Graphical abstract
Keywords: De novo molecule library, De novo drug design, Library profiling, Deep learning
Abbreviations: PCA, Principal components analysis; HTS, High throughput screening; RNN, Recurrent neural networks; VAE, Variational autoencoders; GAN, Generative adversarial networks; FBDD, Fragment-based drug design; DDR1, Discovered potent discoidin domain receptor 1; LSTM, Long short-term memory; SCA, Scaffold-based classification approach; FDR, False discovery rate
Abstract
Various deep learning-based architectures for molecular generation have been proposed for de novo drug design. The flourish of the de novo molecular generation methods and applications has created a great demand for the visualization and functional profiling for the de novo generated molecules. An increasing number of publicly available chemogenomic databases sets good foundations and creates good opportunities for comprehensive profiling of the de novo library. In this paper, we present DenovoProfiling, a webserver dedicated to de novo library visualization and functional profiling. Currently, DenovoProfiling contains six modules: (1) identification & visualization module for chemical structure visualization and identify the reported structures, (2) chemical space module for chemical space exploration using similarity maps, principal components analysis (PCA), drug-like properties distribution, and scaffold-based clustering, (3) ADMET prediction module for predicting the ADMET properties of the de novo molecules, (4) molecular alignment module for three dimensional molecular shape analysis, (5) drugs mapping module for identifying structural similar drugs, and (6) target & pathway module for identifying the reported targets and corresponding functional pathways. DenovoProfiling could provide structural identification, chemical space exploration, drug mapping, and target & pathway information. The comprehensive annotated information could give users a clear picture of their de novo library and could guide the further selection of candidates for chemical synthesis and biological confirmation. DenovoProfiling is freely available at http://denovoprofiling.xielab.net.
1. Introduction
The main objective of drug discovery is to identify a molecule with desired biological properties [1]. Primarily, high throughput screening (HTS) techniques allow a large size of chemical library testing [2], [3]. However, HTS is expensive and with low hit rates, and this technology could be wildly used only in large pharmaceutical companies. Computational-based virtual screening methods can reduce the size of testing molecules. Various ligand-based [4], [5] and structure-based [6] virtual screening methods have been proposed. However, the cost and time consuming for developing a new drug are still demanding [7].
De novo drug design is one of the most promising and scalable approaches to address this issue, particularly, with the advances of deep learning techniques [8], [9], [10]. In the early stage, evolutionary algorithms are used for de novo molecular generation [11], which is commonly based on the combinations of molecular fragments derived from a drug-like library. Over the past years, artificial intelligence algorithms, such as deep learning, reinforcement learning, and transfer learning are proposed in the field of molecule generation, inspired by the wide applications of those methods to generate text, images, video, and music [12], [13]. Recently, several architectures for molecular generation, such as recurrent neural networks (RNN) [1], [14], [15], variational autoencoders (VAE) [16], and generative adversarial networks (GANs) [17] have been developed and proven successfully in generating target-focus molecule library. Furthermore, scaffold-constrained molecular generation methods [18], [19] are developed for lead optimization. Yang et al. also developed linker constraints molecular generation methods using deep conditional transformer neural networks for fragment-based drug design (FBDD) [20]. Zhavoronkov et al. developed a deep generative model with reinforcement learning and discovered potent discoidin domain receptor 1 (DDR1) inhibitors [21]. Yang et al. developed a generative model using long short-term memory (LSTM) neural network and identified a highly potent inhibitor against p300 [22]. These successful cases demonstrated that the deep learning-based de novo molecular design could accelerate the drug discovery process.
The flourish of the de novo molecular generation methods and applications has created a great demand for the visualization and functional profiling for the generated molecules. Generally, the generative models could generate a large chemical library based on sampling criteria and could output with various formats. The following issues, particularly for medicinal chemists, are to visualize, analyze, and select the candidates among the generated molecules. Owing to the development of combinatorial chemistry and high-throughput screening technologies, chemical structures and bioactivity data have rapidly accumulated in the past years and are becoming available in public repositories [23], [24]. There are various well-established cheminformatics and bioinformatics databases available for drug discovery, which provide comprehensive information for bioactive compounds, drugs, targets, pathways, and diseases, such as PDB database [25], PubChem [26], DrugBank [27], ChEMBL [28], and BindingDB [29]. An increasing number of publicly available databases creates good opportunities for comprehensive profiling of the de novo library.
Dealing with chemical libraries is a common practice in drug discovery. Thus, various cheminformatics tools have been developed for chemical library processing and data analysis. Well-known tools for dealing with chemical library are ChemicalToolbox [30], DataWarrior [31], WebMolCS [32], ChemMine [33], CART [34], MONA [35], and CSgator [36]. Those tools mainly focus on specific functionality, such as large library visualization, structure search, or clustering analysis. Even more, some tools are desktop applications, which limited the application. Web-based tools dedicated to de novo generated molecule profiling are rare.
In this work, we present the DenovoProfiling, a webserver for de novo generated molecule library profiling. We aim to provide a user-friendly public webserver, which supports the structure and chemical space visualization, ADMET prediction, molecular alignment, drugs profiling, target & pathway profiling. Cheminformatics tools and databases were integrated to provide comprehensive annotations for the de novo generated molecules. We believe that DenovoProfiling could be an efficient tool for the user to capture the knowledge of de novo generated molecules. DenovoProfiling is freely available at http://denovoprofiling.xielab.net.
2. Materials and methods
2.1. Framework
The framework of DenovoProfiling was outlined in Fig. 1. We integrated the well-known public database PubChem, ChEMBL, DrugBank, and employed open-source cheminformatics toolkits as well as other tools to provide comprehensive information for user-submitted de novo chemical library. The profiling process is fully automatic, in which the user only needs to submit its de novo library files with multiple formats are supported. DenovoProfiling contains 6 modules: identification & visualization, chemical space, ADMET prediction, molecular alignment, drugs mapping, and target & pathway.
Fig. 1.
The framework of the DenovoProfiling web platform.
2.2. Supported formats
Four widely used chemical formats were supported in DenovoProfiling: SDF (structure-data file), SMILES (simplified molecular-input line-entry system), InChI (International Chemical Identifier), and CDX (ChemDraw Exchange). All those formats’ files could be uploaded or be pasted and submitted to the web server, except for the binary CDX format, which cannot be pasted. The Open Babel [37] program was used for chemical file format conversion.
2.3. Modules
Currently, DenovoProfiling provides 6 profiling modules. Each module was functional individually and the user could select the module of interest. The implementations for each module were described as follows.
2.3.1. Identification & visualization
Identification & Visualization module aimed to check whether the de novo structures are already existing and visualize the de novo chemical structures. The submitted de novo molecules were converted into InChIKeys using Open Babel [37]. Subsequently, the InChIKeys were submitted to the PubChem using PubChemPy (https://pubchempy.readthedocs.io), a python package for interacting with PubChem. PubChem is the world's largest collection of freely accessible chemical information with over 109 million compounds [26]. The PubChem compound IDs (CID) were retrieved when de novo molecules were matched. ChemDoodle Web component, a light-weight JavaScript/HTML5 toolkit for chemical graphics, developed by iChemLabs was used for structure visualization [38]. For non-SDF format, Open Babel was used to generate 2D structures for structure visualization. Meanwhile, the drug-like descriptors including molecular weight (MW), ALogP, number of hydrogen bond acceptors (HBA), number of hydrogen bond donors (HBD), number of rotatable bonds (RotBonds), and topological polar surface area (TPSA) were calculated using PaDEL [39] and plotted using Radar Chart.
2.3.2. Chemical space
Chemical space visualization is an efficient way to know the structural similarity or properties similarity of the corresponding molecules through the closeness of the points in this chemical space. Each molecule is defined by a set of numerical descriptors or fingerprints and a set of all molecules corresponded to the points in the same coordinate-based space. Four important approaches including similarity maps, principal components analysis (PCA), drug-like properties distribution, and scaffold analysis were used in DenovoProfiling. The chemical similarity heatmap was generated and interactive, in which the user could move or click the cells of the similarity matrix, and the corresponding structures are visualized beside. The PubChem fingerprints and MACCS fingerprints are supported for similarity calculation. The principal component analysis (PCA) was used to visualize the chemical space based on PubChem fingerprints. The frequency distribution histogram of drug-like descriptors mentioned in Identification & Visualization was also plotted. Scaffold is an important concept for medicinal chemistry when measuring the novelty of a molecule. Generally, for medicinal chemists, a scaffold defines the core structure essential for pharmacological activity, which is dataset dependent and could vary in the different target systems. Bemis and Murcko proposed the Bemis-Murcko (BM) scaffold framework [40], an objective, invariant, and data set independent scaffold representation method. The BM scaffold method dissects molecules into ring systems, linkers, side-chain atoms, and the framework. Scaffold-based classification approach (SCA) [41], an atomic framework of BM scaffold [40] was used here and widely applied in cheminformatics studies [42] and drug discovery projects [43], [44]. The scaffolds were generated for de novo molecules and the number of molecules for each scaffold was calculated and plotted.
2.3.3. ADMET prediction
Early estimation of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) in the discovery phase could reduce the fraction of pharmacokinetics-related and toxicity-related failure in the clinical phases. 13 small molecules ADMET datasets: Caco2, Cell Permeability, P-gp Inhibitors, P-gp Substrates, Biodegradability, CYP1A2, CYP3A4, CYP2D6, CYP2C9, CYP2C19, Human Liver Toxicity, HERG, Rat Acute Oral Toxicity, Carcinogenic Potency were collected from literatures. For each dataset, the structures of the molecules were salt removed and standardized using canonical smiles. The dataset source and the corresponding reference were summarized in Table 1. Caco2 Cell Permeability dataset and P-gp Inhibitors dataset were collected from Wang’s [45] and Chen’s report [46], respectively. P-gp Substrates dataset were collected from reports of Poongavanam [47], Wang [48], and Shaikh [49], Biodegradability and CYP dataset were provided by Cheng, who published their dataset in his previous reports [50], [51]. Human Liver Toxicity dataset were collected from our previous report [52], which compiled from three public databases, including side effect resource (SIDER), OFFSIDES and Comparative Toxicogenomics Database (CTD). HERG, Rat Acute Oral Toxicity, and Carcinogenic Potency dataset were collected from Ji’s report [53]. Deep learning models were constructed using message passing neural network (MPNN) implemented in Chemprop [54]. For each dataset, an 80%/10%/10% training/validation/testing random split was employed and the area under the curve (AUC) for the test set was used as a metric to evaluate the model performance. All experiments were repeated 10 times with a different random seed. The datasets and AUC values were summarized in Table 1. The AUC values of these models range from 0.84 to 0.96. Other model performance metrics: sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were also summarized in Table S1. Y-randomization test was performed using 20 repeats with random labels. The AUC values and balanced metrics, F1 and Matthews correlation coefficient (MCC) of Y-randomization models were compared with the 10 repeated models with real labels (Table S2). These metrics for 13 ADMET dataset decreased dramatically, suggesting the constructed models are not randomly generated. These results mentioned above indicated a good prediction performance and could be used for de novo library ADMET property prediction. The applicability domain was defined using six descriptors [51], [55], Molecular Weight, LogP, TPSA, HBA, HBD, Rotatable Bonds. Descriptors were calculated with RDKit (https://www.rdkit.org), and the distribution of the descriptors were analyzed using quantile R package and summarized in Table S3 in the supporting information. For each descriptor, compounds with values among 1%∼99% values of the dataset were regard as in domain, compounds with values less than 1% value or great than 99% value were regard as out domain.
Table 1.
The collected 13 ADMET datasets and deep learning-based model performance.
| Dataset | Molecules | AUC | SD | Reference |
|---|---|---|---|---|
| Caco2 Cell Permeability | 1946 | 0.92 | 0.018 | [45] |
| P-gp Inhibitors | 4418 | 0.96 | 0.008 | [46] |
| P-gp Substrates | 2100 | 0.85 | 0.021 | [47], [48], [49] |
| Biodegradability | 1604 | 0.91 | 0.023 | [50] |
| CYP1A2 Inhibitors | 14,903 | 0.89 | 0.006 | [51] |
| CYP3A4 Inhibitors | 18,561 | 0.88 | 0.007 | [51] |
| CYP2D6 Inhibitors | 14,741 | 0.86 | 0.015 | [51] |
| CYP2C9 Inhibitors | 14,709 | 0.88 | 0.007 | [51] |
| CYP2C19 Inhibitors | 14,576 | 0.89 | 0.008 | [51] |
| Human Liver Toxicity | 2476 | 0.94 | 0.014 | [52] |
| HERG | 9636 | 0.95 | 0.006 | [53] |
| Rat Acute Oral Toxicity | 12,170 | 0.86 | 0.021 | [53] |
| Carcinogenic Potency | 833 | 0.84 | 0.044 | [53] |
2.3.4. Molecular alignment
When a de novo molecular library was generated, a straightforward point was to align the focused library, in particular scaffold focused library, to compare the structures of molecules. The molecular alignment module was designed to satisfy this demand. The minimum energy conformer for each molecule was obtained using Open Babel Gen3D operations, which concluded geometry optimization with the MMFF94 forcefield and conformational search [56]. Weighted Gaussian Algorithm (WEGA) [57], developed by the previous lab, was used here for molecular alignment. WEGA is an efficient and accurate way for molecular alignment and calculating shape similarity. The shape, pharmacophore, and combined approach in WEGA could be used in DenovoProfiling. The first molecule was used as the template for alignment. After alignment, the user could select the molecules of interest to see or download the alignment results. The three-dimensional conformation alignment was rendered using 3Dmol.js [58].
2.3.5. Drugs mapping
A similar chemical structure may have similar property or activity. Drugs Mapping module was designed to fast retrieve the drugs which are chemically similar to the de novo molecules. The structures and names of drugs were derived from DrugBank [27]. Inorganic molecules, salts, and duplicates were removed using Open Babel. The similarities between the de novo molecule against the drugs were calculated. 2D similarity calculations are based on the atom center fragment [59]. The Tanimoto coefficient was used as a metric to quantify the similarity between two molecules. The similar drugs with similarity over 0.5 against de novo molecules were preserved.
2.3.6. Target & pathway
The bioactivities of targets and corresponding ligands were derived from the ChEMBL database. Duplicates were removed and compounds with multiple binding affinity data, the most potent with minimal value were chosen. After data processing, ligand structures, target, bioactivity data, and corresponding references were obtained and saved in the MySQL database. The target proteins and their bioactivity data for submitted de novo molecules were queried by using the generated InChIKeys. The retrieved results were summarized in an interactive table and a compound target network using a dynamic, browser-based visualization library(vis.js). The targeted proteins were further functional enriched using python client of bioinformatics web service DAVID [60]. The UniProt IDs of the targets retrieved from ChEMBL were used as input for functional enrichment. The enriched KEGG pathways were provided and could be downloaded through an interactive table. For each pathway, the pathway term, gene count, percent, P-value, fold enrichment, Benjamini value, and false discovery rate (FDR) are provided.
2.4. Web server implementation
DenovoProfiling is a publicly accessible platform, which can be accessed through a web browser, such as Chrome (Highly recommended). The D3 library of JavaScript (d3js.org/) was used to illustrate the scatterplots, radical plot, and heatmaps. Storage and management of the submitted job data are implemented by MySQL. The back-end server was developed by the Golang language. The tools used for constructing the DenovoProfiling are summarized in the online tutorial of the help page.
3. Results
To test the functionality of DenovoProfiling, we collected or generated 3 datasets for different purposes (Table 2). The first dataset (Drug Dataset) contains 60 drug molecules randomly selected from DrugBank to check the corrected information retrieved from different profiling database in DenovoProfiling. This dataset aims to verify the utility of identification & visualization, chemical space, drugs mapping, target & pathway. The second dataset (Random Dataset) contains 500 molecules randomly generated using REINVENT [14], an RNN architecture pre-trained on more than one million bioactive structures from ChEMBL. We have developed an interface of REINVENT as a de novo module in our DeepScreening [61]. This dataset aimed to verify the utility of identification & visualization, chemical space, ADMET prediction, drugs mapping, target & pathway. The third dataset (Focused Dataset) contained 50 scaffold-focused de novo molecules based on a scaffold constrained molecular generation approach [18] for verifying the molecular alignment module.
Table 2.
The datasets for testing the functionality of DenovoProfiling.
3.1. Structure identification and visualization of de novo library
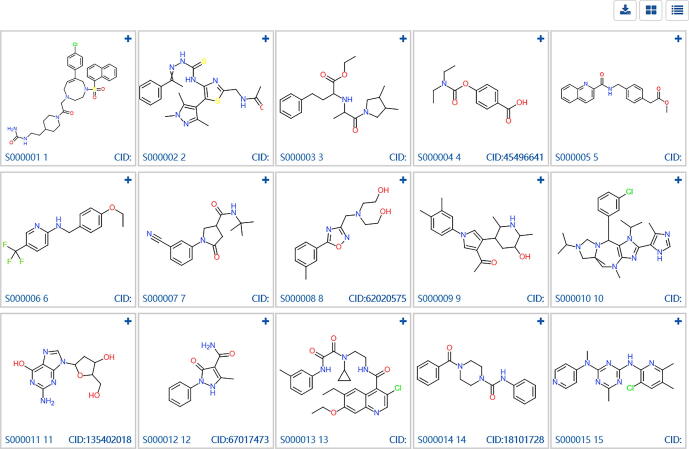
For a de novo generated library, the primary purpose was to visualize the chemical structures and evaluate the structure novelty. The Identification & Visualization module was designed to satisfy this demand. Using the Random dataset as input, the snapshot of this module was shown in Fig. 2. The user could browse the structures with mapped PubChem Compound ID (CID). The properties including molecular weight, LogP, HBA, HBD, number of rotatable bonds, TPSA were given by clicking the upright plus button. The CID was provided at the bottom right and linked the PubChem which provideed more detailed compound information.
Fig. 2.
Structure identification and visualization of de novo library using Random Dataset.
3.2. Chemical space exploration of de novo library
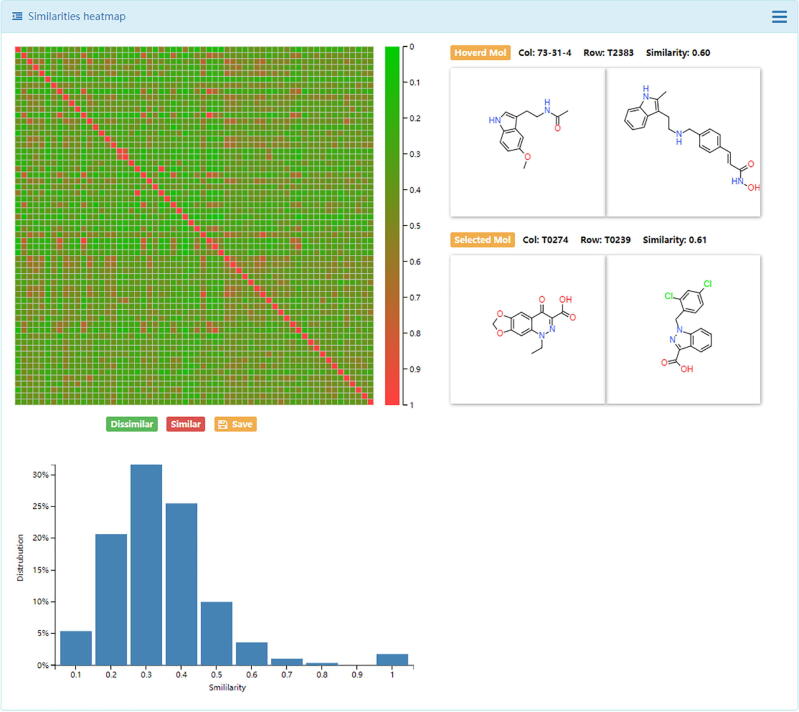
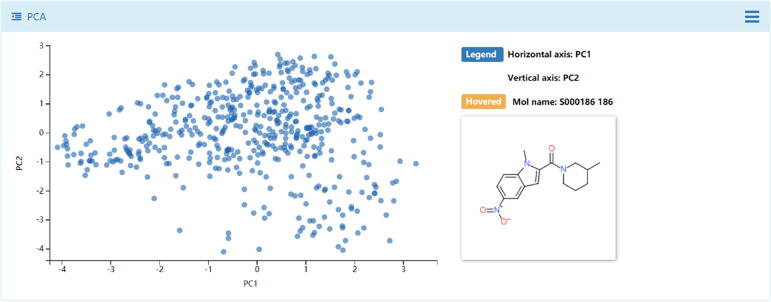
Chemical structures data are sophisticated, in particular for the de novo generated molecular library, and expert knowledge is highly required [62]. In this module, similarity maps, principal components analysis (PCA), drug-like properties distributions, and scaffold analysis were provided in DenovoProfiling. Using Drug Dataset as input, the snapshot of similarity heatmap was shown in Fig. 3. The generated similarity heatmap was interactive, in which the user could move or click the mouse to the target cell, and the corresponding structures, name of molecules, and the similarity value are visualized. Meanwhile, the distribution of the similarities was also plotted. Using the Random Dataset (500 molecules) as input, the snapshots of PCA results were shown in Fig. 4. Each point represents a molecule, and the user could move the mouse to the point, and the corresponding structure and molecule name returned immediately. Meanwhile, the distribution of drug-like properties was also plotted, as shown in Fig. 5. The scaffold is an important concept in drug discovery and medicinal chemistry. Medicinal chemists are seeking chemicals with novel scaffolds for a specific biological target [42]. Using the Random Dataset as input, the snapshots of the scaffold analysis were shown in Fig. 6, the complexity and cyclicity of the scaffolds and statistics of each scaffold were interactive illustrated with scatter plot and histogram plot (Fig. 6). As shown in Fig. 7, the structures of scaffolds and their number of molecules were illustrated in the grid table. The members of molecules for each scaffold could be browsed by clicking the upright plus button. The interactive chemical space exploration could help users capture the structure relations, descriptors landscapes of the de novo library conveniently.
Fig. 3.
Chemical space illustration using similarity heatmap based on Drug Dataset.
Fig. 4.
Chemical space illustration using principal component analysis (PCA) based on Random Dataset.
Fig. 5.
Distribution of drug-like properties based on Random Dataset.
Fig. 6.
Scaffold statistics of chemical scaffolds of de novo library based on Random Dataset.
Fig. 7.
Grid view of the chemical scaffolds of the de novo library based on Random Dataset.
3.3. ADMET profiling of de novo library
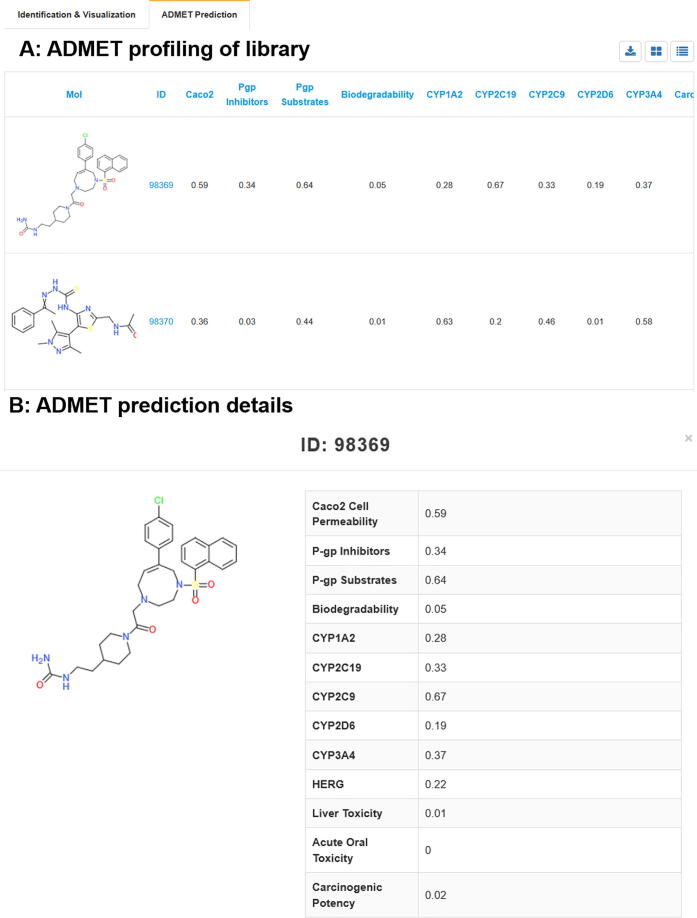
In silico prediction of ADMET properties is an important part of computer-aided drug design in pharmaceutical R&D [63]. 13 ADMET dataset (Caco2 Cell Permeability, P-gp Inhibitors, P-gp Substrates, Biodegradability, CYP1A2, CYP3A4, CYP2D6, CYP2C9, CYP2C19, Human Liver Toxicity, HERG, Rat Acute Oral Toxicity, Carcinogenic Potency) were collected and trained using message passing neural networks and yield good prediction performance models with AUC value ranging from 0.84 to 0.96. These deep learning-based classification models were further used for ADMET profiling in DenovoProfiling. The prediction scores were between 0 and 1 and the value over 0.5 indicates a positive prediction. The ADMET prediction results for Random Dataset were shown in Fig. 8. The user could browse the 13 ADMET properties in a table view and click the molecule in the grid view to get detailed ADMET prediction information. The prediction results also could be downloaded for further filtering in local though click the upright download button. As shown in Fig. 8B, the first molecule in Random Dataset shows good Caco2 cell permeability and could be P-gp substrate, CYP2C9 inhibitor, and non-toxicity based on the prediction results. In the ADMET prediction module, 13 types of ADMET properties were provided. However, machine learning based prediction models relied on experimental data with the limitations. Experimental data were from the molecular level, the cellular level, and the animal level. These different levels of data may not point to the same outcome. Caco2 Cell Permeability and Carcinogenic Potency were all cell-based models, which were limited to cellular activity. P-gp Inhibitors, P-gp Substrates, CYP1A2, CYP3A4, CYP2D6, CYP2C9, CYP2C19, and HERG are molecular level data, which were limited to the single target activity. For Rat Acute Oral Toxicity which was an in vivo toxicity indication. Human Liver Toxicity dataset was based on clinical phenotype and multiple factors could be involved, rather than structures.
Fig. 8.
ADMET prediction snapshot based on Random Dataset. A: Table view of ADMET prediction results for de novo library. B: ADMET prediction details for one molecule.
3.4. Molecular alignment of the scaffold-constrained library
Generally, medicinal chemist starts from drug target, and attempt to generate a target-focused library or a scaffold-constraint library for structural optimization. In this case, shape or pharmacophore features-based molecular alignment was a good apporach to compare the difference of the target-focused or scaffold-constraint de novo library. The shape and pharmacophore combined approach in WEGA was used in DenovoProfiling for molecular alignment. Using Focused Dataset as input, as shown in Fig. 9, DenovoProfiling correctly aligns all the structures to the first structure of the library. The important features, carboxyl, hydroxyl, and benzene ring were corrected recognized, and overlayed. The user could browse the alignment results, and select the molecules of interest to see the alignment result. Stick and line render methods were supported. The user also cloud click the download button to download all the alignment results for local analysis.
Fig. 9.
Molecular alignment of the scaffold-focused de novo library based on Focused Dataset.
3.5. Drugs mapping of de novo library
De novo generated libraries usually are random and cover a larger chemical space. Though, Identification & Visualization, as mentioned above, identify the structures which have been reported. The structural similar drugs against the de novo library were the interest of medicinal chemists. They could fast capture the novelty and pharmacological activities of the de novo compounds when compared with the drug library. Firstly, we used the Drug Dataset as input, DenovoProfiling calculated the similarity between the submitted Drug Dataset and the drug library. The similar drugs for each submitted molecule were returned. As shown in Fig. 10A, the drugs were corrected recognized, and identified 363 similar drugs with similarity values over 0.5. Then, using Random Dataset, DenovoProfiling identified 423 similar drugs with similarity values over 0.5 (Fig. 10B). The grid view (Fig. 10) and the table view (Fig. 11) are provided. For the randomly sampled 500 de novo compounds, as shown in Fig. 11, compounds with a maximal drug similarity over 0.9, and their DrugBank ID were also provided and linked to the original database. Details of this drug information could be obtained directly.
Fig. 10.
Grid view of the drugs mapping. A: Drug Dataset results; B: Random Dataset results.
Fig. 11.
Table view of drugs mapping using Random Dataset.
3.6. Target and pathway profiling for de novo library
The modules we described before were structural annotations for the de novo library. Functional profiling was another important part of user concern for de novo library proofing. Firstly, we used Drug Dataset as input, DenovoProfiling retrieved 365 bioactivity data for the 60 drugs. As shown in Fig. 12, the bioactivity data such as Ki, Kd, IC50, and EC50, and corresponding references are extracted. All those results can be analyzed via a user-friendly table view (Fig. 12). Those results were also can be downloaded for local analysis. The compound target relations were further illustrated using a compound target network (Fig. 12). We further used the Random Dataset as input, DenovoProfiling retrieved 14 bioactivity data for the 500 de novo molecules (Fig. 13). The targets are further enriched to pathways and KEGG pathways are summarized in the table (Fig. 13). DenovoProfiling enriched 115 pathways and 5 pathways for the Drug Dataset and Random Dataset, respectively (Fig. 14).
Fig. 12.
The identified targets in ChEMBL for Drug Dataset and the compound target network.
Fig. 13.
The identified targets in ChEMBL of de novo molecules and the compound target network.
Fig. 14.
The enriched KEGG pathways using the identified targets in ChEMBL. A: Drug Dataset results; B: Random Dataset results.
3.7. Time consuming benchmark test and job management
We submitted these 3 datasets for each module and counted the time consuming for the server to return results. Notably, the first module visualization & identification was required and other modules were optional. The time cost for each module were summarized in Table 3. DenovoProfiling could returns the profiling results from several seconds to several minutes. The chemical space module could take longer time in similarity calculations. The molecular alignment module is time consuming owning to the conformation generation. The ADMET prediction module is library-size depended. The results of the submitted job were saved in DenovoProfiling for 15 days. The user could use job ID to access the profiling results within its validity period.
Table 3.
The time cost for each module (seconds).
| Modules | Drug Dataset (60 mols) |
Focused Dataset (50 mols) |
Random Dataset (500 mols) |
|---|---|---|---|
| Identification & Visualization | 8 | 6 | 10 |
| Chemical Space | 26 | 31 | 124 |
| ADMET Prediction | 61 | 58 | 87 |
| Molecular Alignment | 227 | 25 | 86 |
| Drugs Mapping | 32 | 25 | 73 |
| Target & Pathway | 24 | 18 | 19 |
4. Conclusions
De novo drug design is one of the most promising and scalable approaches to accelerate the drug discovery process. Deep learning-based de novo molecular generation has shown powerful performance in generating de novo target-focused or property-focused libraries. Fast profiling the de novo generated molecules becomes a practical issue in the de novo drug design. To address this issue, we developed DenovoProfiling, a web-based profiling server for de novo generated molecules. DenovoProfiling supports structure identification and visualization, chemical space exploration, ADMET profiling, molecular alignment, drugs profiling, and target & pathway profiling. These functional modules provide structural and functional annotations for de novo molecules generated from various methods. DenovoProfiling provides comprehensive profiling and visualizations tools for de novo molecule library by integrating traditional cheminformatics approaches and state-of-the-art deep learning technologies. However, several potential weaknesses of the present study should be acknowledged. Various de novo molecular generation tools could be integrated into this platform, and user could generate de novo molecules using different methods. Meanwhile, machine learning based bioactivity prediction models could be integrated for evaluating the potency for the generated molecules. Some modules use default values to make it more convenient for users, but resulting in less choice for advanced users. These functional modules could be our future development directions. Overall, we believe this web-based tool could facilitate de novo drug design and accelerate drug discovery.
5. Availability of data and materials
The web platform can be accessible at http://denovoprofiling.xielab.net. The source code for the webserver was deposited in GitHub at https://github.com/nanomolar/DenovoProfiling.
6. Funding
This work was funded by the Guangdong Basic and Applied Basic Research Foundation (2020B1515020046), GDAS Project of Science and Technology Development (Grant No. 2019GDASYL-0103009, 2018GDASCX-0102, 2021GDASYL-20210102003), and National Natural Science Foundation of China (Grant No. 81703416, 81900797, 82072436).
7. Authors’ contributions
LX and JF designed the study. ZL, JD, and BL implemented the web site. The manuscript was written through contributions of all authors and revised by LX and JF. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We also thank professor Jun Xu from Sun Yat-sen University for providing useful suggestions.
Contributor Information
Jiansong Fang, Email: fangjs@gzucm.edu.cn.
Liwei Xie, Email: xielw@gdim.cn.
References
- 1.Segler M.H.S., Kogej T., Tyrchan C., Waller M.P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2018;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayr L.M., Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 3.M.J. Wildey, A. Haunso, M. Tudor, M. Webb, J.H. Connick, High-throughput screening, in: Annu. Rep. Med. Chem., 2017: pp. 149–195. https://doi.org/10.1016/bs.armc.2017.08.004.
- 4.Ripphausen P., Nisius B., Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discov Today. 2011;16:372–376. doi: 10.1016/j.drudis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Zheng M., Liu Z., Yan X., Ding Q., Gu Q., Xu J. LBVS: an online platform for ligand-based virtual screening using publicly accessible databases. Mol Divers. 2014;18:829–840. doi: 10.1007/s11030-014-9545-3. [DOI] [PubMed] [Google Scholar]
- 6.Slater O., Kontoyianni M. The compromise of virtual screening and its impact on drug discovery. Expert Opin Drug Discov. 2019;14:619–637. doi: 10.1080/17460441.2019.1604677. [DOI] [PubMed] [Google Scholar]
- 7.Mullard A. New drugs cost US$2.6 billion to develop. Nat Rev Drug Discov. 2014;13:877. doi: 10.1038/nrd4507. [DOI] [Google Scholar]
- 8.Yang X., Wang Y., Byrne R., Schneider G., Yang S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev. 2019;119:10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 9.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 10.Chen H., Engkvist O., Wang Y., Olivecrona M., Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 11.Devi R.V., Sathya S.S., Coumar M.S. Evolutionary algorithms for de novo drug design – A survey. Appl Soft Comput. 2015;27:543–552. doi: 10.1016/j.asoc.2014.09.042. [DOI] [Google Scholar]
- 12.Schneider G., Clark D.E. Automated De Novo drug design: are we nearly there yet? Angew Chem Int Ed. 2019;58:10792–10803. doi: 10.1002/anie.201814681. [DOI] [PubMed] [Google Scholar]
- 13.Bian Y, Xie X-Q. Generative chemistry: drug discovery with deep learning generative models, 5276 (2020) 1–29. http://arxiv.org/abs/2008.09000. [DOI] [PMC free article] [PubMed]
- 14.Olivecrona M., Blaschke T., Engkvist O., Chen H. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;9:48. doi: 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaschke T., Arús-Pous J., Chen H., Margreitter C., Tyrchan C., Engkvist O., et al. 2.0: an AI tool for de novo drug design. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00915. [DOI] [PubMed] [Google Scholar]
- 16.Blaschke T., Olivecrona M., Engkvist O., Bajorath J., Chen H. Application of generative autoencoder in De Novo molecular design. Mol Inform. 2018;37:1700123. doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimaraes GL, Sanchez-Lengeling B, Outeiral C, Farias PLC, Aspuru-Guzik A. Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models, ArXiv. (2017). http://arxiv.org/abs/1705.10843.
- 18.Langevin M, Minoux H, Levesque M, Bianciotto M. Scaffold-constrained molecular generation, J. Chem. Inf. Model. (2020) acs.jcim.0c01015. https://doi.org/10.1021/acs.jcim.0c01015. [DOI] [PubMed]
- 19.Li Y., Hu J., Wang Y., Zhou J., Zhang L., Liu Z. DeepScaffold: a comprehensive tool for scaffold-based de novo drug discovery using deep learning. J Chem Inf Model. 2020;60:77–91. doi: 10.1021/acs.jcim.9b00727. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y., Zheng S., Su S., Zhao C., Xu J., Chen H. SyntaLinker: automatic fragment linking with deep conditional transformer neural networks. Chem Sci. 2020;11:8312–8322. doi: 10.1039/D0SC03126G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Zhang R., Li Z., Mei L., Wan S., Ding H., et al. Discovery of highly potent, selective, and orally efficacious p300/CBP histone acetyltransferases inhibitors. J Med Chem. 2020;63:1337–1360. doi: 10.1021/acs.jmedchem.9b01721. [DOI] [PubMed] [Google Scholar]
- 23.Lipinski C.A., Litterman N.K., Southan C., Williams A.J., Clark A.M., Ekins S. Parallel worlds of public and commercial bioactive chemistry data. J Med Chem. 2015;58:2068–2076. doi: 10.1021/jm5011308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicola G., Liu T., Gilson M.K. Public domain databases for medicinal chemistry. J Med Chem. 2012;55:6987–7002. doi: 10.1021/jm300501t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. update: improved access to chemical data. Nucleic Acids Res. 2019;47(2019):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray S.A., Lucas X., Kumar A., Grüning B.A. The ChemicalToolbox: reproducible, user-friendly cheminformatics analysis on the Galaxy platform. J Cheminform. 2020;12:40. doi: 10.1186/s13321-020-00442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sander T., Freyss J., Von Korff M., Rufener C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J Chem Inf Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 32.Awale M., Probst D., Reymond J.L. WebMolCS: a web-based interface for visualizing molecules in three-dimensional chemical spaces. J Chem Inf Model. 2017;57:643–649. doi: 10.1021/acs.jcim.6b00690. [DOI] [PubMed] [Google Scholar]
- 33.Backman T.W.H., Cao Y., Girke T. ChemMine tools: An online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:486–491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deghou S., Zeller G., Iskar M., Driessen M., Castillo M., Van Noort V., et al. CART - A chemical annotation retrieval toolkit. Bioinformatics. 2016;32:2869–2871. doi: 10.1093/bioinformatics/btw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilbig M., Rarey M. MONA 2: a light cheminformatics platform for interactive compound library processing. J Chem Inf Model. 2015;55:2071–2078. doi: 10.1021/acs.jcim.5b00292. [DOI] [PubMed] [Google Scholar]
- 36.Park S., Kwon Y., Jung H., Jang S., Lee H., Kim W. CSgator: an integrated web platform for compound set analysis. J Cheminform. 2019;11:17. doi: 10.1186/s13321-019-0339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger M.C. ChemDoodle Web components: HTML5 toolkit for chemical graphics, interfaces, and informatics. J Cheminform. 2015;7:35. doi: 10.1186/s13321-015-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 40.Bemis G.W., Murcko M.A. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 41.Xu J. A new approach to finding natural chemical structure classes. J Med Chem. 2002;45:5311–5320. doi: 10.1021/jm010520k. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z., Ding P., Yan X., Zheng M., Zhou H., Xu Y., et al. ASDB: a resource for probing protein functions with small molecules. Bioinformatics. 2016;32:1752–1754. doi: 10.1093/bioinformatics/btw055. [DOI] [PubMed] [Google Scholar]
- 43.Zhao C., Huang D., Li R., Xu Y., Su S., Gu Q., et al. Identifying novel anti-osteoporosis leads with a chemotype-assembly approach. J Med Chem. 2019;62:5885–5900. doi: 10.1021/acs.jmedchem.9b00517. [DOI] [PubMed] [Google Scholar]
- 44.Guo Q., Zhang H., Deng Y., Zhai S., Jiang Z., Zhu D., et al. Ligand- and structural-based discovery of potential small molecules that target the colchicine site of tubulin for cancer treatment. Eur J Med Chem. 2020 doi: 10.1016/j.ejmech.2020.112328. [DOI] [PubMed] [Google Scholar]
- 45.Wang N.-N., Dong J., Deng Y.-H., Zhu M.-F., Wen M., Yao Z.-J., et al. ADME properties evaluation in drug discovery: prediction of Caco-2 cell permeability using a combination of NSGA-II and boosting. J Chem Inf Model. 2016;56:763–773. doi: 10.1021/acs.jcim.5b00642. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Li Y., Zhao Q., Peng H., Hou T. ADME evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive bayesian classification techniques. Mol Pharm. 2011;8:889–900. doi: 10.1021/mp100465q. [DOI] [PubMed] [Google Scholar]
- 47.Poongavanam V., Haider N., Ecker G.F. Fingerprint-based in silico models for the prediction of P-glycoprotein substrates and inhibitors. Bioorg Med Chem. 2012;20:5388–5395. doi: 10.1016/j.bmc.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z., Chen Y., Liang H., Bender A., Glen R.C., Yan A. P-glycoprotein substrate models using support vector machines based on a comprehensive data set. J Chem Inf Model. 2011;51:1447–1456. doi: 10.1021/ci2001583. [DOI] [PubMed] [Google Scholar]
- 49.Shaikh N., Sharma M., Garg P. Selective fusion of heterogeneous classifiers for predicting substrates of membrane transporters. J Chem Inf Model. 2017;57:594–607. doi: 10.1021/acs.jcim.6b00508. [DOI] [PubMed] [Google Scholar]
- 50.Cheng F., Ikenaga Y., Zhou Y., Yu Y., Li W., Shen J., et al. In silico assessment of chemical biodegradability. J Chem Inf Model. 2012;52:655–669. doi: 10.1021/ci200622d. [DOI] [PubMed] [Google Scholar]
- 51.Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 52.Wu Q., Cai C., Guo P., Chen M., Wu X., Zhou J., et al. In silico identification and mechanism exploration of hepatotoxic ingredients in traditional Chinese Medicine. Front Pharmacol. 2019;10:458. doi: 10.3389/fphar.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji C., Svensson F., Zoufir A., Bender A. eMolTox: prediction of molecular toxicity with confidence. Bioinformatics. 2018;34:2508–2509. doi: 10.1093/bioinformatics/bty135. [DOI] [PubMed] [Google Scholar]
- 54.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., et al. A deep learning approach to antibiotic discovery. Cell. 2020;181:475–483. doi: 10.1016/j.cell.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Klingspohn W., Mathea M., ter Laak A., Heinrich N., Baumann K. Efficiency of different measures for defining the applicability domain of classification models. J Cheminform. 2017;9:44. doi: 10.1186/s13321-017-0230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa N., Hutchison G.R. Fast, efficient fragment-based coordinate generation for Open Babel. J Cheminform. 2019;11:49. doi: 10.1186/s13321-019-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan X., Li J., Liu Z., Zheng M., Ge H., Xu J. Enhancing molecular shape comparison by weighted gaussian functions. J Chem Inf Model. 2013;53:1967–1978. doi: 10.1021/ci300601q. [DOI] [PubMed] [Google Scholar]
- 58.Rego N., Koes D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015;31:1322–1324. doi: 10.1093/bioinformatics/btu829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan X., Gu Q., Lu F., Li J., Xu J. GSA: a GPU-accelerated structure similarity algorithm and its application in progressive virtual screening. Mol Divers. 2012;16:759–769. doi: 10.1007/s11030-012-9403-0. [DOI] [PubMed] [Google Scholar]
- 60.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z., Du J., Fang J., Yin Y., Xu G., Xie L. DeepScreening: a deep learning-based screening web server for accelerating drug discovery. Database. 2019;2019:1–11. doi: 10.1093/database/baz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osolodkin D.I., Radchenko E.V., Orlov A.A., Voronkov A.E., Palyulin V.A., Zefirov N.S. Progress in visual representations of chemical space. Expert Opin Drug Discov. 2015;10:959–973. doi: 10.1517/17460441.2015.1060216. [DOI] [PubMed] [Google Scholar]
- 63.Ferreira L.L.G., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov Today. 2019;24:1157–1165. doi: 10.1016/j.drudis.2019.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The web platform can be accessible at http://denovoprofiling.xielab.net. The source code for the webserver was deposited in GitHub at https://github.com/nanomolar/DenovoProfiling.