Abstract
Pseudomonas putida G7 exhibits chemotaxis to naphthalene, but the molecular basis for this was not known. A new gene, nahY, was found to be cotranscribed with meta cleavage pathway genes on the NAH7 catabolic plasmid for naphthalene degradation. The nahY gene encodes a 538-amino-acid protein with a membrane topology and a C-terminal region that resemble those of chemotaxis transducer proteins. A P. putida G7 nahY mutant grew on naphthalene but was not chemotactic to this aromatic hydrocarbon. The protein NahY thus appears to function as a chemoreceptor for naphthalene or a related compound. The presence of nahY on a catabolic plasmid implies that chemotaxis may facilitate biodegradation.
The ability of bacteria to recognize and swim toward aromatic hydrocarbons is possibly an important prelude to their degradation, but this has not been proven. Recently we reported that naphthalene, a polyaromatic hydrocarbon and a U.S. Environmental Protection Agency priority pollutant (8), is a chemoattractant for Pseudomonas putida G7 (5). Naphthalene is produced in large quantities by the petrochemical industry as a bulk chemical for use in organic syntheses and has been a model compound for studies of biodegradation because it is relatively easily metabolized by soil bacteria (11, 21). The genes required for its complete degradation are typically located on large self-transmissible catabolic plasmids and, in the case of the catabolic plasmid NAH7, have been shown to reside within the boundaries of a defective transposon (17, 20). A mutant derivative of P. putida G7 (G7.C1) that was cured of NAH7 was not chemotactic to naphthalene. Moreover, a 25-kb subclone of the 83-kb NAH7 plasmid conferred on P. putida G7.C1 the ability to grow on naphthalene but did not restore naphthalene chemotaxis to the strain (5). This suggested that an additional gene or genes required for naphthalene chemotaxis might be present on NAH7. Here, we describe work on the molecular basis for chemotaxis to naphthalene and report the identification of a chemotaxis protein transducer gene on the catabolic plasmid NAH7.
Bacterial strains and experimental procedures.
P. putida G7 and its plasmid NAH7-cured derivative, P. putida G7.C1, were obtained from D. T. Gibson at the University of Iowa. P. putida G7.C1(pHG100) has been previously described (5). Cells were grown in Luria broth (1a) or minimal mineral salt medium (7) supplemented with a carbon source. Succinate was used as a growth substrate at a final concentration of 10 mM. Naphthalene was provided as a carbon source by the direct addition of naphthalene crystals to liquid minimal medium, due to the limited solubility of this compound. Liquid cultures were grown at 30°C with shaking at 250 rpm.
The bacterial strains and plasmids used are described in Table 1. Antibiotics were added when necessary at the following concentrations: kanamycin, 100 μg/ml; gentamicin, 10 μg/ml; and tetracycline, 50 μg/ml for P. putida and 25 μg/ml for Escherichia coli. Plasmids were introduced into P. putida from E. coli S17-1 by conjugation. Mating mixtures were spread on minimal medium containing succinate and either kanamycin, gentamicin, or both. In the case of plasmid pHG97, which was used to create the nahY mutant P. putida G7 Y1, exconjugants were screened for sensitivity to gentamicin to ensure that the suicide delivery vector, pSUP102-Gm, had been lost. A gentamicin-susceptible (Gms) kanamycin-resistant (Kmr) colony was then passaged on a minimal medium plate containing succinate and kanamycin for about a week to allow for complete loss of any wild-type NAH7 plasmid that might still be present in the strain.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. putida | ||
| G7 | Wild-type strain containing the NAH7 catabolic plasmid | 3 |
| G7.C1 | NAH7-cured derivative of G7 | D. T. Gibson |
| G7 Y1 | nahY::Km mutant | This work |
| E. coli | ||
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80lacZΔM15 | Gibco-BRL |
| S17-1 | thi pro hdsR hdsM+ recA chromosomal insertion of RP4-2(Tc::Mu Km::Tn7) | 14 |
| Plasmids | ||
| pBBR1-MCS2 | Kmr; multicopy broad-host-range vector | 9 |
| pBBR1-MCS5 | Gmr; multicopy broad-host-range vector | 9 |
| pLAFR1 | Tcr; low-copy-number broad-host-range cosmid | 4 |
| pSUP102::Gm | Gmr Cmr; suicide vector in Pseudomonas | 13 |
| pUC19 | Apr; ColE1 replicon | 19 |
| pHG59 | Kmr; pBBR1-MCS2 containing a 5.9-kb EcoRI fragment from NAH7b | This work |
| pHG60 | Kmr; pBBR1-MCS2 containing a 4-kb SmaI-EcoRI fragment from pHG59 cloned into HincII-EcoRI sites | This work |
| pHG70 | Apr; the insert of pHG60 is cloned into the KpnI-BamHI sites of pUC19, resulting in EcoRI sites on either side of the insert | This work |
| pHG95 | Apr Kmr; pHG70 with the 3′ end of nahY removed with NsiI and replaced with a GenBlock Kmr cassette from Pharmacia that had been digested with PstI | This work |
| pHG97 | Kmr Gmr; the EcoRI insert of pHG95 (containing nahY::Km) cloned into pSUP102::Gm | This work |
| pHG100 | Tcr; pLAFR1 containing a 25-kb EcoRI fragment from NAH7b | 5 |
| pHG125 | Gmr; 1.7-kb BsiWI fragment from pHG59 that contains nahY introduced into the Acc651 (KpnI) site of pBBR1-MCS5 | This work |
Apr, ampicillin resistant; Gmr, gentamicin resistant; Cmr, chloramphenicol resistant, Tcr, tetracycline resistant.
See Fig. 1.
Southern analysis was carried out with the Genius hybridization kit according to the manufacturer’s instructions (Roche Molecular Biochemicals, Inc., Indianapolis, Ind.). Plasmids were isolated from E. coli and P. putida with the Qiagen (Santa Clarita, Calif.) miniprep spin or midiprep system. Nucleotide sequencing was performed by the University of Iowa DNA Core Facility. Sequence data were analyzed with software from Textco, Inc. (West Lebanon, N.H.), Genetics Computer Group (Madison, Wis.), and the National Center for Biotechnology Information (Bethesda, Md.).
Reverse transcriptase PCR (RT-PCR) was used to determine if nahX and nahY were cotranscribed with nahJ, a structural gene of the lower naphthalene degradation pathway. RNA was isolated from naphthalene-grown cells by using the SV Total RNA isolation system from Promega (Madison, Wis.). Cells were sonicated after being resuspended in lysis buffer to improve RNA yield. RT-PCR was performed with the Access RT-PCR system from Promega according to the manufacturer’s instructions. The primers used for RT-PCR were SH-R2 (5′-GCGCCGACTAGCATTAAAAG-3′) and Sal3R-2 (5′-CGCCCAGTTGTACATCCTC-3′) for the region from nahJ to nahX. Primers NahXN (5′-CCATGATCATCTCGACCCTCGAAAC-3′) and NahYN2 (5′-CGTCATAGGCACGCTTTGATTCC-3′) were used for the region from nahX to nahY.
Chemotaxis to naphthalene was tested by a modified capillary assay (5) and also by the agarose-in-plug assay described by Yu and Alam (23). Chemotaxis to salicylate was tested only with the agarose-in-plug assay. Cells used in these assays were grown on naphthalene as a sole source of carbon and energy to mid-logarithmic phase, harvested, and resuspended in chemotaxis buffer (100 mM potassium phosphate [pH 7.0], 20 μM EDTA). Cells were motile in chemotaxis buffer for at least 1 h in the absence of an exogenous energy source. With the capillary method, the open end of a buffer-filled 1-μl capillary tube was packed with finely ground crystals of naphthalene and introduced into a chamber containing a suspension of motile cells. Chemotaxis was determined microscopically and defined as the accumulation of a cloud of cells around the tip of the capillary tube over a period of 10 to 20 min. With the agarose-in-plug method, a blob of melted agarose containing an attractant, either crystals of naphthalene or 50 mM salicylate, was placed between a microscope slide and a glass coverslip that was laid across two strips of plastic to form a bridge. After the agarose solidified to form a plug, a suspension of cells was introduced into the space between the slide and coverslip so that it flooded around the plug. Cells that responded to the naphthalene or salicylate formed a dense band, visible to the naked eye, about 0.5 mm from the plug within a period of 5 min. Cells did not respond to a plug of agarose prepared with chemotaxis buffer alone. Soft agar swarm plates were used to assess chemotaxis to succinate, 4-hydroxybenzoate, and benzoate, as described previously (2).
The NAH7 plasmid contains a gene or genes necessary for chemotaxis to naphthalene.
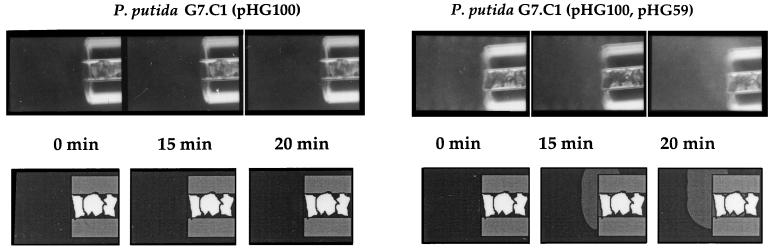
To determine whether plasmid NAH7 included genes that were required for chemotaxis to naphthalene, we started with a strain [P. putida G7.C1(pHG100)] that we had previously constructed which was cured of the NAH7 plasmid but could grow on naphthalene because it carried all of the upper and most of the lower naphthalene pathway genes on pHG100, a subclone of NAH7 (5) (Fig. 1). We presume that ortho cleavage genes located on the chromosome allowed the strain to circumvent the need for all of the lower pathway genes. This strain did not exhibit chemotaxis to naphthalene. We then introduced additional subclones of NAH7 into strain G7.C1(pHG100) on a compatible plasmid and tested for naphthalene chemotaxis. Naphthalene chemotaxis was restored by a subclone (pHG59) that contained a 5.9-kb EcoRI fragment that mapped immediately adjacent to the 25-kb EcoRI fragment used to make pHG100 (Fig. 2). Introduction of the vector pBBR1-MCS2, used to construct pHG59, into strain G7.C1(pHG100) did not restore chemotaxis to naphthalene. Strain G7.C1 carrying only pHG59 sometimes exhibited a weak chemotactic response to naphthalene. However, a consistently strong response was seen only when pHG100 was present together with pHG59 in strain G7.C1.
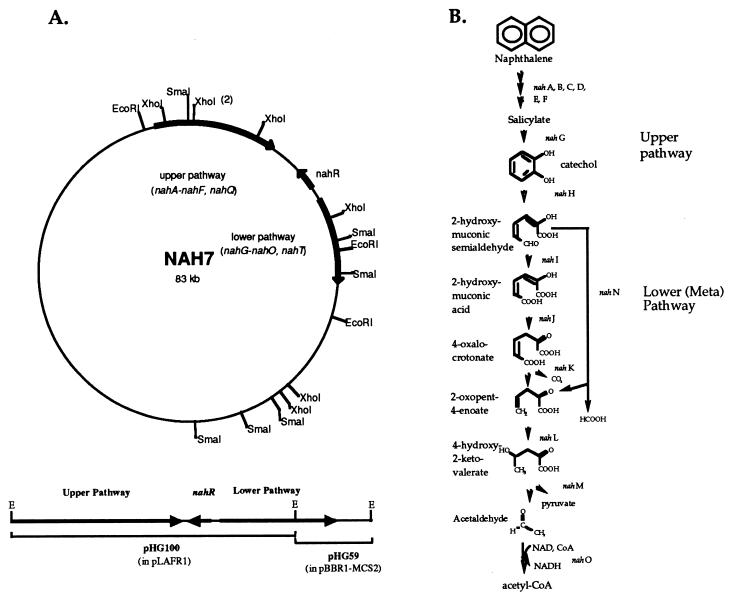
FIG. 1.
The naphthalene catabolic plasmid and degradation pathway in P. putida G7. (A) NAH7. The region of NAH7 involved in naphthalene degradation is indicated by arrows. Two subclones made from NAH7, pHG100 and pHG59, are shown. Only EcoRI sites in or around the naphthalene degradation genes are shown (E, EcoRI). (B) Naphthalene catabolic pathway. The meta pathway, which converts catechol to acetyl-coenzyme A (acetyl-CoA) and pyruvate, is encoded on the NAH7 plasmid.
FIG. 2.
Chemotactic responses of Pseudomonas strains to naphthalene in modified capillary assays. Naphthalene chemotaxis was restored when pHG59 was introduced into P. putida G7.C1(pHG100). Cells were grown on naphthalene. The results are shown in both photographic (top) and schematic (bottom) form. Naphthalene crystals are visible inside the mouths of the capillary tubes. The accumulation of a cloud of cells around the mouth of a capillary tube over time indicates a chemotactic response.
Genes present on the chemotaxis-complementing clone.
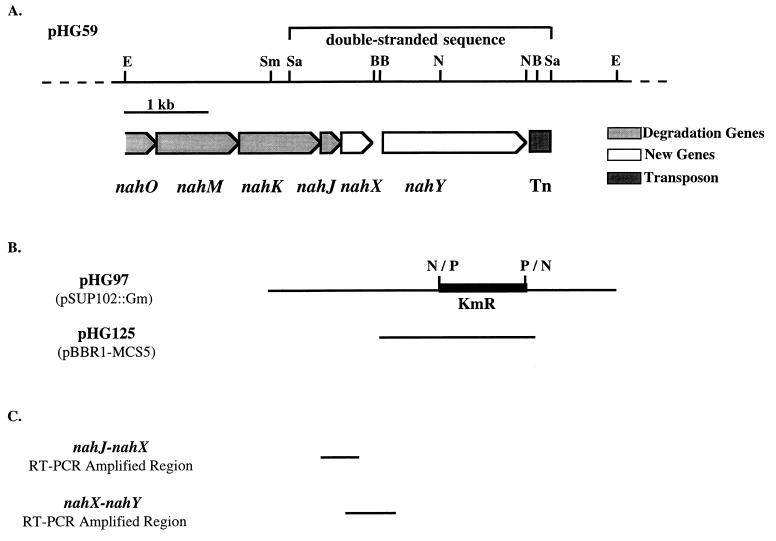
Nucleotide sequencing of pHG59 revealed several genes on the 5.9-kb EcoRI insert that had not previously been described (Fig. 3). As expected, several lower naphthalene pathway genes required for the degradation of the intermediate salicylate via a meta cleavage pathway were found, their locations having previously been roughly mapped to the region of the 5.9-kb EcoRI fragment (21). The order of the genes nahOMKJ was slightly different from that previously reported based on an analysis of Tn5 mutations in this region (21). The sequences of the nahO and nahM genes, encoding acetaldehyde dehydrogenase and 4-hydroxy-2-oxovalerate aldolase, respectively, from the naphthalene catabolic plasmid pWW60-22 had already been reported (10). The deduced amino acid sequences of the nahK and nahJ genes show a high level of identity (about 90%) to those of homologous meta pathway genes involved in phenol and toluene degradation (6, 12). nahJ encodes 4-oxalocrotonate tautomerase, and nahK encodes 4-oxalocrotonate decarboxylase.
FIG. 3.
Plasmid pHG59. (A) Restriction and gene map of pHG59. Sufficient sequencing was done to determine the locations of the starts and stops of all the genes illustrated. B, BsiWI; E, EcoRI; N, NsiI; P, PstI; Sa, SalI; Sm, SmaI. Not all SalI or NsiI sites are shown. (B) Subclones of pHG59. Plasmid pHG97 (see Table 1 for construction details) was derived from a 4-kb SmaI-EcoRI fragment of pHG59. A kanamycin resistance cassette was introduced into the NsiI sites of this 4-kb fragment. The second clone, pHG125, contains nahY and was used for complementation analysis. (C) Transcript analysis of nahJ, nahX, and nahY. The intergenic regions between nahJ and nahX and between nahX and nahY that were amplified by RT-PCR are shown.
Downstream of nahOMKJ were two new genes that we have termed nahX and nahY. nahX is predicted to encode a 140-amino-acid protein that resembles (38% amino acid identity) a Sphingomonas sp. protein (CmpX) of undefined function. It may be noteworthy that CmpX is encoded by an open reading frame that is also located among plasmid-borne meta cleavage genes (22).
nahY shows 28% deduced amino acid identity with PctB, a membrane-bound transducer protein from Pseudomonas aeruginosa that functions in chemotaxis to amino acids (16). The 538-amino-acid predicted NahY protein has several features that are conserved among chemotaxis transducer proteins, also known as methyl-accepting chemotaxis proteins (MCPs). These include two predicted membrane-spanning regions, one near amino acid 10 and the other near amino acid 200.We assume, based on homology with other MCPs, that the intervening sequence is located in the cellular periplasm. Residues 375 to 425 of NahY are about 70% identical to the chemotaxis-signaling domain of transducer proteins from E. coli and Salmonella typhimurium (15). This region, predicted to be in the cytoplasm, is highly conserved among chemotaxis transducer proteins from diverse bacteria. The C-terminal cytoplasmic regions of MCPs have two domains on either side of the signaling domain that contain glutamate or glutamine residues which are either methylated, demethylated (glutamate), or deamidated (glutamine) as part of the adaptation phase of chemotaxis (15). In the region corresponding to the methylation region designated K1 in the enteric transducers, NahY has two glutamine residues, one at position 291 and the other at position 296, that align to within one amino acid residue of the first two methylation sites of the E. coli transducers Tsr, Tar, and Trg. NahY has a glutamate at position 304 that aligns exactly with a glutamine that has been shown to serve as a site of methylation in each of the E. coli MCPs. There appears to be only one potential methylation site in the portion of NahY that corresponds to the R1 methylation region. This is a glutamate at position 506 that aligns with glutamates that are methylated in the E. coli MCPs. The last five amino acids (NWETF) of the major E. coli and S. typhimurium chemotaxis transducers serve as a methyltransferase binding sequence (18). NahY lacks this sequence.
Immediately downstream of nahY was a region of DNA that was almost identical to the left inverted repeat of Tn4655, a transposon previously reported to be part of the NAH7 plasmid (17).
RT-PCR showed that nahJ, nahX, and nahY were cotranscribed in naphthalene-grown cells. This suggests that nahX and nahY may be part of the lower naphthalene pathway operon of NAH7.
Construction of a nahY mutant.
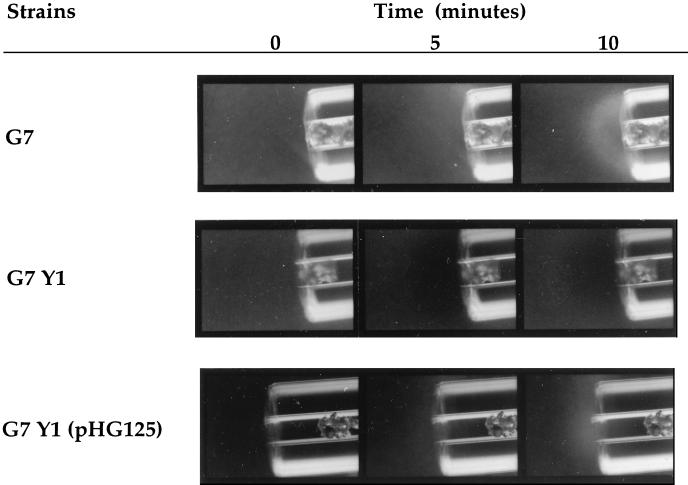
The deduced structural characteristics of NahY suggested that it might serve as a catabolic plasmid-encoded chemoreceptor for naphthalene. To test this, we constructed a mutant strain (G7 Y1) in which the 3′ end of nahY, including the domain that is highly conserved in transducer proteins, was replaced with a kanamycin resistance cassette (Table 1). Southern hybridization analysis was used to confirm the strain construction and to show that P. putida G7 Y1 contained no detectable wild-type copies of nahY. The nahY mutant strain grew on naphthalene at the same rate as its wild-type parent, but naphthalene-grown cells of the mutant were not attracted to naphthalene (Fig. 4). The chemotaxis defect appears to be specific to the attractant naphthalene, as the nahY mutant had a wild-type chemotactic response to succinate, benzoate, salicylate, and 4-hydroxybenzoate (data not shown). The unstimulated-swimming patterns of the nahY mutant were the same as those of wild-type cells, as judged by microscopic observation. Neither the wild type nor the mutant increased its swimming speed in response to the addition of any of the attractants tested. Wild-type nahY, supplied on a broad-host-range plasmid, complemented the naphthalene chemotaxis phenotype of P. putida G7 Y1 in trans (Fig. 4).
FIG. 4.
NahY is necessary for naphthalene chemotaxis, as determined by capillary assay. The mutant P. putida G7 Y1 is not chemotactic to naphthalene, but the mutation is complemented by the introduction of pHG125, a clone containing nahY. The wild-type strain also exhibited a positive response to naphthalene by the agarose-in-plug method, whereas the nahY mutant strain G7 Y1 did not (data not shown). Cells were grown on naphthalene.
NahY is a catabolic plasmid-encoded chemoreceptor.
The phenotype of the nahY mutant and the deduced amino acid sequence of NahY suggest that this protein is likely to function by binding naphthalene or a related compound on its periplasmic face to initiate chemosensory signaling in a manner analogous to that of other bacterial transducer proteins. Members of this family of proteins have been best studied in E. coli and S. typhimurium, where they bind amino acids or sugars and initiate sensory signal transduction by altering the activity of CheA, an associated histidine kinase. CheA-P phosphorylates a response regulator protein, CheY, that interacts with rotational “switch” proteins in the flagellar motor. This causes a change in swimming behavior such that cells migrate towards chemoattractants (15). A cluster of genes whose products are homologous to five of the six soluble proteins required for chemotaxis in E. coli have recently been identified in P. putida (2), suggesting that the two species process sensory information to effect chemotaxis in similar ways.
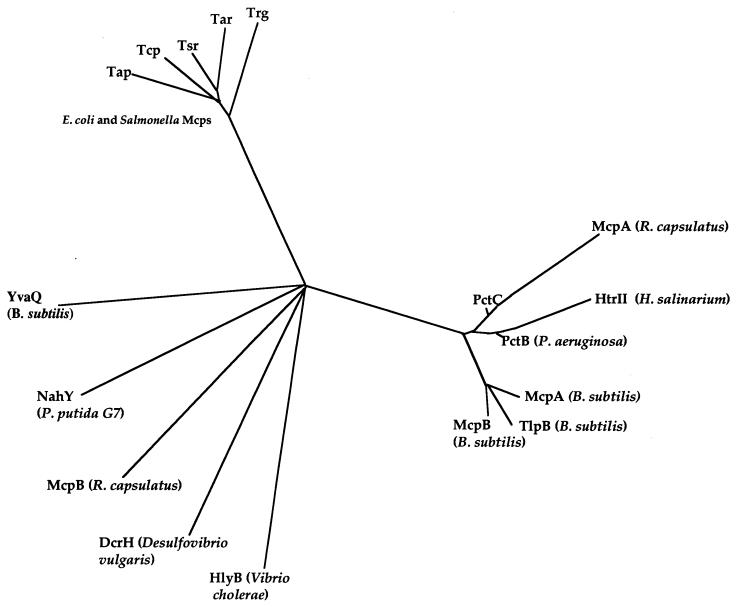
Although NahY has a signaling domain, potential methylation sites, and a membrane topology typical of chemotaxis transducer proteins, it does not show a high degree of overall amino acid identity with other MCPs. This is particularly evident if one compares the predicted N-terminal periplasmic domains of transducers and transducer-like proteins (Fig. 5). The presumed periplasmic sensing domain of NahY is clearly an outlier, rather distantly related to the N-terminal sensing domains of other bacterial and archaeal transducer proteins. This raises the possibility that NahY may have distinct chemosensing characteristics. In fact, several features of NahY chemoreceptor function remain to be explored. We still do not know, for example, exactly what NahY senses and whether it functions alone or in concert with NahX. We have been unable to consistently demonstrate naphthalene chemotaxis in a P. putida strain that carries the nahY gene in trans without naphthalene degradation genes also being present. This may suggest that a metabolite of naphthalene, rather than naphthalene itself, is the chemoattractant. If this is the case, then such a metabolite is likely to be an intermediate of the upper naphthalene degradation pathway, since the nahY mutant is not defective in chemotaxis to salicylate, the starting compound of the lower pathway.
FIG. 5.
Phylogenetic tree of the predicted N-terminal periplasmic domains of selected bacterial transducer proteins constructed by the AllAll program of the Computational Biochemistry Research Group (1). The proteins included, sources, and National Center for Biotechnology Information Entrez protein accession numbers are as follows: Tap, MCP IV (dipeptide chemoreceptor) from E. coli, 2506839; Tcp, methyl-accepting chemotaxis citrate transducer from S. typhimurium, 400235; Tsr, MCP I (serine chemoreceptor) from E. coli, 400233; Tar, MCP II (aspartate chemoreceptor protein) from E. coli, 2506837; Trg, MCP III (ribose and galactose chemoreceptor protein) from E. coli, 2506838; PctC, chemotaxis transducer protein (amino acid chemoreceptor) from P. aeruginosa, 2626833; PctB, chemotaxis transducer protein (amino acid chemoreceptor) from P. aeruginosa, 2626836; McpA, methyl-accepting chemoreceptor (presumed) from Rhodobacter capsulatus, 2126470; HtrII, sensory rhodopsin II transducer (methyl-accepting phototaxis protein II) from Halobacterium salinarium, 3023997; McpA, MCP (amino acid chemoreceptor) from Bacillus subtilis, 730002; McpB, MCP (amino acid chemoreceptor) from B. subtilis, 730003; TlpB, MCP (amino acid chemoreceptor) from B. subtilis, 730959; YvaQ, methyl-accepting chemotaxis-like protein from B. subtilis, 2635882; McpB, methyl-accepting chemoreceptor (presumed) from R. capsulatus, 2126471; DcrH, methyl-accepting chemoreceptor (presumed) from Desulfovibrio vulgaris, 887858; HlyB, hemolysin secretion protein precursor from Vibrio cholerae, 123206; NahY, chemotaxis transducer protein for naphthalene from P. putida, AF100302.
That nahY is located on the NAH7 catabolic plasmid and cotranscribed with genes for naphthalene degradation suggests that chemotaxis may be an important adjunct to biodegradation. The availability of nahY mutants should facilitate work to critically test the possibility that chemotaxis to aromatic hydrocarbons may enhance their biodegradation in natural environments.
Nucleotide sequence accession number.
The nahKJXY sequences have been assigned GenBank accession no. AF100302.
Acknowledgments
This work was supported by grant MCB9603551 from the National Science Foundation.
REFERENCES
- 1.Computational Biochemistry Research Group.http://cbrg.inf.ethz.ch /section3_1.html. Swiss Federal Institute of Technology, Zurich, Switzerland. [15 September 1998, last date accessed.]
- 1a.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 2.Ditty J L, Grimm A C, Harwood C S. Identification of a chemotaxis gene region from Pseudomonas putida. FEMS Microbiol Lett. 1998;159:267–273. doi: 10.1111/j.1574-6968.1998.tb12871.x. [DOI] [PubMed] [Google Scholar]
- 3.Dunn N W, Gunsalus I C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 5.Grimm A C, Harwood C S. Chemotaxis of Pseudomonas spp. to the polyaromatic hydrocarbon naphthalene. Appl Environ Microbiol. 1997;63:4111–4115. doi: 10.1128/aem.63.10.4111-4115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harayama S, Rekik M. Comparison of the nucleotide sequences of the meta-cleavage pathway genes of TOL plasmid pWW0 from Pseudomonas putida with other meta-cleavage genes suggests that both single and multiple nucleotide substitutions contribute to enzyme evolution. Mol Gen Genet. 1993;239:81–89. doi: 10.1007/BF00281605. [DOI] [PubMed] [Google Scholar]
- 7.Harwood C S, Nichols N N, Kim M-K, Ditty J L, Parales R E. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol. 1994;176:6479–6488. doi: 10.1128/jb.176.21.6479-6488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keith L H, Telliard W A. Priority pollutants. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 9.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–801. [PubMed] [Google Scholar]
- 10.Platt A, Shingler V, Taylor S C, Williams P A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology. 1995;141:2223–2233. doi: 10.1099/13500872-141-9-2223. [DOI] [PubMed] [Google Scholar]
- 11.Resnick S M, Lee K, Gibson D T. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Ind Microbiol. 1996;17:438–457. [Google Scholar]
- 12.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon R, O’Connell M, Labes M, Pühler A. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 14.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–789. [Google Scholar]
- 15.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 16.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 17.Tsuda M, Iino T. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol Gen Genet. 1990;223:33–39. doi: 10.1007/BF00315794. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 19.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 20.Yen K-M, Gunsalus I C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci USA. 1982;79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen K-M, Serdar C M. Genetics of naphthalene catabolism in pseudomonads. Crit Rev Microbiol. 1988;15:247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]
- 22.Yrjala K, Paulin L, Romantschuk M. Novel organization of catechol meta-pathway genes in Sphingomonas sp. HV3 pSKY4 plasmid. FEMS Microbiol Lett. 1997;154:403–408. doi: 10.1111/j.1574-6968.1997.tb12674.x. [DOI] [PubMed] [Google Scholar]
- 23.Yu H S, Alam M. An agarose-in-plug bridge method to study chemotaxis in the Archaeon Halobacterium salinarum. FEMS Microbiol Lett. 1997;156:265–269. doi: 10.1111/j.1574-6968.1997.tb12738.x. [DOI] [PubMed] [Google Scholar]