Abstract
Kidney pathophysiology is often complex, nonlinear and heterogeneous, which limits the utility of hypothetical-deductive reasoning and linear, statistical approaches to diagnosis and treatment. Emerging evidence suggests that artificial intelligence (AI)-enabled decision support systems — that use algorithms based on learned examples —may have an important role in nephrology. Contemporary AI applications can accurately predict the onset of acute kidney injury before notable biochemical changes occur; identify modifiable risk factors for chronic kidney disease onset and progression; match or exceed human accuracy in recognizing renal tumors on imaging studies and may augment prognostication and decision-making following renal transplantation. Future AI applications have the potential to make real-time, continuous recommendations for discrete actions and yield the greatest probability of achieving optimal kidney health outcomes. Realizing the clinical integration of AI applications will require cooperative, multidisciplinary commitment to ensure algorithm fairness, overcome barriers to clinical implementation, and build an AI-competent workforce. AI-enabled decision support should preserve the preeminence of wisdom and augment rather than replace human decision-making by anchoring intuition with objective predictions and classifications, and should favor clinician intuition when it is honed by experience.
Introduction
The complex, nonlinear, and heterogeneous nature of kidney pathophysiology limits the utility of hypothetical-deductive reasoning and linear, statistical approaches for disease diagnosis and treatment. Many off the ~68,000 diagnostic codes in the 10th revision of the International Statistical Classification of Diseases (ICD) system represent manifestations of kidney disease or systemic diseases that interact with kidney function. Moreover, an individual patient may have a combination of diseases, each influenced by behavioral, social, environmental, and genetic determinants of health. Thus, the complexity of kidney disease can reache or exceed the limits of human cognition and additive modeling.
Artificial intelligence (AI)-enabled decision support has the potential to mitigate these challenges and improve clinical care and research in nephrology. Whereas traditional clinical decision support systems conform to rules, AI models learn from examples and may therefore more accurately identify complex processes, such as kidney disease.1, 2 However, high-level evidence that supports the efficacy of AI-enabled decision support in nephrology is scarce, and few AI models have been deployed in the clinical setting. Realizing the potential of AI to transform nephrology care and research requires a robust understanding of AI fundamentals, the promises and perils of algorithm fairness, barriers and solutions to its clinical implementation, and pathways toward the development of an AI-competent workforce. This Review endeavors to impart understanding of these elements by providing an overview of the state-of-the-art of AI-enabled decision support systems in nephrology.
Fundamentals of AI in health care
“[AI is] about making computers that can help us — that can do the things that humans can do but our current computers can’t.” — Dr. Yoshua Bengio, deep learning pioneer and recipient of the Turing Award (2018)3
Scientific foundation for AI algorithms in health care
In 1966, Warner Slack used a transistorized computer, called the Laboratory INstrument Computer (LINC), to conduct the very first direct patient interview using a computer,4 — in this instance, regarding the patients’ history of allergy-related conditions. Among the fifty patients interviewed, physicians did not detect any allergic condition that was missed by LINC, whereas LINC detected 22 conditions that were missed by a physician. Remarkably, among the 30 patients who expressed a preference in the approach used to obtain their medical history, 18 preferred the computer-based approach over the physician. This system is one of the earliest successful examples of AI in medicine.
Broadly, the types of challenges that AI research aims to address include knowledge reasoning, planning, machine learning, natural language processing, computer vision, and robotics. The LINC system developed by Slack and colleagues reflects early efforts in AI to capture and encode expert knowledge within computer systems — referred to as “expert systems.” However, advances in machine learning — the subfield of AI that uses algorithms to learn rules and relationships among variables derived directly from data — have largely displaced other AI subfields given the superior ability of machine learning to effectively solve challenges that previously required expert systems. Thus, modern use of the term “AI” is almost synonymous with machine learning.5 The ability of AI algorithms to learn rules and relationships directly from data has highlighted the potential of AI and data mining in health care. Similarly, deep learning —a subfield of machine learning in which computer systems learn to represent data by adjusting the weights of associations among input variables across a multi-layered hierarchy of nodes that together comprise an artificial neural network —has also demonstrated utility in the health-care space, most notably in enabling computer vision analyses of medical images. In the 1980s, the development of convolutional neural network (CNN) that mimicked human sight inspired substantial advances in computer vision that are now embedded in imaging applications in fields such as pathology, ophthalmology, cardiology, radiology and nephrology.6 For example, computer vision can survey X-rays and stratify their priority for radiologist review according to the severity of findings.7 Similarly, computer vision can prioritize specimen reviews by pathologists by identifying normal and abnormal cells and tissues.8
Although newer deep learning methods have proven to be particularly effective in many health-care contexts, they have several drawbacks. Specifically, they are most beneficial in the context of a high signal-to-noise ratio (that is, when the outcome can be consistently predicted by experts); they require a much larger sample size than traditional statistical approaches, and high-level evidence that supports their efficacy in the clinical setting is currently sparse.9 However, greater adoption of electronic health records (EHRs),10 genomic sequencing11 and wearable devices12 has led to broader availability of multimodal health-oriented data. Thus the application of AI approaches may become more feasible for in-hospital and community health-care settings, and offer opportunities to build a stronger evidence base for AI-enabled decision support.
The common tasks for which AI can be applied in health care fall into three broad categories: unsupervised, supervised, and reinforcement learning (Box 1). An unsupervised learning algorithm learns the underlying relationships between variables in a dataset. These relationships can be used to assign groups of observations to clusters, or to reduce high-dimensional data, such as genomic data for which there are hundreds or thousands of variables representing single cells or patients, to lower-dimensional representations. These algorithms are considered to be unsupervised as there is typically no “gold standard” against which the algorithms are trained or judged. By contrast, supervised learning involves the training of models to predict one or more outcomes using a set of predictor variables or features. The goal of supervised learning is to learn generalizable relationships between predictors and the outcome without overfitting the model to the training dataset. Although models that are flexible and highly parameterized can often better fit complex relationships than supervised learning approaches that are less flexible, they often underperform in health-care settings.13 A model that establishes excellent fit to complex relationships in training data by leveraging high parameterization (overfitting) may perform poorly when applied to another dataset in which the data are slightly different, as often occurs in health-care settings.
Box 1. Summary of common AI algorithms and applications in healthcare.
| Types of Health Data | Types of AI | Algorithms | Applications |
|---|---|---|---|
| Structured data - Demographics - Laboratory tests - Medications - Diagnoses - Procedures Unstructured data - Clinical notes - Waveform data - Images - Videos |
Unsupervised learning Supervised learning Reinforcement learning |
Generalized linear models Discriminant analysis Naive Bayes Support vector machine Decision trees Random forest Gradient boosting machines Neural networks - Convolutional neural networks - Recurrent neural networks |
Biomarker discovery Drug discovery Disease diagnosis - CheXNet - Diabetic retinopathy - Skin cancer - Breast cancer nodal metastasis Patient risk stratification Treatment recommendation systems |
Problems to which supervised learning approaches are applied are commonly divided into regression problems (with continuous outcomes) and classification problems (with binary or multinomial outcomes). In reality, however, many distinct combinations of these problems exist. As a consequence, problems to which supervised learning approaches are applied are often considered from the viewpoint of a loss function that quantifies the quality of a model’s predictions by calculating differences between model predictions and the ground truth.
Reinforcement learning is a process that can be used to learn optimal actions from data. The researcher defines rewards for desirable actions and costs for undesirable ones. The algorithm is designed to learn an optimal strategy that maximizes the overall rewards. In some settings, such as robotics, reinforcement learning often involves physical actions (for example, moving the robot) to learn the consequences of the action in real-time. However, the implementation of such actions is often impossible or unethical in health-care settings when the proposed action (or inaction) lacks equipoise. Thus, reinforcement learning in health care is often used to learn optimal actions from retrospective data alone.
Although generalized linear models continue to have a major role in tabular biomedical data (for example, sets of laboratory and vital sign values), modelling of unstructured data (for example, natural language or imaging data) particularly benefits from deep learning approaches, Hence, deep neural networks have emerged as the dominant mechanism by which unstructured data are modelled. In this approach, information from the variables being used to predict the outcome enters an input layer of neurons, whereupon it is assigned initial, arbitrary weights that change over time as the model learns associations between inputs and the outcome. The information moves forward through hidden layers to the final output layer that represents the outcome of interest. After calculating the loss function, weights are optimized with backpropagation to optimize the association between inputs and the outcome of interest. Such network architectures have been developed for specific types of data. For example, CNNs are useful for modelling imaging data, recurrent neural networks (and its variants) are useful for modelling sequential data, and generative adversarial neural networks are useful for modelling synthetic data.
Examples of AI algorithms in health care
Unsupervised, supervised, and reinforcement learning algorithms have been applied effectively in many health-care settings. For example, unsupervised hierarchical clustering has been used to discover patient behavioral and clinical trajectory patterns.14, 15 Supervised algorithms have demonstrated efficacy in estimating probabilities of researcher-defined outcomes such as physiologic deterioration, sepsis, diabetic retinopathy, and pathologic imaging findings,demonstrating the potential of this approach to augment clinical diagnostics and decision-making.16–22 Reinforcement learning approaches based on retrospective analyses have demonstrated the potential to facilitate medication and intravenous fluid dosing decisions.23–25 In resource-limited settings, AI systems may prove useful when alternative means of diagnosis are unavailable.26 In resource-rich settings, AI systems might help physicians to make diagnoses more quickly and miss fewer rare abnormalities.27
AI and the learning health system
AI-driven clinical interventions can be viewed through the lens of a learning health systems approach that uses cycles of learning to assess and improve care (Figure 1). To support health-care decisions, AI models must integrate within EHRs and prospectively generate patient data that can be used to assess the impact of the intervention to the health problem of interest (that is, a practice to data approach). Models that are trained on the gathered data must then be evaluated for their ability to generate accurate predictions and classifications according to a specific clinical context (that is, converting data to knowledge). Finally, accurate predictions should be linked to interventions that are tailored to the patient needs (that is, converting knowledge to practice). The efficacy of AI-driven interventions is evaluated through the analysis of prospective data and through clinical trials;28 these implementations often follow a continuous lifecycle (Figure 2).
Fig. 1: Artificial intelligence systems through the lens of a learning healthcare system.
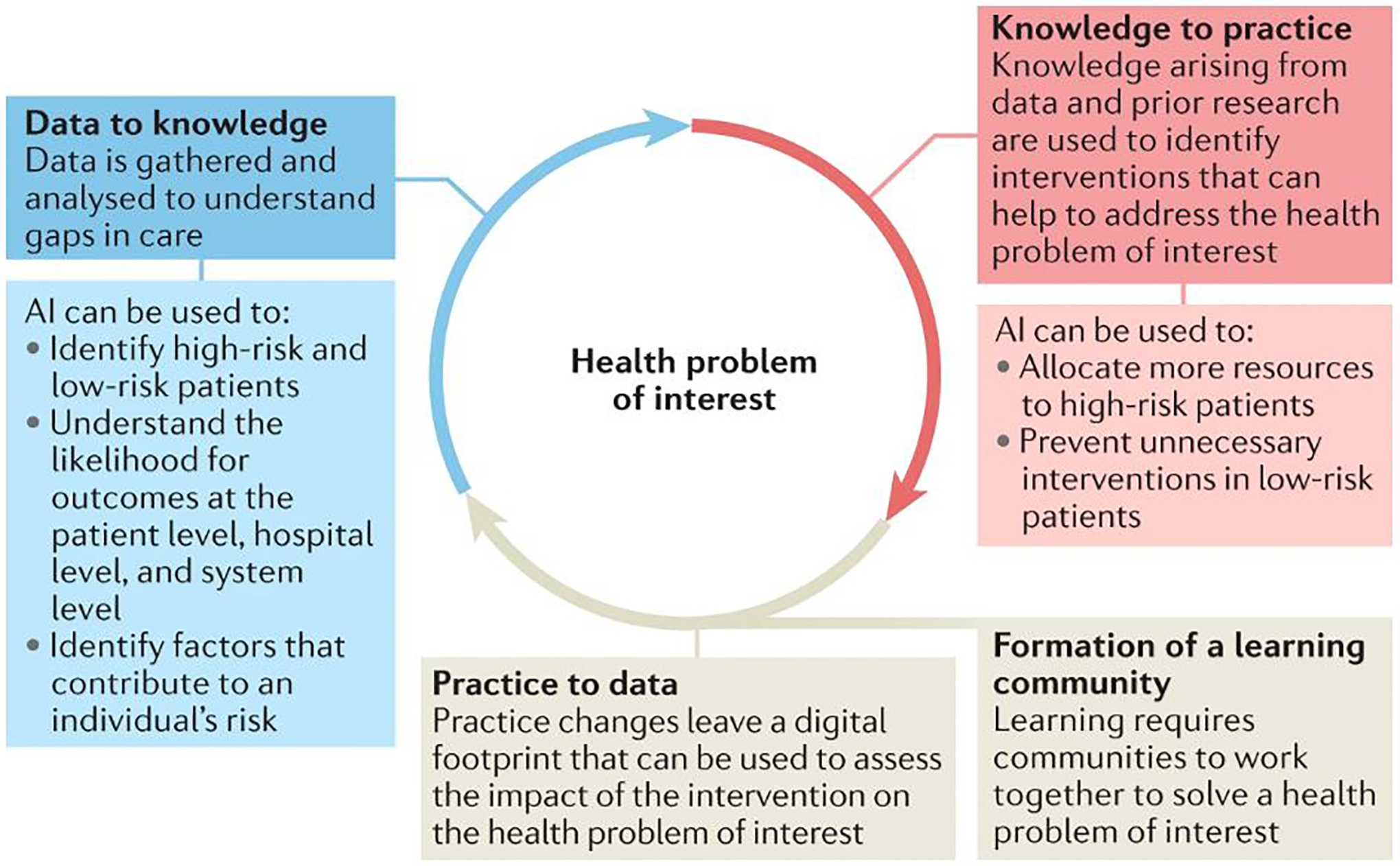
From: Artificial intelligence-enabled decision support in nephrology
The learning cycle framework describes how communities learn. Communities use data to generate locally relevant knowledge and use that knowledge to inform changes in practice. This process generates data that is used to assess changes in the quality of care.
Fig. 2: Lifecycle of artificial intelligence for decision support.
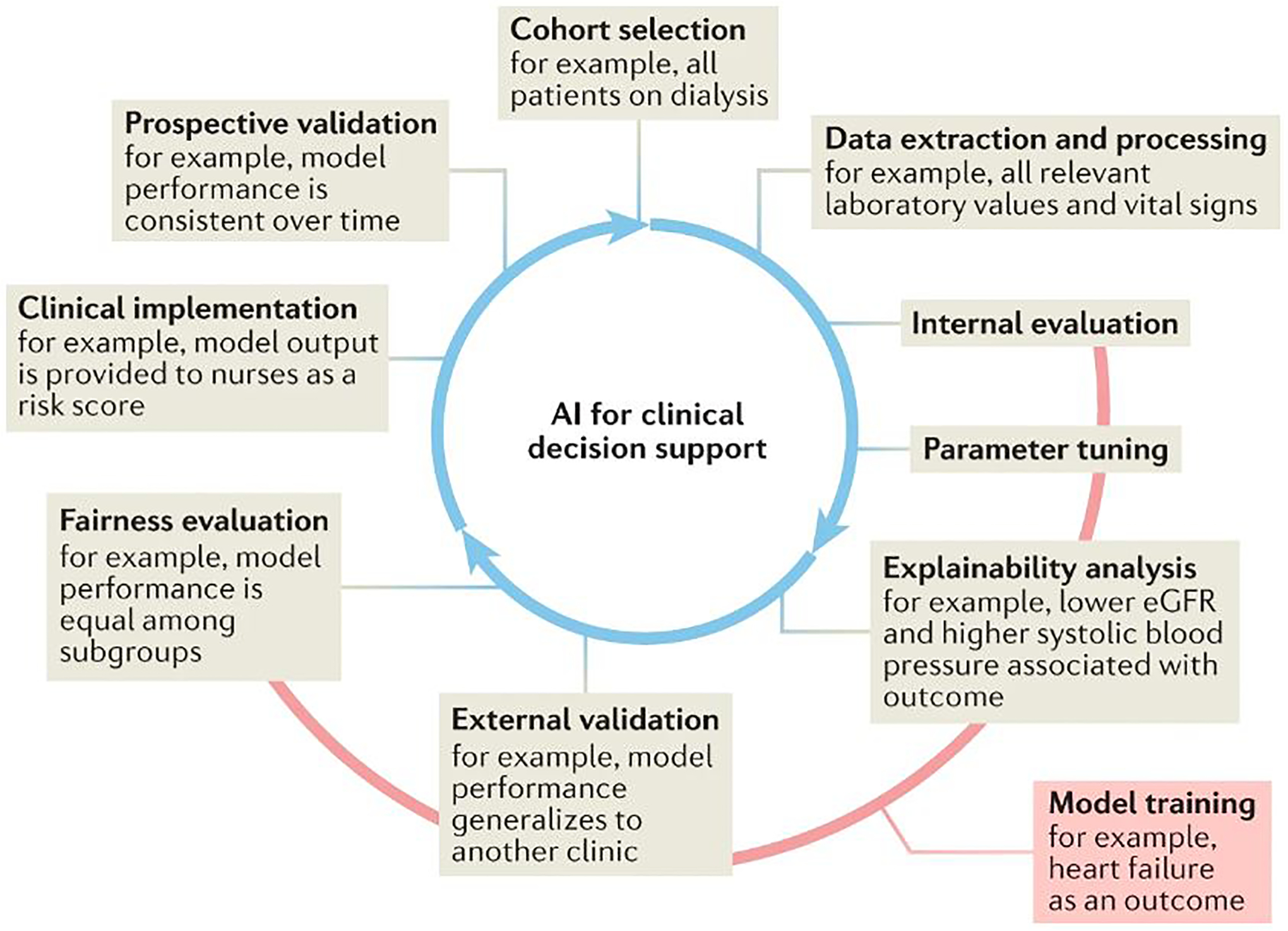
From: Artificial intelligence-enabled decision support in nephrology
The lifecycle of artificial intelligence (AI) for clinical decision support is an integrated, iterative and complex series of processes. Building a model for clinical decision support often relies on the use of retrospective data. First, the cohort of interest must be investigated using inclusion and exclusion criteria. Second, relevant data elements must be extracted and their quality and conformation to modelling requirements ensured. Training a model to predict or classify an outcome between cases and controls requires several steps. Internal validation refers to the process of refining a model on a training subset of the cohort through such processes such as cross-validation. During the model refinement stage, model parameters (such as weights and biases) are altered to optimize the associations between inputs and outputs. Model explainability or interpretability can be explored at this stage to elucidate the relative importance of inputs. External validation is then performed to assess the generalizability and reproducibility of the model. Model fairness, or its equity in performance across sociodemographic factors, is critical for safe and effective implementation. Operationalizing a model requires attention to implementation science best practices. It is imperative that the performance of an established model is continually monitored in prospective deployment to safeguard against population drifts or data shifts that may result in deteriorating performance over time.
Despite evidence that the implementation of AI-driven tools has improved clinical outcomes in some areas of medicine,29 the evaluation of AI models in the field of nephrology remains mainly in silico. This incremental progress largely reflects challenges in integrating AI models within EHRs and in the linking of model results to meaningful interventions.30 However, the maturation of AI-enabled decision-support systems in nephrology could be facilitated by the initiating greater numbers of AI-driven clinical trials that are conducted in accordance with relevant guidelines to ensure that findings are robust and reproducible.31, 32
State-of-the-art of AI in nephrology
AI techniques have the potential to enable earlier detection and define more granular, patient-specific representations of kidney diseases and their treatments while avoiding the inherent risks of invasive testing. The scope of AI applications in nephrology will expand as data standardization, integration practices and workflows evolve alongside advances in AI techniques. To date, emerging AI technologies have demonstrated potential for the early detection and accurate representation of acute kidney injury (AKI) and chronic kidney disease (CKD), renal cell carcinoma (RCC) and renal allograft dysfunction (Table 1), although most applications to date have been described in retrospective studies and require prospective validation.33 Below we summarize sentinel articles that highlight the state-of-the-art of AI in nephrology by disease area.
Table 1.
Examples of artificial intelligence applications in major domains of nephrology
| Domain | Patient Data | Sample Size | Models | Findings | Ref |
|---|---|---|---|---|---|
| AKI | Demographics, diagnoses, procedures, laboratory results, medications, orders, vital signs, health factors, clinical note titles | 703,782 patients | Recurrent neural network | Deep learning predicted 55.8% of inpatient AKI episodes and 90.2% of all AKI requiring dialysis up to 48 h in advance with two false alerts for every true alert (AUROC 0.921) | Tomašev et al. (2019)52 |
| CKD | Risk factors (age, sex, ethnicity, diabetes, hypertension); retinal images | 11,758 patients; 23,516 retinal images | Convolutional neural network | CKD prediction using risk factors alone outperformed retinal images alone (AUROC 0.916 vs. 0.911). Hybrid models integrating both data sources yielded maximum accuracy (AUROC 0.938). | Sabanayagam et al. (2020)57 |
| eGFR and CKD | US images | 1,299 patients; 4,505 US images | Convolutional neural network pre-trained with ImageNet, gradient boosting trees | The model predicted CKD status with accuracy greater than that of experienced nephrologists (0.86 vs. 0.60–0.80) | Kuo et al. (2019)58 |
| Kidney failure | Demographics, chronic conditions, diagnoses, procedures, medications, medical costs, episode counts | 550,000 CKD patients; 10,000,000 medical insurance claims | Word embeddings (Word2Vec), gradient boosting trees | Accurate prediction of kidney failure diagnosis within 6 months (AUROC 0.93). Influential factors included chronic and ischemic heart disease comorbidities, age, and number of hypertensive episodes |
Segal et al. (2020)60 |
| Mortality in CRRT patients | Demographics, mechanical ventilation, comorbidities, vital signs, laboratory results | 1,571 CRRT patients | Random forest | Random forest modeling had similar or greater performance in predicting mortality compared with existing illness severity scoring systems (AUROC 0.784 vs. 0.722 [Mosaic score], 0.677 [SOFA], 0.611 [APACHE II]). | Kang et al. (2020)62 |
| DKD | Laboratory tests, profiles, medications, diagnoses, free text (medical examinations, nutrition consultations) | 64,059 patients with T2DM | Convolutional autoencoder, logistic regression | The model accurately predicted 180-day DKDaggravation (AUROC 0.743; accuracy 71%) | Makino et al. (2019)63 |
| Renal cell carcinoma | Clinical variables (gender, age, tumour volume); MRI (T1C, T2WI) | 1,162 MRI | Convolutional neural network pretrained with ImageNet | Model performance was similar to or greater than experts for accuracy (0.70 vs. 0.60, P=0.053), sensitivity (0.92 vs. 0.80, P=0.017), and specificity (0.41 vs. 0.35, P=0.450) | Xi et al. (2020)66 |
| Kidney transplant graft survival | Demographics, kidney failure cause, KRT modality, ABO type, comorbidities, immunologic factors | 3,117 kidney transplant patients | Decision tree bagging ensemble | A decision tree ensemble had a superior prediction of 1-year graft failure; serum creatinine levels 3 months after transplant were an important risk factor | Yoo et al. (2017)67 |
AKI, acute kidney injury; APACHE: acute physiology and chronic health evaluation; AUROC: area under the receiver operating characteristic curve, CKD: chronic kidney disease, CRRT: continuous renal replacement therapy, DKD, diabetic kidney disease; eGFR: estimated glomerular filtration rate, ESRD: end-stage renal disease, KRT; ICU: intensive care unit; kidneyl replacement therapy, SOFA: sequential organ failure assessment; T2DM, type 2 diabetes mellitus; US, ultrasound.
Acute kidney injury
AI may be particularly useful for time-sensitive applications in nephrology, such as in diagnosing, forecasting, and suggesting treatments for AKI.34, 35 Patients with AKI often present with non-specific symptoms, suggesting there may be utility in AI tools that can automatically extract AKI-associated patterns from high-throughput patient data to predict the onset of AKI.36 Indeed, the development of such AI algorithms has demonstrated efficacy in several clinical contexts, including in patients with hospital-acquired AKI,37 postoperative AKI,38–41 and in patients who develop AKI as a consequence of cancer,42 traumatic injury,43 or critical illness settings.44–48 To optimize their clinical efficacy, AKI prediction tools must represent temporalities, such as the time span of the input data and/or prediction horizons. For example, one study developed logistic regression and random forest models from EHR data from 1 year prior to hospital admission to 48 hours after admissionto stratify the risk of developing multi-stage hospital-acquired AKI more than 48 hours after admission37. This approach identified features early in disease progression — such as use of nonsteroidal anti-inflammatory drugs — that were associated with AKI risk. Use of causal inference models or randomized control trials are needed to determine whether such features are causative and could represent preventative or therapeutic targets.
In attempts to develop tools that enable earlier, more dynamic AKI prediction and monitoring, several studies have implemented models that output AKI predictions periodically (for example, every 15 minutes,49, 50 one hour,51 six hours,52 or twelve hours,53, 54) or whenever a particular clinical descriptor (for example, serum creatinine level) changes in value.42, 55, 56 One study53 used nonoverlapping 12-hour intervals of EHR data with gradient boosted machine learning algorithms to continuously predict the onset of stage 2 AKI within the following 24 hours (area under the receiver operating characteristic curve (AUROC) 0.90; 95% CI: 0.90–0.90) and 48 hours (AUROC 0.87; 95% CI: 0.87–0.87), and the receipt of kidney replacement therapy within 48 hours (AUROC 0.96; 95% CI: 0.96–0.96). The clinical implementation of such AKI monitoring tools therefore also requires a careful balance between sensitivity and specificity for maximizing the discovery of positive cases and minimizing alarm fatigue. Indeed, a comparison of these critical predictive metrics at various predicted probability thresholds, ranging from 0.004 (18,932 patients; sensitivity 0.96; specificity 0.61; positive predictive value (PPV) 0.043; negative predictive value (NPV) 0.998) to 0.125 (2,362 patients; sensitivity 0.44; specificity: 0.98, PPV: 0.327, NPV: 0.992) demonstrated that different predicted probability thresholds have substantial effects on model performance.53 Nonetheless, the ability to predict AKI onset prior to notable elevations in serum creatinine level — a metric that is commonly used to identify AKI —implies potential clinical benefits for the real-time implementation of time-sensitive AKI forecasting.
Most AI-enabled AKI prediction systems involve conventional classifiers, such as logistic regression,55, 56 random forests,41, 45, 49 gradient boosting,51, 53, 54 k-nearest neighbors,43 or a comparison of multiple such algorithms. Alternatively, deep learning methods derive more hierarchical and robust health representations that may yield greater accuracy and clinical utility than conventional classifiers.50, 52 For example, the development of a continuous AKI prediction framework using variations of recurrent neural networks to predict AKI every 6 hours after hospital admission, enabled AKI events to be predicted up to 48 hours in advance of an AKI episode, with an AUROC of 0.92.52 This approach correctly forecasted subsequent increases in biochemical laboratory measurements of renal function with 88.5% probability. These studies indicate that identifying imminent AKI before biochemical changes occur is feasible and could enable early preventative and therapeutic measures that may improve patient outcomes.
Chronic kidney disease and kidney failure
Worldwide,CKD and kidney failure are associated with approximately 1.2 million deaths per year, and substantial health-care costs, suggesting a need for earlier detection and more accurate prediction of CKD trajectory. In response to this need, researchers57 have used CNNs to develop an automated, noninvasive screening tool for the detection of CKD from two-field retinal photographic images. Internal validation demonstrated superior classification performance (AUROC: 0.938 [95% CI 0.917 – 0.959], Sensitivity: 0.84, Specificity: 0.85) when retinal images were combined with existing clinical risk factors, compared with the use of risk factors (AUROC: 0.916 [0.891 – 0.941], Sensitivity: 0.82, Specificity: 0.84) or retinal images (AUROC: 0.911 [0.886 – 0.936], Sensitivity: 0.83, Specificity: 0.83]) alone. However, the added value of retinal photography was not replicated in two external validation experiments. Of note, NPV for the hybrid approach was high (0.96 – 0.99); however, PPV was low (0.09 – 0.57), with wide variance partially attributable to population and CKD prevalence differences, suggesting that the model was effective in identifying the absence of CKD, but not its presence. Nevertheless, this study57 remains an important step towards the goal of AI-enabled CKD screening. A second study58 developed a similar autonomous deep learning framework using a residual Network (ResNet)59 CNN architecture to predict estimated glomerular filtration rate (eGFR) — a key component for the diagnosis and staging of CKD — from kidney ultrasonography images at a single institution. CKD prediction performance varied across eGFR thresholds of 30 ml/min/1.73m2 (AUROC: 0.8036, Sensitivity: 0.7027, Specificity: 0.7791), 45 ml/min/1.73m2 (AUROC: 0.8326, Sensitivity: 0.8077, Specificity: 0.7321), and 60 ml/min/1.73m2 (AUROC: 0.9036, Sensitivity: 0.9213, Specificity: 0.6061), with superior accuracy, precision, recall, and F1 score compared with that of four experienced nephrologists. The observed lower performance at lower eGFR may be attributable to an uneven distribution of eGFRs in the dataset, limitations in ultrasonography in detecting all pathophysiologic factors affecting CKD, or a combination of these factors. Another study60 that analyzed insurance claims data from more than 20 million patients used a gradient boosting tree61 algorithm to predict the probability of an individual developing kidney failure six months later. Their model implemented a code embedding procedure inspired by natural language processing techniques, resulting in excellent predictive performance (AUROC: 0.930 [95% CI: 0.916 – 0.943], Sensitivity: 0.715, Specificity: 0.958, PPV: 0.517, NPV: 0.981). The most important predictors of kidney failure were age, high CKD stage, number of hypertensive events, and newly diagnosed hypertension, consistent with clinical experience. A separate study of patients receiving kidney replacement therapy62 demonstrated that random forest modeling performed similarly or better in predicting mortality than existing scoring systems, such as the sequential organ failure assessment (SOFA) score and the acute physiology and chronic health evaluation (APACHE) II score. Thus AI-enabled prediction of CKD onset and progression to kidney failure offers the potential for accurate screening and prognostication without the need for invasive testing.
Diabetic kidney disease
Diabetic kidney disease affects approximately 1 in 3 patients living with type 1 or type 2 diabetes in the USA; early diagnosis has important implications for treatment success. One study developed a deep learning approach based on integrated EHR data from 64,059 patients with diabetes that comprised longitudinal time series of laboratory tests, structured billing codes, and disease concepts extracted from unstructured clinical notes to predict worsening diabetic kidney disease six months prior to clinical diagnosis63. This work used convolutional autoencoders to extract information from multivariate longitudinal time series and implemented disease name matching and topic modeling to convert text notes into a structured format. This multi-modal approach yielded greater predictive performance compared with that achieved by an approach that used patient characteristics only (AUROC: 0.743 versus 0.562, accuracy: 0.701 versus 0.548). A separate study64 approached the diagnosis of diabetic kidney disease from a pathology and computer vision perspective, using glomerular immunofluorescent images (of IgG, IgA, IgM, C3, C1q and fibrinogen staining) from 885 patients to train a CNN to diagnose kidney disease. The tool demonstrated high accuracy (98.23 +/− 2.81) on a limited test dataset of six patients. Although these studies are in their early stages, findings to date suggest an important role for early, AI-enabled diagnosis and prognostication of diabetic kidney disease.
Renal cell carcinoma
RCC is the most common form of renal cancer, with forecasts of more than 55,000 new diagnoses per year by 2050.65 Automated computer vision approaches based on computed tomography, magnetic resonance and histopathology images have potential to aid the diagnosis and characterization of RCC, to enable earlier and more accurate non-invasive screening and diagnosis. One study66 developed an ensemble deep learning model that integrated clinical variables and magnetic resonance images to distinguish benign from malignant renal tumors, yielding diagnostic accuracy comparable to that achieved by four expert radiologists (0.70 vs. 0.60, respectively, P=0.053), demonstrating the utility of this imaging-based approach in diagnosing renal cell carcinoma. Similar to computer vision models developed for CKD screening, the framework used in this study was also based on the ResNet deep learning architecture. An internally validated retrospective study highlighted the importance of integrating data derived from clinical variables such as age, gender, and tumor volume with imaging data from T1C-weighted and T2-weighted magnetic resonance images, with an integrated approach demonstrating better performance (AUROC: 0.73, Sensitivity: 0.92, Specificity: 0.41, PPV: 0.67) than that achieved by clinical variables alone (AUROC: 0.43, Sensitivity: 0.83, Specificity: 0.12, PPV: 0.55), T1C-weighted images alone (AUROC: 0.62, Sensitivity: 0.87, Specificity: 0.35, PPV: 0.63), or T2-weighted images alone (AUROC: 0.70, Sensitivity: 0.90, Specificity: 0.41, PPV: 0.66). Furthermore, the fully automated deep learning ensemble outperformed conventional machine learning techniques that used handcrafted radiomic features such as shape, intensity, and texture (AUROC: 0.59, Sensitivity: 0.79, Specificity: 0.39), highlighting the power and potential of deep learning for accurate, autonomous renal tumor classification applications.
Renal allograft function
AI harbors potential to augment prognostication and decision-making following renal transplantation. One study67 developed an AI framework for predicting allograft survival, using clinical variables and immunological factors as inputs to generate a survival decision-tree model. The model demonstrated high predictive accuracy (AUROC: 0.77 – 0.82) and establishedimportant associations between early rejection and eventual graft failure. Another preduction system, called the iBox risk score68 integrates functional, histological and immunological features, such as HLA antibody profiles, to predict renal allograft failure using a multivariate Cox proportional hazards regression model. The inclusion of histology information improved the overall discrimination of the model compared with that achieved by individual feature subsets. iBox was validated at several time points after kidney transplantation and in three randomized clinical trial datasets, suggesting that this model represents a state-of-the-art tool for predicting allograft survival.
Algorithm fairness
Generating fair datasets
AI-enabled decision support can exacerbate implicit bias and discrimination if trained on data that mirrors the health-care disparities experienced by groups defined by race, ethnicity, gender, sexual orientation, socioeconomic status or geographic location.69 Unfair datasets can potentiate minority bias (for example, by failing to include representative data from a sufficient number of patient groups to enable the model to learn accurate, representative patterns that serve all patients) and potentiate missing data bias (for example, if a dataset lacks data from select groups in a non-random fashion). To mitigate these pitfalls, AI algorithms must be trained on fair datasets that include and accurately represent social, environmental, and economic factors that influence health. Gaps created by missing sociodemographic informationfrom the EHRs that are used for algorithm training, can be filled by linking to publicly available datasets.70, 71 This approach can improve adherence to FAIR principles (https://www.go-fair.org/fair-principles/),72 but cannot account for systemic and structural bias that arises from two sources: label bias in which a subpopulation is misdiagnosed frequently, and the nonexistence of training data representing individuals with inadequate access to health-care resources.73 Models themselves can identify and correct label bias. However, other than data simulations guided by census data and expert opinion, we are unaware of any robust, data-driven solution for developing models that represent individuals who have no opportunity to leave a data trail.74
Implications of including race and social determinants of health in training data
Including race in training data must be done with care to avoid inadvertent harm from the modelling of racial inequities, which are common among patients with kidney disease. Black individuals are at increased risk of developing more severe and rapidly progressive CKD. The vast majority of this risk may be largely a consequence of structural racism, resulting in suboptimal access to high-quality health care, housing, education, and employment opportunities.75 These associations have important implications for the inclusion of race in AI algorithms. For example, incorporating the observation that Black patients are at increased risk of mortality after coronary artery bypass in a decision-making algorithm could reduce the likelihood that Black patients will garner the benefits of an indicated procedure, especially if the observed mortality risk is mainly attributable to inadequate access to health-care resources.69, 76, 77, 78 Thus, associations between race and health-care outcomes often reflect inequitable social conditions rather than genetic differences. For predictions or classifications in which a plausible biologic mechanism exists by which race is associated with the outcome of interest – for example, the higher risk of kidney disease attributable to APOL1 risk variants, which are most often seen in people of West African, excluding race could reduce model accuracy. However, in the absence of a plausible biologic mechanism, the inclusion of race as a variable in an AI algorithm may introduce implicit bias. The Kidney Donor Risk Index (KDRI) evaluates organ quality of potential donor kidneys using ten variables, one of which is race. Black kidney donor race is associated with increased risk of allograft loss by as much as 20%.78 This increased risk may be partially attributable to the higher incidence of APOL1 variants among Black donors compared to white donors in the USA, and may be partially attributable to more general structural racial inequities in health-care. However, the inclusion of race in the KDRI may decrease the predicted quality of organs from Black donors and could contribute to disparities in kidney transplant donation by Black donors, even though the prevalence of APOL1 among Black individuals is only 10–22%.79
For some applications, it may be preferable to exclude race from AI learning algorithms. For example, although Black and non-Black categories have long been included in commonly used eGFR equations, a growing academic consensus has questioned the biological rationale for the inclusion of race in these equations and highlighted the potential for harm from this approach. These events have led to the development of. new eGFR equations that do not include race.80,81,82
Race also has important interactions with social determinants of health (SDOH) that represent non-medical factors (that is, the conditions under which we live, grow and work) that contribute to health outcomes.83 SDOH are classified into five, key elements: health-care access and quality; education access and quality; social and community context; economic stability; and neighborhood and built environment.84 These elements are associated with disparities in the incidence, progression, and treatment of CKD.84–87 Information about these five SDOH elements are often missing from EHRs of patients from vulnerable populations, and the absence of this information can potentiate bias in algorithms that attempt to include SDOH elements and race in the model.88 Even when information on SDOH is collected uniformly across populations, differences in access to health care across social groups may worsen the under-representation of SDOH data for vulnerable populations. Finally, SDOH derived from census data are subject to inaccuracies from the infrequent collection of census data (every 10 years in the USA).
Best practices for fair algorithms
To mitigate potential harms of algorithm bias, we recommend several best practices. The multiple etiologies of bias in AI-decision support require a comprehensive, multi-faceted approach to ensuring algorithm fairness.across the phases of algorithm design; training and development; and assessment and deployment (Figure 3). These best practices for the development of fair algorithms are consistent with American Medical Association policy recommendations that aim to promote the development of high-quality, clinically validated AI support systems that are transparent, can identify and mitigate bias, and that avoid introducing or exacerbating health disparities, particularly for new AI tools that are tested or deployed in vulnerable populations.89
Fig. 3: Key elements in algorithm fairness.

From: Artificial intelligence-enabled decision support in nephrology
To ensure that the algorithms used in artificial intelligence (AI)-enabled decision support represents and serves all patients equitably, several potential sources of bias must be considered and mitigated at each phase of algorithm development and deployment. PROBAST, Prediction model Risk Of Bias ASsessment Tool.
Study design phase
Researchers must ensure that patients included in studies for AI model development are representative of the patients for whom the model will be applied. For example, models that are trained and tested on a cohort in which women are under-represented — as might be expected in studies that use US Department of Veterans Affairs datasets, in which approximately 6% of participants are female — may not produce effective decision support applications for use in female patients.52 Researchers who use retrospective data for AI model development should consider the possibility that training data may be affected by systemic and structural health-care disparities that could introduce bias, and communicate these weaknesses to the patients and providers who might use the resulting AI-enabled decision support. In addition, researchers must ensure that data collection occurs uniformly across patient subpopulations to avoid scenarios whereby training data disproportionately represent certain sociodemographic groups, which could compromise the efficacy of the decision support system when applied to under-represented populations.
Training and development phase
Incorporating information about person context-level SDOH is essential to ensure equity in AI tools, methods and health-care applications. Careful consideration of each input feature is needed to minimize bias in model outputs. Researchers should consider whether the associations between input features, including race and SDOH, are the result of true biologic mechanisms or bias. In the absence of knowledge about biologic mechanisms, excluding those SDOH from model feature sets may promote fairness. In addition, researchers must consider the effect of confounding on associations between SDOH and the health-care outcomes that are forecasted by AI-enabled decision support. For example, if an AI model “learns” that low socioeconomic status is associated with poor outcomes after an otherwise effective disease-specific treatment but the association is actually driven by interactions between SDOH and outcomes rather than interactions between the treatment and outcomes, then the AI model may recommend against providing an effective treatment for patients with low socioeconomic status. Use of different cut-off values for different patient subgroups has been proposed as a strategy to adjust for such confounders; however, this approach raises questions as to the identification of an equitable means to determine the optimal cut-off value and requires further investigation.90
Assessment and deployment phase
Following the development of a model, researchers should ensure that the deployment phase variables represent the same concepts and values as their training phase counterparts. For example, a urine output variable must use the same measurement units and time interval in both the training and deployment phases. Standardized reporting guidelines can promote transparency in the training process by documenting the association of SDOH variables with model outputs and recommendations that correspond to model outputs.91 Before their deployment, models should be assessed with validated tools — such as the Prediction model Risk Of Bias ASsessment Tool (PROBAST) — to determine their risk of bias,. Following deployment, researchers and independent oversight committees should ensure that the models provide equal clinical benefit, predictive performance, and resource allocation for all patient subgroups.74, 92
Clinical implementation
Despite great potential for AI-enabled decision support systems to augment care, few have been implemented clinically.93, 94 However, as described below, solutions for the various barriers to clinical implementation are often available (Table 2).
Table 2.
Barriers and solutions to clinical implementation of AI-enabled decision support
| Challenges | Solutions |
|---|---|
| Lack of data standardization impairs multi-centre validation and dissemination | Use common data models or federated learning to maintain data security while sharing models |
| Patients and clinicians mistrust “black-box” models and fear the possibility of egregious errors | Implement model interpretability, explainability and uncertainty estimation mechanisms |
| Models applied outside of their training environment and patient population could cause harm | Perform technology readiness assessments, perform model stress testing with simulated data |
| Manual data entry requirements and additional work drive clinician apathy | Integrate automated decision-support systems with existing clinical and digital workflows |
| AI models cannot incorporate some subjective finding and the wisdom of experience | Preserve human intuition in decision-making processes that are augmented by recommendations from AI models |
| Accountability for errors associated with AI-enabled decision support remains challenging | Guide the development and implementation of AI models toward social benefit with altruism, creativity and clinical expertise |
AI, artificial intelligence.
Data standardization
Dissemination and clinical implementation of AI models across health-care systems requires data structures to be standardized across centres. For example, serum creatinine measurements must have a single, common variable name or label. We suggest that variables used in AI algorithms should be mapped to a single, interoperable scheme, such as the open-source OMOP (Observational Medical Outcomes Partnership) or PCORnet (National Patient-Centered Clinical Research Network) common data models. An advantage of AI decision support models that are built using standardized labels is that they can be shared between different institutions by translating the model into an interoperable programming language (for example, Predictive Model Markup Language) followed by retraining and/or testing of the model on interoperable data from different practice settings. Such sharing of an interoperable model obviates the need for data sharing and the ensuing potential for compromising the security of patients’ private health data. Alternatively, collaborative modelling without data sharing can be accomplished by federated learning in which local models train separately and send gradients or coefficients to a centralized, global model. This approach — privacy by design — ensures that the models can be generalized across participating institutions without the need to share patient data, and offers the potential advantage of learning from a greater number of rare scenarios that are represented sparsely in smaller, single-institution datasets. Accurate representation of these rare scenarios is critical for building AI-enabled decision-support for rare kidney diseases.
Uncertainty and mistrust
Mistrust hinders clinical implementation of AI-enabled decision support through skepticism regarding opaque, “black box” model outputs and recognition that AI models can make egregious errors.95–97 Such skepticism can in part bemitigated by the adoption of mechanisms that aid interpretation of the model output by communicating the relative importance or weight of input variables that determine the model outputs and describe objectively what the model has learned. Such mechanisms are most useful when their results correlate with logic, scientific evidence, domain knowledge and effective interventions.98, 99 For example, a model that predicts the development of AKI could also identify the most important modifiable risk factors for individual patients, such as nephrotoxic medications that could be discontinued, contrast imaging studies that could be avoided, or renal perfusion deficits that could be corrected with intravascular volume expansion. These techniques, while potentially useful, fail to address valid concerns regarding model uncertainty. Among model types, computer vision models are, arguably, the most successful in performing health care prediction and classification tasks. Yet, even computer vision models can fail catastrophically at seemingly simple tasks.97, 100 For an AI model that yields 95% accuracy, one may — and perhaps should — fear that a particular output is among the 5% that are incorrect. This fear may be alleviated by conveying the probability that a given model output is inaccurate, termed here as model uncertainty.99 Common statistical measures of spread, such as standard deviations and interquartile ranges, cannot convey model uncertainty because they are undefined for point predictions, which are the outputs of a risk prediction model. Instead, one proposed approach to estimate uncertainty in deep learning prediction models is to generate sets of predictions using a series of models that are modified slightly by dropping different sets of neurons each time data are passed101. In this scenario, high variance (that is, poor precision) across predictions made by the model variants suggests high overall model uncertainty, incicating that any single prediction made by the model is likely to be inaccurate. Mechanistic models that use mathematical expressions of pathophysiology or pragmatic, clinical truths (for example, patients requiring mechanical ventilation or vasopressor support require admission to an intensive care unit) can simulate data to account for unique, heterogeneous patient populations and practice patterns, and can infer causal relationships by changing individual variables and appraising their effects on outcomes. Of note, further exploration of these approaches is required to establish efficacy for medical applications.102, 103
Technology readiness
Clinical application of AI-enabled decision support in nephrology should occur only after high-level evidence of safety and efficacy has been validated by a robust peer-review process. To avoid harm, models should be subjected to assessments of technology readiness, similar to those adopted by the National Aeronautics and Space Administration (NASA) after the Challenger tragedy.104 Following the development and validation of models using datasets that are sufficiently large to contain an adequate number of patients with and without the medical condition being modeled, the implementation of AI models should mimic Phase 1 and 2 clinical trials. That is, small, prospective studies should be performed under close surveillance and high scrutiny.105 Approaches to mimic traditional phase 3 and 4 clinical trials for definitive validation of AI decision support are typically hindered by their cost and requirement for substantial research infrastructure resources. An alternative approach to understand the performance of a model in different clinical practice environments and with different patient populations may require investigators to determine how their models respond to simulations of different and rare input data structures. For example, the open-source What-If Tool allows investigators to vary input features, such as patient age or proportions of vulnerable populations, and perform interactive model reassessments under these simulated, hypothetical circumstances.106 These technology readiness assessments could avoid large-scale failures and facilitate realistic expectations regarding real-world model performance, before AI-enabled decision support is deployed in clinical settings.
Integration with clinical and digital workflows
Within six years of the Health Information Technology for Economic and Clinical Health Act of 2009,107 which incentivized adoption of EHR systems, more than four of five US hospitals had adopted EHRs. This large-scale adoption of EHR systems is largely responsible for the massive volumes of data available today that are necessary for optimal AI algorithm training.107–109 Of note, contemporary data management systems allow the use of automated, real-time EHR data as model input features, obviating the manual data entry requirements that incite apathy toward clinical decision support systems.110 In practical terms, the automated inputting of real-time EHR can be achieved by linking EHR data with census data to incorporate data representing SDOH information, and by passing the merged dataset through validated preprocessing algorithms for handling outliers, missing values, normalization, and resampling. For large-scale real-time data, we suggest implementing a modular platform using open-source tools (for example, Spark Streaming or Cassandra), that dynamically scale computing resources according to real-time workloads by linking and coordinating multiple computer servers.110, 111
The integration of AI models with clinical and digital workflows may also decrease deployment costs associated with the organizational effort and resources required for clinical implementation.111 Finally, the clinical integration process should reflect the reality that AI models describe relationships and associations rather than causality, with the caveat that in silico randomized trials can infer causality. When causal relationships and uncertainty have not yet been established, which is true of nearly all contemporary AI-enabled decision support models, outputs that are provided to patients, caregivers or clinicians should represent estimations and recommendations that enrich rather than replace human decision-making and preserve the pre-eminence of human wisdom and intuition.
Heuristics and intuition
Under time constraints and uncertainty, human decision-making is influenced by heuristics (cognitive shortcuts), that can lead to cognitive errors and patient harm.112–114 For example, incorrect drug doses are the most common cause of adverse drug events; patients with kidney dysfunction are particularly vulnerable to drug dosing errors.115, 116 AI models have the theoretical advantages of objectivity and formality grounded in mathematical expressions of logic, suggesting utility for AI-enabled decision support systems to serve as a bulwark against cognitive errors. For experienced clinicians, heuristics can be advantageous. When rewards or penalties follow life experiences, limbic system neuronal architectures adapt so intuitive positive or negative emotions are recalled when similar experiences are subsequently encountered.117, 118 Intuition yields performance advantages in controlled settings and can identify life-threatening medical conditions that are underrepresented by objective clinical parameters alone.119–121 Therefore, clinical applications of AI-enabled decision-support should seek to anchor intuition with objective predictions and classifications, and should favor clinician intuition when it is honed by experience.
Legal governance
Unlike ethical considerations, legal governance is explicit and audited externally.122 From a medico-legal perspective, in the USA, AI-enabled decision support in nephrology fits the Software as Medical Device (SaMD) category generated and maintained by the US Food and Drug Administration.121 Guidelines proposed by the SaMD Working Group provide frameworks for aiding health care software developers in creating, evaluating and implementing SaMD safely and effectively in clinical settings. Judicial oversight and precedents for SaMD are not defined rigidly. Hence, assigning accountability for medical errors and complications associated with SaMD remains challenging, especially when legal governance overlaps with ethical considerations. For example, an AI model that predicts the short-term and long-term utility of continuous renal replacement therapy (CRRT) for a critically ill patient may support the decision to use CRRT resources for that patient, which decreases CRRT resource availability for other patients, regardless of whether they have or will develop a greater need for CRRT relative to the index patient. The dynamic, complex nature of these dilemmas hinders the development of universal rules and policies for legal governance of medical AI. Instead, we suggest that AI-enabled decision support in nephrology is guided by altruism, creativity and clinical expertise towards the greatest possible and most equitable net social benefit.
Workforce development
Experiences from non-medical industries
AI has transformed non-medical industries by automating repetitive, time-consuming tasks and providing decision support. Automation enables human workers to shift their focus from mundane tasks toward higher-level critical thinking and problem-solving. AI can organize data into information, and translate information into knowledge; presently, applying that knowledge to complex decision-making paradigms requires the wisdom of human experience. In non-medical contexts, developments in AI-enabled automation have shifted workforce expertise away from assembly-line type skillsets toward the skills necessary to interpret and leverage AI applications in achieving work-related objectives. Businesses are reframing multidisciplinary teams to include domain experts, business experts, and AI experts. To fuel these initiatives, such businesses are providing reskilling, upskilling, cross training, and mentorship programs to bridge knowledge gaps between subgroups in multidisciplinary teams.123 These programs, often delivered online by large corporations such as Amazon, AT&T, and Microsoft, generate customized learning paths from digital knowledge assessments.124–127 Experiences from non-medical industries in AI workforce development can inform the development and implementation of similar programs in medical fields.
Deficiencies in current medical training paradigms
Medical training is perhaps one of the longest and most extensive of all professional training pathways. The field of medicine is undergoing extensive transformation as a consequence of digitization. However, medical education has not kept pace with these transformations and remains largely based on traditional curricula that lack adequate exposure to the concepts and applications of AI.128 The Accreditation Council for Graduating Medical Education (ACGME) in the USA focuses medical education on the following domains: patient care; medical knowledge; interpersonal and communication skills; practice-based learning and improvement; professionalism; and systems-based practice.129 However, in practice this approach often translate into participants memorizing medical facts and then applying them to patient care.130 There is almost no systematic commitment to the teaching of probabilistic thinking or the basics of AI, and little motivation to addend these traditions since they are not tested in medical training or professional licensing exams.131 The lack of curricula to ensure that emerging AI technologies are part of routine medical education, means that AI skillsets are disproportionately available to individuals who seek training or to select institutions with contemporary training programs that introduce medical AI concepts and applications.131
The second major deficiency in the current paradigm is a lack of education relating to the quality assessment and optimization of EHR data at both graduate and postgraduate levels. Training on the use of EHRs often consists of brief introductory courses that teach the basic skills necessary for clinical work and rarely address data quality, the impact of biased EHR data, and methods for maximizing EHR use for enriching – rather than encroaching – the patient-physician relationship.132 This gap in training is a major problem, since most medical AI systems use EHR data and physicians are a primary source of EHR data entry. Fortunately, deficiencies in current medical training paradigms are somewhat self-limited, as many high schools and colleges in the USA and elsewhere now offer courses in AI and data science. Similarly, postgraduate training should adapt toward building an AI-competent medical workforce.
Building an AI-competent medical workforce
An understanding of data-driven medicine, probabilistic thinking, and key aspects of AI applications should be an integral part of graduate, postgraduate, and continuing medical education. Although a comprehensive set of recommendations for achieving these goals is beyond the scope of this Review, we suggest a framework for the creation of an AI-competent medical workforce. First, both didactic and practical education should be embedded throughout all phases of medical education and standardized testing (Figure 4). Second, multidisciplinary teams with expertise in data and implementation science should integrate with learners at all levels. Finally, we must collaborate with our ethics and humanity colleagues to ensure that computational skills are balanced with and complement person-centered aspects of medicine, including communication, empathy, creativity and shared decision-making. In the data-driven age of medicine, the personal aspects of medicine will take even greater precedence as we ask patients to trust not the algorithms, but rather the clinicians using the algorithms.
Fig. 4: A framework for building an AI-competent medical workforce.
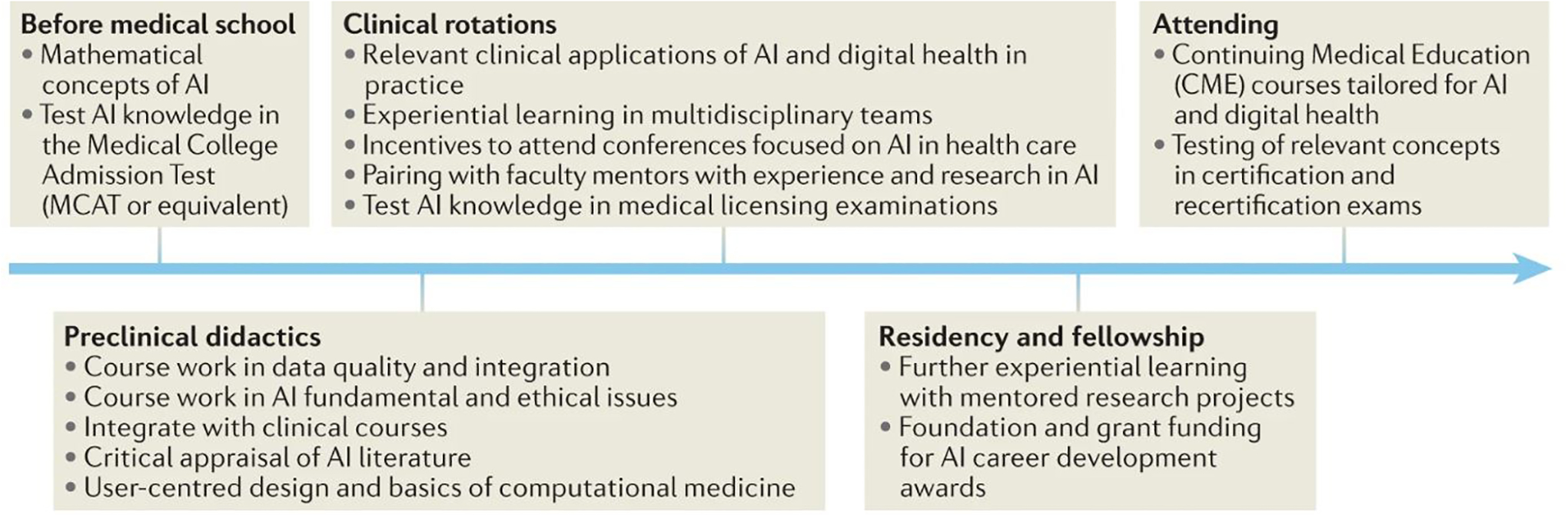
From: Artificial intelligence-enabled decision support in nephrology
Didactic and practical training activities apply at various stages of pre-medical, medical, graduate and continuing medical education, building on previous stages of development. Theoretical and practical exposure to applications of artificial intelligence (AI) in healthcare has the potential to develop and sustain an AI-competent medical workforce.
Conclusions
Artificial intelligence (AI)-enabled decision support offers opportunities to understand and manage the complex, non-linear, and heterogeneous pathophysiology of kidney disease by learning from examples rather than conforming to rules. AI models can already predict the onset of AKI before biochemical changes occur, identify modifiable risk factors for CKD development and progression, and match or exceed human accuracy in recognizing renal tumors on imaging studies. In the future, decision support applications could leverage reinforcement learning technologies to offer real-time, continuous recommendations for discrete actions that yield the greatest probability of achieving optimal kidney health outcomes. Realizing these potential advantages will require that clinicians, data scientists and administrators work together to build an AI-competent workforce and overcome substantial barriers to ensure the efficient and effective clinical implementation of fair algorithms while preserving the preeminence of wisdom and intuition by augmenting rather than replacing human decision-making.
Key points.
Hypothetical-deductive reasoning and linear, statistical approaches to diagnosis and treatment often fail to adequately represent the complex, non-linear, and heterogeneous nature of kidney pathophysiology.
Artificial intelligence (AI)-enabled decision support systems use algorithms that learn from examples to accurately represent complex pathophysiology, including kidney pathophysiology, offering opportunities to enhance patient-centered diagnostic, prognostic, and treatment approaches.
Contemporary AI applications can accurately predict kidney injury before the development of measurable biochemical changes, identify modifiable risk factors, and match or exceed human accuracy in recognizing kidney pathology on imaging studies.
Advances in the past few years suggest that AI models have potential to make real-time, continuous recommendations for discrete actions that yield the greatest probability of achieving optimal kidney health outcomes.
Optimizing the clinical integration of AI-enabled decision-support in nephrology will require multidisciplinary commitment to ensure algorithm fairness and the building of an AI-competent medical workforce.
AI-enabled decision support should preserve the preeminence of human wisdom and intuition in clinical decision-making by augmenting rather than replacing interactions between patients, caregivers, clinicians, and data.
Acknowledgements
T.J.L. was supported by the National Institute of General Medical Sciences (NIGMS) of the NIH under Award Number K23 GM140268. T.O.B. was supported by K01 DK120784, R01 DK123078, and R01 DK121730 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), R01 GM110240 from NIGMS, R01 EB029699 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and R01 NS120924 from the National Institute of Neurological Disorders and Stroke (NINDS). B.S.G was supported by R01MH121923 from the National Institute of Mental Health (NIMH), R01AG059319 and R01AG058469 from the National Institute for Aging (NIA), and 1R01HG011407–01Al from the National Human Genome Research Institute (NHGRI). G.N.N is supported by R01 DK127139 from NIDDK and R01 HL155915 from the National Heart Lung and Blood Institute (NHLBI). L.C. was supported by K23 DK124645 from the NIDDK. A.B. was supported R01 GM110240 from NIGMS, R01 EB029699 and R21 EB027344 from NIBIB, R01 NS120924 from NINDS, and by R01 DK121730 from NIDDK.
Competing interests
B.S.G has received consulting fees from Anthem AI and consulting and advisory fees from Prometheus Biosciences. K.S. has received grant funding from Blue Cross Blue Shield of Michigan and Teva Pharmaceuticals for unrelated work, and serves on a scientific advisory board for Flatiron Health. G.N.N. has received consulting fees from AstraZeneca, Reata, BioVie, Siemens Healthineers, and GLG Consulting; grant funding from Goldfinch Bio and Renalytix; financial compensation as a scientific board member and advisor to Renalytix; owns equity in Renalytix and Pensieve Health as a cofounder and is on the advisory board of Neurona Health. The other authors declare no competing interests.
Glossary
- Nodes
Computational units in a neural network. Each node has a weight that is influenced by other nodes and affects predictions made by the neural network
- Computer vision
An AI subfield in which deep models use pixels from images and videos as inputs
- Convolutional neural networks
A type of neural network that assembles patterns of increasing complexity to avoid overfitting (fitting too closely on inputs), which can compromise predictive performance when the model is applied to new, previously unseen data. Convolutional neural networks are commonly used in imaging applications
- Loss function
A mathematical function that calculates errors. AI algorithms are typically designed to minimize loss as the algorithm learns associations between input variables and outcomes
- Deep neural networks
Neural networks with several layers of nodes between the input layer and final output layer
- Generative adversarial neural networks
Two neural networks that compete with and learn from one another, offering the ability to generate synthetic data
- Hierarchical clustering
Forming groups of elements that are similar to one another and different than others by iteratively merging points according to pair-wise distances
- Random forest
A type of AI model that assembles outputs from a set of decision trees and uses the majority vote or average prediction of the individual trees to produce a final prediction
- Gradient boosting
An AI technique for iteratively improving predictive performance by ensuring that the next permutation of the AI model, when combined with the prior permutation, offers a performance improvement
- Sensitivity
The true positive rate; the percentage of patients with a disease for whom a model or test predicted a positive result, also known as recall. Sensitivity indicates the ability of a model or test to identify subjects who have a condition
- Specificity
The true negative rate; the percentage of patients without a disease for whom a model or test predicted a negative result. Specificity indicates the ability of a model or test to identify subjects who do not have a condition
- Positive predictive value
The probability that a positive prediction made by a model or test is correct according to the gold standard or ground truth. Positive predictive value is also known as precision
- Negative predictive value
The probability that a negative prediction made by a model or test is correct according to the gold standard or ground truth
- K-nearest neighbors clustering
Forming groups of elements that are similar to one another and different than others by using distances between points to assign elements to one of “K” groups, where “K” is the total number of groups and is assigned by the investigator
- F1 score
A measurement of accuracy that considers both precision, which is also known as positive predictive value, and recall, which is also known as sensitivity
- Convolutional autoencoder
A convolutional neural network variant that learns which filters should be used to detect features of interest among model inputs, which are usually imaging data
- Topic modeling
An AI technique for detecting groups of text data that are similar to one another and different than others
- Ensemble model
A model that assembles outputs from multiple algorithms to achieve predictive performance that is greater than that of individual algorithms
- FAIR principles
The findability, accessibility, interoperability, and reuse principles of digital assets for scientific investigation are intended to optimize the reuse of data
- Prediction model Risk Of Bias ASsessment Tool
An instrument for assessing the risk of bias associated with a prediction model that provides diagnostic or prognostic information
- Observational Medical Outcomes Partnership (OMOP)
An organization that designed a common data model that standardizes the way medical information is captured across health-care institutions and provides metadata tables describing relationships among data elements
- National Patient-Centered Clinical Research Network (PCORnet)
An organization that designed a common data model that standardizes the way medical information is captured across health-care institutions and is widely adopted by institutions participating in the Patient Centered Outcomes Research Institute
- Predictive Model Markup Language
A programming language that standardizes methods for describing predictions models, which may facilitate sharing models among investigator groups
- Federated learning
A technique for generating a central AI model that is built with information from several local AI models that train on local data. This approach has the potential advantage of training AI models on data from multiple centers without sharing data across centers, thereby promoting data security and privacy
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Schwartz WB, Patil RS, Szolovits P. Artificial intelligence in medicine. Where do we stand? N Engl J Med. Mar 12 1987;316(11):685–8. doi: 10.1056/nejm198703123161109 [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto DA, Rosman G, Rus D, Meireles OR. Artificial Intelligence in Surgery: Promises and Perils. Ann Surg. Jul 2018;268(1):70–76. doi: 10.1097/sla.0000000000002693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millar J, Barron B, Hori K, Finlay R, Kotsuki K, Kerr I. Accountability in AI. Promoting Greater Social Trust. https://cifar.ca/cifarnews/2018/12/06/accountability-in-ai-promoting-greater-social-trust/ 2018.
- 4.Slack WV, Hicks GP, Reed CE, Van Cura LJ. A computer-based medical-history system. N Engl J Med. Jan 27 1966;274(4):194–8. doi: 10.1056/nejm196601272740406 [DOI] [PubMed] [Google Scholar]
- 5.Beam AL, Kohane IS. Big Data and Machine Learning in Health Care. Jama. Apr 3 2018;319(13):1317–1318. doi: 10.1001/jama.2017.18391 [DOI] [PubMed] [Google Scholar]
- 6.Esteva A, Chou K, Yeung S, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. Jan 8 2021;4(1):5. doi: 10.1038/s41746-020-00376-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Titano JJ, Badgeley M, Schefflein J, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med. Sep 2018;24(9):1337–1341. doi: 10.1038/s41591-018-0147-y [DOI] [PubMed] [Google Scholar]
- 8.Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. Aug 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. Dec 22 2014;14:137. doi: 10.1186/1471-2288-14-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apathy NC, Holmgren AJ, Adler-Milstein J. A decade post-HITECH: Critical access hospitals have electronic health records but struggle to keep up with other advanced functions. J Am Med Inform Assoc. Aug 13 2021;28(9):1947–1954. doi: 10.1093/jamia/ocab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. Oct 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.About one-in-five Americans use a smart watch or fitness tracker. https://www.pewresearch.org/fact-tank/2020/01/09/about-one-in-five-americans-use-a-smart-watch-or-fitness-tracker.
- 13.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. Jun 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 14.Doshi-Velez F, Ge Y, Kohane I. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. Jan 2014;133(1):e54–63. doi: 10.1542/peds.2013-0819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K, Choudhry NK, Krumme AA, et al. A concept-wide association study to identify potential risk factors for nonadherence among prevalent users of antihypertensives. Pharmacoepidemiol Drug Saf. Oct 2019;28(10):1299–1308. doi: 10.1002/pds.4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh K, Valley TS, Tang S, et al. Evaluating a Widely Implemented Proprietary Deterioration Index Model among Hospitalized Patients with COVID-19. Ann Am Thorac Soc. Jul 2021;18(7):1129–1137. doi: 10.1513/AnnalsATS.202006-698OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escobar GJ, Liu VX, Kipnis P. Automated Identification of Adults at Risk for In-Hospital Clinical Deterioration. Reply. N Engl J Med. Feb 4 2021;384(5):486. doi: 10.1056/NEJMc2034836 [DOI] [PubMed] [Google Scholar]
- 18.Tarabichi Y, Cheng A, Bar-Shain D, et al. Improving Timeliness of Antibiotic Administration Using a Provider and Pharmacist Facing Sepsis Early Warning System in the Emergency Department Setting: A Randomized Controlled Quality Improvement Initiative. Crit Care Med. Aug 20 2021;doi: 10.1097/ccm.0000000000005267 [DOI] [PubMed] [Google Scholar]
- 19.Gargeya R, Leng T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology. Jul 2017;124(7):962–969. doi: 10.1016/j.ophtha.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 20.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. Jama. Dec 13 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 21.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. Nov 2018;15(11):e1002686. doi: 10.1371/journal.pmed.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Kohlberger T, Norouzi M, et al. Artificial Intelligence-Based Breast Cancer Nodal Metastasis Detection: Insights Into the Black Box for Pathologists. Arch Pathol Lab Med. Jul 2019;143(7):859–868. doi: 10.5858/arpa.2018-0147-OA [DOI] [PubMed] [Google Scholar]
- 23.Yu C, Liu J, Nemati S. Reinforcement learning in healthcare: A survey. arXiv preprint 2019;arXiv:1908.08796 [Google Scholar]
- 24.Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nature Medicine. 2018/11/01 2018;24(11):1716–1720. doi: 10.1038/s41591-018-0213-5 [DOI] [PubMed] [Google Scholar]
- 25.Gottesman O, Johansson F, Komorowski M, et al. Guidelines for reinforcement learning in healthcare. Nat Med. Jan 2019;25(1):16–18. doi: 10.1038/s41591-018-0310-5 [DOI] [PubMed] [Google Scholar]
- 26.Tanakasempipat P Google launches Thai AI project to screen for diabetic eye disease. https://www.reuters.com/article/us-thailand-google-idUSKBN1OC1N2. Reuters. 2018;
- 27.Steiner DF, MacDonald R, Liu Y, et al. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am J Surg Pathol. Dec 2018;42(12):1636–1646. doi: 10.1097/pas.0000000000001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz LI, Kuznetsova M, Jones SA. Creating a Learning Health System through Rapid-Cycle, Randomized Testing. N Engl J Med. Sep 19 2019;381(12):1175–1179. doi: 10.1056/NEJMsb1900856 [DOI] [PubMed] [Google Scholar]
- 29.Escobar GJ, Liu VX, Schuler A, Lawson B, Greene JD, Kipnis P. Automated Identification of Adults at Risk for In-Hospital Clinical Deterioration. N Engl J Med. Nov 12 2020;383(20):1951–1960. doi: 10.1056/NEJMsa2001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challener DW, Prokop LJ, Abu-Saleh O. The Proliferation of Reports on Clinical Scoring Systems: Issues About Uptake and Clinical Utility. Jama. Jun 25 2019;321(24):2405–2406. doi: 10.1001/jama.2019.5284 [DOI] [PubMed] [Google Scholar]
- 31.Cruz Rivera S, Liu X, Chan AW, Denniston AK, Calvert MJ. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Lancet Digit Health. Oct 2020;2(10):e549–e560. doi: 10.1016/s2589-7500(20)30219-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. Sep 2020;26(9):1364–1374. doi: 10.1038/s41591-020-1034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Huang S, Chen T, et al. Machine Learning for Prediction and Risk Stratification of Lupus Nephritis Renal Flare. Am J Nephrol. 2021;52(2):152–160. doi: 10.1159/000513566 [DOI] [PubMed] [Google Scholar]
- 34.Kashani K, Herasevich V. Sniffing out acute kidney injury in the ICU: do we have the tools? Curr Opin Crit Care. Dec 2013;19(6):531–6. doi: 10.1097/mcc.0000000000000024 [DOI] [PubMed] [Google Scholar]
- 35.Rahman M, Shad F, Smith MC. Acute kidney injury: a guide to diagnosis and management. Am Fam Physician. Oct 01 2012;86(7):631–9. [PubMed] [Google Scholar]
- 36.Lewington AJ, Cerda J, Mehta RL. Raising awareness of acute kidney injury: a global perspective of a silent killer. Kidney Int. Sep 2013;84(3):457–67. doi: 10.1038/ki.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cronin RM, VanHouten JP, Siew ED, et al. National Veterans Health Administration inpatient risk stratification models for hospital-acquired acute kidney injury. J Am Med Inform Assoc. Sep 2015;22(5):1054–71. doi: 10.1093/jamia/ocv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thottakkara P, Ozrazgat-Baslanti T, Hupf BB, et al. Application of Machine Learning Techniques to High-Dimensional Clinical Data to Forecast Postoperative Complications. PLoS One. 2016;11(5):e0155705. doi: 10.1371/journal.pone.0155705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HC, Yoon HK, Nam K, et al. Derivation and Validation of Machine Learning Approaches to Predict Acute Kidney Injury after Cardiac Surgery. J Clin Med. Oct 2018;7(10)doi: 10.3390/jcm7100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HC, Yoon SB, Yang SM, et al. Prediction of Acute Kidney Injury after Liver Transplantation: Machine Learning Approaches vs. Logistic Regression Model. J Clin Med. Nov 2018;7(11)doi: 10.3390/jcm7110428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adhikari L, Ozrazgat-Baslanti T, Ruppert M, et al. Improved predictive models for acute kidney injury with IDEA: Intraoperative Data Embedded Analytics. PLoS One. 2019;14(4):e0214904. doi: 10.1371/journal.pone.0214904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park N, Kang E, Park M, et al. Predicting acute kidney injury in cancer patients using heterogeneous and irregular data. PLoS One. 2018;13(7):e0199839. doi: 10.1371/journal.pone.0199839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran NK, Sen S, Palmieri TL, et al. Artificial intelligence and machine learning for predicting acute kidney injury in severely burned patients: A proof of concept. Burns. 09 2019;45(6):1350–1358. doi: 10.1016/j.burns.2019.03.021 [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Yao L, Mao C, Srivastava A, Jiang X, Luo Y. Early Prediction of Acute Kidney Injury in Critical Care Setting Using Clinical Notes. Proceedings (IEEE Int Conf Bioinformatics Biomed). Dec 2018;2018:683–686. doi: 10.1109/bibm.2018.8621574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flechet M, Falini S, Bonetti C, et al. Machine learning versus physicians’ prediction of acute kidney injury in critically ill adults: a prospective evaluation of the AKIpredictor. Crit Care. Aug 16 2019;23(1):282. doi: 10.1186/s13054-019-2563-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun M, Baron J, Dighe A, et al. Early Prediction of Acute Kidney Injury in Critical Care Setting Using Clinical Notes and Structured Multivariate Physiological Measurements. Stud Health Technol Inform. Aug 21 2019;264:368–372. doi: 10.3233/shti190245 [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Ho KM, Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit Care. Apr 08 2019;23(1):112. doi: 10.1186/s13054-019-2411-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmerman LP, Reyfman PA, Smith ADR, et al. Early prediction of acute kidney injury following ICU admission using a multivariate panel of physiological measurements. BMC Med Inform Decis Mak. Jan 31 2019;19(Suppl 1):16. doi: 10.1186/s12911-019-0733-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiofolo C, Chbat N, Ghosh E, Eshelman L, Kashani K. Automated Continuous Acute Kidney Injury Prediction and Surveillance: A Random Forest Model. Mayo Clin Proc. 05 2019;94(5):783–792. doi: 10.1016/j.mayocp.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 50.Rank N, Pfahringer B, Kempfert J, et al. Deep-learning-based real-time prediction of acute kidney injury outperforms human predictive performance. NPJ Digit Med. 2020;3:139. doi: 10.1038/s41746-020-00346-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohamadlou H, Lynn-Palevsky A, Barton C, et al. Prediction of Acute Kidney Injury With a Machine Learning Algorithm Using Electronic Health Record Data. Can J Kidney Health Dis. 2018;5:2054358118776326. doi: 10.1177/2054358118776326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomasev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. Aug 2019;572(7767):116–119. doi: 10.1038/s41586-019-1390-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koyner JL, Carey KA, Edelson DP, Churpek MM. The Development of a Machine Learning Inpatient Acute Kidney Injury Prediction Model. Crit Care Med. Jul 2018;46(7):1070–1077. doi: 10.1097/ccm.0000000000003123 [DOI] [PubMed] [Google Scholar]
- 54.Churpek MM, Carey KA, Edelson DP, et al. Internal and External Validation of a Machine Learning Risk Score for Acute Kidney Injury. JAMA Netw Open. Aug 3 2020;3(8):e2012892. doi: 10.1001/jamanetworkopen.2020.12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kate RJ, Pearce N, Mazumdar D, Nilakantan V. A continual prediction model for inpatient acute kidney injury. Comput Biol Med. 01 2020;116:103580. doi: 10.1016/j.compbiomed.2019.103580 [DOI] [PubMed] [Google Scholar]
- 56.Simonov M, Ugwuowo U, Moreira E, et al. A simple real-time model for predicting acute kidney injury in hospitalized patients in the US: A descriptive modeling study. PLoS Med. 07 2019;16(7):e1002861. doi: 10.1371/journal.pmed.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabanayagam C, Xu D, Ting DSW, et al. A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet Digit Health. 06 2020;2(6):e295–e302. doi: 10.1016/S2589-7500(20)30063-7 [DOI] [PubMed] [Google Scholar]
- 58.Kuo CC, Chang CM, Liu KT, et al. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ Digit Med. 2019;2:29. doi: 10.1038/s41746-019-0104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016:770–778. [Google Scholar]
- 60.Segal Z, Kalifa D, Radinsky K, et al. Machine learning algorithm for early detection of end-stage renal disease. BMC Nephrol. Nov 27 2020;21(1):518. doi: 10.1186/s12882-020-02093-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen T, Guestrin C. XGBoost: A scalable tree boosting system, in proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’16). San Francisco, CA. 2016;785:794. [Google Scholar]
- 62.Kang MW, Kim J, Kim DK, et al. Machine learning algorithm to predict mortality in patients undergoing continuous renal replacement therapy. Crit Care. 02 06 2020;24(1):42. doi: 10.1186/s13054-020-2752-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makino M, Yoshimoto R, Ono M, et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci Rep. Aug 14 2019;9(1):11862. doi: 10.1038/s41598-019-48263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitamura S, Takahashi K, Sang Y, Fukushima K, Tsuji K, Wada J. Deep Learning Could Diagnose Diabetic Nephropathy with Renal Pathological Immunofluorescent Images. Diagnostics (Basel). Jul 9 2020;10(7)doi: 10.3390/diagnostics10070466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santoni M, Piva F, Porta C, et al. Artificial Neural Networks as a Way to Predict Future Kidney Cancer Incidence in the United States. Clin Genitourin Cancer. Apr 2021;19(2):e84–e91. doi: 10.1016/j.clgc.2020.10.008 [DOI] [PubMed] [Google Scholar]
- 66.Xi IL, Zhao Y, Wang R, et al. Deep Learning to Distinguish Benign from Malignant Renal Lesions Based on Routine MR Imaging. Clin Cancer Res. 04 15 2020;26(8):1944–1952. doi: 10.1158/1078-0432.CCR-19-0374 [DOI] [PubMed] [Google Scholar]
- 67.Yoo KD, Noh J, Lee H, et al. A Machine Learning Approach Using Survival Statistics to Predict Graft Survival in Kidney Transplant Recipients: A Multicenter Cohort Study. Sci Rep. 08 21 2017;7(1):8904. doi: 10.1038/s41598-017-08008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loupy A, Aubert O, Orandi BJ, et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: international derivation and validation study. Bmj. Sep 17 2019;366:l4923. doi: 10.1136/bmj.l4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson-Mann CN, Loftus TJ, Bihorac A. Equity and Artificial Intelligence in Surgical Care. JAMA Surg. Jun 1 2021;156(6):509–510. doi: 10.1001/jamasurg.2020.7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.US Census Bureu. https://www.census.gov/data/data-tools.html. Accessed NOv 10, 2021.
- 71.US DHHS. https://health.gov/healthypeople/objectives-and-data/data-sources-and-methods/data-sources. Accessed Nov 10, 2021.
- 72.Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Scientific data. 2016;3:160018–160018. doi: 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajkomar A, Hardt M, Howell MD, Corrado G, Chin MH. Ensuring Fairness in Machine Learning to Advance Health Equity. Ann Intern Med. Dec 18 2018;169(12):866–872. doi: 10.7326/m18-1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang G. Identifying and Correcting Label Bias in Machine Learning. 2019.
- 75.Eneanya ND, Boulware LE, Tsai J, et al. Health inequities and the inappropriate use of race in nephrology. Nat Rev Nephrol. Nov 8 2021:1–11. doi: 10.1038/s41581-021-00501-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vyas DA, Eisenstein LG, Jones DS. Hidden in Plain Sight - Reconsidering the Use of Race Correction in Clinical Algorithms. N Engl J Med. Aug 27 2020;383(9):874–882. doi: 10.1056/NEJMms2004740 [DOI] [PubMed] [Google Scholar]
- 77.Shahian DM, Jacobs JP, Badhwar V, et al. The Society of Thoracic Surgeons 2018 Adult Cardiac Surgery Risk Models: Part 1-Background, Design Considerations, and Model Development. Ann Thorac Surg. May 2018;105(5):1411–1418. doi: 10.1016/j.athoracsur.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 78.Tesi RJ, Deboisblanc M, Saul C, Frentz G, Etheredge E. An increased incidence of rejection episodes. One of the causes of worse kidney transplantation survival in black recipients. Arch Surg. 1997. Jan;132(1):35–9; discussion 40. doi: 10.1001/archsurg.1997.01430250037007. [DOI] [PubMed] [Google Scholar]
- 79.Limou S, Nelson GW, Kopp JB, Winkler CA. APOL1 kidney risk alleles: population genetics and disease associations. Advances in chronic kidney disease. 2014;21(5):426–433. doi: 10.1053/j.ackd.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delgado C, Baweja M, Burrows NR, et al. Reassessing the Inclusion of Race in Diagnosing Kidney Diseases: An Interim Report From the NKF-ASN Task Force. Am J Kidney Dis. Jul 2021;78(1):103–115. doi: 10.1053/j.ajkd.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inker LA, Eneanya ND, Coresh J, et al. New Creatinine- and Cystatin C-Based Equations to Estimate GFR without Race. N Engl J Med. Nov 4 2021;385(19):1737–1749. doi: 10.1056/NEJMoa2102953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delgado C, Baweja M, Crews DC, et al. A Unifying Approach for GFR Estimation: Recommendations of the NKF-ASN Task Force on Reassessing the Inclusion of Race in Diagnosing Kidney Disease. Am J Kidney Dis. Feb 2022;79(2):268–288 e1. doi: 10.1053/j.ajkd.2021.08.003 [DOI] [PubMed] [Google Scholar]
- 83.Hall YN. Social Determinants of Health: Addressing Unmet Needs in Nephrology. Am J Kidney Dis. Oct 2018;72(4):582–591. doi: 10.1053/j.ajkd.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 84.Patzer RE, Amaral S, Wasse H, Volkova N, Kleinbaum D, McClellan WM. Neighborhood poverty and racial disparities in kidney transplant waitlisting. J Am Soc Nephrol. Jun 2009;20(6):1333–40. doi: 10.1681/asn.2008030335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volkova N, McClellan W, Klein M, et al. Neighborhood poverty and racial differences in ESRD incidence. J Am Soc Nephrol. Feb 2008;19(2):356–64. doi: 10.1681/asn.2006080934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grace BS, Clayton P, Cass A, McDonald SP. Socio-economic status and incidence of renal replacement therapy: a registry study of Australian patients. Nephrol Dial Transplant. Nov 2012;27(11):4173–80. doi: 10.1093/ndt/gfs361 [DOI] [PubMed] [Google Scholar]
- 87.Edmonds A, Braveman P, Arkin E, D. J Making the case for linking community development and health. http://www.buildhealthyplaces.org/content/uploads/2015/10/making_the_case_090115.pdf. Robert Wood Johnson Foundation. Accessed December 4, 2017.
- 88.Garg A, Boynton-Jarrett R, Dworkin PH. Avoiding the Unintended Consequences of Screening for Social Determinants of Health. Jama. Aug 23–30 2016;316(8):813–4. doi: 10.1001/jama.2016.9282 [DOI] [PubMed] [Google Scholar]
- 89.Association AM. AMA passes first policy recommendations on augmented intelligence. Accessed at www.amaassn.org/ama-passes-first-policy-recommendations-augmented-intelligence. Accessed July 6, 2018.
- 90.Barocas S, Hardt M, Narayanan A. FAIRNESS AND MACHINE LEARNING Limitations and Opportunities.
- 91.McCradden MD, Joshi S, Mazwi M, Anderson JA. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digit Health. May 2020;2(5):e221–e223. doi: 10.1016/s2589-7500(20)30065-0 [DOI] [PubMed] [Google Scholar]
- 92.Wolff RF, Moons KGM, Riley RD, et al. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann Intern Med. Jan 1 2019;170(1):51–58. doi: 10.7326/m18-1376 [DOI] [PubMed] [Google Scholar]
- 93.Peterson ED. Machine Learning, Predictive Analytics, and Clinical Practice: Can the Past Inform the Present? JAMA. Nov 22 2019;doi: 10.1001/jama.2019.17831 [DOI] [PubMed] [Google Scholar]
- 94.Loftus TJ, Tighe PJ, Filiberto AC, et al. Artificial Intelligence and Surgical Decision-making. JAMA Surg. Feb 1 2020;155(2):148–158. doi: 10.1001/jamasurg.2019.4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stubbs K, Hinds PJ, Wettergreen D. Autonomy and common ground in human-robot interaction: A field study (vol 22, pg 42, 2007). Ieee Intell Syst. May-Jun 2007;22(3):3–3. [Google Scholar]
- 96.Linegang MP, Stoner HA, Patterson MJ, et al. Human-Automation Collaboration in Dynamic Mission Planning: A Challenge Requiring an Ecological Approach. Proceedings of the Human Factors and Ergonomics Society Annual Meeting. 2006;50(23):2482–2486. doi: 10.1177/154193120605002304 [DOI] [Google Scholar]
- 97.Rosenfeld A, Zemel R, Tsotsos J. The Elephant in the Room. arXiv:1808.03305 [cs.CV]. 2018; [Google Scholar]
- 98.Miller T Explanation in artificial intelligence: Insights from the social sciences. Artif Intell. Feb 2019;267:1–38. doi: 10.1016/j.artint.2018.07.007 [DOI] [Google Scholar]
- 99.Tonekaboni S, Joshi S, McCradden MD, Goldenberg A. What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use. presented at: Proceedings of the 4th Machine Learning for Healthcare Conference; 2019; Proceedings of Machine Learning Research. http://proceedings.mlr.press [Google Scholar]
- 100.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. Feb 02 2017;542(7639):115–118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gal Y, Ghahramani Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. presented at: Proceedings of The 33rd International Conference on Machine Learning; 2016; Proceedings of Machine Learning Research. http://proceedings.mlr.press/v48/gal16.html [Google Scholar]
- 102.Prosperi M, Guo Y, Bian J. Bagged random causal networks for interventional queries on observational biomedical datasets. J Biomed Inform. Mar 2021;115:103689. doi: 10.1016/j.jbi.2021.103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Castro DC, Walker I, Glocker B. Causality matters in medical imaging. Nat Commun. Jul 22 2020;11(1):3673. doi: 10.1038/s41467-020-17478-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fleuren LM, Thoral P, Shillan D, Ercole A, Elbers PWG, Right Data Right Now C. Machine learning in intensive care medicine: ready for take-off? Intensive Care Med. May 12 2020;doi: 10.1007/s00134-020-06045-y [DOI] [PubMed] [Google Scholar]
- 105.Shortliffe EH, Sepulveda MJ. Clinical Decision Support in the Era of Artificial Intelligence. JAMA. Dec 4 2018;320(21):2199–2200. doi: 10.1001/jama.2018.17163 [DOI] [PubMed] [Google Scholar]
- 106.Wexler J, Pushkarna M, Bolukbasi T, Wattenberg M, Viegas F, Wilson J. The What-If Tool: Interactive Probing of Machine Learning Models. Ieee T Vis Comput Gr. Jan 2020;26(1):56–65. doi: 10.1109/Tvcg.2019.2934619 [DOI] [PubMed] [Google Scholar]
- 107.Birkhead GS, Klompas M, Shah NR. Uses of electronic health records for public health surveillance to advance public health. Annu Rev Public Health. Mar 18 2015;36:345–59. doi: 10.1146/annurev-publhealth-031914-122747 [DOI] [PubMed] [Google Scholar]
- 108.Adler-Milstein J, Holmgren AJ, Kralovec P, Worzala C, Searcy T, Patel V. Electronic health record adoption in US hospitals: the emergence of a digital “advanced use” divide. J Am Med Inform Assoc. Nov 01 2017;24(6):1142–1148. doi: 10.1093/jamia/ocx080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stanford Medicine 2017 Health Trends Report: Harnessing the Power of Data in Health. Accessed 23 Feb 2019. Available at: http://med.stanford.edu/content/dam/sm/sm-news/documents/StanfordMedicineHealthTrendsWhitePaper2017.pdf.
- 110.Leeds IL, Rosenblum AJ, Wise PE, et al. Eye of the beholder: Risk calculators and barriers to adoption in surgical trainees. Surgery. Aug 24 2018;164(5):1117–1123. doi: 10.1016/j.surg.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.110i. Apache Software Foundation. Spark streaming makes it easy to build scalable fault-tolerant streaming applications. https://spark.apache.org/streaming. Accessed Mar 2, 2022.
- 112.111i. Apache Cassadra. Open Source NoSQL Database. https://cassandra.apache.org/_/index.html. Accessed Mar 2, 2022.; Morse KE, Bagley SC, Shah NH. Estimate the hidden deployment cost of predictive models to improve patient care. Nat Med. Jan 2020;26(1):18–19. doi: 10.1038/s41591-019-0651-8 [DOI] [PubMed] [Google Scholar]
- 113.Blumenthal-Barby JS, Krieger H. Cognitive biases and heuristics in medical decision making: a critical review using a systematic search strategy. Med Decis Making. May 2015;35(4):539–57. doi: 10.1177/0272989X14547740 [DOI] [PubMed] [Google Scholar]
- 114.Ludolph R, Schulz PJ. Debiasing Health-Related Judgments and Decision Making: A Systematic Review. Med Decis Making. Jun 1 2017:272989X17716672. doi: 10.1177/0272989X17716672 [DOI] [PubMed] [Google Scholar]
- 115.Goldenson RM. The encyclopedia of human behavior; psychology, psychiatry, and mental health. 1st ed. Doubleday; 1970. [Google Scholar]
- 116.Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. Jan 22–29 1997;277(4):301–6. [PubMed] [Google Scholar]
- 117.Bates DW, Cullen DJ, Laird N, et al. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE Prevention Study Group. JAMA. Jul 5 1995;274(1):29–34. [PubMed] [Google Scholar]
- 118.Sanfey AG, Rilling JK, Aronson JA, Nystrom LE, Cohen JD. The neural basis of economic decision-making in the Ultimatum Game. Science. Jun 13 2003;300(5626):1755–8. doi: 10.1126/science.1082976 [DOI] [PubMed] [Google Scholar]
- 119.Kahneman D. Thinking, fast and slow. 1st pbk. ed. Farrar, Straus and Giroux; 2013:499 p. [Google Scholar]
- 120.Van den Bruel A, Thompson M, Buntinx F, Mant D. Clinicians’ gut feeling about serious infections in children: observational study. BMJ. Sep 25 2012;345:e6144. doi: 10.1136/bmj.e6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.U.S.Food & Drug Administration. Software as a Medical Device (SaMD). https://www.fda.gov/medical-devices/digital-health-center-excellence/software-medical-device-samd. Accessed Mar 2, 2022.
- 122.Van den Bruel A, Haj-Hassan T, Thompson M, Buntinx F, Mant D, European Research Network on Recognising Serious Infection i. Diagnostic value of clinical features at presentation to identify serious infection in children in developed countries: a systematic review. Lancet. Mar 6 2010;375(9717):834–45. doi: 10.1016/S0140-6736(09)62000-6 [DOI] [PubMed] [Google Scholar]
- 123.Bechara A, Damasio H, Tranel D, Damasio AR. Deciding advantageously before knowing the advantageous strategy. Science. Feb 28 1997;275(5304):1293–5. [DOI] [PubMed] [Google Scholar]
- 124.Theodorou A, Dignum V. Towards ethical and socio-legal governance in AI. Nature Machine Intelligence. 2020/01/01 2020;2(1):10–12. doi: 10.1038/s42256-019-0136-y [DOI] [Google Scholar]
- 125.Hamoni R, Lin O, Matthews M, Taillon PJ. Building Canada’s Future AI. Workforce: In the Brave New (Post-Pandemic) World. https://www.ictc-ctic.ca/wp-content/uploads/2021/03/ICTC_Report_Building_ENG.pdf. Accessed Nov 10, 2021.
- 126.Caminiti S AT&T’s $1 billion gambit: Retraining nearly half its workforce for jobs of the future. https://www.cnbc.com/2018/03/13/atts-1-billion-gambit-retraining-nearly-half-its-workforce.html. CNBC.com. Accessed Nov 10, 2021.
- 127.Moritz B Preparing everyone, everywhere, for the digital world, PricewaterhouseCoopers. https://www.pwc.com/gx/en/issues/upskilling/everyone-digital-world.html?WT.mc_id=CT1-PL50-DM2-TR1-LS40-ND30-TTA3-CN_JoeAtkinsonDigitalTransformationBlog-&eq=CT1-PL50-DM2-CN_JoeAtkinsonDigitalTransformationBlog. Accessed Nov 10, 2021.
- 128.Amazon Staff. Our Upskilling 2025 programs, Amazon. October 2020. https://www.aboutamazon.com/news/workplace/our-upskilling-2025-programs. Accessed Nov 10, 2021.
- 129.Smith Brad. Microsoft launches initiative to help 25 million people worldwide acquire the digital skills needed in a COVID-19 economy, Microsoft,
- 130.June 2020. https://blogs.microsoft.com/blog/2020/06/30/microsoft-launches-initiative-to-help-25-million-people-worldwide-acquire-the-digital-skills-needed-in-a-covid-19-economy/. Accessed Nov 10, 2021.
- 131.Pfeifer CM. A progressive three-phase innovation to medical education in the United States. Med Educ Online. Dec 2018;23(1):1427988. doi: 10.1080/10872981.2018.1427988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nasca TJ, Philibert I, Brigham T, Flynn TC. The next GME accreditation system--rationale and benefits. N Engl J Med. Mar 15 2012;366(11):1051–6. doi: 10.1056/NEJMsr1200117 [DOI] [PubMed] [Google Scholar]
- 133.Wartman SA, Combs CD. Medical Education Must Move From the Information Age to the Age of Artificial Intelligence. Acad Med. Aug 2018;93(8):1107–1109. doi: 10.1097/acm.0000000000002044 [DOI] [PubMed] [Google Scholar]
- 134.Paranjape K, Schinkel M, Nannan Panday R, Car J, Nanayakkara P. Introducing Artificial Intelligence Training in Medical Education. JMIR Med Educ. Dec 3 2019;5(2):e16048. doi: 10.2196/16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wald HS, George P, Reis SP, Taylor JS. Electronic health record training in undergraduate medical education: bridging theory to practice with curricula for empowering patient- and relationship-centered care in the computerized setting. Acad Med. Mar 2014;89(3):380–6. doi: 10.1097/acm.0000000000000131 [DOI] [PubMed] [Google Scholar]
Related links
- FAIR principles: https://www.go-fair.org/fair-principles/
- Observational Medical Outcomes Partnership: https://www.ohdsi.org/
- National Patient-Centered Clinical Research Network: https://pcornet.org
- What-If Tool: https://pair-code.github.io/what-if-tool/
- Spark Streaming: https://spark.apache.org/streaming/
- Cassandra: https://cassandra.apache.org/_/index.html


