With immunotherapies and inhibitors of metabolic enzymes in clinical development, the altered lipid metabolism and immune response in African-American men provides potential therapeutic opportunities to attenuate racial disparities in prostate cancer.
Abstract
African-American (AA) men are more likely to be diagnosed with and die from prostate cancer than European American (EA) men. Despite the central role of the androgen receptor (AR) transcription factor in prostate cancer, little is known about the contribution of epigenetics to observed racial disparities. We performed AR chromatin immunoprecipitation sequencing on primary prostate tumors from AA and EA men, finding that sites with greater AR binding intensity in AA relative to EA prostate cancer are enriched for lipid metabolism and immune response genes. Integration with transcriptomic and metabolomic data demonstrated coinciding upregulation of lipid metabolism gene expression and increased lipid levels in AA prostate cancer. In a metastatic prostate cancer cohort, upregulated lipid metabolism associated with poor prognosis. These findings offer the first insights into ancestry-specific differences in the prostate cancer AR cistrome. The data suggest a model whereby increased androgen signaling may contribute to higher levels of lipid metabolism, immune response, and cytokine signaling in AA prostate tumors. Given the association of upregulated lipogenesis with prostate cancer progression, our study provides a plausible biological explanation for the higher incidence and aggressiveness of prostate cancer observed in AA men.
Significance:
With immunotherapies and inhibitors of metabolic enzymes in clinical development, the altered lipid metabolism and immune response in African-American men provides potential therapeutic opportunities to attenuate racial disparities in prostate cancer.
Introduction
African-American (AA) men are more likely to be diagnosed with prostate cancer, to be diagnosed at a younger age, to have more aggressive disease, and to die from prostate cancer compared with European American (EA) men (1, 2). While socioeconomic and psychosocial factors contribute, a growing body of literature also supports biological differences underlying these disparities, including ancestry-specific genetic risk and somatic variants, RNA expression, tumor microenvironment, and androgen levels between AA and East Asian (EA) men (3–7). Comparatively less is known about the role of epigenetics.
Several studies suggest that aberrant DNA methylation, a repressive epigenetic mark, may contribute to prostate cancer aggressiveness in AA men (8–10). Transcription factors, proteins that bind specific DNA sequences to dynamically regulate gene transcription, are strongly implicated in prostate cancer development (11–14). We previously demonstrated that the androgen receptor (AR) cistrome is extensively reprogrammed in prostate cancer tumorigenesis and disease progression (11, 12). These findings provided important insights into events that drive normal prostate epithelium to transform into prostate cancer and established epigenetic reprogramming of the AR cistrome as central to prostate tumorigenesis. These data, however, were generated exclusively in samples from EA men. How or if the prostate cancer AR cistrome differs in AA men and whether this contributes to observed racial disparities is not known.
In this study, we present the first description of the AR cistrome in primary prostate cancer from AA men. Our data suggest that differential AR binding in AA and EA prostate cancer may contribute to distinct transcriptional programs, including biological processes known to be dysregulated in prostate tumors in AA men, such as lipid metabolism, immune response, and cytokine signaling.
Materials and Methods
Tissue cohort
Fresh-frozen radical prostatectomy specimens were selected from the Dan L Duncan Comprehensive Cancer Center Human Tissue Acquisition and Pathology Core at Baylor College of Medicine (Houston, TX) and the Dana-Farber Cancer Institute (Boston, MA) Gelb Center biobank. A genitourinary pathologist reviewed slides stained with hematoxylin and eosin from each case and isolated areas enriched for prostate tumor tissue (≥50% tumor cellularity) or normal prostate epithelium. 23 subjects were selected for chromatin immunoprecipitation sequencing (ChIP-seq) analysis. Informed consent was obtained from all subjects whose samples were included in the study. This study was approved by the Baylor College of Medicine and the Dana-Farber Cancer Institutional Review Boards (IRB).
ChIP-seq data generation and analysis
Using a 2-mm2 core needle, one core was extracted from frozen regulatory potential (RP) tissue blocks in the areas marked on the corresponding slide. Frozen cores were pulverized using the Covaris CryoPrep system. The tissue was then fixed using 2 mmol/L disuccinimidyl glutarate (DSG) for 10 minutes followed by 1% formaldehyde buffer for 10 minutes and quenched with glycine. Chromatin was sheared to 300 to 500 bp using the Covaris E220 ultrasonicator. The resulting chromatin was incubated overnight with 5 μg of antibody to AR (RB Anti-AR PAb, Spring Bioscience, REF: E2724, REF: 05300886001, LOT: 170118LVA) bound to protein A and protein G beads (Life Technologies). Five percent of the sample was not exposed to antibody and used as control. The samples were then de–cross-linked, treated with RNase and proteinase K, and DNA was extracted (Qiagen). DNA sequencing libraries were prepared using the ThruPLEX-FD Prep Kit (Rubicon Genomics). Libraries were sequenced on an Illumina HiSeq 4000 to generate 150-bp paired-end reads (Novogene).
ChIP-seq reads were aligned to the human genome build hg19 using the Burrows-Wheeler Aligner (BWA) version 0.7.15 (15). Nonuniquely mapped/redundant reads were discarded. MACS v2.1.1.20140616 was used for ChIP-seq peak calling (q < 0.01) and for the generation of the BigWig and BED files (16). ChIP-seq data quality was evaluated by a variety of measures, including total peak number, fraction of reads in peak (FRiP) score, number of high-confidence peaks, and percent of peak overlap with DNase I hypersensitive site peaks derived from the ENCODE project. ChIP-seq peaks were assessed for overlap with gene features and CpG islands using annotatr (17). IGV was used to visualize normalized ChIP-seq read counts at specific genomic loci (18).
Heatmap clustering, principal component analysis, and identification of subgroup-specific binding sites were performed using Mapmaker (https://bitbucket.org/cfce/mapmaker), a ChIP-seq analysis pipeline implemented with Snakemake (19). Read counts for each peak were normalized to the total number of mapped reads for each sample. Quantile normalization was applied to this matrix of normalized read counts. Using DESeq2 (20), tumor-specific peaks (T-ARBS) and normal tissue-specific peaks (N-ARBS) were identified at the indicated FDR-adjusted P and log2 fold change cut-offs (Padjusted < 0.01, log2 fold change > 1) in the AA prostate samples. The same DESeq2 comparison was applied to compare AA and EA prostate tumors (Padjusted < 0.01, log2 fold change > 1).
Unsupervised hierarchical clustering was performed based on Spearman correlation between samples. Principal component analysis was performed using the prcomp R function. Enriched de novo motifs in differential peaks were detected using HOMER version 4.11 (21). The top nonredundant motifs were ranked by adjusted P. The GREAT tool was used to assess for enrichment of and MSigDB perturbation annotations among genes near differential ChIP-seq peaks, assigning each peak to the nearest gene within 500 kb (22). CISTROME-GO (23) was used to assess the gene-level regulatory potential scores and enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in AA tumor-specific ARBS (compared with EA tumors). All differential peaks identified by DESeq2 were used as input (Padjusted < 0.01, log2-fold change > 1). A half-decay distance of 10.0 kb was set. Single-sample gene set enrichment analysis (ssGSEA; ref. 24) was applied to the quantile normalized matrix of AR ChIP-seq read counts to compute Hallmark (h) and KEGG (c2.cp.kegg) gene set enrichment in each individual AR ChIP-seq sample. Pathway-level z-scores were calculated and used for heatmap plotting.
RNA sequencing data generation and analysis
Tissue samples were obtained from the Human Tissue Acquisition and Pathology Core of the Dan L. Duncan Comprehensive Cancer Center at Baylor College of Medicine and were collected from fresh radical prostatectomy specimens after obtaining informed consent under an IRB-approved protocol. Cancer samples contained a minimum of 70% cancer and benign tissues were free of cancer on pathologic examination. RNAs were extracted using Qiagen DNA/RNA Mini kit according to manufacturer's instruction. The isolated total RNA was assessed for quantity and degradation on an RNA 6000 Nano chip ran on a 2100 BioAnalyzer (Agilent). RNAs with RIN number ≥7 were chosen for RNA sequencing (RNA-seq) analysis. Sequencing libraries are prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina, Inc). Briefly, rRNA was depleted from total RNA and the remaining RNA purified, fragmented appropriately, and primed for cDNA synthesis. Blunt-ended cDNA was generated after first and second strand synthesis. Adenylation of the 3′ blunt-ends was followed by adapter ligation prior to the enrichment of the cDNA fragments. Final library quality control was carried out by evaluating the fragment size on a DNA1000 chip ran on a 2100 BioAnalyzer (Agilent). The concentration of each library was determined by qPCR by the KAPA Library Quantification Kit for Next Generation Sequencing (KAPA Biosystems) prior to sequencing.
Libraries were normalized to 2 nmol/L in 10 mmol/L Tris-Cl, pH8.5 with 0.1% Tween 20 and then pooled evenly. The pooled libraries were denatured with 0.1N NaOH and diluted to 20 pmol/L. Cluster generation of the denatured libraries was performed according to the manufacturer's specifications (Illumina, Inc) utilizing the appropriate HiSeq paired-end cluster chemistry and flow cells. Libraries were clustered appropriately with a 1% PhiX spike-in. Sequencing-by-synthesis (SBS) was performed on a HiSeq2500 utilizing the appropriate chemistry with paired-end 101-bp reads. Sequence read data were processed and converted to FASTQ format for downstream analysis by Illumina BaseSpace analysis software, FASTQ Generation v1.0.0.
FASTQ files were processed using the VIPER workflow (25). Read alignment to human genome build hg19 was performed with STAR (26). Cufflinks was used to assemble transcript-level expression data from filtered alignments (27). Differential gene expression analysis was performed using DESeq2 (20). For downstream analyses (GSEA), the RNA-seq matrix was further trimmed mean of M-values (TMM)-normalized using the edgeR package.
RNA-seq/ChIP-seq correlation analysis
To check for concordance between RNA-seq and AR ChIP-seq data, GSEA was used to run the following two analyses (24):
First, the top 500 most genes with the highest DESeq2 log-fold change in AA versus EA prostate tumors were used as a custom reference gene set. The normalized AR ChIP-seq matrix of AA and EA prostate tumors was used as “expression dataset” input after assigning every peak to the gene with the closest transcriptional start site. Enrichment of AA differential expression genes in regions of AA-specific AR ChIP-seq signal was tested. Permutation analysis (1,000 permutations) was used to generate FDR-corrected P.
Second, the top 500 most differentially marked genomic regions based on DESeq2 log–fold change (AA versus EA prostate tumors) were used as a custom gene set, after being assigned to the gene with the closest transcriptional start site. The TMM-normalized RNA-seq matrix of AA and EA prostate tumors was used as “expression dataset” input. Enrichment of AA differentially marked AR ChIP-seq sites around AA differential expression genes was tested. Permutation analysis (1,000 permutations) was used to generate FDR-corrected P values.
Ancestry score calculation
Genetic ancestry was inferred using common polymorphisms called from off-target and on-target sequencing reads. Germline variant imputation was performed using the STITCH imputation software applied to AR ChIP-seq BAM files for the immunopreciptiation product (merged with the corresponding input control BAMs when available) across all tumor samples (28). This method leverages ultralow coverage read data together with the 1000 Genomes reference panel to infer probabilistic germline calls for the autosomal chromosomes. Analysis was restricted to variants with imputation INFO > 0.4 and variant allele frequency (VAF) > 0.01. Ancestry components were inferred for each individual by linear projection using publicly available weights computed by the SNPWEIGHTS software (29), which had been trained on European, AA, and EA individuals in the 1000 Genomes project (30). The projection was performed using the imputed dosages and the PLINK2--score function to compute the African ancestry component in each sample.
Tissue microarrays
Tissue microarrays (TMA) were previously described (31). Briefly, TMA were constructed from radical prostatectomy tissues from AA and EA patients operated on at the Michael E. DeBakey VA Medical Center between 1995 and 2013. Patients provided written informed consent for the use of tissues under an IRB-approved protocol. Areas of cancer and benign tissue were identified by pathologic examination and 1 mm cores of cancer and matched benign tissues from each prostatectomy were used to construct TMAs.
IHC
IHC for FAS was carried out on a Leica BOND III autostainer using online heat treatment with ER1 antigen retrieval solution (citrate pH 6.0) for 20 minutes. Primary antibody incubation was carried out using anti-FAS rabbit mAb (Cell Signaling Technology C20G5) at 1:50 dilution for 30 minutes. Detection was carried out using a Bond Polymer Refine Detection Kit (Leica) for 16 minutes followed by chromogen for 5 minutes. Counterstain was hematoxylin.
Metabolomic profiling
The study cohort has been described previously (32). Briefly, samples from the Dana-Farber Cancer Institute/Harvard Cancer Center SPORE Prostate Cancer Cohort were used. Fresh-frozen radical prostatectomy specimens from 124 patients were used, with matched normal prostate tissue for 105 out of the 124. Specimens were received fresh from the operating room, inked, sliced, formalin-fixed, paraffin embedded (FFPE), and embedded in optimal cutting temperature (OCT) compound and stored in liquid nitrogen. A total of 5-μm–thick sections cut from FFPE and OCT blocks were stained with hematoxylin and eosin and examined histologically. Gleason score was assigned based on a representative tumor focus in the corresponding tissue block.
For each individual in the study, approximately 1 mg of tissue was sent to Metabolon, Inc. Metabolon prepared the frozen tissue cores and serum samples for analysis using their proprietary solvent extraction method and internal standards were added to each sample for normalization and quality control. Additional details are described previously (33).
Metabolites with more than 50% missing values were excluded from the analysis. Data were further corrected for batch effects using median-scaling, normalized using the probabilistic quotient approach (34), and log2-transformed. Missing values were imputed using a knn-based approach (35). The preprocessed data included 273 metabolites measured in 124 samples (110 EA and 14 AA). For the differential analysis, only tumor samples from EA and AA patients were used. Differential metabolite abundances were estimated using a linear model (met ∼ Race), and the corresponding p-values were corrected for multiple testing using the Benjamini–Hochberg method (FDR < 0.2).
Clinical cohort
We used publicly available data on patients with metastatic prostate adenocarcinoma treated with AR signaling inhibitors (ARSI; ref. 36). We restricted the analysis to patients who had both clinical and transcriptomic data available (81 patients). We applied ssGSEA to normalized RNA-seq data and extracted sample-level enrichment scores for the Hallmark Fatty Acid Metabolism gene set. The cohort was then split into two groups: top quartile, and lower three quartiles based on ssGSEA enrichment scores. Overall survival (OS) and time to treatment failure (TTF) were compared between the two groups. For OS, patients who were alive were censored at the date of last follow-up. For TTF, patients who were alive without progression and were still on treatment with the same ARSI were censored at the date of last follow-up. The distributions of OS and TTF were estimated with the Kaplan–Meier method along with the corresponding HRs between the two groups, as well 95% confidence intervals (CI). All tests were two-tailed; statistical significance was defined as P < 0.05. Survival analyses were performed using the “survival” and “survminer” R packages.
Cell culture
LNCaP cells were cultured in RPMI (Gibco #11875-093) medium supplemented with 10% FBS (Gibco #10483) and 1% Pen-Strep. PCa2b Cells were cultured with BRFF-HPC1 media (Athenaes, catalog no. 0403) supplemented with 20% FBS (Gibco #10483) and 1% Pen-Strep. Cell lines were authenticated by short tandem repeat (STR) fingerprinting and routine PCR-based Mycoplasma testing was carried out using Mycoplasma detection kit, Alfa Aesar (VWR, catalog no. 10067-040).
qRT-PCR
For qRT-PCR experiment, 350K LNCaP cells /well were seeded in a 12-well plate with RPMI 1640 phenol-red free media supplemented with 10% charcoal stripped FBS and 1% Pen-Strep. For PCa2b cells, 350K cells/well were seeded in a 12-well plate in BRFF-HPC1 media with 20% FBS and 1 x Pen-Strep with fibronectin, collagen, and albumin coating. After 48 hours of seeding, cells were transferred to DMEM (Gibco #11965-092) + 0.1% FBS + 1 X Pen-Strep and treated after 24 hours of culturing cells in DMEM + 0.1% FBS + 1 X Pen-Strep. After 24 hours of seeding, cells were treated with 10 nmol/L of R1881 or EtOH (as vehicle control) and after 48 hours of seeding, a subset was treated with 20 μmol/L of enzalutamide. Cells were harvested 48 hours later and processed for RNA isolation using RNeasy Micro Kit (Qiagen #74004). cDNA was made from 500 ng of total RNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific #4368814). Gene expression of specific target genes were analyzed using the primers mentioned below and Power SYBR Green PCR Master Mix (Applied Biosystems #4367659) with Quantstudio3 Real-Time PCR System (Applied Biosystems). qPCR data were analyzed by the 2–∆∆Ct method using 18S rRNA as the reference transcript and gene expression of the treatment groups were represented as the fold change in comparison with the vehicle treated cells. Primer sequences were as follows: 18S (forward: ACCGCAGCTAGGAATAATGGA; reverse: GCCTCAGTTCCGAAAACCA), KLK3 (forward: ACCTGCACCCGGAGAGCT; reverse: TCACGGACAGGGTGAGGAAG), FASN (forward: TTCTACGGCTCCACGCTCTTCC; reverse: GAAGAGTCTTCGTCAGCCAGGA). To examine expression of FASN mRNA in patient tumor tissues, we carried out qRT-PCR (Q-RT-PCR on an Applied Biosystems StepOne; Life Technologies). Following total RNA extraction, cDNA was synthesized using an iScript cDNA Synthesis kit (Bio-Rad) with OligodT in a PTC-200 thermocycler (5 minutes at 25°C; 30 minutes at 42°C; 5 minutes at 85°C). FASN, and β-actin TaqMan probes (ABI) were utilized. PCR conditions were set using standard two-step manufacturer's protocol. Differences in mRNA levels were analyzed using the 2−ΔΔCT method normalized to β-actin expression. Each measurement point was repeated at least in duplicate.
Data availability
The ChIP-seq data for patient samples has been deposited in Gene Expression Omnibus (GEO; GSE181440 and GSE181441). The RNA-seq, and metabolomic data for patient samples that support the findings of this study are available upon request from the corresponding authors (M.L. Freedman and S. Kaochar) to comply with institutional ethics regulations to protect patient privacy. All requests for raw and analyzed data will be promptly reviewed to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Data Transfer Agreement.
Results
The AR cistrome is reprogrammed in prostate tumorigenesis in AA men
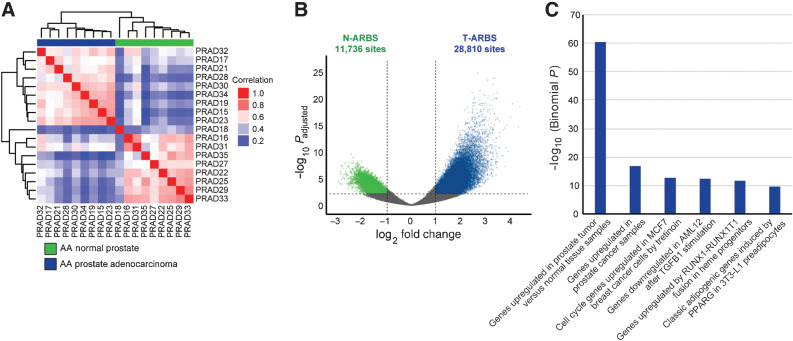
We generated and analyzed AR ChIP-seq data on 23 human prostate tissue specimens including 9 prostate cancer and histologically normal prostate samples from AA men and 5 prostate cancer samples from EA men (Materials and Methods; Supplementary Table S1). Self-reported ancestry was confirmed through genotyping (Supplementary Fig. S1). The AA prostate AR cistrome undergoes extensive reprogramming during tumorigenesis, similar to what we previously observed in EA men (Fig. 1A; ref. 11). As expected, AR binding sites (ARBS) with greater intensity across AA tumors relative to normal specimens were highly enriched for genes upregulated in prostate cancer (Fig. 1B and C).
Figure 1.
The AR cistrome is reprogrammed in prostate tumorigenesis in AA men. A, Unsupervised pairwise correlation of the AR cistromes from AA prostate tumor and normal specimens. Hierarchical clustering demonstrates the relatedness of each AR cistrome. B, Volcano plot of ARBS enriched in AA tumor versus normal specimens (T-ARBS; N = 28,810) and normal versus tumor specimens (N-ARBS; N = 11,736) with an FDR-adjusted P < 0.01 and log2-fold change > 1. C, MSigDB perturbation pathways enriched in the 28,810 T-ARBS using the GREAT tool (22).
Differences in the prostate cancer AR cistrome between AA and EA men associate with distinct RNA expression programs
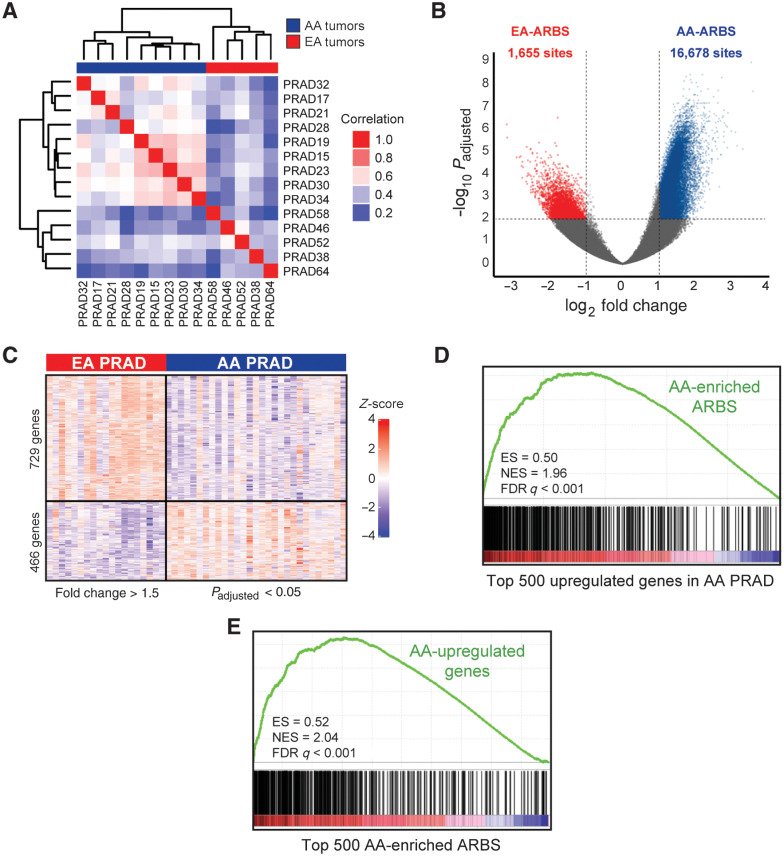
To interrogate whether tumor AR binding patterns differ by ancestry, we performed an unsupervised analysis of the prostate cancer AR cistromes, which clustered clearly into AA and EA groups (Fig. 2A). 118,467 and 115,584 ARBS were identified in AA and EA prostate cancer, respectively. Although the total number of ARBS was similar, 16,678 demonstrated significantly greater binding intensity in AA relative to EA prostate cancer (AA-ARBS), while only 1,655 demonstrated greater binding intensity in EA prostate cancer (EA-ARBS; Fig. 2B; Materials and Methods).
Figure 2.
Differences in the prostate cancer AR cistrome between AA and EA men associate with distinct RNA expression programs. A, Unsupervised pairwise correlation of the AR cistromes from AA and EA prostate tumors. Hierarchical clustering demonstrates the relatedness of each AR cistrome. B, Volcano plot of ancestry-enriched ARBS. The 16,678 ARBS were enriched in AA relative to EA (AA-ARBS) prostate tumors and 1,655 ARBS in EA relative to AA prostate tumors (EA-ARBS) with an FDR-adjusted P < 0.01 and log2-fold change > 1. C, Differential gene expression analysis of 30 AA and 19 EA paired tumor-normal prostate specimens. The 466 genes were upregulated in AA and 729 genes were upregulated in EA prostate tumors with an FDR-adjusted P < 0.05 and log2-fold change > 1.5. D, GSEA of AA-ARBS are enriched for genes upregulated in AA relative to EA prostate tumors (NES = 1.96; P < 0.001). E, GSEA of genes upregulated in AA prostate tumors are enriched for AA-ARBS (NES = 2.04; P < 0.001). ES, enrichment score.
We next sought to evaluate whether ancestry-enriched AR binding associates with differential RNA expression. We performed RNA-seq on an independent set of paired tumor-normal prostate specimens from 30 AA men and 19 EA men, identifying 466 genes upregulated and 729 downregulated in AA relative to EA prostate cancer (Fig. 2C; Supplementary Tables S2–S4). We observed a strong overall correlation between epigenomic and transcriptomic data. Using GSEA, transcripts nearest AA-ARBS were enriched for genes upregulated in AA relative to EA prostate cancer in the RNA-seq data [normalized enrichment score (NES) = 1.96; P < 0.001; Fig. 2D]. Likewise, genes with higher expression in AA than EA tumors were enriched for AA-ARBS (NES = 2.04; P < 0.001; Fig. 2E), suggesting that a significant portion of differential gene expression between AA and EA prostate cancer may be driven by ancestral differences in AR binding.
The AR prostate cancer cistrome associates with lipid metabolism and immune response
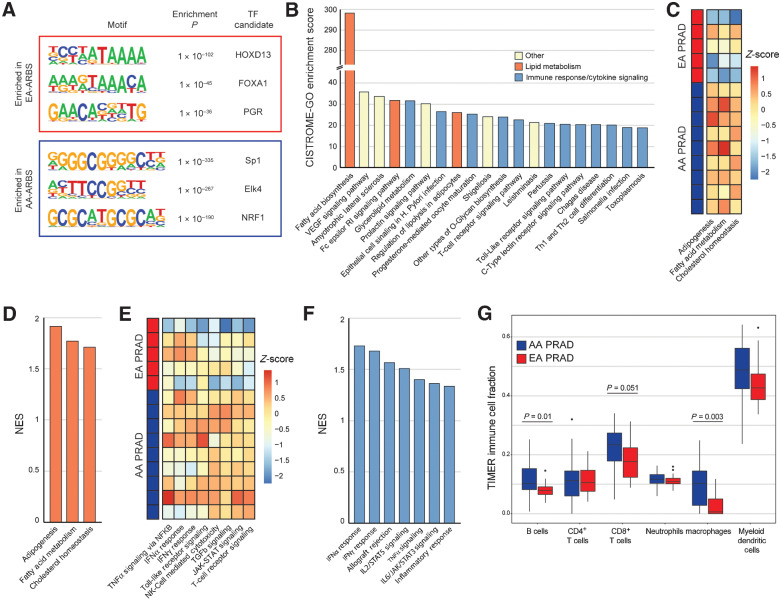
To investigate biological processes associated with ancestral differences in the prostate cancer AR cistromes, we first performed motif analysis on the EA-ARBS and AA-ARBS (Fig. 3A). Motifs for the transcription factors (TF) HOXD13, FOXA1, and progesterone receptor (PGR) were enriched in EA-ARBS. HOXB13, which shares a nearly identical motif with HOXD13, and FOXA1 are known to colocalize to ARBS in prostate cancer in EA men (11). Motifs for Sp1, Elk4, and NRF1 were enriched in AA-ARBS. Sp1 is a TF reported to colocalize with AR and regulate de novo lipogenesis and proliferation in prostate cancer cells (37, 38). NRF1 is a TF reported to be a coactivator of AR and regulates key metabolic genes to cellular growth (39). Elk4 is an ETS family TF that is highly expressed in a subset of prostate cancer and is involved in promoting cell growth (40).
Figure 3.
The AR prostate cancer cistrome associates with lipid metabolism, immune response, and cytokine signaling. A, Three most significantly enriched nucleotide motifs present in AA-ARBS and EA-ARBS by de novo motif analysis. B, Pathway enrichment of the AA-ARBS identified using CISTROME-GO (23). C and E, AR binding intensity in each AA and EA prostate tumor for Hallmark lipid metabolism (C) and immune response and cytokine signaling (E) gene sets using ssGSEA analysis (24). D and F, Differential expression analysis in our RNA-seq data identifies upregulation of Hallmark lipid metabolism (D) and immune response and cytokine signaling (F) gene sets in AA (n = 30) versus EA (n = 19) prostate tumors. G, Estimation of tumor infiltrate immune populations demonstrates greater signal for B cells (P = 0.01) and macrophages (P = 0.003), and a trend towards CD8+ T cells (P = 0.051) in AA versus EA prostate cancer (41). NK, natural killer.
We next analyzed the AA-ARBS using CISTROME-GO, a tool that performs functional enrichment analysis of TF ChIP-seq peaks (23). Three of the top 10 most enriched gene sets, including the top overall gene set, pertained to lipid metabolism (Fig. 3B). Using ssGSEA, we observed greater and more consistent AR binding intensity at KEGG lipid metabolism gene sets across AA compared with EA prostate cancer samples (Fig. 3C). Consistent with the strong global correlation between ancestry-enriched ARBS and RNA expression, we observed that greater AR binding intensity at lipid metabolism genes was associated with transcriptional upregulation of these pathways in our independent RNA-seq dataset (Fig. 3D; Supplementary Tables S3 and S4). Notably, immune response and cytokine signaling were also strongly represented in the list of pathways enriched in AA-ARBS, comprising 12 of the 20 top gene sets (Fig. 3B). ssGSEA demonstrated greater and more consistent AR binding intensity at KEGG immune gene sets across AA compared with EA prostate cancer samples (Fig. 3E). In our independent RNA-seq dataset, several immune response gene sets were significantly upregulated in AA versus EA prostate cancer (Fig. 3F). Further, applying TIMER2.0, which provides quantitative estimates of six tumor-infiltrating immune subsets, to our RNA-seq demonstrated a significantly higher signal for B cells (P = 0.01) and macrophages (P = 0.003), and a trend towards CD8+ T cells (P = 0.051; Fig. 3G; ref. 41). These findings are concordant with previously published differential gene expression analyses comparing AA and EA prostate tumors, most of which identified upregulation of lipid metabolism, immune response, and/or cytokine signaling gene sets in AA versus EA prostate cancer (4, 42–48). Our data implicate differential AR binding as a potential driver of these distinct transcriptional programs. Results of CISTROME-GO analysis of the EA-ARBS are shown in Supplementary Table S5.
Multiomic analysis demonstrates upregulated lipid metabolism in AA prostate cancer
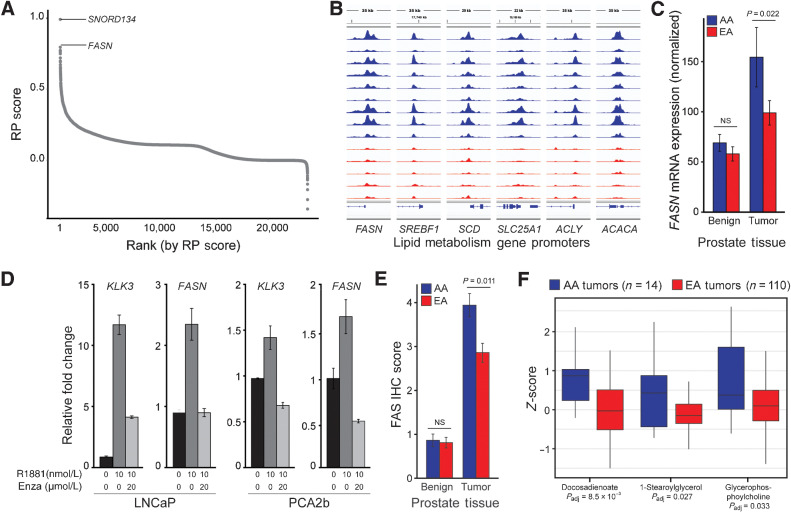
We next assessed differential AR binding at individual genes. Each gene was assigned a RP score: a quantitative value reflecting the likelihood that a set of TF ChIP-seq peaks are a direct regulator of a given gene (23). The gene with the second highest RP score in the AA-ARBS was FASN, which encodes fatty acid synthase (FAS), a critical catalytic enzyme in fatty acid synthesis whose expression is associated with aggressive prostate cancer (Fig. 4A; Supplementary Table S6; refs. 49–51). The gene with the highest RP score was SNORD134, an uncharacterized small nucleolar RNA embedded in intron 11 of the FASN gene. Visualization of AR binding at the FASN locus in the 14 prostate tumors clearly demonstrates greater binding intensity at the FASN promoter in prostate tumors from AA men (Fig 4B). Notably, several additional genes encoding key lipid metabolism proteins, including sterol regulatory element binding transcription factor 1 (SREBF1), stearoyl-CoA desaturase (SCD), citrate transport protein (SLC25A1), ATP citrate lyase (ACLY), and acetyl-CoA carboxylase alpha (ACACA) exhibited greater AR binding intensity at their gene promoters in AA versus EA prostate cancer (Fig. 4B).
Figure 4.
AR binding associates with FASN and other lipid metabolism genes, which are regulated by AR in vitro. A, Gene-level RP score in the 16,678 AA-ARBS identifies FASN as the gene with the greatest difference in AR binding intensity in AA versus EA prostate cancer (23). B, AR binding intensity is greater in AA than EA prostate tumors at the FASN promoter, as well as several other genes that encode key lipid metabolism enzymes. Each track depicts ChIP-seq AR binding intensity in each sample. C, Normalized FASN mRNA expression in paired normal prostate tissue and prostate cancer from 24 AA and 24 EA men. Error bars, SE. D, RNA expression for FASN and KLK3 in LNCaP cells and PCA2b cells treated with vehicle, R1881 for 72 hours, or R1881 72 hours and enzalutamide for 48 hours. Expression values for cells treated with R1881 or R1881 plus enzalutamide were relative to vehicle-treated cells (black). Error bars, SE. E, FAS protein expression in 492 prostate tissue specimens from AA (102 tumor and 112 normal) and EA men (150 tumor and 128 normal) demonstrating significantly higher FAS expression in AA than EA prostate tumors. Error bars, SE. F, Metabolomic analysis of 94 lipids in 14 AA and 110 EA prostate tumors identified lipids and fatty acids present at significantly higher levels in AA prostate tumors. Box plots are displayed with a median center line, box range from the 25th to 75th percentile and whiskers extending to the most extreme observation within 1.5 times the interquartile range. NS, not significant; Padj, Padjusted.
Consistent with published transcriptomic data and greater AR binding intensity at the FASN promoter in AA prostate cancer, an independent cohort of 48 samples demonstrated higher FASN expression in AA (n = 24) than EA tumors (n = 24; P = 0.022; Fig. 4C; ref. 4). FASN expression was higher in prostate cancer compared with normal prostate tissue in both ancestral groups. To evaluate the regulatory relationship between AR and FASN expression, we treated two cell lines–LNCaP and PCA2b, the only AA-derived prostate cancer cell line–with or without the synthetic androgen R1881. As expected, we observed R1881-mediated upregulation of KLK3, a canonical AR-regulated gene, in both cell lines (Fig. 4D). In response to androgen stimulation, FASN mRNA levels also increased in both cell lines. Subsequent treatment with the AR antagonist enzalutamide abrogated androgen-induced upregulation of both genes (Fig. 4D), confirming that AR regulates FASN gene expression in vitro.
Whether enhanced FASN expression translates into higher FAS protein expression in AA prostate cancer is not known. To characterize FAS protein levels, we performed IHC on an independent set of 492 prostate tissue samples, including 102 tumor and 112 normal specimens from AA men and 150 tumor and 128 normal specimens from EA men. We observed significantly greater FAS protein levels in tumors compared with normal specimens for both ancestral groups (Fig. 4E). Comparison of AA versus EA tumors demonstrated significantly higher FAS protein levels in AA prostate cancer (P = 0.0011). Notably, there was no difference in FASN RNA or FAS protein levels in normal prostate tissue from AA and EA men.
Based on results of this integrated analysis, we speculated that upregulation of FAS would translate to higher lipid levels in AA prostate cancer. We therefore performed metabolic profiling in an independent cohort of 14 AA and 110 EA prostate tumors. 65 (69%) of 94 lipids, including 26 (79%) of 33 fatty acids, demonstrated numerically higher levels in AA than EA prostate cancer; 9 lipids, including 4 fatty acids, were present at significantly higher levels (Fig. 4F). These data add to a growing literature demonstrating an altered lipid metabolic profile in AA prostate cancer (52, 53). This may be biologically and clinically relevant as prostate cancer cells upregulate de novo lipogenesis to support rapid cellular division, increased uptake of exogenous lipids is linked to prostate cancer aggressiveness, and high-fat diets and obesity are associated with prostate cancer incidence and progression (54).
Upregulated lipid metabolism is associated with aggressive prostate cancer
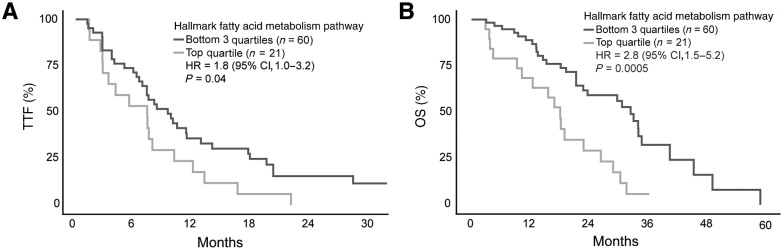
While previous studies have demonstrated an association between altered lipid metabolism and prostate cancer aggressiveness in preclinical models, its relation to clinical outcomes in men with prostate cancer is not well defined (55, 56). To investigate the clinical implications of upregulated lipogenesis, we analyzed publicly available transcriptomic and clinical data from men with metastatic prostate cancer. We applied ssGSEA to normalized RNA-seq data and extracted sample-level enrichment scores for the Hallmark Fatty Acid Metabolism gene set in 81 men with metastatic prostate cancer (36). Greater expression of the 160 genes in the Hallmark Fatty Acid Metabolism gene set was associated with significantly shorter progression-free survival to first-line ARSIs, abiraterone or enzalutamide, (HR = 1.8; 95% CI, 1.0–3.2; P = 0.04) and OS (HR = 2.8; 95% CI, 1.5–5.2; P = 0.0005; Fig. 5A and B). This is the first demonstration in a contemporary metastatic prostate cancer cohort that high expression of lipid metabolism genes is associated with poor response to ARSIs and shorter survival.
Figure 5.
Upregulated lipid metabolism is associated with worse prostate cancer outcomes. Kaplan–Meier survival curves for time to treatment failure (A) and OS (B) on abiraterone or enzalutamide for 81 men with metastatic castration-resistant prostate cancer based on lipid metabolism activity (36). Lipid metabolism scores were generated by applying ssGSEA to normalized RNA-seq data and extracting sample-level enrichment scores for the Hallmark Fatty Acid Metabolism gene set. Outcomes were compared between men in the top quartile versus the lower three quartiles based on ssGSEA enrichment scores.
Discussion
This study presents the first description of the AR cistrome in primary prostate cancer from AA men, resulting in important biological and clinical observations. There are three key findings of the present research. First, we demonstrate clear differences in the prostate cancer AR cistrome between AA and EA men. Second, our data suggest that these epigenetic differences may contribute to distinct RNA expression programs in AA and EA prostate cancer. The third key finding is that sites with greater AR binding intensity in AA prostate cancer are enriched for lipid metabolism genes with concomitant upregulation of lipogenic activity. Given the substantial association of upregulated lipid metabolism with prostate cancer aggressiveness, this provides a plausible role for differential AR binding in contribution to prostate cancer disparities.
Epigenetics provide a potential link between ancestry, environment, and cancer biology. The finding that approximately 75% of variation in DNA methylation is explained by genomic ancestry suggests that environmental factors not captured by ancestry also contribute to epigenomic variation (57). Indeed, compelling data supports that factors experienced differently across ancestral groups, such as diet, can modify the epigenome (58). The evolving understanding of epigenetics at the intersection of ancestry, environment, and biology highlights the importance of understanding epigenetic differences across ancestral groups for mitigating prostate cancer racial disparities.
The role of the AR in prostate cancer initiation and progression is well-established, yet how the prostate cancer AR cistrome differs across men of different ancestry is not known. We previously demonstrated that the AR cistrome is extensively reprogrammed in prostate cancer tumorigenesis and disease progression (11, 12). Further, differences in the AR cistrome across states generated novel mechanistic insights into events driving normal prostate cells to undergo malignant transformation. Reflective of a broader issue of underrepresentation of samples from minority populations in molecular cancer research, this data was generated exclusively in EA men. Herein, we report that the prostate cancer AR cistrome is also extensively reprogrammed in AA men. This finding has two important implications. First, it solidifies epigenetic reprogramming as central to human prostate tumorigenesis, irrespective of ancestry. Second, it provides a novel opportunity to identify differences in prostate cancer biology between AA and EA men. Comparative analyses across several molecular features have identified ancestry-specific genetic risk and somatic variants, and differences in RNA expression, tumor microenvironment, and androgen levels between AA and EA men (3–7). This paper provides the first insights into how differences in AR binding may contribute to observed racial disparities and identify novel therapeutic strategies to improve prostate cancer outcomes for AA men.
We observed that differences in AR binding between AA and EA men is associated with distinct RNA expression programs. Consistent with its known role as a direct regulator of gene transcription, we observed global upregulation of genes in AA prostate cancer near AA-ARBS as well as greater AR binding intensity near genes upregulated in AA prostate cancer. These results imply that a significant portion of differential gene expression between AA and EA prostate cancer may be driven by ancestral differences in AR binding. This novel finding provides the first suggestion that divergence in the AR cistrome, and likely other epigenetic features, may underlie differences in prostate cancer biology between AA and EA men. This idea is further supported by the observation that the top two biological processes enriched in AA-ARBS, lipid metabolism and immune response/cytokine signaling, are upregulated in AA versus EA prostate cancer in prior comparative transcriptomic analyses (4, 42–48). Notably, a recent study reported greater expression of genes involved in adipogenesis in EA than AA prostate cancer (43). However, the effect size was modest and this report comprised a single dataset. In comparison, we observed evidence of upregulated lipid metabolism in AA prostate cancer across independent epigenomic, transcriptomic, and metabolomic datasets.
Higher expression of lipid metabolism genes in our RNA-seq dataset and lipids and fatty acids in our metabolomic dataset in prostate cancer from AA versus EA men adds to the literature supporting an altered lipid metabolic profile in prostate cancer in AA men (4, 52, 53). Further, we report three novel findings that add to our understanding of differences in the lipid metabolism pathway in AA prostate cancer and its potential clinical implications. The first is that AA prostate cancer exhibits greater AR binding intensity at lipid metabolism genes. The role of AR in upregulating aberrant lipogenesis is supported by our in vitro experiments demonstrating a regulatory relationship between AR signaling and FASN expression in an AA prostate cancer cell line. The second discovery pertains to FASN, which encodes FAS, a critical catalytic enzyme in lipid metabolism that is associated with aggressive prostate cancer (49–51). FASN has been previously shown to be upregulated in prostate cancer in AA versus EA men, but whether this correlates with increased FAS protein expression was not known (4). We show for the first time that FAS protein levels are significantly elevated in prostate cancer in AA versus EA men. It's notable that while FASN RNA and FAS protein levels were significantly higher in AA than EA prostate cancer, there was no difference in either in normal prostate tissue between ancestral groups. The absence of difference in normal prostate tissue suggests that FAS upregulation in AA prostate cancer is likely independent of external factors, such as differences in diet, that may be experienced differently by AA and EA men.
The third observation in this study pertains to the clinical implications of upregulated lipid metabolism. While preclinical models demonstrate its association with aggressive prostate cancer, upregulated lipid metabolism is not known to impact clinical outcomes in men with prostate cancer (55, 56). Our data suggest that higher expression of lipid metabolism genes is associated with poor response to ARSIs and shorter survival in a contemporary metastatic prostate cancer cohort. This finding further strengthens the rationale for inhibiting lipid metabolism as a novel therapeutic approach in men with prostate cancer and suggests that AA men may be more likely to benefit from these drugs. We previously demonstrated that the selective FAS inhibitor IPI-9119 reduces tumor growth in castration-resistant prostate cancer (CRPC) preclinical models and human organoids (49). With drugs targeting FAS and other lipid metabolism enzymes in clinical development, we urge clinical trials to enroll diverse patient populations. In addition, correlative studies to identify biomarkers, such as FAS protein levels and/or lipid metabolism gene expression, will be critical to identify patients most likely to benefit from this treatment approach.
In addition to lipid metabolism, ARBS with greater intensity in prostate cancer in AA men demonstrated a strong enrichment for immune response and cytokine signaling genes. While epigenetic regulation of immune response is well-established, our data is the first to suggest that differences in the prostate cancer tumor microenvironment between AA and EA men may be driven in part by differential AR binding (59). This novel finding was concordant with greater expression of immune response and cytokine signaling gene sets in our independent RNA-seq dataset, which is consistent with results of several comparative transcriptomic analyses (4, 42–48). Provocative data suggests a potential clinical correlation of this differential immune response between AA and EA men with prostate cancer. Sipuleucel-T is an autologous cellular immunotherapy demonstrated to prolong survival in men with metastatic prostate cancer (60). Analysis of the PROCEED Registry demonstrated that AA men treated with Sipuleucel-T lived significantly longer than EA men (61). Our RNA-seq data also demonstrated greater intratumoral activity of macrophages, B cells, and CD8+ T-cells, all of which are involved in the immune response to Sipuleucel-T (62). The prostate tumor microenvironment may have clinical implications beyond response to immunotherapy. A recent report suggests that tumor-associated macrophages promote castration-resistance by contributing cholesterol for intratumoral androgen production (63). In light of our findings, further studies are warranted to explore the relationship between tumor microenvironment, lipid metabolism, and androgen signaling in prostate cancer across men of different ancestry.
While the present results strongly support the conclusions discussed herein, it is appropriate to recognize potential limitations. First, the small sample size of the AR ChIP-seq cohort is modest. In addition, due to limitations in tissue availability, we were unable to perform DNA-seq or RNA-seq on the same samples that underwent AR ChIP-seq analysis. This precluded our ability to correlate the AR ChIP-seq results with other molecular features that can alter AR signaling, such as ERG fusion status and CAG repeats, which are known to differ between AA and EA men. However, the results of the differential AR binding analysis identified biological processes corroborated by our independent RNA-seq and metabolomic cohorts, as well as published data, strongly supports the epigenetic findings. Another limitation is that AR was the only epigenetic feature evaluated in this study. Other TFs, such as FOXA1 and HOXB13, and histone modifications clearly play a role in prostate cancer development (13, 14). It will be important that future studies integrate comprehensive epigenetic profiling with RNA-seq and DNA sequencing data. Performing these analyses within the same samples will provide clarity on how genomic alterations influence epigenetics, the regulatory relationship between epigenetic features and RNA expression, and how these differences contribute to prostate cancer racial disparities.
In summary, our data suggest a model whereby differential androgen signaling may contribute to higher levels of lipid metabolism, immune response, and cytokine signaling in AA prostate tumors. Given the substantial association of upregulated lipogenesis with prostate cancer progression, our study provides a plausible biological explanation of the higher incidence and aggressiveness of prostate cancer observed in AA men (1, 2, 64, 65). With inhibitors of key lipid metabolism enzymes as well as immunotherapies in clinical development, our findings suggest a potential therapeutic opportunity to target and attenuate racial disparities in prostate cancer. Further exploration of these treatment approaches in preclinical AA prostate cancer models and enrollment of diverse patient populations in future clinical trials is warranted. In conclusion, this study offers the first insights into ancestry-specific differences in the prostate cancer AR cistrome. More broadly, our study demonstrates the utility of epigenomic approaches to gain insight into the biological differences underlying cancer disparities.
Supplementary Material
Acknowledgments
J.E. Berchuck. is supported by the Department of Defense (grant no. W81XWH-20-1-0118). S.C. Baca is supported by a Young Investigator Award from the American Society of Clinical Oncology and a fellowship from the PhRMA Foundation and the Kure It Cancer Research Foundation. D. Lowder and C. McColl are supported by the National Institute of General Medical Sciences of the NIH under award number T32GM136554 and T32GM008231. D. Lowder is also supported by the PhRMA Foundation. M. Loda is supported by the NIH (grant no. RO1CA131945), Department of Defense (grant nos. PC160357, PC180582, P50CA211024), and the Prostate Cancer Foundation. M.L. Freedman is supported by the Claudia Adams Barr Program for Innovative Cancer Research, the H.L. Snyder Medical Research Foundation, and the Cutler Family Fund for Prevention and Early Detection. N. Mitsiades, M. Ittmann, and S. Kaochar are supported by the Prostate Cancer Foundation Young Investigator and Challenge Awards, the NCI (grant no. U54-CA233223), and the Department of Defense (grant nos. PCRP IDA W81XWH-18-1-0288 and PCRP HDRA W81XWH2110253).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
J.E. Berchuck reports grants from Dana-Farber Cancer Institute and Department of Defense during the conduct of the study and personal fees from Digital Science Press and from Genome Medical outside the submitted work. D. Lowder reports grants from NIH during the conduct of the study. C. McColl reports grants from NIH during the conduct of the study. N. Mitsiades reports grants from NCI during the conduct of the study. M.L. Freedman reports other support from Precede Bio outside the scope of the submitted work. S. Kaochar reports grants from NCI, Prostate Cancer Foundation, Baylor College of Medicine, and Department of Defense during the conduct of the study; other support from Department of Defense and FGH BioTech outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
J.E. Berchuck: Conceptualization, data curation, formal analysis, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing. E. Adib: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. S. Abou Alaiwi: Conceptualization, data curation, formal analysis, investigation, writing–review and editing. A.K. Dash: Conceptualization, data curation, investigation. J.N. Shin: Conceptualization, data curation, investigation. D. Lowder: Conceptualization, data curation, investigation. C. McColl: Conceptualization, data curation, investigation. P. Castro: Resources, data curation, supervision. R. Carelli: Conceptualization, formal analysis, investigation. E. Benedetti: Conceptualization, formal analysis, investigation. J. Deng: Formal analysis, investigation. M. Robertson: Formal analysis. S.C. Baca: Formal analysis, supervision. C. Bell: Data curation, investigation. H.M. McClure: Investigation. T. El Zarif: Investigation. M.P. Davidsohn: Data curation, investigation. G. Lakshminarayanan: Data curation, investigation. K. Rizwan: Formal analysis, investigation. D.G. Skapura: Conceptualization, formal analysis, investigation. S.L. Grimm: Formal analysis. C.M. Davis: Formal analysis, investigation. E.A. Ehli: Formal analysis, investigation. K.M. Kelleher: Data curation. J.-H. Seo: Supervision, investigation. N. Mitsiades: Resources. C. Coarfa: Formal analysis. M.M. Pomerantz: Conceptualization, resources, supervision. M. Loda: Conceptualization, resources, data curation, supervision, methodology, writing–review and editing. M. Ittmann: Conceptualization, resources, data curation, supervision. M.L. Freedman: Conceptualization, resources, formal analysis, supervision, funding acquisition, methodology, writing–review and editing. S. Kaochar: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–review and editing.
References
- 1. Farkas A, Marcella S, Rhoads GG. Ethnic and racial differences in prostate cancer incidence and mortality. Ethn Dis 2000;10:69–75. [PubMed] [Google Scholar]
- 2. Fowler JE, Bigler SA, Bowman G, Kilambi NK. Race and cause specific survival with prostate cancer: influence of clinical stage, Gleason score, age and treatment. J Urol 2000;163:137–42. [DOI] [PubMed] [Google Scholar]
- 3. Mahal BA, Alshalalfa M, Kensler KH, Chowdhury-Paulino I, Kantoff P, Mucci LA, et al. Racial differences in genomic profiling of prostate cancer. N Engl J Med 2020;383:1083–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Powell IJ, Dyson G, Land S, Ruterbusch J, Bock CH, Lenk S, et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomarkers Prev 2013;22:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaston KE, Kim D, Singh S, Ford OH, Mohler JL. Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J Urol 2003;170:990–3. [DOI] [PubMed] [Google Scholar]
- 6. Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A 2006;103:14068–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, Chou A, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet 2021;53:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang B-D, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res 2010;16:3539–47. [DOI] [PubMed] [Google Scholar]
- 9. Devaney JM, Wang S, Furbert-Harris P, Apprey V, Ittmann M, Wang B-D, et al. Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men. Epigenetics 2015;10:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rubicz R, Zhao S, Geybels M, Wright JL, Kolb S, Klotzle B, et al. DNA methylation profiles in African American prostate cancer patients in relation to disease progression. Genomics 2019;111:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47:1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pomerantz MM, Qiu X, Zhu Y, Takeda DY, Pan W, Baca SC, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet 2020;52:790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parolia A, Cieslik M, Chu S-C, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019;571:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline Mutations in HOXB13 and Prostate-Cancer Risk. N Engl J Med 2012;366:141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cavalcante RG, Sartor MA. annotatr: genomic regions in context. Bioinformatics 2017;33:2381–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Köster J, Rahmann S. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics 2012;28:2520–2. [DOI] [PubMed] [Google Scholar]
- 20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S, Wan C, Zheng R, Fan J, Dong X, Meyer CA, et al. Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks. Nucleic Acids Res 2019;47:W206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cornwell M, Vangala M, Taing L, Herbert Z, Köster J, Li B, et al. VIPER: visualization pipeline for RNA-seq, a snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinf 2018;19:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davies RW, Flint J, Myers S, Mott R. Rapid genotype imputation from sequence without reference panels. Nat Genet 2016;48:965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C-Y, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics 2013;29:1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong M, Bierman Y, Pettaway C, Kittles R, Mims M, Jones J, et al. Comparative analysis of p16 expression among African American and European American prostate cancer patients. Prostate 2019;79:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Penney KL, Tyekucheva S, Rosenthal J, El Fandy H, Carelli R, Borgstein S, et al. Metabolomics of prostate cancer gleason score in tumor tissue and serum. Mol Cancer Res 2021;19:475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cacciatore S, Zadra G, Bango C, Penney KL, Tyekucheva S, Yanes O, et al. Metabolic profiling in formalin-fixed and paraffin-embedded prostate cancer tissues. Mol Cancer Res 2017;15:439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 2006;78:4281–90. [DOI] [PubMed] [Google Scholar]
- 35. Do KT, Wahl S, Raffler J, Molnos S, Laimighofer M, Adamski J, et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics 2018;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tewari AK, Yardimci GG, Shibata Y, Sheffield NC, Song L, Taylor BS, et al. Chromatin accessibility reveals insights into androgen receptor activation and transcriptional specificity. Genome Biol 2012;13:R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu S, Archer MC. Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int J Cancer 2010;126:416–25. [DOI] [PubMed] [Google Scholar]
- 39. Schultz MA, Hagan SS, Datta A, Zhang Y, Freeman ML, Sikka SC, et al. Nrf1 and Nrf2 transcription factors regulate androgen receptor transactivation in prostate cancer cells. PLoS One 2014;9:e87204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 2009;69:2734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2. 0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res 2008;68:927–36. [DOI] [PubMed] [Google Scholar]
- 43. Rayford W, Beksac AT, Alger J, Alshalalfa M, Ahmed M, Khan I, et al. Comparative analysis of 1152 African-American and European-American men with prostate cancer identifies distinct genomic and immunological differences. Commun Biol 2021;4:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kinseth MA, Jia Z, Rahmatpanah F, Sawyers A, Sutton M, Wang-Rodriguez J, et al. Expression differences between African American and Caucasian prostate cancer tissue reveals that stroma is the site of aggressive changes. Int J Cancer 2014;134:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hardiman G, Savage SJ, Hazard ES, Wilson RC, Courtney SM, Smith MT, et al. Systems analysis of the prostate transcriptome in African-American men compared with European-American men. Pharmacogenomics 2016;17:1129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rahmatpanah F, Robles GD, Lilly M, Keane T, Kumar V, Mercola D, et al. RNA expression differences in prostate tumors and tumor-adjacent stroma between Black and White Americans. Oncotarget 2021;12:1457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Awasthi S, Berglund A, Abraham-Miranda J, Rounbehler RJ, Kensler K, Serna A, et al. Comparative genomics reveals distinct immune-oncologic pathways in African American men with prostate cancer. Clin Cancer Res 2021;27:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yuan J, Kensler KH, Hu Z, Zhang Y, Zhang T, Jiang J, et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet 2020;16:e1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zadra G, Ribeiro CF, Chetta P, Ho Y, Cacciatore S, Gao X, et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc Natl Acad Sci U S A 2019;116:631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Epstein JI, Carmichael M, Partin AW. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology 1995;45:81–6. [DOI] [PubMed] [Google Scholar]
- 51. Bastos DC, Ribeiro CF, Ahearn T, Nascimento J, Pakula H, Clohessy J, et al. Genetic ablation of FASN attenuates the invasive potential of prostate cancer driven by Pten loss. J Pathol 2021;253:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou X, Mei H, Agee J, Brown T, Mao J. Racial differences in distribution of fatty acids in prostate cancer and benign prostatic tissues. Lipids Health Dis 2019;18:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Figiel S, Pinault M, Domingo I, Guimaraes C, Guibon R, Besson P, et al. Fatty acid profile in peri-prostatic adipose tissue and prostate cancer aggressiveness in African-Caribbean and Caucasian patients. Eur J Cancer 2018;91:107–15. [DOI] [PubMed] [Google Scholar]
- 54. Bader DA, McGuire SE. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat Rev Urol 2020;17:214–31. [DOI] [PubMed] [Google Scholar]
- 55. Lounis MA, Péant B, Leclerc-Desaulniers K, Ganguli D, Daneault C, Ruiz M, et al. Modulation of de novo lipogenesis improves response to enzalutamide treatment in prostate cancer. Cancers 2020;12:3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Han W, Gao S, Barrett D, Ahmed M, Han D, Macoska JA, et al. Reactivation of androgen receptor-regulated lipid biosynthesis drives the progression of castration-resistant prostate cancer. Oncogene 2018;37:710–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Galanter JM, Gignoux CR, Oh SS, Torgerson D, Pino-Yanes M, Thakur N, et al. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife 2017;6:e20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun 2018;9:3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Q, Cao X. Epigenetic regulation of the innate immune response to infection. Nat Rev Immunol 2019;19:417–32. [DOI] [PubMed] [Google Scholar]
- 60. Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 2010;363:411–22. [DOI] [PubMed] [Google Scholar]
- 61. Sartor O, Armstrong AJ, Ahaghotu C, McLeod DG, Cooperberg MR, Penson DF, et al. Survival of African-American and Caucasian men after sipuleucel-T immunotherapy: outcomes from the PROCEED registry. Prostate Cancer Prostatic Dis 2020;23:517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Madan RA, Antonarakis ES, Drake CG, Fong L, Yu EY, McNeel DG, et al. Putting the pieces together: completing the mechanism of action jigsaw for sipuleucel-T. J Natl Cancer Inst 2020;112:562–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. El-Kenawi A, Dominguez-Viqueira W, Liu M, Awasthi S, Abraham-Miranda J, Keske A, et al. Macrophage-derived cholesterol contributes to therapeutic resistance in prostate cancer. Cancer Res 2021;81:5477–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. De Piano M, Manuelli V, Zadra G, Otte J, Edqvist P-HD, Pontén F, et al. Lipogenic signalling modulates prostate cancer cell adhesion and migration via modification of Rho GTPases. Oncogene 2020;39:3666–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yue S, Li J, Lee S-Y, Lee HJ, Shao T, Song B, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab 2014;19:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq data for patient samples has been deposited in Gene Expression Omnibus (GEO; GSE181440 and GSE181441). The RNA-seq, and metabolomic data for patient samples that support the findings of this study are available upon request from the corresponding authors (M.L. Freedman and S. Kaochar) to comply with institutional ethics regulations to protect patient privacy. All requests for raw and analyzed data will be promptly reviewed to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Data Transfer Agreement.