083. S. lividans TK21, pIJ486, and pIJ4083 were kindly provided by D. A. Hopwood (John Innes Centre, Norwich, United Kingdom). DNA manipulations in Escherichia coli and Streptomyces were performed as described by Sambrook et al. (20) and Hopwood et al. (7), respectively.
Sequence of the varS gene.
To identify a gene cotranscribed with barB, 1.95 kbp of the barB downstream region was sequenced on both strands by the dideoxy chain termination method with a BcaBEST dideoxy sequencing kit (Takara Shuzo Co.) or a Thermo sequencing kit (Amersham Pharmacia Biotech, Tokyo, Japan) and an ALF DNA sequencer (Amersham Pharmacia Biotech, Tokyo, Japan) (Fig. 1). Frame analysis (3) of the nucleotide sequence revealed a 1,554-bp open reading frame (ORF) transcribed in the same direction as barB and flanked by a typical Shine-Dalgarno sequence (GGGAGG) 6 bp upstream of the TTG initiation codon and a perfectly matched inverted repeat sequence 10 bp downstream of the TGA stop codon. The inverted repeat sequence was judged to form a strong secondary structure (ΔG = −37.4 kcal/mol), as evident from the complete termination of further DNA sequencing. Only by using a minimized template of 100 bp on an M13 phage and an extension reaction with BcaBEST DNA polymerase at 65°C were we able to determine the nucleotide sequence of the corresponding region. The ORF started 161 bp downstream of the barB stop codon, and the intergenic region contained several pairs of hexanucleotides that resembled typical −10 sequences for Streptomyces promoters (4), suggesting that the ORF may be transcribed monocistronically in addition to the bicistronic transcription with barB (described in more detail below).
FIG. 1.
Nucleotide and deduced amino acid sequences for the varS locus. varS spans nucleotides 285 to 1838. The deduced amino acids are shown below the nucleotides as one-letter notations. The putative ribosome-binding site and the −35 and −10 sequences are underlined and marked as SD, −35, and −10, respectively. The transcriptional start site is boxed and marked as +1. Inverted repeat sequences in the varS 3′ region are indicated by arrows. The oligonucleotide used for primer extension analysis is indicated by a broken arrow. The 14 TMS of VarS are indicated by shading.
Characterization of the deduced ORF product.
From the nucleotide sequence, the ORF was deduced to encode a hydrophobic 518-amino-acid protein (Mr, 52,191) containing multiple potential transmembrane domains. Database searches revealed that the ORF product likely belongs to a superfamily of integral membrane proteins that act as drug resistance proteins by exporting toxic compounds from cells with the aid of transmembrane electrochemical gradients (data not shown). Very high homology (68 to 73% identity and 78 to 84% similarity) was observed with RifP (1) of Amycolatopsis mediterranei and Ptr (22) of S. pristinaespiralis, while moderate homology (31 to 37%) was observed with several proteins, such as ActVA.1 (5) of S. coelicolor, QacA (18) of Staphylococcus aureus, and TcmA (6) of S. glaucescens. All the homologous proteins were found to belong to family 1 of Paulsen et al. (16) and were classified as drug (resistance) transporters having 14 transmembrane segments (TMS). Because both the hydropathy plot and the sequence alignment (data not shown) indicated the probable presence of TMS in the ORF product, the ORF was named varS (for S. virginiae antibiotic resistance).
Transcriptional analysis of the varS gene.
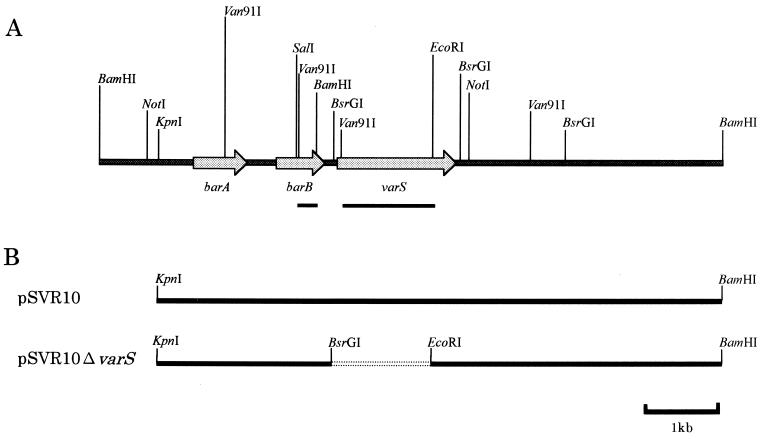
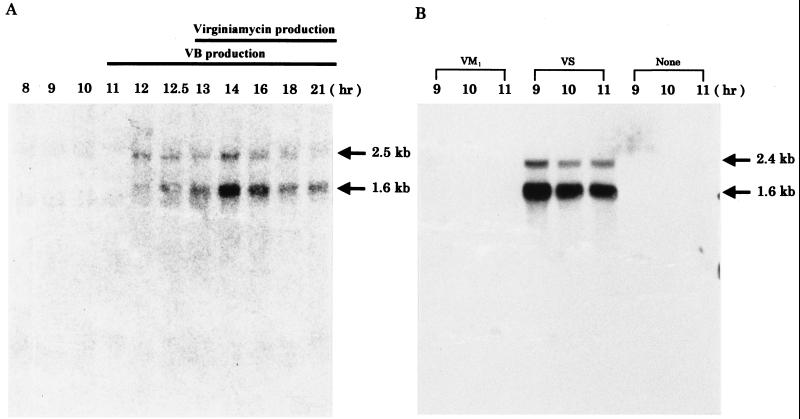
To elucidate the transcriptional pattern of varS, we carried out a Northern blot analysis by using a varS probe (Van91I-EcoRI fragment [Fig. 2A]) against mRNA samples collected from an 8- to 21-h culture of S. virginiae by the method of Kirby et al. (10) with modifications by Hopwood et al. (7) (Fig. 3A). Two different varS transcripts (1.6 and 2.5 kb) were detected at 12 h of cultivation, 1 h before the production of virginiamycin. As previously reported (9), a barB probe (SalI-BamHI fragment [Fig. 2A]) also hybridized to the large varS transcript (data not shown), confirming that varS was cotranscribed with the upstream barB gene. This fact indicates that both barB and varS are under the transcriptional control of the BarA-VB system. Because the presence of VB (at 11 h of cultivation [Fig. 3A]) leads to virginiamycin production in S. virginiae at 13 h of cultivation, although via a still-unknown pathway, the occurrence of varS transcription prior to virginiamycin production is rational if VarS participates in antibiotic resistance.
FIG. 2.
(A) Restriction map of an 8.2-kb BamHI fragment containing varS and the upstream and downstream regions. ORFs corresponding to barA, barB, and varS are indicated by shaded arrows. Probes (1.19-kb Van91I-EcoRI fragment for varS and 262-bp SalI-BamHI fragment for barB) used for Northern blot hybridization are indicated by filled boxes below the arrows. (B) Schematic representation of the inserts in pSVR10 and pSVR10ΔvarS used for the in vivo functional analysis of varS. Inserts were first constructed in pUC18, recovered as HindIII-XbaI fragments by use of the corresponding flanking restriction sites of pUC18, and then ligated into HindIII-XbaI-digested pIJ486. Broken lines indicate the deletion of varS.
FIG. 3.
Northern blot hybridization analysis of the varS transcripts during cultivation of S. virginiae (A) and for virginiamycin-induced mRNAs (B). (A) Total RNA was extracted from cells cultivated for the indicated times (hours) at 28°C. RNA (10 μg) was loaded in each lane, electrophoresed on a 1.2% agarose gel, and transferred to Hybond-N+ (Amersham Pharmacia Biotech) according to the manufacturer’s recommendations. Hybridization was carried out at 65°C for 20 h with the varS probe (Fig. 2A). VB and virginiamycin production under the experimental conditions started at 11 and 13 h of cultivation, respectively. (B) RNAs from cells without any addition or with either VM1 (10 μg/ml) or VS (10 μg/ml) added at 8 h and harvested at 9, 10, or 11 h of cultivation were analyzed. Probe and hybridization conditions were the same as those used for panel A.
In addition to the bicistronic transcription with barB, varS seemed to be transcribed independently from barB, as evident from the presence of the 1.6-kb transcript, which agreed well with the size of varS alone (1,554 bp).
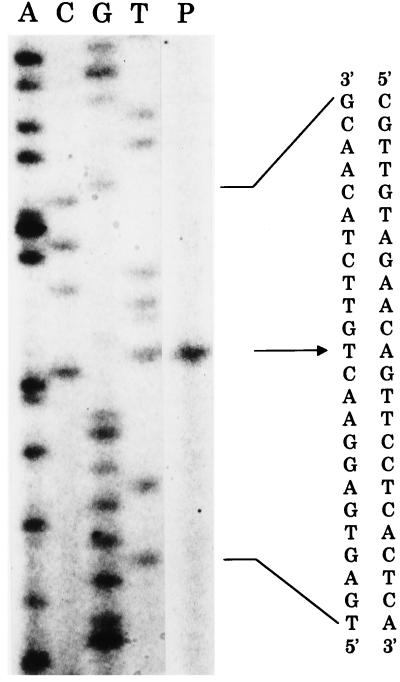
To confirm that varS has its own promoter, primer extension analysis was performed. A 26-mer primer (5′-GCGCCTGGGGTTGCGGGCCGGTACAG-3′) complementary to positions +54 to +28 relative to the putative varS start codon was 5′ end labeled with [γ-32P]ATP and hybridized with RNA from a 14-h culture. The hybrid was extended with reverse transcriptase as described by Sambrook et al. (20). The extended product suggested that varS has a single transcriptional start site at an A situated 29 bp upstream from the TTG initiation codon (Fig. 4). Furthermore, the presence of a functional promoter was confirmed in S. lividans with the aid of promoter-probe vector pIJ4083 (data not shown). The transcriptional start site was consistent with the presence of typical −35 (TTGTAC) and −10 (TACGTT) sequences that showed a high degree of similarity to the consensus sequence of the Streptomyces G2 promoter (4). However, the presence of the promoter raised the possibility that an additional mechanism regulates the monocistronic promoter. For the barB-varS bicistronic operon, BarA bound to the barB promoter sequence and repressed transcription (9). For the varS promoter region, no BarA-binding sequence was present, nor was any binding of BarA detected by surface plasmon resonance analysis (unpublished data), suggesting that a factor(s) other than BarA is involved.
FIG. 4.
Primer extension analysis for the varS transcriptional start site. The primer extension reaction was carried out with total RNA prepared from a 14-h culture of S. virginiae. Lanes A, C, G, and T, DNA sequencing ladder obtained with the same primer; lane P, primer extension reaction. The varS transcriptional start site is indicated by an arrow.
Because VarS seemed to be involved in antibiotic resistance, we used Northern blot analysis to investigate the possibility that either VM1 or VS influences the synthesis of the monocistronic transcript (Fig. 3B). RNA was prepared from cells with added VM1 or VS at 8 h of culturing, at which time neither internal virginiamycin nor internal VB was present. The 1.6-kb monocistronic varS transcript was detected only in the RNA sample with added VS, indicating that VS, not VM1, induced the synthesis of the monocistronic varS transcript. A 2.4-kb transcript was also observed. Because the barB probe did not show any sign of the corresponding signal on the same membrane (data not shown), we conclude that the 2.4-kb transcript is a minor transcript covering varS and the downstream region, rather than a barB-varS bicistronic transcript, although the transcript sizes are similar (2.5 kb for the barB-varS transcript and 2.4 kb for the varS-downstream region transcript). Therefore, the large varS transcript in Fig. 3A, especially from 14 h of cultivation, should be considered to contain both the barB-varS bicistronic transcript and the varS-downstream region transcript.
In vivo functional analysis of the varS gene.
To confirm the function of VarS in vivo, we first attempted to introduce a 2.0-kbp BamHI-NotI fragment containing varS alone into S. lividans by using pIJ486. However, no S. lividans transformant harboring intact varS was obtained, suggesting that the overexpression of VarS is toxic to the cells, probably because of the very hydrophobic nature of the VarS protein. Next, a 7.5-kbp fragment containing both a 2.2-kbp fragment from the upstream region and a 3.8-kbp fragment from the region downstream of varS (pSVR10 [Fig. 2B]) was introduced into S. lividans TK21. Transformants were readily available, but the reason for this result is unknown. As a control, pSVR10ΔvarS lacking only varS was used. Both constructs were used to determine susceptibility to several antibiotics. S. lividans harboring pSVR10 was 16 times more resistant to VS (Table 1) than S. lividans harboring pSVR10ΔvarS, while no difference between the strains was observed with VM1, erythromycin, tylosin, gramicidin, polymyxin, streptomycin, kanamycin, gentamicin, rifampin, lincomycin, chloramphenicol, or tetracycline. Because VM1 is known to enhance synergistically the antibacterial activity of VS (2), the susceptibility of both strains to the synergistic mixture of VM1 plus VS (VM1/VS ratio, 7:3) was measured. S. lividans containing pSVR10 was 3.3 times more resistant than S. lividans harboring pSVR10ΔvarS (Table 1). These results, together with the VS-dependent increase of varS transcription, indicated that varS encodes a VS-specific resistance protein which presumably transports VS from S. virginiae cells.
TABLE 1.
Virginiamycin resistance conferred by varS in S. lividans TK21
| Plasmid | MICa (μg/ml) of:
|
||
|---|---|---|---|
| VM1 | VS | VM1 + VS | |
| pIJ486 | 40 | 5 | 3 |
| pSVR10ΔvarS | 40 | 5 | 3 |
| pSVR10 | 40 | 80 | 10 |
The MIC was determined as the concentration lethal for cell growth after incubation for 3 days at 28°C.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper has been submitted to the GenBank/DDBJ data bank under accession no. AB019519.
Acknowledgments
This study was supported in part by the Research for the Future Program of the Japan Society for the Promotion of Science.
REFERENCES
- 1.August P R, Tang L, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffmann D, Kim C G, Zhang X, Hutchinson C R, Floss H G. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 2.Barriere J C, Berthaud N, Beyer D, Dutka-Malen S, Paris J M, Desnottes J F. Recent developments in streptogramin research. Curr Pharm Design. 1998;4:155–180. [PubMed] [Google Scholar]
- 3.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Bourn W R, Babb B. Computer assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 1995;23:3696–3703. doi: 10.1093/nar/23.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caballero J L, Martinez E, Malpartida F, Hopwood D A. Organization and function of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol Gen Genet. 1991;230:401–412. doi: 10.1007/BF00280297. [DOI] [PubMed] [Google Scholar]
- 6.Guilfoile P G, Hutchinson C R. Sequence and transcriptional analysis of the Streptomyces glaucescens tcmAR tetracenomycin C resistance and repressor gene loci. J Bacteriol. 1992;174:3651–3658. doi: 10.1128/jb.174.11.3651-3658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopwood D A, Bibb M J, Chater K F, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 8.Kim H S, Nihira T, Tada H, Yanagimoto M, Yamada Y. Identification of binding protein of virginiae butanolide C, an autoregulator in virginiamycin production, from Streptomyces virginiae. J Antibiot. 1989;42:769–778. doi: 10.7164/antibiotics.42.769. [DOI] [PubMed] [Google Scholar]
- 9.Kinosita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J Bacteriol. 1997;179:6986–6993. doi: 10.1128/jb.179.22.6986-6993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirby T, Fox-Carter E, Guest M. Isolation of deoxyribonucleic acid and ribosomal ribonucleic acid from bacteria. Biochem J. 1967;104:258–262. doi: 10.1042/bj1040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo K, Higuchi Y, Sakuda S, Nihira T, Yamada Y. New virginiae butanolides from Streptomyces virginiae. J Antibiot. 1989;42:1873–1876. doi: 10.7164/antibiotics.42.1873. [DOI] [PubMed] [Google Scholar]
- 12.Murakami T, Anzai H, Imai S, Satoh A, Nagaoka K, Thompson C J. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning and characterization of the gene cluster. Mol Gen Genet. 1986;205:42–50. [Google Scholar]
- 13.Nakano H, Takehara E, Nihira T, Yamada Y. Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J Bacteriol. 1998;180:3317–3322. doi: 10.1128/jb.180.13.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto S, Nakajima K, Nihira T, Yamada Y. Virginiae butanolide binding protein from Streptomyces virginiae. J Biol Chem. 1995;270:12319–12326. doi: 10.1074/jbc.270.20.12319. [DOI] [PubMed] [Google Scholar]
- 15.Paris J M, Barriere J C, Smith C, Bost P E. The chemistry of pristinamycin. In: Lukacs G, Ohno M, editors. Recent progress in the chemical synthesis of antibiotics. Berlin, Germany: Springer-Verlag KG; 1990. pp. 183–248. [Google Scholar]
- 16.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson W R, Lipman D J. Imported tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2446. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 19.Sakuda S, Yamada Y. Stereochemistry of butyrolactone autoregulators from Streptomyces. Tetrahedron Lett. 1991;32:1817–1820. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Bioeng. 1989;68:170–173. [Google Scholar]
- 22.Veronique B, Bey K J, Folcher M, Tompson C J. Molecular characterization and transcriptional analysis of a multidrug resistance gene cloned from the pristinamycin-producing organism, Streptomyces pristinaespiralis. Mol Microbiol. 1995;17:989–999. doi: 10.1111/j.1365-2958.1995.mmi_17050989.x. [DOI] [PubMed] [Google Scholar]
- 23.Ward J M, Janssen G R, Kieser T, Bibb M J, Buttner M J, Bibb M J. Construction and characterisation of a series of multi-copy promotor-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 25.Yamada Y, Nihira T, Sakuda S. Butyrolactone autoregulators, inducers of virginiamycin in Streptomyces virginiae: their structures, biosynthesis, receptor proteins, and induction of virginiamycin biosynthesis. In: Strohl W R, editor. Biotechnology of antibiotics. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 63–79. [Google Scholar]