Hybrid-capture, targeted deep sequencing of lung cancer mutational burden in cell-free BAL fluid identifies more tumor-derived mutations with increased allele frequencies compared with plasma cell-free DNA.
Abstract
Genomic profiling of bronchoalveolar lavage (BAL) samples may be useful for tumor profiling and diagnosis in the clinic. Here, we compared tumor-derived mutations detected in BAL samples from subjects with non–small cell lung cancer (NSCLC) to those detected in matched plasma samples. Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) was used to genotype DNA purified from BAL, plasma, and tumor samples from patients with NSCLC. The characteristics of cell-free DNA (cfDNA) isolated from BAL fluid were first characterized to optimize the technical approach. Somatic mutations identified in tumor were then compared with those identified in BAL and plasma, and the potential of BAL cfDNA analysis to distinguish lung cancer patients from risk-matched controls was explored. In total, 200 biofluid and tumor samples from 38 cases and 21 controls undergoing BAL for lung cancer evaluation were profiled. More tumor variants were identified in BAL cfDNA than plasma cfDNA in all stages (P < 0.001) and in stage I to II disease only. Four of 21 controls harbored low levels of cancer-associated driver mutations in BAL cfDNA [mean variant allele frequency (VAF) = 0.5%], suggesting the presence of somatic mutations in nonmalignant airway cells. Finally, using a Random Forest model with leave-one-out cross-validation, an exploratory BAL genomic classifier identified lung cancer with 69% sensitivity and 100% specificity in this cohort and detected more cancers than BAL cytology. Detecting tumor-derived mutations by targeted sequencing of BAL cfDNA is technically feasible and appears to be more sensitive than plasma profiling. Further studies are required to define optimal diagnostic applications and clinical utility.
Significance:
Hybrid-capture, targeted deep sequencing of lung cancer mutational burden in cell-free BAL fluid identifies more tumor-derived mutations with increased allele frequencies compared with plasma cell-free DNA.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide and the majority of patients are diagnosed with metastatic disease that is generally incurable. Survival for lung cancer remains poor in large part due to detection at late stages (1–3). Genotyping is now standard of clinical care for treating lung cancer, but this can be difficult to perform due to issues with the amount and quality of sample acquired during clinical evaluation.
Therapies that target specific oncogenic alterations in lung cancers have been approved for clinical use (4, 5). Appropriate use of these therapies requires molecular profiling of lung tumor tissue. The diagnosis and genomic profiling of lung cancer is achieved by biopsy or surgical resection for patients with early-stage disease (localized or regional disease) or biopsy alone for those with advanced disease (widespread). Unfortunately, biopsy samples are difficult to obtain in a significant subset of patients and/or contain insufficient material for mutation profiling. As an example, for bronchoscopic biopsies of peripheral lung lesions, diagnostic yield is 50% (6, 7) and adequate molecular profiling of diagnostic biopsies ranges from 60% to 100% (8, 9). Patients are therefore frequently subjected to repeat sampling via multiple procedures, or clinicians are left to render treatment decisions with incomplete data.
Chest practitioners have routinely performed bronchoalveolar lavage (BAL) for decades at academic and community medical centers across the world with little change in practice and few new assays developed to increase utility (10). BAL is simple to perform, easily obtained, relatively inexpensive to process, and safely retrieved during bronchoscopy for lung cancer evaluation, but it has a poor yield for cancer diagnosis by cytology alone (11). Currently, there is no role for BAL fluid to molecularly profile lung cancer, despite evidence that it might provide complementary information to tissue sampling (7).
Genomic profiling from a routine blood draw is a convenient and promising approach to noninvasively profile lung cancer (12), yet its utility for detecting and profiling early tumors is limited by the low quantities of circulating tumor DNA (ctDNA) shed in patients with low tumor burden (13).
Studying proximal fluids such as pleural fluid, urine, ascites, or cerebrospinal fluid can enhance tumor detection since they are often enriched for molecular markers of the cancer of interest. Molecular analyses of biofluids to detect tumors and their associated mutation profiles is therefore an important and emerging approach for solid cancer evaluation (14–16). For lung cancer, new paradigms of biofluid analysis for genomic analysis could reduce patient morbidity, decrease costs of care, shorten time to treatment, and increase treatment efficacy.
We hypothesized that BAL fluid from patients with early-stage lung cancer is enriched for lung tumor–derived DNA when compared with blood. To explore this question, we utilized Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) to identify mutations in regions of the genome that are commonly altered in lung cancer (12, 17–19). We identified putative tumor mutations in BAL fluid and then compared these results to tumor tissue and plasma profiling. We demonstrate that BAL fluid obtained during routine bronchoscopy frequently contains lung tumor-derived mutant DNA and could add value to routine BAL cytology.
Materials and Methods
Patient enrollment
Our goal was to explore whether molecular profiling of BAL fluid might have utility for genotyping and detection of lung cancer by utilizing the CAPP-Seq platform (Supplementary Fig. S1). We performed routine bronchoscopy and phlebotomy in patients undergoing clinical evaluation for lung cancer to collect BAL and plasma samples from 2015 to 2017 at Stanford Health Care (Stanford, CA). We enrolled a separate cohort of patients without cancer who underwent lung cancer screening from 2016 to 2017 at Vanderbilt Health (Nashville, TN) to identify field cancerization and to then develop a BAL genome classifier. Additional details on sample collection, DNA extraction, library preparation, and statistical analyses of biofluids are provided in the Supplementary Methods.
Written informed consent was obtained from the patients in this study in accordance with ethical guidelines put forth by the Declaration of Helsinki at each participating center. Protocols were approved by the institutional review board at each center prior to initiation of the study.
CAPP-Seq analysis
Targeted capture and sequencing analysis of all samples was performed using CAPP-Seq (17). We employed a 302-kb CAPP-Seq selector targeting 771 noncontiguous regions of the human genome, spanning 276 genes (18). A maximum of 32 ng DNA was input into sequencing library preparation. For plasma and BAL fluid samples with less than 32 ng of isolated cell-free DNA (cfDNA), all the extracted cfDNA was used for library preparation, down to a minimum of 16 ng. Samples were sequenced using 2 × 100 or 2 × 150 reads on an Illumina HiSeq 2500 or 4000. Sequencing data were processed using a previously described bioinformatics pipeline (12, 17). SNVs and indels were genotyped in all samples (17).
Mutation identification pipeline
Tumor informed
Mutational profiling of primary tumor biopsy samples in 34 patients was performed in the Stanford pathology department using the Solid Tumor Actionable Mutation Panel (STAMP), a Clinical Laboratory Improvement Amendments (CLIA)–certified tumor genotyping assay (20). Variants that were identified in tumor tissue were then compared to the corresponding biofluids from these patients (18). We limited our analysis to genomic positions targeted by both STAMP and the lung cancer-focused CAPP-Seq selector. 62 genes (53.5 kb total) overlapped between the STAMP (150 total genes) and CAPP-Seq (276 total genes) panels. Overlapping positions were identified to generate patient-specific tumor variant lists after removing germline variants that were identified by sequencing germline DNA from plasma depleted whole blood. Sequencing results from corresponding plasma, BAL cfDNA and BAL cell pellets were then queried for the presence of single-nucleotide variants (SNV) and small structural variants (indels) that were identified in matched tumor tissue using our previously described Monte-Carlo–based ctDNA detection index (12, 14, 21, 22). We refer to variants detected in tumor and biofluids as tumor-derived.
Tumor-naïve
BAL is usually performed during a biopsy procedure or prior to surgical resection, so determining whether BAL mutation detection is useful without a priori knowledge of a tumor's molecular profile is relevant for clinical utility assessment. To study this question, we applied an adaptation of a previously described tumor-naïve genotyping strategy developed by our group to analyze BAL cfDNA (19). SNVs for patients with cancer and noncancer patients for all sequenced plasma cfDNA and BAL cfDNA were identified without using primary biopsy data after comparing to matched germline DNA as previously described (12, 17, 19).
BAL genomic classifier model development
We also performed an exploratory analysis to determine whether BAL fluid profiling might be useful for diagnosis of lung cancer. In order to classify benign controls versus lung cancer cases, the following set of features was defined to summarize the mutations identified in a BAL sample: (i) the mean variant allele frequency (VAF) across all the mutations identified (mVAF), (ii) the total number of mutations (n), (ii) the maximum allele frequency (mxVAF), (iv) the number of mutations identified that were observed in ≥1 cancer cases observed in the COSMIC database (CosmicGenomeScreens v85; nCOSMIC1), (v) the number of mutations identified that were observed in ≥1 lung cancer cases in the COSMIC database (CosmicGenomeScreens v85; nCOSMICL1), (vi) the number of mutations identified that were observed in ≥10 lung cancer cases observed in the COSMIC database (CosmicGenomeScreens v85; nCOSMICL10), (vii) the number of mutations in canonical lung cancer driver genes (lung_driver; ref. 23), (viii) the mean allele frequency of nonsynonymous mutations (ns mVAF), (ix) the number of nonsynonymous mutations (nns), (x) the fraction of mutations present in matched leukocyte DNA (nGp), and (xi) fraction of mutations with a control-compared empirical P ≤ 0.05 or nCOSMICL1 > 0 (nEp).
Using these 11 features a Random Forest model with n = 1,000 trees was then trained, and a classifier was generated through a leave-one-out cross-validation framework. An ROC curve and its AUC were used to summarize model performance. Diagnostic sensitivity and specificity were calculated per standard methods based on the derived ROC curves. R packages randomForest and pROC were used for the analysis. To evaluate individual feature importance for the proposed classifier, we used the ‘mean decrease in accuracy’ metric and summarized this metric across the models generated in the leave-one-out cross-validation. A comparison with BAL cytology was performed by AUC analysis, where BAL cytology was dichotomized as not diagnostic for cancer for atypical, suspicious or no malignancy detected and diagnostic for cancer if definitive malignant cells were reported.
Data availability
The data generated in this study are available within the article and its supplementary data files. Additional data generated not reported herein are available upon request from the corresponding author. Raw data for this study were generated at Stanford University and are available from the corresponding author upon request if permitted by the local institutional review board.
Results
Cohort
In total, we analyzed 200 samples from 59 participants that included 38 subjects with lung cancer and 21 high-risk controls without cancer (Supplementary Table S1). Controls were patients who had nodules detected on CT scan that ultimately were diagnosed as not having cancer (n = 11) or were undergoing lung cancer screening based on age and tobacco history and did not have cancer (n = 10). 34 patients with lung cancer had their primary tumor sequenced by STAMP (Supplementary methods). For each subject we profiled matching BAL fluid, plasma, and plasma depleted whole blood (PDWB) by CAPP-Seq (17, 19). Most BAL specimens from patients with cancer were adequate for library preparation and sequencing after extraction (35 of 38, 92%). All 21 control subjects at risk for lung cancer had adequate cfDNA isolated from BAL fluid and plasma to carry forward for sequencing.
Characteristics of BAL cellular DNA
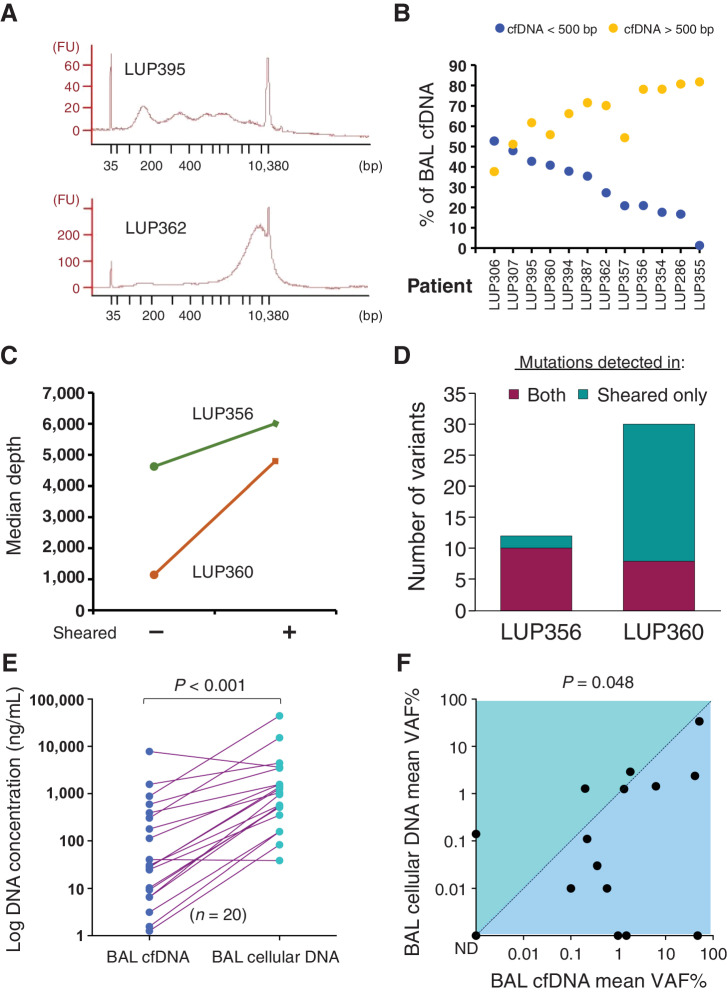
Because sequencing of BAL fluid is not well characterized in the literature (24–26), we assessed the quality of DNA and sequencing between the BAL cell pellet (BAL cellular DNA) and supernatant (BAL cfDNA). DNA fragments from BAL cfDNA were consistently larger in size when compared with plasma fragments that have a stereotyped size distribution with a mode fragment size of approximately 167 bp (Fig. 1A and B; ref. 27). We therefore sequenced BAL cfDNA before and after DNA shearing to understand the impact of fragment size on our ability to genotype tumor-derived mutations in BAL cfDNA. Sheared BAL cfDNA samples yielded more unique, “deduplicated” DNA reads and detected more mutations than unsheared BAL cfDNA (Fig. 1C and D). Based on these results, all subsequent BAL cfDNA samples analyzed were sheared before library preparation and sequencing.
Figure 1.
Characteristics of BAL DNA from lung cancer. A, cfDNA fragment size distribution for two representative BAL cfDNA samples. FU, fluorescence units. B, DNA fragment size for BAL cfDNA stratified by a 500-bp threshold (orange, >500 bp; blue, <500 bp) for n = 12 patients (x-axis). C, Median sequencing depth obtained from BAL cfDNA samples with and without shearing. D, Number of mutations detected in both sheared and unsheared samples or in sheared samples alone for the two patients from C. A total of 42 mutations were detected in sheared samples versus 18 in unsheared (P < 0.001). E, BAL DNA concentrations in 20 patients with lung cancer for BAL cfDNA and BAL cellular DNA. F, Comparison of mean VAF% for BAL cfDNA and BAL cellular DNA for 15 patients. ND, not detected. P is displayed above the plot.
In 20 patients with lung cancer, BAL cellular DNA concentrations ranged from 38.5 to 44,213 ng/mL (median, 1,007 ng/mL), which was 34 times higher compared with BAL cfDNA (range, 1.3–7,795 ng/mL; median, 29.3 ng/mL; P < 0.001; Fig. 1E). Despite more DNA being present in cellular BAL, the median VAF of tumor-derived mutations was significantly lower in BAL cellular DNA (0.11%, range, 0%–34%) compared with BAL cfDNA (0.99%, range, 0%–51%; n = 15; P = 0.048; Fig. 1F; Supplementary Table S2). Furthermore, 87% of BAL cfDNA samples contained tumor-derived DNA as determined by tumor-informed monitoring using a cut-off of P < 0.05 to identify variants compared with only 60% in cellular BAL. Finally, the mean VAF of all mutations detected in BAL cfDNA samples was higher in 11 of the 15 BAL samples that analyzed both BAL cfDNA and BAL cellular DNA. Taken together, these results suggest that BAL cfDNA best captures tumor-derived fragments, and we therefore focused on sequencing BAL cfDNA for our subsequent analyses.
Cell-free BAL fluid contains a higher concentration of tumor DNA than plasma
DNA was extracted from a median of 4.7 mL of cell-free BAL [interquartile range (IQR): 4.1–7.0 mL] and 4.0 mL plasma (IQR: 3.8–4.9 mL) from 38 cancer subjects to input for library preparation and sequencing (Supplementary Table S3). DNA quantity extracted from BAL cfDNA (P = 0.71) and plasma (P = 0.29) did not differ stratified by stage I/II versus III/IV lung cancer. Median deduplicated read depth per sample for BAL cfDNA and plasma cfDNA samples was 2,228 (IQR: 1,366–3,240) and 3,612 (IQR: 2,666–4,903) respectively (P < 0.001; Supplementary Fig. S2). Read depth did not differ by tumor stage for BAL cfDNA (P = 0.46) or plasma cfDNA (P = 0.30).
We identified a median of four mutations per tumor. Mutations affecting TP53 and KRAS were the most frequent alterations detected (41% and 35% of tumors respectively; Supplementary Table S4). Patient smoking status (ever vs. never) was associated with more tumor mutations (P = 0.015), but age (>65 years, P = 0.66), gender (P = 0.30) and stage (I/II vs. III/IV, P = 0.33) were not.
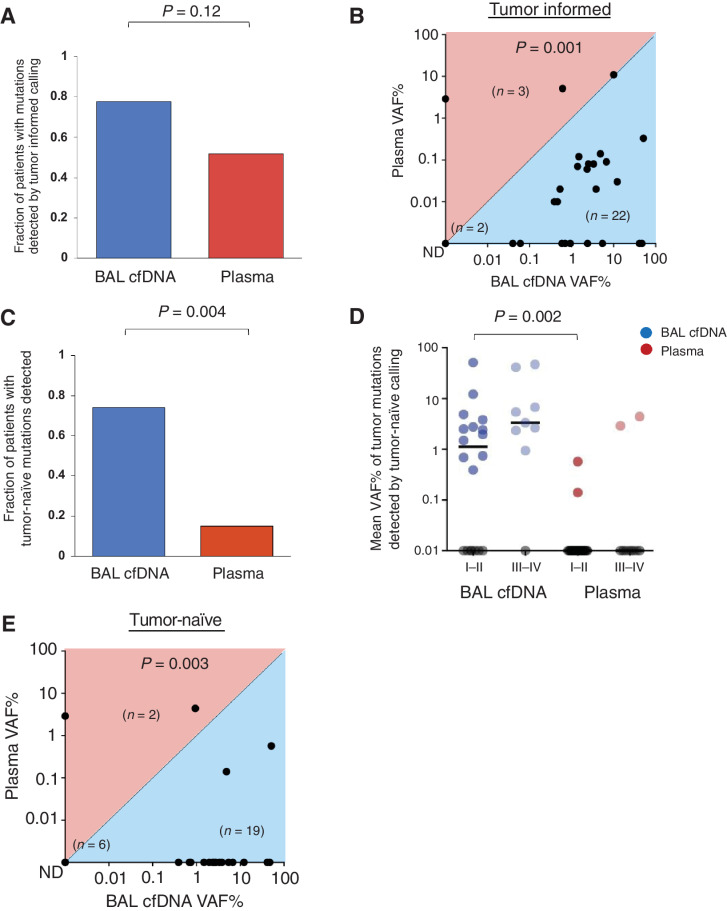
Using tumor-informed analysis (which leverages prior knowledge of somatic mutations from tumor tissue sequencing), we identified tumor-derived variants (SNVs and indels) in 81% of BAL cfDNA samples and 47% of plasma cfDNA samples (P = 0.016; Table 1). There was no significant association between stage and the number of tumor-derived mutations identified in BAL cfDNA (I–II vs. III–IV, P = 0.96; Supplementary Table S5). SNVs were identified in 21 of 27 BAL cfDNA samples (78%) compared with 14 of 27 plasma cfDNA samples (52%, P = 0.12; Fig. 2A; Supplementary Table S5). Furthermore, the tumor-derived mean VAF% of SNVs was higher in BAL cfDNA than for plasma cfDNA in 22 of 27 patients (P = 0.001; Fig. 2B).
Table 1.
Tumor mutation characteristics by fluid type.
| Variant statistic | Tumor, n = 34 | BAL cfDNA, n = 31 | Plasma, n = 34 |
|---|---|---|---|
| Tumor variants detecteda | 34 (100%) | 25 (81%) | 16 (47%) |
| Mean number of variants | 5.1 | 1.9 | 0.91 |
| Median number of variants | 3.5 | 1.0 | 0.0 |
| Mean VAF% | 12.9 | 6.6 | 2.5 |
| Median VAF% | 4.2 | 2.4 | 0.09 |
| Mean VAF%, drivers only | 17.4 | 9.0 | 3.3 |
| Median VAF%, drivers only | 8.9 | 2.4 | 0.08 |
aIncluding indels, see Supplementary Table S5; P < 0.05 level using a Monte-Carlo approach described in Materials and Methods.
Figure 2.
BAL identifies more tumor mutations than plasma. A, Fraction of patients with tumor-derived mutations identified in BAL cfDNA (blue) compared with plasma cfDNA (red) using the tumor-informed approach for 27 patients (Supplementary Table S5). B, Mean VAF% in BAL and plasma cfDNA using the tumor-informed approach to identify tumor-derived variants. Each dot represents one patient. Numbers of patients in each half of the plot are shown in parentheses within the plot along with the number of samples with no tumor-derived mutations detected. C, Driver mutations identified in BAL cfDNA (blue) compared with plasma cfDNA (red) using the tumor-naïve calling approach for 27 patients (Supplementary Table S7). D, Mean VAF% of mutant DNA detected in BAL cfDNA (blue) or plasma cfDNA (red) by stage using the tumor-naïve calling strategy (P = 0.002). Each dot represents one patient and samples with no mutations detected are shown on the x-axis in gray. E, Mean VAF% in BAL and plasma cfDNA using the tumor-naïve approach. Each dot represents one patient. Numbers of patients in each half of the plot are shown in parentheses within the plot along with the number of samples with no tumor-derived mutations detected. ND, not detected.
We then applied a tumor-naïve calling approach that would approximate a clinical scenario where tumor mutation data was not available, such as in the diagnostic setting (Supplementary Tables S6 and S7). We again observed that analysis of BAL cfDNA identified more tumor-associated variants than analysis of plasma (SNVs and indels, P = 0.004). Using the tumor-naïve approach, we detected at least one tumor-derived mutation in 20 of 27 (74%) patients using BAL cfDNA but only 4 of 27 (15%) patients using plasma cfDNA (Fig. 2C; Supplementary Fig. S3A). In addition, among early-stage patients BAL cfDNA harbored more variants (12/18, 67% vs. 2/18, 11%) with a higher median VAF% (0.74% vs. 0.0%; P = 0.002) than plasma (Fig. 2D). 19 of the 27 BAL samples with variants identified by tumor-naïve calling had higher mean VAF% in BAL cfDNA compared with plasma cfDNA (P = 0.003, Fig. 2E).
As expected, fewer tumor-derived mutations were detected in both BAL cfDNA and plasma cfDNA using a naïve calling strategy (n = 27 subjects) when compared with the informed approach. This is due to the fact that tumor-informed CAPP-Seq analysis decreases multiple hypothesis testing by only interrogating positions known to be mutant in the matching tumor and achieves an ≥10-fold lower detection limit (∼0.01% vs. ∼0.1%–0.5%; refs. 12, 17). However, due to the higher concentrations of tumor-derived DNA, analysis of BAL cfDNA identified more mutations than plasma cfDNA (P < 0.001; Supplementary Fig. S3B). Importantly, statistically significant decreases between the two approaches were noted for plasma cfDNA (P < 0.001) but not for BAL cfDNA. When focusing on cancer driver genes only, the proportion of mutations in these genes detected in BAL cfDNA was again higher compared with plasma cfDNA (P < 0.001) and not significantly different between the two approaches (Supplementary Fig. S3C).
Development of a diagnostic classifier from tumor-derived mutations in BAF
A common indication for bronchoscopy and BAL is for diagnosis of lung cancer in patients with lung nodules. Therefore, we wished to explore if BAL cfDNA analysis might aid in distinguishing patients with lung cancer from at-risk controls. One potential complication for such an approach is field cancerization, which refers to the acquisition of somatic mutations in morphologically normal appearing tissues (28). Because the lung is susceptible to field cancerization (29–31), we aimed to compare results of tumor-naïve BAL cfDNA analysis in 35 patients with cancer and 21 noncancer controls at risk for lung cancer who underwent bronchoscopy as part of research studies at two medical centers (Supplementary Methods).
The 21 non-lung cancer controls consisted of 7 subjects with benign nodules or masses who underwent bronchoscopy for lung cancer evaluation and 14 subjects that were current or ex-smokers from a lung cancer screening program, 4 of whom had lung nodules detected on their screening CT. Gender (P = 0.72), and smoking status (P = 0.53) were not significantly different between cases and controls with age showing a trend towards difference (P = 0.07). The concentration of cfDNA and sequencing depth did not differ in the two groups (P = 0.79 and P = 0.21 respectively; Supplementary Table S3). While the mean VAF% of detected mutations (Fig. 3A) and the frequency of lung cancer driver mutations (Fig. 3B) was significantly lower in both biofluids for at-risk controls compared with patients with cancer, mutations in lung cancer driver genes were identified in 4/21 (19%) of BAL cfDNA controls (Fig. 3C; Supplementary Table S8). A total of 38 driver mutations were detected in BAL cfDNA among the patients with lung cancer, 25 of which were also present in matched tumor tissue (concordance 66%) This indicates that the presence of mutations in cancer driver genes alone is insufficiently specific for distinguishing between controls and patients with lung cancer using BAL cfDNA.
Figure 3.
Tumor mutation profiling of BAL fluid in at risk controls identifies field cancerization. A, Mean VAF% (y-axis, log10 scale) for mutations detected in lung cancer patients compared with risk-matched controls for BAL cfDNA (P < 0.001) and plasma cfDNA (P = 0.002) by the tumor-naïve calling approach (Supplementary Table S8). Box plots depict median values, 25% to 75% IQR, and minimal and maximal values. The number of variants represented in the scatter plots are denoted in the x-axis labels. Blue, BAL cfDNA. Red, Plasma. P values are displayed above the plot. B, Fraction of patients with cancer and controls with driver mutations detected in BAL (blue) or plasma cfDNA (red). NS, not significant. C, Oncoprint of lung cancer driver genes in 27 lung cancer cases and 21 risk-matched controls for BAL cfDNA profiles (Supplementary Table S8). Tumor DNA results are shown for patients with cancer. Each column denotes one patient and each row a driver mutation. For mutations found in tumors, BAL cfDNA mutations are only shown if they are identical. Clinical characteristics [tumor histology, tumor stage according to American Joint Committee on Cancer (AJCC) VIII guidelines, smoking status, and age in years] are displayed at the top of the oncoprint. For diagnosis, a black box represents a cancer patient and a white box a noncancer patient. NSCLC, non–small cell lung cancer.
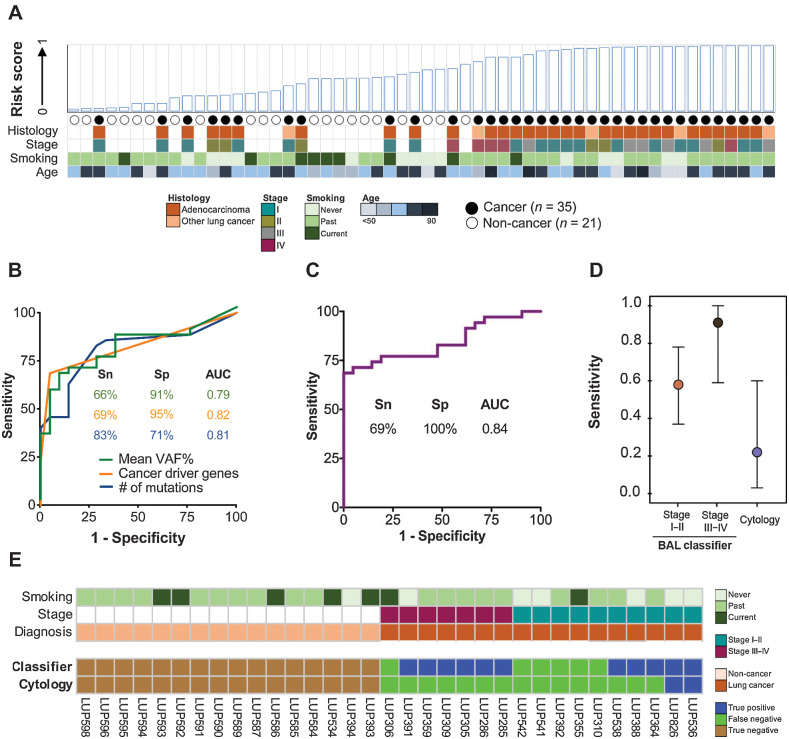
We therefore performed an exploratory analysis to test if it is possible to develop a machine learning-based classifier to distinguish between the two groups of patients. Specifically, we trained a multivariable BAL genomic classifier using a random forest model on 56 samples (35 cases and 21 controls) and eleven gene features (Supplementary Table S9). Performance was evaluated using leave-one-out cross-validation (Fig. 4A). The three features with the largest impact on model performance were mean VAF% of detected mutations, number of cancer driver mutations detected, and number of total single nucleotide variants detected (Fig. 4B; Supplementary Fig. S4A). The genomic classifier achieved an AUC of 0.84 with all 11 features incorporated (Fig. 4C), with 69% sensitivity at 100% specificity. Notably, the classifier outperformed BAL cytology (P = 0.001) for both early- and late-stage patients (Fig. 4D), and it was not associated with patient age, gender, and smoking status (Supplementary Fig. S4B). Of the 17 lung cancer cases profiled with BAL cytology, 2 (12%) were diagnosed with lung cancer by cytology, compared with 11 (65%) using the BAL genomic classifier (Fig. 4E).
Figure 4.
Derivation and performance of a BAL cfDNA genomic classifier for lung cancer diagnosis. A, BAL risk score based on 11 genomic features (Supplementary Table S9; see Materials and Methods). Case–control status and relevant clinicopathologic variables are indicated. NSCLC, non–small cell lung cancer. B, Individual performance of the top three features contributing to the BAL cfDNA classifier at the optimal cut point (Youden value). Mean VAF% (green line), cancer driver genes (orange line), and number of mutations (blue line). C, Performance of the BAL cfDNA classifier using all 11 features at the optimal cut-off point (Supplementary Table S9). D, Sensitivity of the BAL genomic classifier and BAL cytology at a specificity of 100% for diagnosing lung cancer stratified by stage. Point estimates are represented by a circle and 95% confidence intervals by whiskers. Analysis based on 17 patients with cancer and 16 controls who had both cytology and BAL genomics scores available. E, Patient-level comparison of BAL cytology and BAL genomic classifier risk scores using a 100% specificity threshold. Seventeen patients with cancer and 16 noncancer patients that had both cytology and a risk score assigned to them are arranged on the x-axis by stage and cancer status.
Discussion
Here, we compared BAL cfDNA and plasma cfDNA as two different sources of tumor derived DNA and found that BAL cfDNA analysis is more sensitive than plasma for identifying lung cancer-derived mutations. We also explored the potential of tumor-naïve BAL cfDNA analysis for detection of lung cancer. Our results suggest that BAL cfDNA analysis could have clinical utility for identification of mutations in lung cancer patients and, potentially, for the diagnosis of lung cancer.
Although previous studies have demonstrated the feasibility of tumor genotyping via BAL (25, 26, 32), we directly compared BAL with plasma cfDNA. We observed that tumor DNA concentrations were significantly higher in BAL than in plasma and more likely to be above our assay's detection limit. Using tumor-informed analysis, we found higher concentrations of tumor-derived DNA and an increased sensitivity for identifying tumor mutations in BAL cfDNA samples (17/22, 77%) compared with plasma cfDNA samples 10/22 (45%) in early stage I to II disease. Tumor-naïve analysis, which is clinically relevant in the diagnostic setting when tumor profiling is not available, also demonstrated higher ctDNA concentrations and better performance in BAL cfDNA (12/18, 67%) than plasma cfDNA in stage I to II disease (2/18, 11%). Only a subset of stage I to II lung cancers can be detected using ultrasensitive plasma cfDNA profiling methods (19, 33, 34). In addition, our study adds to an emerging literature that has demonstrated successful high throughput genome sequencing of proximal fluids in several cancer types including cerebrospinal fluid for gliomas (35–37), urine for genito-renal cancers (14, 38), and lavage or ascites for gynecologic cancers (39–41).
Several groups have demonstrated that gene expression profiling of histologically normal bronchial mucosal tissue can identify patients at high risk for developing lung cancer, due to a process called field cancerization (29, 30, 42). Genomic analysis of brushing specimens and single cell analysis has recently confirmed this effect at the DNA level (31, 43). Our data are consistent with these findings. Specifically, our observation of mutations in cancer driver genes in BAL cfDNA that were not found in the matching tumor specimens and were also detected in at-risk patients without lung cancer (Fig. 3C) supports the existence of field cancerization and less likely tumor heterogeneity. We speculate that these mutations reflect clonal mutagenesis of airway epithelial cells, analogous to the clonal mutations observed in tissues such as blood, esophagus, skin, and uterus (44–47). Finally, our exploratory analysis developing a BAL cfDNA-based classifier suggests that it may be possible to leverage field cancerization to diagnose lung cancer in patients with nondiagnostic bronchoscopic biopsies.
Strengths of our work include benchmarking of alterations identified in plasma cfDNA and BAL cfDNA to those present in matched tumor samples, sequencing of matched leukocytes to remove germline alterations and alterations arising due to clonal hematopoiesis, use of risk-matched controls to account for field cancerization, and utilization of a validated and ultrasensitive sequencing method to detect tumor derived mutations from cfDNA.
Our study has a number of limitations. First, we performed this work on patients enrolled in an observational study of BAL biomarkers without predefined assay criteria, since this was a proof-of-concept and method development study. Second, the majority of patients had adenocarcinoma and we therefore did not have sufficient power to examine the impact of tumor histology on the ability to detect tumor-derived variants in BAL. Third, we developed our BAL genomics classifier using a cross-validation framework, and therefore validation in an independent cohort will be required. Finally, collection protocols were not standardized between the two centers.
Before we can realistically consider BAL genomics for mutational profiling or detection of lung cancer in the clinic, further studies are required to enable a better understanding of how preanalytic variables influence tumor DNA identification in BAL. Standardization of collection methods will facilitate robust genomic profiling of BAL in larger cohorts of patients. In addition, it will be important to investigate alternative tumor DNA detection strategies such as whole-genome sequencing or DNA methylation analysis (34, 48, 49) and to compare these with our mutation-based approach. Finally, it will be informative to explore if features such as histology, grade, and location in the lung are associated with shedding of tumor DNA in BAL. These studies will more fully elucidate the clinical utility of BAL genomic profiling during lung cancer evaluation as a complementary liquid biopsy approach to blood-based analyses (50).
Supplementary Material
Acknowledgments
This work was supported with grants from the Canary Foundation (to V.S. Nair), the Stanford Department of Radiology (to V.S. Nair), The Fred Hutchinson Cancer Research Center Support Grant (grant no. P30CA0115704 to V.S. Nair), The Tobacco Related Disease Research Program (to V.S. Nair and M. Diehn), NCI (grant no. U01CA253166 to V.S. Nair; grant nos. R01CA188298 and R01CA254179 to M. Diehn and A.A. Alizadeh), the NIH Director's New Innovator Award Program (grant no. 1-DP2-CA186569 to M. Diehn), the Virginia and D.K. Ludwig Fund for Cancer Research (to M. Diehn and A.A. Alizadeh), and the CRK Faculty Scholar Fund (to M. Diehn). This article used the Genome Sequencing Service Center by the Stanford Center for Genomics and Personalized Medicine, supported by the grant award NIH S10OD020141. This paper is dedicated to the late Sanjiv “Sam” Gambhir and Pierre P. Massion, each of whom were remarkable mentors and visionaries in their fields. They made everyone around them better, and their loss leaves a void that will not be easily filled. The authors thank the patients who agreed to participate in this research.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
V.S. Nair reports grants from NIH NCI and grants from Roche Diagnostics during the conduct of the study. J.J. Chabon reports personal fees from Foresight Diagnostics outside the submitted work and patent filings related to cancer biomarkers and ownership interest in Foresight Diagnostics. B.Y. Nabet reports a patent for Immunmodulatory RNA pending and a patent for Liquid biopsy markers of immune checkpoint inhibitor outcomes pending; in addition, B.Y.Nabet is currently an employee and stock holder of Genentech/Roche. A.A. Chaudhuri reports personal fees and nonfinancial support from Roche; grants and personal fees from Tempus; other support from Geneoscopy, Droplet Biosciences; personal fees from NuProbe, Daiichi Sankyo, AstraZeneca, AlphaSights, Guidepoint, Dava Oncology; and other support from LiquidCell Dx outside the submitted work. N.S. Lui reports grants from Intuitive Foundation outside the submitted work. L.M. Backhus reports personal fees from Johnson & Johnson, Genentech and personal fees from Bristol-Myers Squibb outside the submitted work. A.A. Alizadeh reports nonfinancial support and other support from Roche and from Foresight Diagnostics during the conduct of the study; nonfinancial support and other support from Cibermed outside the submitted work; in addition, A.A. Alizadeh reports personal fees and other support from Roche, Gilead, Chugai; other support from Genentech, Celgene, Janssen, FortySeven, CiberMed Inc., Foresight Diagnostics, Pharmacyclics; and personal fees and other support from grants from BMS during the conduct of the study; in addition, A.A. Alizadeh has a patent 20210214437 issued, licensed, and with royalties paid from FortySeven; a patent 20210172022 pending and licensed to Foresight; a patent 20210033608 pending to MARIA; a patent 10167514 issued and licensed to CiberMed; a patent 10633450 issued; a patent 20190338364 pending to CiberMed; a patent 9605320 issued to Idiotype Vaccines; and a patent 20140296081 issued, licensed, and with royalties paid from Roche. M. Diehn reports grants and personal fees from AstraZeneca; personal fees and nonfinancial support from Illumina; grants and personal fees from Genentech; personal fees from Novartis, Gritstone Oncology, BioNTech, Boehringer Ingelheim, Roche Sequencing Solutions; other support from Foresight Diagnostics; and other support from CiberMed outside the submitted work; in addition, M. Diehn has a patent for ctDNA methods issued, licensed, and with royalties paid from Roche; a patent for ctDNA methods pending, licensed, and with royalties paid from Foresight Diagnostics; and a patent for single cell profiling methods issued, licensed, and with royalties paid from Celgene. No disclosures were reported by the other authors.
Authors’ Contributions
V.S. Nair: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. A.B.-Y. Hui: Data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.J. Chabon: Data curation, formal analysis, supervision, investigation, visualization, writing–original draft, writing–review and editing. M. Shahrokh Esfahani: Data curation, software, formal analysis, methodology, writing–original draft, writing–review and editing. H. Stehr: Data curation, software, formal analysis, methodology, writing–original draft, writing–review and editing. B.Y. Nabet: Data curation, software, formal analysis, visualization, methodology. L. Zhou: Data curation, methodology. A.A. Chaudhuri: Formal analysis, visualization, writing–original draft, writing–review and editing. J.A. Benson: Resources, data curation, writing–review and editing. K. Ayers: Resources, data curation. H. Bedi: Data curation, patient enrollment. M.C. Ramsey: Data curation, patient enrollment. R. Van Wert: Data curation, patient enrollment. S. Antic: Data curation, supervision, patient enrollment. N.S. Lui: Data curation, patient enrollment. L.M. Backhus: Data curation, patient enrollment. M.F. Berry: Data curation. A.W. Sung: Conceptualization, resources, data curation, patient enrollment. P.P. Massion: Resources, data curation, patient enrollment. J.B. Shrager: Resources, data curation, supervision, project administration, writing–review and editing. A.A. Alizadeh: Resources, data curation, supervision, project administration. M. Diehn: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P, et al. The international association for the study of lung cancer staging project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol 2009;4:792–801. [DOI] [PubMed] [Google Scholar]
- 2. Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE collaborative group. J Clin Oncol 2008;26:3552–9. [DOI] [PubMed] [Google Scholar]
- 3. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 6. Folch EE, Pritchett MA, Nead MA, Bowling MR, Murgu SD, Krimsky WS, et al. Electromagnetic navigation bronchoscopy for peripheral pulmonary lesions: one-year results of the prospective, multicenter NAVIGATE study. J Thorac Oncol 2019;14:445–58. [DOI] [PubMed] [Google Scholar]
- 7. Ost DE, Ernst A, Lei X, Kovitz KL, Benzaquen S, Diaz-Mendoza J, et al. Diagnostic yield and complications of bronchoscopy for peripheral lung lesions. results of the AQuIRE registry. Am J Respir Crit Care Med 2016;193:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lim JH, Kim MJ, Jeon SH, Park MH, Kim WY, Lee M, et al. The optimal sequence of bronchial brushing and washing for diagnosing peripheral lung cancer using non-guided flexible bronchoscopy. Sci Rep 2020;10:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tajarernmuang P, Ofiara L, Beaudoin S, Gonzalez AV. Bronchoscopic tissue yield for advanced molecular testing: are we getting enough? J Thorac Dis 2020;12:3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baughman RP. Technical aspects of bronchoalveolar lavage: recommendations for a standard procedure. Semin Respir Crit Care Med 2007;28:475–85. [DOI] [PubMed] [Google Scholar]
- 11. Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest 2013;143Suppl 5:e142S–e65S. [DOI] [PubMed] [Google Scholar]
- 12. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med 2011;3:109ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dudley JC, Schroers-Martin J, Lazzareschi DV, Shi WY, Chen SB, Esfahani MS, et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov 2019;9:500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Li L, Douville C, Cohen JD, Yen TT, Kinde I, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med 2018;10:eaap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Springer SU, Chen CH, Rodriguez Pena MDC, Li L, Douville C, Wang Y, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7:e32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020;580:245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang SR, Lin CY, Stehr H, Long SR, Kong CS, Berry GJ, et al. Comprehensive genomic profiling of malignant effusions in patients with metastatic lung adenocarcinoma. J Mol Diagn 2018;20:184–94. [DOI] [PubMed] [Google Scholar]
- 21. Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AF, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016;8:364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buttitta F, Felicioni L, Del Grammastro M, Filice G, Di Lorito A, Malatesta S, et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin Cancer Res 2013;19:691–8. [DOI] [PubMed] [Google Scholar]
- 25. Ryu JS, Lim JH, Lee MK, Lee SJ, Kim HJ, Kim MJ, et al. Feasibility of bronchial washing fluid-based approach to early-stage lung cancer diagnosis. Oncologist 2019;24:e603–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roncarati R, Lupini L, Miotto E, Saccenti E, Mascetti S, Morandi L, et al. Molecular testing on bronchial washings for the diagnosis and predictive assessment of lung cancer. Mol Oncol 2020;14:2163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. [DOI] [PubMed] [Google Scholar]
- 28. Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat Rev Cancer 2018;18:19–32. [DOI] [PubMed] [Google Scholar]
- 29. Kadara H, Fujimoto J, Yoo SY, Maki Y, Gower AC, Kabbout M, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst 2014;106:dju004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361–6. [DOI] [PubMed] [Google Scholar]
- 31. Kadara H, Sivakumar S, Jakubek Y, San Lucas FA, Lang W, McDowell T, et al. Driver mutations in normal airway epithelium elucidate spatiotemporal resolution of lung cancer. Am J Respir Crit Care Med 2019;200:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeng D, Wang C, Mu C, Su M, Mao J, Huang J, et al. Cell-free DNA from bronchoalveolar lavage fluid (BALF): a new liquid biopsy medium for identifying lung cancer. Ann Transl Med 2021;9:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol 2021;32:1167–77. [DOI] [PubMed] [Google Scholar]
- 35. De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martinez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015;6:8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, et al. Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol 2016;34:2404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panditharatna E, Kilburn LB, Aboian MS, Kambhampati M, Gordish-Dressman H, Magge SN, et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res 2018;24:5850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sin MLY, Mach KE, Sinha R, Wu F, Trivedi DR, Altobelli E, et al. Deep sequencing of urinary RNAs for bladder cancer molecular diagnostics. Clin Cancer Res 2017;23:3700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, Vergote I, et al. Lavage of the uterine cavity for molecular detection of mullerian duct carcinomas: a proof-of-concept study. J Clin Oncol 2015;33:4293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shah RH, Scott SN, Brannon AR, Levine DA, Lin O, Berger MF. Comprehensive mutation profiling by next-generation sequencing of effusion fluids from patients with high-grade serous ovarian carcinoma. Cancer cytopathology 2015;123:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, Schumacher CA, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med 2016;13:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med 2015;373:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshida K, Gowers KHC, Lee-Six H, Chandrasekharan DP, Coorens T, Maughan EF, et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 2020;578:266–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019;565:312–7. [DOI] [PubMed] [Google Scholar]
- 45. Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, et al. Somatic mutant clones colonize the human esophagus with age. Science 2018;362:911–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moore L, Leongamornlert D, Coorens THH, Sanders MA, Ellis P, Dentro SC, et al. The mutational landscape of normal human endometrial epithelium. Nature 2020;580:640–6. [DOI] [PubMed] [Google Scholar]
- 48. Zviran A, Schulman RC, Shah M, Hill STK, Deochand S, Khamnei CC, et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med 2020;26:1114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rolfo C, Mack P, Scagliotti GV, Aggarwal C, Arcila ME, Barlesi F, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the international association for the study of lung cancer. J Thorac Oncol 2021;16:1647–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Additional data generated not reported herein are available upon request from the corresponding author. Raw data for this study were generated at Stanford University and are available from the corresponding author upon request if permitted by the local institutional review board.



![Figure 3. Tumor mutation profiling of BAL fluid in at risk controls identifies field cancerization. A, Mean VAF% (y-axis, log10 scale) for mutations detected in lung cancer patients compared with risk-matched controls for BAL cfDNA (P < 0.001) and plasma cfDNA (P = 0.002) by the tumor-naïve calling approach (Supplementary Table S8). Box plots depict median values, 25% to 75% IQR, and minimal and maximal values. The number of variants represented in the scatter plots are denoted in the x-axis labels. Blue, BAL cfDNA. Red, Plasma. P values are displayed above the plot. B, Fraction of patients with cancer and controls with driver mutations detected in BAL (blue) or plasma cfDNA (red). NS, not significant. C, Oncoprint of lung cancer driver genes in 27 lung cancer cases and 21 risk-matched controls for BAL cfDNA profiles (Supplementary Table S8). Tumor DNA results are shown for patients with cancer. Each column denotes 1 patient and each row a driver mutation. For mutations found in tumors, BAL cfDNA mutations are only shown if they are identical. Clinical characteristics [tumor histology, tumor stage according to American Joint Committee on Cancer (AJCC) VIII guidelines, smoking status, and age in years] are displayed. NSCLC, non–small cell lung cancer.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/938e/9379362/55fc21225348/2838fig3.jpg)