Abstract
Hepatitis C virus mRNA contains an internal ribosome entry site (IRES) that mediates end‐independent translation initiation, requiring a subset of eukaryotic initiation factors (eIFs). Biochemical studies revealed that direct binding of the IRES to the 40S ribosomal subunit places the initiation codon into the P site, where it base pairs with eIF2‐bound Met‐tRNAiMet forming a 48S initiation complex. Subsequently, eIF5 and eIF5B mediate subunit joining, yielding an elongation‐competent 80S ribosome. Initiation can also proceed without eIF2, in which case Met‐tRNAiMet is recruited directly by eIF5B. However, the structures of initiation complexes assembled on the HCV IRES, the transitions between different states, and the accompanying conformational changes have remained unknown. To fill these gaps, we now obtained cryo‐EM structures of IRES initiation complexes, at resolutions up to 3.5 Å, that cover all major stages from the initial ribosomal association, through eIF2‐containing 48S initiation complexes, to eIF5B‐containing complexes immediately prior to subunit joining. These structures provide insights into the dynamic network of 40S/IRES contacts, highlight the role of IRES domain II, and reveal conformational changes that occur during the transition from eIF2‐ to eIF5B‐containing 48S complexes and prepare them for subunit joining.
Keywords: hepatitis C virus IRES, ribosome, eIF2, eIF5B, translation initiation
Subject Categories: Structural Biology, Translation & Protein Quality
Cryo‐EM structures of translation initiation intermediates show a dynamic network of contacts between the HCV IRES, the ribosomal 40S subunit, initiation factors and initiator tRNA.

Introduction
The canonical initiation process begins with formation of the 43S preinitiation complex (PIC) comprising the 40S ribosomal subunit, the eIF2·GTP/Met‐tRNAi Met ternary complex (eIF2‐TC), eIF1, eIF1A, and eIF3 (Jackson et al, 2010). The 43S PIC attaches to the capped 5′‐terminal region of mRNA and then scans to the initiation codon in a favorable nucleotide context (containing A/G and G at the −3 and +4 positions relative to the AUG, respectively) where it stops and forms the 48S initiation complex (IC) with established codon–anticodon base pairing. Attachment is mediated by eIFs 4A, 4B, and eIF4F, which cooperatively unwind the cap‐proximal region allowing attachment and also assist 43S PIC scanning. eIF1, in cooperation with eIF1A, stabilizes an “open” scanning‐competent conformation of the 43S PIC and monitors the fidelity of initiation codon selection (Pestova et al, 1998a; Pestova & Kolupaeva, 2002; Passmore et al, 2007; Hussain et al, 2014). Establishment of codon–anticodon base pairing in the 48S IC leads to dissociation of eIF1 and eIF5‐induced hydrolysis of eIF2‐bound GTP, and thereby switches the 40S subunit to the “closed” conformation (Unbehaun et al, 2004; Maag et al, 2005). After that, eIF5B, in its GTP‐bound form, displaces residual eIF2·GDP (Pisarev et al, 2006) and promotes the joining of the 60S subunit (Pestova et al, 2000). Interaction of eIF5B with eIF1A enhances eIF5B's subunit joining activity and the hydrolysis of eIF5B‐bound GTP, leading to the coupled release of eIF5B·GDP and eIF1A from the assembled 80S ribosome (Marintchev et al, 2003; Acker et al, 2006; Nag et al, 2016).
A number of viral mRNAs contain internal ribosomal entry sites (IRESs), structured RNA regions that mediate cap‐independent initiation of translation using only a subset of the eIFs that are required by canonical initiation. All IRES‐mediated initiation mechanisms are based on noncanonical interactions of IRESs with canonical components of the translation apparatus (Jackson et al, 2010). The ∼300‐nt‐long hepatitis C virus (HCV) IRES is located in the 5′‐terminal region of the viral genome and epitomizes a class of related RNA elements. HCV‐like IRESs occur in the genomes of pestiviruses (e.g., classical swine fever virus (CSFV)), some pegiviruses and caliciviruses, and numerous members of Picornaviridae (Arhab et al, 2020, 2022). The HCV IRES comprises two major domains (II–III), with domain III divided into several subdomains (Fig 1A). Ribosomal recruitment of HCV and HCV‐like IRESs occurs by direct binding of the IRES to the 40S subunit and does not involve scanning, group 4 eIFs, or eIF1 (Pestova et al, 1998b). Domain III binds at the back of the 40S subunit, whereas the long, bent domain II loops out and reaches into the E site. The sites of interaction with the 40S subunit include domains IIIa and IIIc (which bind to eS1, eS26, and eS27), the apex of domain IIId (which base pairs to expansion segment (ES) 7 of 18S rRNA), domain IIIe (which interacts with helix (h) 26 of 18S rRNA), and the apex of domain II, which interacts with uS7 and eS25 in the head and uS11 on the platform of the 40S subunit, intercalating into the mRNA‐binding channel, and causing tilting of the head and forcing the 40S subunit to adopt the open conformation (Kolupaeva et al, 2000; Kieft et al, 2001; Hashem et al, 2013; Malygin et al, 2013a, 2013b; Matsuda & Mauro, 2014; Quade et al, 2015; Yamamoto et al, 2015; Angulo et al, 2016; Yokoyama et al, 2019).
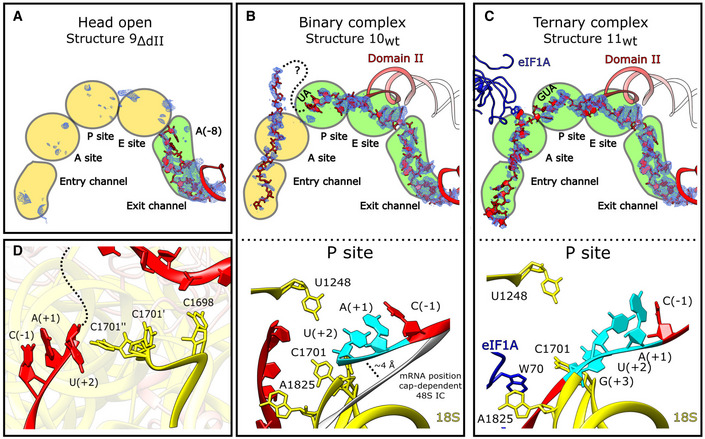
Figure 1. Overview of HCV IRES‐mediated initiation complexes.
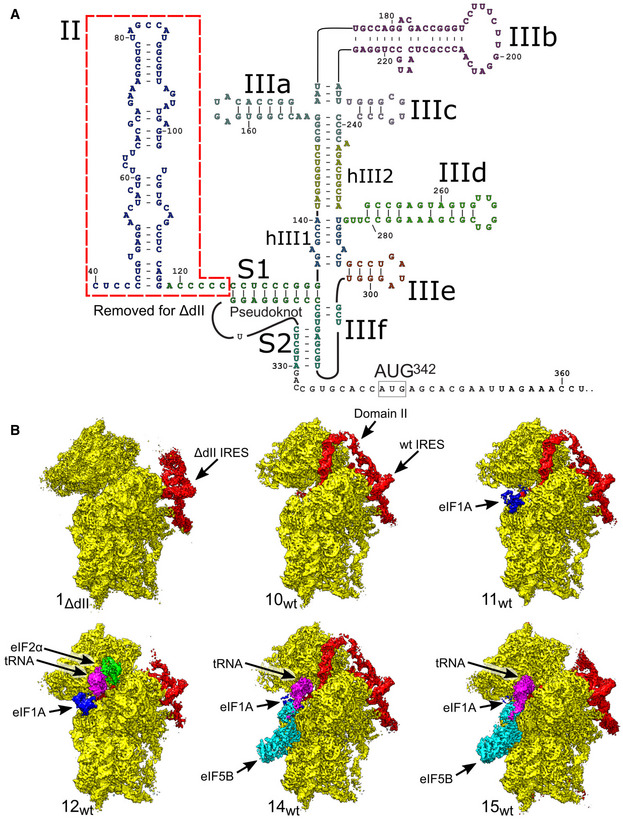
-
ASecondary structure of the HCV IRES, annotated to show individual elements.
-
BSegmented maps of the indicated ribosomal complexes assembled on the wt or ΔdII HCV IRES, showing the 40S subunit (yellow), IRES (red), eIF1A (blue), Met‐tRNAi Met (magenta), and initiation factors eIF2 (green) or eIF5B (cyan).
Although domain II does not contribute to the affinity of the HCV IRES to the 40S subunit (e.g., Kieft et al, 2001; Spahn et al, 2001), the open conformation of the 40S subunit promoted by domain II facilitates loading of the region containing the initiation codon into the mRNA‐binding channel, accounting for the stimulatory role of domain II during initiation on HCV‐like IRESs (Honda et al, 1996; Reynolds et al, 1996; Odreman‐Macchioli et al, 2001; Filbin & Kieft, 2011). Upon binding to the 40S subunit, the initiation codon is placed in the immediate vicinity of the P site, where it base pairs with the anticodon of Met‐tRNAi Met as a part of the eIF2‐TC, leading to the formation of the 48S IC (Pestova et al, 1998b). After that, eIF5 and eIF5B mediate the subunit joining step to complete the formation of the elongation‐competent 80S ribosome (Locker et al, 2007; Pestova et al, 2008; Terenin et al, 2008). Notably, in 80S complexes assembled on the HCV IRES, the P‐site Met‐tRNAi Met, and eIF5B·GTP, which correspond to the last stage in the initiation process prior to the formation of an elongation‐competent ribosome, the tilt of the 40S subunit is reversed, and the apex of domain II is released from its position on the head of the 40S subunit (Yamamoto et al, 2014). Remarkably, when levels of active eIF2 are reduced due to stress‐induced phosphorylation, Met‐tRNAi Met can be recruited by eIF5B instead of the IRES/40S complexes (Pestova et al, 2008; Terenin et al, 2008). In both eIF2‐ and eIF5B‐mediated pathways, eIF1A enhances 48S complex formation (de Breyne et al, 2008; Jaafar et al, 2016), whereas eIF1 inhibits the process and even induces dissociation of preassembled 48S ICs, but this inhibition can be alleviated by deletion of domain II (Pestova et al, 2008).
In addition to 40S subunits, HCV and related IRESs also bind to eIF3 via their apical IIIa and IIIb domains (Sizova et al, 1998; Pestova et al, 1998b; Ji et al, 2004; Hashem et al, 2013). Strikingly, in 40S/IRES/eIF3 complexes, HCV‐like IRESs displace eIF3 from its ribosomal position (Hashem et al, 2013), usurping eIF3's key ribosomal contacts involving eS1, eS26, and eS27 (des Georges et al, 2015). Moreover, the ribosome‐binding surface of eIF3 is now involved in interaction with the IRES (Hashem et al, 2013). In in vitro reconstituted initiation reactions, eIF3 only modestly enhances 48S complex formation on HCV‐like IRESs (Pestova et al, 1998b; Hashem et al, 2013), which led to the suggestion that in vivo, the role of the eIF3/IRES interaction is likely to relieve the competition between the IRES and eIF3 for a common ribosomal binding site, and to reduce the formation of 43S PICs, thereby favoring translation of viral mRNAs (Hashem et al, 2013).
Cryo‐EM studies have been indispensable in providing insights into the architecture and interactions of HCV and HCV‐like IRES ribosomal complexes, and the mechanism of the IRES function, initially through low‐resolution 40S/HCV IRES and 80S/HCV IRES structures (Spahn et al, 2001; Boehringer et al, 2005) and continuing with sub‐nanometer resolution structures of 40S/eIF3/CSFV IRES and 80S/Met‐tRNAi Met/eIF5B·GMPPNP/HCV IRES functional complexes (Hashem et al, 2013; Yamamoto et al, 2014), and the more recent near‐atomic resolution reconstructions of 80S·HCV IRES complexes (Quade et al, 2015; Yamamoto et al, 2015; Yokoyama et al, 2019). However, despite these advances, the structures of 48S ICs assembled on the HCV IRES, and the transitions between different states in the initiation pathways and accompanying conformational changes have remained unknown. To fill these gaps, we present cryo‐EM structures of HCV IRES ribosomal complexes up to 3.5 Å resolution that cover all major stages of IRES‐mediated initiation pathways, from IRES binding to the 40S subunit through eIF2‐containing 48S ICs to the final eIF5B‐containing 48S ICs immediately prior to the joining of the 60S subunit. Individually, these structures also provide detailed insights into the dynamic network of contacts between the IRES and the 40S subunit, highlight the role of IRES domain II, and importantly, include the first structure of eIF5B bound to the 40S subunit, prior to subunit joining.
Results
Overview of cryo‐EM analysis of initiation complexes assembled on the wt and the ΔdII HCV IRES with eIF2 or eIF5B
To capture discrete states in the eIF2‐ and the eIF5B‐mediated initiation pathways on the HCV IRES and to visualize the role of IRES domain II in these processes, initiation complexes were reconstituted in vitro by incubating the wt or the ΔdII mutant IRES (Fig 1A) with individual purified translation components. To follow the eIF2‐mediated pathway, assembly mixtures contained the wt or the ΔdII IRES, 40S subunits, eIF2, eIF3, eIF1A, and Met‐tRNAi Met, and to follow the eIF5B‐mediated pathway, eIF2 was replaced by eIF5B, thus yielding four discrete sample types (Appendix Table S1). Cryo‐EM grids of each complex were imaged at 300 kV producing high‐contrast micrographs with easily identifiable 40S ribosomal particles (Appendix Fig S1A–E and Table S2). The images were processed using maximum‐likelihood classification techniques implemented in Relion 3.1 (Scheres, 2012, 2016; Zivanov et al, 2018, 2019), yielding 18 structures containing different sets of components at resolutions as high as 3.5 Å (Figs 1B and EV1A, Appendix Figs S1F and G, and S2, Table S3). Although some flexible regions had a poor local resolution (e.g., eS12 in the 40S head or IRES domain IIIb), most of the ribosomes, all IRES‐ribosome contacts, and all initiation factors present had resolutions, between 3–7 Å (Appendix Figs S2 and S3A–F), that allowed modeling of these components. None of the structures obtained contained eIF3. During initiation on HCV‐like IRESs, eIF3 interacts with the apical region of IRES domain III rather than with the 40S subunit (Hashem et al, 2013). This interaction is sensitive to the process of grid preparation and is more stable when grids have thicker ice, so that imaging complexes that contain eIF3 require the intentional selection of regions with sufficiently thick ice (e.g., Hashem et al, 2013; Neupane et al, 2020); however, our study aimed to determine the details of ribosomal interactions with the IRES, initiation factors, and Met‐tRNAi Met at high resolution, which relies on imaging in regions with thinner ice. Employing an imaging strategy that is unlikely to capture eIF3‐containing states does not affect data interpretation because all complexes studied can be assembled efficiently without eIF3 (Pestova et al, 1998b, 2008) and because the structure of the IRES is not affected by the binding of eIF3 (Sizova et al, 1998).
Figure EV1. All HCV IRES‐mediated initiation complexes.
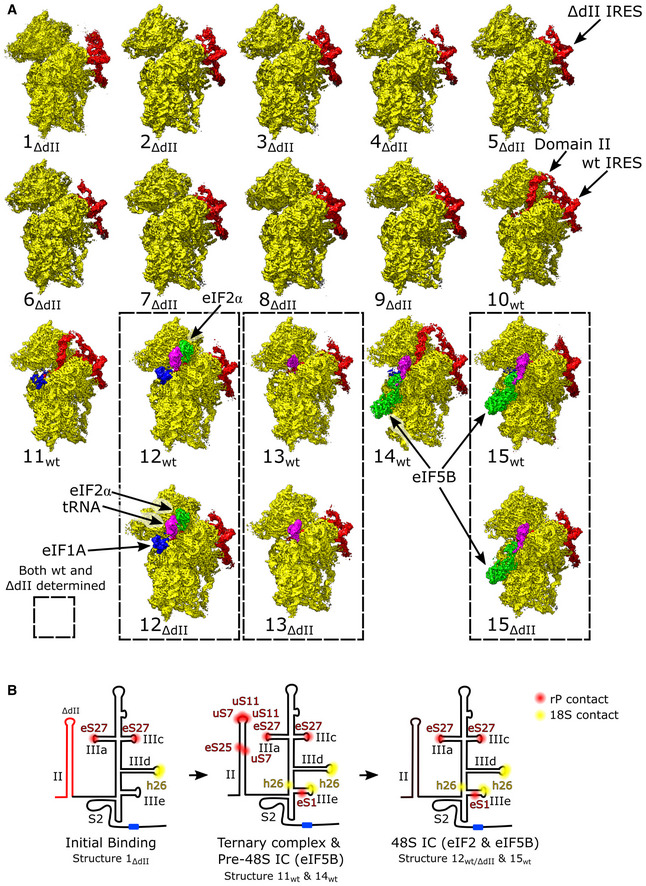
-
ASegmented maps of the indicated 40S ribosomal complexes assembled on the wt or ΔdII HCV IRES, showing the 40S subunit (yellow), IRES (red), eIF1A (blue), Met‐tRNAi Met (magenta), and initiation factors eIF2 (green) or eIF5B (cyan). Complexes assembled on the wt or ΔdII HCV IRES that share identical 40S subunit conformation and factor composition are enclosed by dashed lines.
-
BInteraction between the IRES and the 40S subunit at different stages of initiation. Contacts with ribosomal proteins (red) and 18S rRNA (yellow), and the AUG codon (blue) are marked for the indicated complexes. See Appendix Table S4 for additional information.
The structures obtained comprise the 40S/IRESΔdII binary complex in various conformational states (structures 1ΔdII‐9ΔdII); the 40S/IRESwt binary complex in a single conformational state (structure 10wt); the 40S/eIF1A/IRESwt ternary complex (structure 11wt); 48S ICs containing 40S subunits in the closed conformation, eIF2, Met‐tRNAi Met, eIF1A, and the wt or the ΔdII IRES (structures 12wt and 12ΔdII); 48S complexes containing the wt or the ΔdII IRES base‐paired with the P‐site Met‐tRNAi Met but lacking eIF2 and thus likely mimicking the stage after eIF2 dissociation following hydrolysis of GTP (structures 13wt and 13ΔdII); the pre‐48S IC containing 40S subunits in the open conformation, eIF5B, eIF1A, and the wt IRES base‐paired with P‐site Met‐tRNAi Met and IRES dII inserted into the E site (structure 14 wt); and 48S ICs containing 40S subunits in the closed conformation, eIF5B, eIF1A, Met‐tRNAi Met, and the wt or the ΔdII IRES (structures 15wt and 15ΔdII; Figs 1B and EV1A).
Thus, the structures obtained cover the entire initiation pathway, from the initial binding of the IRES to the 40S subunit to the formation of the eIF5B‐containing 48S complex prior to subunit joining.
Stepwise binding of the IRES to the 40S subunit
All previously published structures of the IRES showed domains IIIa/IIIc/IIId/IIIe/IIIf in an identical conformation on the platform side of the 40S subunit, regardless of the presence or absence of domain II (Spahn et al, 2001; Quade et al, 2015; Yamamoto et al, 2015; Weisser et al, 2017; Yokoyama et al, 2019). Although domain II is required for efficient translation (e.g, Reynolds et al, 1996; Filbin & Kieft, 2011) its contribution to IRES binding to the ribosome is minor and binding occurs with similar affinity for both the wt and ΔdII IRES (Kieft et al, 2001; Ji et al, 2004). For both wt and ΔdII IRES, their structures show binding to the 40S subunit platform involves multiple contacts formed by regions of the IRES (domains IIIa, IIIc, IIId, IIIe, and helix (h) S2) and ribosomal proteins eS1, eS27, and eS28, and h26 in ES7 of 18S rRNA (e.g., Quade et al, 2015; Weisser et al, 2017; Yamamoto et al, 2015; Fig EV1B). Deletion or mutation of these domains (IIIa‐f) impairs ribosomal binding of the IRES to different extents, reflecting their cumulative importance (Kieft et al, 2001; Ji et al, 2004). In contrast to all previous structures of the HCV IRES, we observed a small proportion of 40S/IRES binary complexes formed on the ΔdII IRES with conformational differences in the individual positions of domains IIIa/IIIc/IIId/IIIe/IIIf (structures 1ΔdII‐6ΔdII). Examination of these complexes (structures 1ΔdII‐5ΔdII) revealed the ΔdII IRES undergoing a transition from a minimally associated state (structure 1ΔdII) to the canonically bound conformation observed here (structures 6ΔdII and 7ΔdII‐15wt/ΔdII) and elsewhere (e.g., Spahn et al, 2001; Quade et al, 2015; Yamamoto et al, 2015; Weisser et al, 2017; Yokoyama et al, 2019). By ordering structures 1ΔdII‐6ΔdII based on the number of IRES‐40S subunit contacts we inferred a putative sequence of binding events between the ΔdII IRES and the 40S subunit (Fig 2A and Appendix Table S4).
Figure 2. Initial events during binding of the HCV IRES to the 40S subunit.
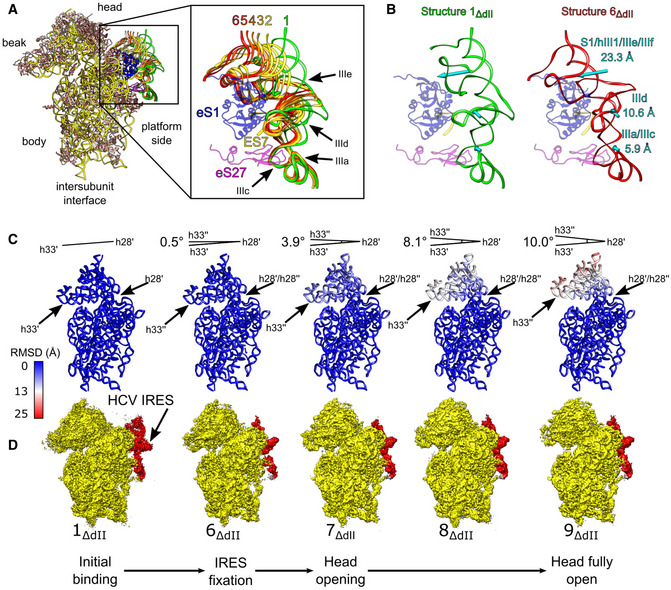
-
AIRES models from structures 1ΔdII‐6ΔdII aligned on the 40S subunit. IRES domains and ribosomal proteins eS1 (blue), eS27 (magenta), and 18S rRNA ES7 (yellow) are indicated (inset).
-
BComparison between minimally bound (structure 1ΔdII) and fully bound (structure 6ΔdII) IRES complexes showing the displacement of the indicated IRES domains (cyan arrows).
-
CRMSD (Å) of 18S rRNA for complexes shown directly underneath in (D) relative to the minimally bound state (structure 1ΔdII), color‐coded as in the inset key. The angle formed between helix 33 (h33′) and helix 28 (h28′) in structure 1ΔdII and helix 33 (h33′′) in other complexes is indicated.
-
DSegmented maps for the indicated complexes showing the 40S subunit (yellow) and IRES (red) organized in a putative sequence showing the minimally bound state (structure 1ΔdII), canonical IRES binding (structure 6ΔdII), and the induction of opening of the head of the 40S subunit (structures 7ΔdII‐9ΔdII).
Comparison between the least‐ and the most‐bound states (structures 1ΔdII and 6ΔdII) shows that the IRES domains undergo displacement of varying extents during IRES binding (Fig 2B). Across all structures, the most uniform regions of the IRES are the linked domains IIIa and IIIc (Fig 2A and B), which are known to contact eS27 via nt. 163 and 233 (Quade et al, 2015; Yamamoto et al, 2015). Given that we identified no complexes in which domain IIIa/IIIc showed major conformational differences regardless of the conformation of the rest of the IRES, the impairment of IRES activity by nucleotide substitutions in these domains (e.g., Tang et al, 1999; Kieft et al, 2001), suggests that the observed interactions between domain IIIa/IIIc and eS27 are critical for correct IRES binding to the 40S subunit and hence for function. Other IRES domains are more dynamic. Thus, to transition from the least‐ to the most‐bound states involves the translation of domain IIId by 10.6 Å toward the intersubunit face and domains hIII1/IIIe/IIIf by 23.3 Å toward the mRNA exit channel, whereas domains IIIa/IIIc move by 5.9 Å (Fig 2B). The relative differences in the displacement of these domains show how the IRES attaches to the ribosome in a zipper‐like fashion by building consecutive contacts from the stable domain IIIa/IIIc region to the terminal domains hIII1/IIIe/IIIf.
The local resolution was sufficient to identify specific 40S/IRES contacts and to follow their dynamics in structures 1ΔdII‐6ΔdII. The IRES fully bound to 80S ribosomes form six interactions with ES7: four Watson‐Crick base pairs between ES7 nt. U1115 and C1116–1118 and IRES domains hIII1 (A136) and IIId (GGG266–268), respectively; a reverse Hoogsteen base pair between ES7 nt. U1114 and domain IIIe (A296); and a stacking interaction between ES7 nt. U1114 and domain IIIe (G295; Quade et al, 2015; Yamamoto et al, 2015). Although this network of interactions is present in structures 6ΔdII and 7ΔdII‐15wt/ΔdII, the full complement was not observed during the early‐stage association of the IRES with the 40S subunit (structures1ΔdII‐5ΔdII; Fig 2B and Appendix Table S4). Initially, structure 1ΔdII showed only two of the six canonical interactions with ES7, namely the base pairs between domain IIId (G267–268) and ES7 nt. C1116–1117. The position of domain IIId in structure 1ΔdII also allows some interaction between this domain and α‐helix 6 in eS1, a region that contains Lys199, a residue that interacts with domain IIIe in the fully bound IRES (Quade et al, 2015; Yamamoto et al, 2015). These contacts between ES7 and domain IIId, along with the domain IIIa/IIIc interactions with eS27 are the only bonds between the IRES and the 40S subunit in this complex. Structure 2ΔdII maintains all of these contacts, but the repositioning of domain IIId by 4.4 Å relative to structure 1ΔdII allows for the formation of the third Watson‐Crick base pair, between ES7 (C1118) and domain IIId (G266). In the following complex (structure 3ΔdII), domain IIId moves closer to the 40S platform by 2.8 Å; hS1/hS2/hIII1/IIIe/IIIf moves by 5.0 Å, whereas the position of the stable domain IIIa/IIIc changes by only 1.5 Å. This repositioning breaks none of the contacts observed in structure 2ΔdII and allows the formation of a stacking interaction between domain IIIe G295 and ES7 U1114. The subsequent complex, structure 4ΔdII, maintains all previous contacts and forms the fifth interaction with ES7, between domain IIIe (A296) and ES7 (U1114). Globally, the IRES domains continue to move closer to their canonically bound positions, with domain IIId shifting by 2.8 Å and domains hS1/hIII1/IIIe/IIIf moving by 4.7 Å compared with structure 3ΔdII. Although structures 5ΔdII and 6ΔdII have both formed the final canonical contact, a base pair between domain hIII1 (A136) and ES7 (U1115), these complexes differ by the position of hS1 and the degree of accommodation of hS2/IIIf, with structure 6ΔdII resembling the fully accommodated state (e.g., structures 7ΔdII‐15wt/ΔdII). Structures 5ΔdII and 6ΔdII are also the first complexes in which the IRES is in a position to form a hydrogen bond between Asn147 in eS1 and the phosphate backbone of GG300–301, an interaction that is maintained in all subsequent complexes.
Taken together, this series of structures (1ΔdII‐6ΔdII) indicates that domains IIIa/IIIc, the first elements of the IRES to form canonical contacts with the 40S subunit, act as a pivot to complete the docking of domain IIId and hIII1/IIIe/IIIf onto ES7. These structures (1ΔdII‐6ΔdII) may potentially represent transient states in the binding of both the ΔdII and wt IRES, which we were able to capture in samples prepared using the ΔdII IRES because the altered kinetic landscape of the initiation pathway in the absence of domain II allowed them to accumulate.
An important corollary of IRES binding is the conformational changes that occur in the 40S subunit. Whereas complexes with an incompletely accommodated IRES (structures 1ΔdII‐5ΔdII) contain 40S subunits in the same closed conformation, ribosomal structures with the full complement of IRES/40S contacts (structure 6ΔdII‐9ΔdII) show striking differences in the position of the head, from the closed conformation in structure 6ΔdII (matching the head position in structures 1ΔdII‐5ΔdII) to the fully open state in structure 9ΔdII (Fig 2C and D). Although structures 7ΔdII‐9ΔdII have a canonically bound IRES with domain III contacting eS1, eS27, and ES7 as in structure 6ΔdII, they show large‐scale conformational changes to the head as the 40S subunit transitions from semi‐closed (structure 7ΔdII) to fully open (structure 9ΔdII) states. Thus, structure 7ΔdII opens by 3.9°, structure 8ΔdII by 8.1°, and structure 9ΔdII by 10.0° compared with the conformation of the 18S rRNA in structures 1ΔdII‐6ΔdII. These global changes to the position of the 40S head are reflected in changes in the P site as the distance between U1248 and C1701 increases from 7.3 Å, to 9.9 Å and finally to 11 Å in structures 7ΔdII, 8ΔdII, and 9ΔdII, respectively.
Thus, even in the absence of domain II, the establishment of the full complement of IRES/40S contacts results in the transition of the 40S subunit from the closed to the open state, which is required for the accommodation of the initiation codon and surrounding regions in the mRNA‐binding channel.
Accommodation of the IRES in the mRNA‐binding channel
In contrast to 40S/IRES binary complexes assembled on the ΔdII IRES, which showed differences in the conformation of the 40S subunit head and IRES position, binary complexes assembled on the wt IRES yielded a uniform structure that was refined to 3.8 Å resolution from 119,320 particles (structure 10wt; Fig 1B). Both the conformation of the 40S subunit and its contacts with IRES domains IIIa/IIIc/IIId/IIIe/IIIf in structure 10wt are identical to those in structure 9ΔdII (Fig EV2A and B). Structure 10wt was assembled using the wt IRES, and so, domain II is present and inserted into the 40S subunit E site, in a conformation identical to that observed in IRES/80S complexes (Quade et al, 2015; Yamamoto et al, 2015). Superposition of structure 10wt and a closed‐state 40S subunit shows that in the latter, steric clashes between uS7 and domain II would prohibit insertion of this domain into the E site. Thus, domain II prevents the 40S subunit from returning to the closed state.
Figure EV2. Conformational changes in the 40S subunit induced by association with the wt or ΔdII HCV IRES.
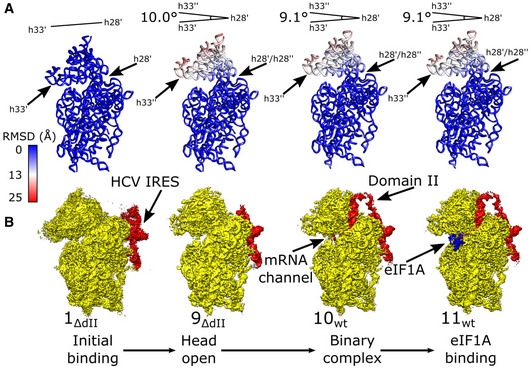
-
ARMSD (Å) of 18S rRNA for complexes shown directly underneath in (B) relative to the minimally bound state (structure 1ΔdII), color‐coded as in the inset key. The angle formed between helix 33 (h33′) and helix 28 (h28′) in structure 1ΔdII and helix 33 (h33′′) in other complexes is indicated.
-
BSegmented maps for the indicated complexes showing 40S subunit (yellow), IRES (red), and eIF1A (blue) organized in a putative sequence showing the minimally bound state (structure 1ΔdII), fully opened binary complex containing ΔdII IRES (structure 9ΔdII), binary complex containing the wt IRES (structure 10wt), and the eIF1A‐containing ternary complex (structure 11wt).
For the wt IRES, we also obtained the structure of the 40S·IRES·eIF1A ternary complex that was refined to 3.8 Å resolution from 204,782 particles (structure 11wt; Fig 1B). The conformation of the 40S subunit and the position of the IRES were identical to those in the 40S/IRESwt binary complex (structure 10wt; Figs 1B and EV2B). The complex clearly shows density for eIF1A located between 18S rRNA h44 and the ribosomal proteins eS30 and uS12, allowing us to model the OB domain and the C‐terminal subdomain of eIF1A (residues 22–122). Although structure 11wt lacks tRNA and the 40S subunit is in the open state, the position and conformation of eIF1A on the ribosome are identical to those in canonical 48S complexes (Brito Querido et al, 2020; Simonetti et al, 2020).
As expected, binary complexes assembled on the ΔdII IRES and containing the 40S subunit in the closed conformation (1ΔdII‐6ΔdII) do not have mRNA in the mRNA‐binding channel. However, although structures 9ΔdII, 10wt, and 11wt all have 40S subunits in the identical open conformation that is required for loading mRNA into the channel, they differ strongly in the degree of accommodation of the initiation codon and surrounding regions (Fig EV2A and B). In the 40S/IRESΔdII binary complex (structure 9ΔdII) there is clear density for mRNA only in the exit portion of the channel up to the (−8) position (HCV nt 334; Fig 3A). The wt 40S/IRES binary complex (structure 10wt), on the other hand, shows density corresponding to mRNA from the exit portion, through the E site, where it is stabilized by domain II, to AU342‐3 in the P site (Fig 3B). The (+3) nucleotide, G344, could not be identified due to disorder in the map. Although there is additional mRNA density in the mRNA channel ∼8 Å from the (+3) P‐site nucleotide, the identity of these nucleotides does not correspond to those that immediately follow the start codon, and this density extends 20 Å out from the mRNA channel, suggesting that after the P site, the mRNA is looped out. Thus, the presence of domain II results in accommodation of the additional nine nucleotides from the mRNA exit to the P site (i.e., from G335 to U343), while density corresponding to mRNA located downstream from the P site is likely a mixture of different sequence registers of mRNA. Strikingly, the presence of eIF1A in ribosomal complexes (structure 11wt) resulted in the accommodation of mRNA along the entire mRNA‐binding cleft (Fig 3C). Examination of critical P‐site nucleotides for all complexes shows that in each case C1701 of 18S rRNA is in a single conformation, except for the binary complex prepared with the wt IRES (structure 10wt), where it is present in two states as determined by examination of the Coulomb potential around that nucleotide (Fig 3D). In the conformation of the second state, C1701 contacts the upstream mRNA base U343(+2), possibly contributing to stabilizing the mRNA when it has not been completely accommodated in the mRNA channel at the P site. The highly conserved C1698 (Prince et al, 1982) contacts downstream mRNA and may act as a sensor to stimulate C1701 adopting the second conformation in which it can stabilize the incompletely loaded mRNA.
Figure 3. The roles of IRES domain II and eIF1A in loading mRNA into the mRNA channel.
-
A‐D(A and upper panels of B and C) The mRNA channel spanning the entry and exit channels, A, P, and E sites viewed through the ribosome head toward the body for the indicated complexes. The IRES (red), Coulomb potential (blue mesh), and eIF1A (blue) are shown. (Lower panels of B and C) P site showing mRNA interactions for the (B) binary complex (structure 10wt) and the (C) eIF1A‐containing ternary complex (structure 11wt). 18S rRNA (yellow), eIF1A (blue), IRES mRNA (red) with start codon (cyan) are all marked. (B) The position of mRNA modeled in the canonical cap‐dependent 48S IC (PDB: 6ZMW) is shown in gray. (D) Position of key nucleotides in the P site in structure 10wt showing interactions between incompletely loaded mRNA and C1698 resulting in C1701 sampling dual conformations near the (+2) position.
Taken together, the structures of binary complexes assembled on ΔdII and wt IRESs, and the eIF1A‐containing complex assembled on the wt IRES provide structural insight into the roles of domain II and eIF1A in sequentially loading the mRNA channel. Even though the ΔdII IRES induces the open conformation of the 40S subunit that is required for the accommodation of mRNA in the mRNA‐binding channel, accommodation in this case is only partial, and the 40S subunit is not stably present in the open conformation. This is consistent, on one hand, with the ability of the ΔdII IRES to function in initiation, but also, on the other hand, with it being less active than the wt IRES (e.g., Reynolds et al, 1996). The presence of domain II results in accommodation of the initiation codon and the upstream region in the mRNA‐binding channel, thereby enhancing the efficiency of initiation (Reynolds et al, 1996), whereas eIF1A promotes mRNA fixation downstream from the P site, accounting for its stimulatory role (Jaafar et al, 2016).
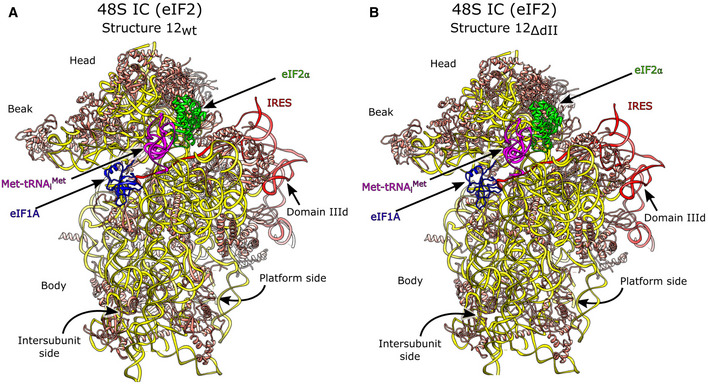
The structure of eIF2‐containing 48S complexes assembled on the HCV IRES
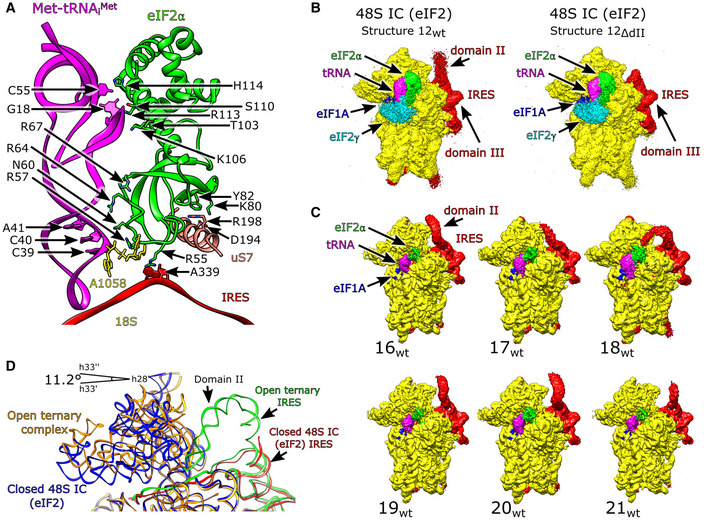
The structures of eIF2‐containing 48S ICs assembled on the wt and ΔdII IRES were refined from 46,904 and 103,813 particles to resolutions of 3.6 and 3.5 Å, respectively (structures 12wt and 12ΔdII; Figs 1B and EV1A, and Appendix Fig S1). Both wt and ΔdII IRES 48S ICs form identical complexes with respect to the 40S subunit's closed conformation, the positions of Met‐tRNAi Met, eIF2, and eIF1A, and the established P‐site codon–anticodon base pairing (Figs 1B, and EV3A and B). The IRES‐containing 48S IC is also structurally identical to the canonical 48S complex formed by cap‐dependent initiation (e.g., Simonetti et al, 2020) with respect to the global conformation of the 40S subunit and the placement of Met‐tRNAi Met, eIF2α, and eIF1A. Thus, the E site‐associated eIF2α contacts the highly conserved uS7/Asp194 directly and interacts with Met‐tRNAi Met via Thr103, Arg67, and His114 (Fig 4A). As with canonical 48S ICs (Hussain et al, 2014; Simonetti et al, 2020), eIF2α forms hydrogen bonds with the (−3) adenosine via Arg55, a contact that enhances codon selection in the scanning mode of initiation, presumably by stabilizing the arrested 48S IC (Pisarev et al, 2006; Thakur et al, 2020). Contacts between eIF2α Arg57 and the tRNA acceptor stem loop (ASL) and the 18S rRNA that occur in canonical 48S ICs (Simonetti et al, 2020) are also present (Fig 4A). The unsharpened map also contained density corresponding to eIF2γ (Fig 4B) identical to that seen in cap‐dependent 48S ICs (Simonetti et al, 2020), but it had a low local resolution at the acceptor end of Met‐tRNAi Met and was not modeled after map sharpening. The occupancy of the mRNA‐binding channel for 48S ICs prepared on both wt and ΔdII IRESs (Structures 12wt/ΔdII) is identical, with mRNA density present from the exit channel through the E, P, and A sites to the entry region. The observation that eIF2α interacts via Arg55 with the (−3) context nucleotide, as in the canonical initiation process, is consistent with its importance in HCV IRES function (Ma et al, 2018).
Figure EV3. HCV IRES·eIF2‐containing 48S initiation complexes.
-
A, BOverview of (A) the wt IRES eIF2‐containing 48S IC (structure 12wt) and (B) the ΔdII IRES eIF2‐containing 48S IC (structure 12ΔdII).
Figure 4. The HCV IRES·eIF2‐containing 48S initiation complex.
-
AContacts between eIF2α and the HCV IRES, Met‐tRNAi Met, and the 40S subunit.
-
BUnsharpened maps for 48S ICs formed on the wt IRES (structure 12wt), left, and the ΔdII IRES (structure 12ΔdII), right. Comparison with canonical cap‐dependent initiation complexes (PDB: 6YAL) identifies the presence of eIF2γ (cyan).
-
CIRES domain II occupies multiple positions in 48S ICs formed on the wt IRES.
-
DConformation of 18S rRNA in the open ternary complex (structure 11wt) and the eIF2‐containing 48S IC (structure 12wt).
The position of domain II differs substantially between the 48S IC and the corresponding 40S/IRES binary and 40S/eIF1A/IRES ternary complexes assembled on the wt IRES (structures 12wt, 10wt, and 11wt, respectively). In both the binary and ternary complexes, it is inserted into the E site, a position that is incompatible with the binding of eIF2. Thus, in the 48S IC, domain II is oriented away from the subunit interface, toward the solvent side of the 40S subunit, and shows an attenuation of density so that a model of domain II is not fully enclosed by the map. Focused classification of this region revealed that domain II is flexible and occupies multiple conformations oriented away from the E site (Fig 4C and Appendix Table S5). Interestingly, eIF2α Arg55 contacting the mRNA at A(−3) in 48S ICs is in the same position as C83 in IRES domain II in the 40S/eIF1A/IRES ternary complex, indicating that this location in the E site is important for stabilizing the mRNA regardless of the conformation of the 40S subunit and of differences in the position of mRNA at other locations along the mRNA‐binding channel.
Compared with the open binary or ternary complexes (structures 10wt and 11wt), the head of the 40S subunit in 48S ICs formed on both wt and ΔdII IRESs is in a closed position, having moved by 11.2° relative to the open states (Fig 4D). This position is in an even more closed conformation than in closed binary complexes (structures 1ΔdII‐6ΔdII), in which the position of the head differed from that in structures 10wt and 11wt by ∼9.0° (Fig EV2B). This indicates that the formation of the codon–anticodon interaction, seen in structures 12wt/ΔdII, causes the ribosome to enter a distinct closed conformation matching that is observed for both yeast and mammalian canonical cap‐dependent 48S ICs (e.g., Llácer et al, 2015; Simonetti et al, 2020). This closed state likely serves as a signal that an appropriate start codon has been identified so that subsequent initiation steps can proceed (Ogle et al, 2002).
We also obtained 40S complexes containing platform‐bound wt or ΔdII IRES and P‐site Met‐tRNAi Met but lacking all initiation factors (structures 13wt and 13ΔdII; Fig EV1A). These closed complexes, refined from 15,906 and 15,598 particles to 4.6 and 4.4 Å, respectively, showed clear density for codon–anticodon base pairing (Appendix Fig S3G and Table S3). It is unclear whether initiation factors dissociated due to slow eIF5‐independent hydrolysis of eIF2‐bound GTP (Unbehaun et al, 2004), or through denaturation at the air‐water interface and/or due to the shear forces associated with blotting (d'Imprima et al, 2019; Glaeser, 2021). In any case, this complex likely mimics an intermediate state immediately after eIF2 dissociation and prior to the binding of eIF5B.
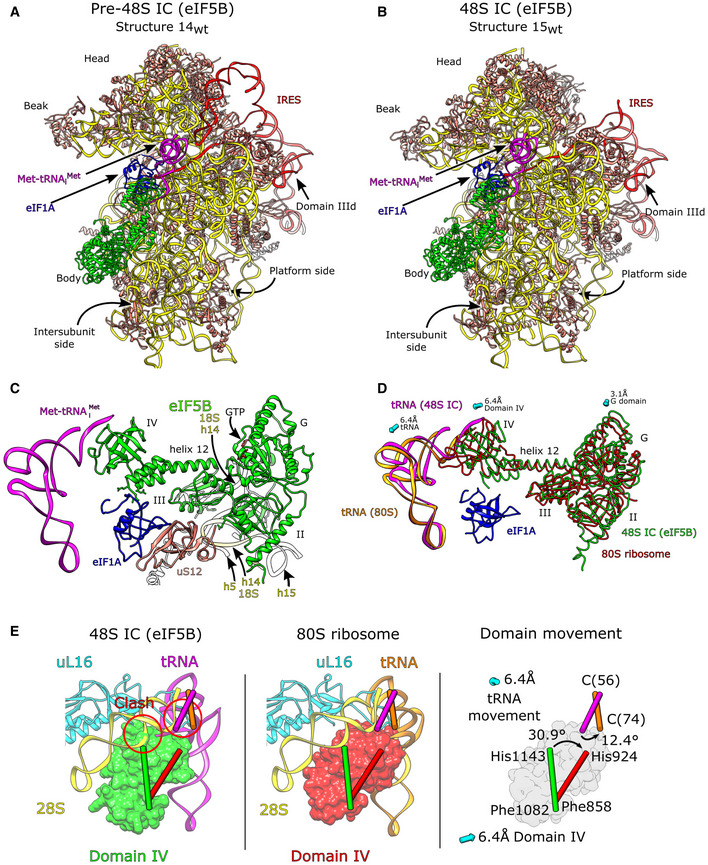
The structure of eIF5B‐containing 48S complexes assembled on the IRES
Classification of eIF5B‐containing ribosomal complexes yielded two structures formed on the wt IRES (structures 14wt and 15wt) and one structure (15ΔdII) assembled on the ΔdII IRES, which were refined to 3.8, 3.7, and 3.7 Å resolution from 60,578, 133,782, and 61,648 particles, respectively (Figs 1B and EV1A). All structures contain eIF5B, eIF1A, and the P‐site Met‐tRNAi Met base‐paired with the initiation codon. Structures 15wt and 15ΔdII were in the same closed conformation (Fig EV4A and B, and Appendix Fig S4A), identical to that in 48S ICs assembled on canonical cap‐dependent mRNA (Simonetti et al, 2020), and were accordingly classified as 48S ICs. Thus, in eIF5B‐ and eIF2‐containing 48S ICs, 18S rRNA nucleotides C1701 and U1248 are separated by ∼3.5 Å, and the contacts of the P‐gate nucleotides G1639 and A1640 in the 18S rRNA and Arg146 in uS9, with the tRNA anticodon arm on the opposite side to the anticodon, are also present in both 48S ICs. Similar to eIF2‐containing 48S ICs, in eIF5B‐containing ICs assembled on the wt IRES (structure 15wt), domain II is oriented away from the subunit interface, toward the solvent side of the 40S subunit, and shows an attenuation of density so that the model of domain II is not fully enclosed by the map. Focused classification of this region revealed that domain II is flexible and occupies multiple conformations oriented away from the E site (Appendix Fig S4B and Table S5).
Figure EV4. The HCV IRES·eIF5B‐containing 48S initiation complex.
-
A, BOverview of (A) the wt IRES eIF5B‐containing pre‐48S IC (structure 14wt) and (B) the wt IRES eIF5B‐containing 48S IC (structure 15wt).
-
CGlobal position of eIF5B bound to the intersubunit face of the 40S ribosomal subunit. Interactions between 18S rRNA (yellow), uS12 (salmon), and eIF1A (blue) are shown.
-
DPosition of eIF5B and tRNA in the 48S and 80S initiation complexes. Domains II, G, and III of eIF5B undergo relatively little movement between the 48S (green) and 80S stages (red), whereas domain IV translates and rotates (see E) causing movement in the tRNA between the two complexes (magenta and orange, respectively). Arrows show displacement for labeled domains or components between the eIF5B‐containing 48S IC and the pre‐elongation 80S ribosome.
-
EThe position of eIF5B domain IV in the 48S IC (green, left panel) would clash with 28S rRNA H89 and uL16 in the 60S subunit (left panel). To avoid this clash, in the 80S complex, domain IV (red, middle panel) translates by 6.4 Å toward the platform side of the 40S subunit and rotates by 30.9° causing the tRNA to rotate by 12.4° and translate by 6.4 Å toward the head (right panel).
However, in contrast to structures 15wt/ΔdII, the 40S subunit in structure 14wt is in an open conformation with domain II inserted into the E site, which led us to designate this structure as pre‐48S IC (Fig 1B). In the eIF5B‐containing pre‐48S IC, the separation between C1701 and U1248 is increased to ∼11 Å, and tRNA does not contact the P gate or uS9. In all structures, eIF1A occupies its usual position over h44 and eS30 and uS12, whereas eIF5B resides on the intersubunit face of the 40S subunit as in 80S ribosomal complexes (Yamamoto et al, 2014; Huang & Fernández, 2020; Wang et al, 2020). Both eIF5B‐containing pre‐48S ICs (structure 14wt) and 48S ICs (structure 15wt/ΔdII) showed full occupancy of the entire mRNA‐binding channel, from the exit portion through the E, P, and A sites to the entry region.
All structures show clear density for eIF5B residues 592–1,218, corresponding to all major domains (Fig EV4C). This is the first high‐resolution structure of mammalian eIF5B and also the first structure of the 40S/eIF5B subunit complex prior to subunit joining. Although we used full‐length native eIF5B, extensive 3D classification and processing did not reveal any structure corresponding to the N‐terminal region, suggesting that it is highly disordered, an observation that is supported by the structure of full‐length eIF5B predicted using AlphaFold (Jumper et al, 2021). In all structures, the G domain and domains II and III form the central core of eIF5B that connects via domain III and helix 12 to the tRNA acceptor stem‐binding domain IV (Fig EV4C). All complexes also contained eIF5B‐bound GTP (Appendix Fig S4C), and consistently, showed switch 1, switch 2, and the β9‐β10 loop in domain II in the GTP‐bound conformation, identical to that in fungal GTP‐bound eIF5B (Kuhle & Ficner, 2014; Appendix Fig S4D).
We identified multiple contacts between eIF5B domains II and III and 18S rRNA h5, h14, and h15, and an interaction between eIF5B domain II and uS12 (Fig EV4C and Appendix Table S6) and noted that the β13‐β14 loop (Appendix Fig S4E, green) could interfere with the transition of switch 1 from GTP‐bound to GDP‐bound (Appendix Fig S4E, blue and cyan). However, on the yeast 80S ribosome, the β13‐β14 loop of GTP‐bound eIF5B contacts A415 in 18S rRNA h14 and is positioned away from the path that switch 1 might take as it changes to the GDP‐bound conformation (Wang et al, 2020; Appendix Fig S4E, red). Although there is no contact between the equivalent nucleotide (A464) and β13‐β14 in our complexes, h14 is accessible. These observations suggest that the 60S subunit might stimulate the interaction between A415 and the β13‐β14 loop to reposition this loop away from switch 1 so that a transition from the GTP‐ to the GDP‐bound conformation is possible. In both pre‐48S ICs and 48S ICs, the position of domain IV also allows it to contact eIF1A via interactions between His112 in the helical subdomain of eIF1A (Battiste et al, 2000) and the extreme C‐terminal region of eIF5B, and between the eIF1A L23 β‐turn (near Gly54) and Arg1186 in domain IV of eIF5B (Fig 5A and B). These interactions are distinct from the previously reported binding of eIF1A's extreme C‐terminal DDIDI sequence to the h12/h13 loop in eIF5B domain IV (Marintchev et al, 2003; Zheng et al, 2014) and of the eIF1A L45 loop to eIF5B domain III (Nag et al, 2016). Previously reported interactions involved isolated polypeptides and were detected by NMR (Marintchev et al, 2003; Nag et al, 2016) or X‐ray crystallography (Zheng et al, 2014), whereas the interactions reported here were observed in functional 48S complexes.
Figure 5. The HCV IRES·eIF5B‐containing 48S initiation complex.
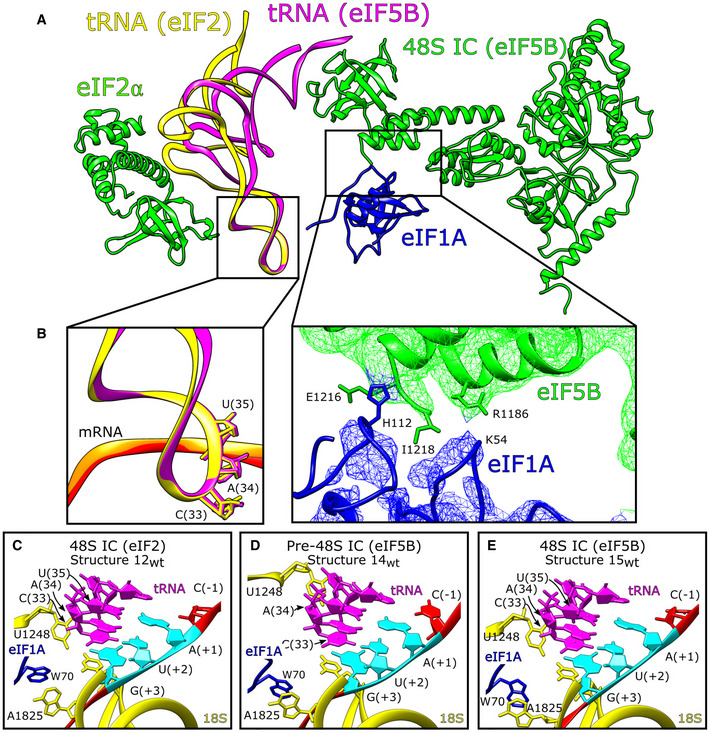
-
AChanges to the position of P site tRNA depending on the presence of either eIF2 or eIF5B. Upon binding of eIF5B, the T‐arm, D‐arm, and acceptor stem of Met‐tRNAi Met move by 15.6 Å and 14o relative to their positions in the eIF2‐containing 48S complex.
-
BInset showing the unchanged conformation of the tRNA acceptor stem loop in eIF2‐ and eIF5B‐containing 48S complexes (left), and contacts between eIF5B domain IV and eIF1A (right).
-
C–EP site for (C) eIF2‐containing 48S IC (structure 12wt), (D) eIF5B‐containing pre‐48S IC (structure 14wt), and (E) eIF5B‐containing 48S IC (structure 15wt) showing 18S rRNA (yellow), Met‐tRNAi Met (magenta), eIF1A (blue), and IRES mRNA (red) with the start codon (cyan) marked.
Comparison of the position of the P‐site Met‐tRNAi Met in the eIF2‐containing 48S IC (Simonetti et al, 2020) and in the 80S ribosome (Yamamoto et al, 2014; Wang et al, 2020) shows that in the latter, tRNA has rotated 14° toward the 40S subunit body and the T‐loop has moved by ∼15 Å to allow placement of the acceptor stem into the P site of the 60S subunit. Comparison of eIF2‐ and eIF5B‐ containing 40S ribosomal complexes (structures 12wt/ΔdII, 14wt, and 15wt/ΔdII) revealed that in eIF5B‐containing complexes, Met‐tRNAi Met has rotated by ∼14° and has moved by 15 Å from the head of the 40S subunit to a position that matches the orientation seen in 80S structures (Wang et al, 2020; Fig 5A). This repositioning of tRNA was observed for all structures that contained eIF5B, indicating that eIF5B re‐orients Met‐tRNAi Met on the 40S subunit prior to subunit joining. However, despite the differences in the orientation of the acceptor arm of the P‐site tRNA and the conformation of the 40S subunit, the anticodon loop in eIF2‐containing 48S ICs, and eIF5B‐containing 48S ICs and pre‐48S ICs is positioned identically (Fig. 5A and B). Interestingly, although the overall position of eIF5B‐Met‐tRNAi Met in eIF5B‐containing 48S ICs and in 80S ribosomes is generally similar, upon binding of a 60S subunit, domain IV of eIF5B undergoes a 33° rotation toward the platform side of the 40S subunit, and a translation by 6.4 Å parallel to the mRNA channel toward the platform of the 40S subunit, which result in repositioning of the tRNA acceptor stem by 6.4 Å toward the ribosome head (i.e., toward ribosomal protein eS25) without changing the position of the ASL and with only minor movement of eIF5B helix 12 (Fig EV4D). Such repositioning of domain IV and the tRNA acceptor stem upon binding of a 60S subunit would avert steric clashes of domain IV with H84 of 28S rRNA and between uL16 and the tRNA acceptor stem (∼A74; Fig EV4E). Similar repositioning of IF2 domain C2 (equivalent to eIF5B domain IV) to avoid analogous steric clashes was observed in bacteria upon joining of 50S subunits to 30S ICs (Hussain et al, 2016; Kaledhonkar et al, 2019). Adjustment of the orientation of initiator tRNA prior to subunit joining is a critical step in initiation, and the mechanism by which it is mediated, by rotation and translation of domain IV of the universally‐conserved initiation factor IF2/eIF5B, likely appeared early in evolution.
Dynamics of ribosomal contacts of mRNA along the mRNA‐binding channel throughout the initiation process
We next compared all complexes containing mRNA in the mRNA‐binding channel (structures 9ΔdII‐15wt/ΔdII) to determine how the conformational state of the 40S subunit and the presence of different translational components correlate with ribosomal interactions of defined mRNA regions along the channel and their consequent stabilization (Fig 6A and B). The open‐state binary 40S/IRES complexes (structures 9ΔdII and 10wt) both show clear mRNA density at the exit portion of the mRNA‐binding channel up to the (−8) position, likely due to stabilizing contacts of eS28 residues Arg31 and Arg66 with the (−13)/(−11) and (−8) nucleotides, respectively. In addition, mRNA in the wt complex (structure 10wt) also forms interactions at the (−7) and (−5) nucleotides with eS28 Arg69 and uS11 Arg66. Although in the unsharpened map, mRNA is visible from the exit through to the P site, the position of each nucleotide was clearly defined only in the (−13) to (−5) nt region where mRNA contacts eS28 and uS11, whereas the downstream region up to position (+2) was more heterogenous and the quality of the sharpened map declined. In the open 40S/IRES/eIF1A ternary complexes (structure 11wt), association with eIF1A resulted in mRNA accommodation in the entire mRNA‐binding channel, but it did not influence the mRNA‐40S interactions observed in the binary complexes and also did not stabilize the mRNA region from the E to A sites. Establishment of the P‐site codon–anticodon interaction in the open eIF5B‐containing pre‐48S IC (structure 14wt) led to stabilization of the P‐site codon, resulting in clear density‐forming Watson‐Crick base pairs with the tRNA anticodon (Fig 5D), but preserved mRNA interactions with eS28 and uS11 and its overall position and conformation observed in the 40S/IRES/eIF1A ternary complex. As in the ternary complex (structure 11wt), the presence of eIF1A at high occupancy in pre‐48S ICs was not associated with stabilization of the (+4) to (+7) region of mRNA.
Figure 6. Ribosomal contacts and stability of mRNA along the mRNA‐binding channel at different stages of initiation on the HCV IRES.
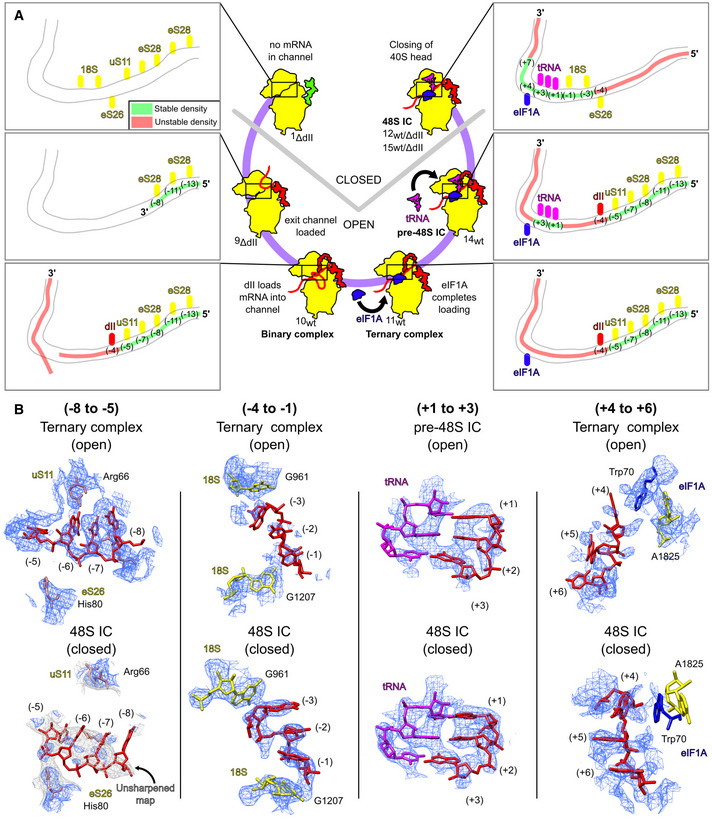
-
AOccupancy of the mRNA channel for the indicated complexes showing the identity of ribosomal components (yellow), tRNA (magenta), eIF1A (blue), or IRES domain II (red) that interact with mRNA in the mRNA‐binding channel. The stability of mRNA in the channel was assessed as either stable (green) or unstable (red) depending on quality of density attributable to each nucleotide.
-
BComparison of density maps and models for specific areas of mRNA in the mRNA‐binding channel in the indicated open and closed ribosomal complexes.
The closed‐state eIF2‐ and eIF5B‐containing 48S ICs (structure 12wt/ΔdII, and 15wt/ΔdII) show identical mRNA conformations and stability patterns along the mRNA‐binding channel that differ from those observed in the open‐state complexes. Due to the head closure, eS28 and uS11 can no longer make contacts with the (−13), (−11), and (−5) nucleotides, and mRNA has moved by ∼6 Å toward the body so that eS26 His80 contacts the (−5) nucleotide phosphate backbone. In contrast to the open‐state complexes, the new position of mRNA is characterized by the poorly defined region upstream of the (−5) nucleotide, whereas the (−3) to (−1) region is stabilized, forming a stacking triple sandwiched between 18S rRNA G961 and G1207. In all 48S ICs, the P‐site codon shows clear density interacting with the tRNA anticodon (Fig 5C and E). In contrast to the pre‐48S IC, the presence of eIF1A influences the stability of the (+4) to (+7) nucleotides in 48S ICs. In complexes with high eIF1A occupancy (structures 12ΔdII, 15wt), the (+4) nucleotide forms a stacking triple with eIF1A Trp70 and 18S rRNA A1825, and the (+4) to (+7) region is stable. Such stabilization was not observed in complexes with lower eIF1A occupancy (structures 12wt, 15ΔdII).
Thus, our data indicate that transition from the open to the closed states is accompanied by changes in mRNA interactions and stability patterns along the mRNA‐binding channel. Movement of the 40S subunit head in 48S ICs results in the loss of the interaction between the mRNA region upstream of the (−5) nucleotide with eS28 and uS11 and subsequent destabilization of this region compared with the open‐state complexes. On the other hand, the 40S subunit closure leads to stabilization of the (−3) to (−1) mRNA region between 18S rRNA bases. Notably, in closed 48S ICs, but not in open ribosomal complexes, the (+4) to (+7) mRNA region can be stabilized by eIF1A.
Comparison of eIF5B‐containing pre‐48S initiation complexes with eIF2‐containing scanning 43S complexes
In contrast to initiation on the HCV IRES, eIF5B cannot substitute for eIF2 in recruiting Met‐tRNAi Met to the 40S subunit in the canonical scanning mechanism. Canonical initiation also requires eIF1, which binds to the 40S subunit below the mRNA channel at the P site between h24 and the region connecting h44 to h45 and, in cooperation with eIF1A, stabilizes the open conformation of the 40S subunit (Passmore et al, 2007; Llácer et al, 2015), thereby promoting ribosomal attachment to mRNA, scanning and initiation codon selection (e.g., Pestova & Kolupaeva, 2002). Comparison of the eIF5B‐containing pre‐48S ICs (structure 14wt) and canonical scanning, eIF2‐containing 43S complexes that also contained eIF1 (Brito Querido et al, 2020) revealed that the positions of mRNA and conformations of the anticodon loop of Met‐tRNAi Met in the P site in both complexes are very similar (Fig 7A), and the anticodon in the scanning 43S complex is also in a position that enables the establishment of codon–anticodon base pairing (Fig 7B, center panel). However, whereas the conformations of the anticodon in the P site in both complexes are similar, in the eIF5B‐containing complex, the T‐arm and the acceptor stem are shifted toward the body of the 40S subunit by bending of the anticodon loop region (Fig 7A). Consequently, the contacts between the ASL of Met‐tRNAi Met and 18S rRNA nucleotides GA1639‐40 and the N‐terminal region of uS9 in the head of the 40S subunit, which are present in all eIF2‐containing complexes, including mRNA‐free 43S complexes, scanning 43S complexes and 48S ICs (e.g., Hussain et al, 2014; Brito Querido et al, 2020; Simonetti et al, 2020; Fig 7B, center panel), do not exist in the eIF5B‐containing pre‐48S ICs and only form after ribosomal closure in 48S ICs (Fig 7B, left and right panels). Thus, interaction with eIF2 allows tRNA to maintain the contacts with the head in all 40S subunit conformations, from fully open to fully closed, and the position of the anticodon loop that allows it to inspect mRNA in the scanning 43S complexes is determined by the rotation of the head to a position that is intermediate between fully open and fully closed states. In contrast, in the open pre‐48S IC, tRNA is stabilized by contacts between eIF5B domain IV and the acceptor stem, codon–anticodon base pairing, and by stacking of the aromatic rings of ASL C33 with C1701 of 18S rRNA (Fig 7B, left panel). Interestingly, in the scanning 43S complex, the C33 ribose instead forms a lone pair‐π interaction with U1248 of 18S rRNA (Fig 7B, middle panel), whereas in canonical 48S ICs (e.g., Simonetti et al, 2020), and in eIF5B‐containing 48S ICs (structure 15wt), C33 is involved in both interactions (Fig 7B, right panel). Thus, whereas some aspects of eIF5B‐containing pre‐48S complexes are analogous to those of eIF2‐containing scanning 43S complexes, the overall orientation and the specific interactions of tRNA in them differ.
Figure 7. Position of eIF5B‐bound P site Met‐RNAi Met in open ribosomal complexes is not compatible with eIF1‐mediated scanning.
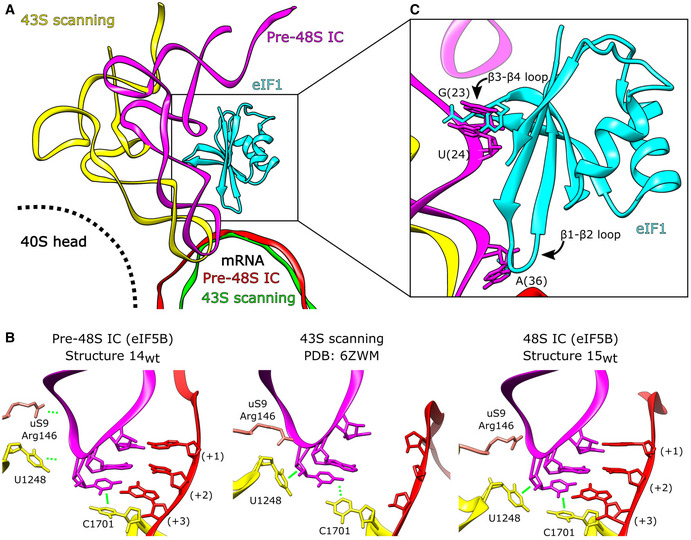
-
APosition of the P site Met‐tRNAi Met in the scanning 43S complex (PDB: 6ZMW; yellow), and the pre‐48S IC (structure 14wt; magenta). mRNA from the 43S scanning complex (green) and pre‐48S IC (red) is shown. eIF1 associated with the scanning 43S complex is in cyan.
-
BContacts between tRNA (magenta), the initiation codon (red), and the P site (yellow) for the indicated complexes.
-
CThe β1‐β2 and β3‐β4 loops of eIF1 bound as in the 43S scanning complex (PDB: 6ZMW) would clash with the position of P site Met‐tRNAi Met in eIF5B‐containing pre‐48S ICs.
During eIF2‐dependent canonical initiation, the open, scanning‐competent conformation of the 40S subunit is determined by the binding of eIF1. We therefore analyzed whether the binding of eIF1 would be compatible with the positions of eIF5B and Met‐tRNAi Met in the open pre‐48S ICs. The position of eIF1 placed into such complexes suggests that it would clash with tRNA. Thus, the repositioning of tRNA in eIF5B‐containing complexes causes the AAC38‐40 nucleotides of Met‐tRNAi Met to move toward the 40S subunit body by ∼3.0 Å, so that the binding of eIF1 as in the 43S scanning complex would create a clash between A36 of Met‐tRNAi Met and the β1‐β2 loop (Fig 7C). Accommodation of eIF1 would require either reorganization of this loop or displacement of the P‐site tRNA. Examination of human (Fletcher et al, 1999) and yeast (Reibarkh et al, 2008) solution NMR structures of eIF1 did not identify any conformations of the β1‐β2 loop that would allow a clash with the anticodon stem of tRNA to be avoided in the eIF5B‐containing pre‐48S and 48S ICs. Moreover, the eIF1 β3‐β4 loop would also clash with tRNA nucleotides GU23‐24, and a clash between Phe113 and tRNA nucleotide G25 is also possible (Fig 7C). These observations suggest that even if eIF5B were able to bind Met‐tRNAi Met with high affinity and recruit it to the 40S subunit efficiently, the structure of the resulting complexes would not be compatible with the binding of eIF1 and hence, with the scanning mechanism of initiation. On the other hand, the stabilizing interaction of the acceptor arm of Met‐tRNAi Met with domain IV of eIF5B in the closed 48S complexes following dissociation of eIF1 and eIF2·GDP would lock the complex, preventing leaky scanning from occurring. The incompatibility of Met‐tRNAi Met and eIF1 on eIF5B‐containing pre‐48S complexes also likely explains why eIF1 disrupts 48S complexes prepared using wt but not the ΔdII variant of the HCV‐like CSFV IRES (Pestova et al, 2008). Domain II has the propensity to insert into the E site (Quade et al, 2015; Yamamoto et al, 2015; this report), stabilizing the open conformation of the 40S subunit, and if eIF1 binds to this complex, then the insertion of its β1‐β2 loop into the mRNA channel creates steric hindrance between mRNA and tRNA in the P site, thereby dislodging the tRNA. We did not identify complexes formed on the ΔdII IRES in the open conformation, suggesting that closure is fast in the absence of domain II, so that eIF1 is unable to gain access to such complexes and destabilize them.
Discussion
Here we present the most comprehensive structural overview of the HCV IRES‐mediated initiation pathway to date (Fig 8) that shows the sequential conformational and compositional changes that occur during this process. Initially, the IRES binds to the 40S subunit through domains IIIa/IIIc and then pivots onto its platform side where it establishes the complete set of contacts (structures 1ΔdII‐6ΔdII). Formation of the full set of canonical contacts between the IRES domains IIIa/IIIc/IIId/IIIe/IIIf and the 40S subunit platform riboproteins eS1, eS27, and 18S rRNA ES7 induces the head of the 40S subunit to open (structures 8ΔdII‐9ΔdII). Although head opening can occur in the absence of IRES domain II, such complexes are nevertheless characterized by remarkable heterogeneity in the position of the 40S subunit head. In contrast, 40S/IRES binary complexes assembled on the wt IRES yield a uniform structure, in which the 40S subunit is in the open conformation, and domain II is inserted into the E site (structure 10wt). Importantly, in the absence of domain II, mRNA density was clearly seen only in the exit portion of the channel up to the (−8) position of mRNA (structure 9ΔdII). In the wt 40S/IRES binary complex, mRNA nucleotides could be identified at the exit channel through the E site, where it is stabilized by domain II, to AU342‐3 located in the P site (structure 10wt), and eIF1A promotes further accommodation of the mRNA in the entire mRNA‐binding channel (structure 11wt). Thus, complexes (1ΔdII‐11wt) provide structural insights into the functions of multiple IRES domains, including IIIa/IIIc in establishing the initial ribosome contacts, IIId and hIII1/IIIe/IIIf in fixing the IRES to the 40S subunit and inducing ribosomal head opening, and II in imposing the open conformation and promoting fixation of mRNA in and upstream of the P site. Our analysis also revealed the role of eIF1A in completing mRNA accommodation, as prior to eIF1A binding the mRNA is incompletely loaded into the mRNA channel (structure 10wt) and loading is complete only after eIF1A binding (structure 11wt).
Figure 8. The HCV IRES‐mediated initiation pathway.
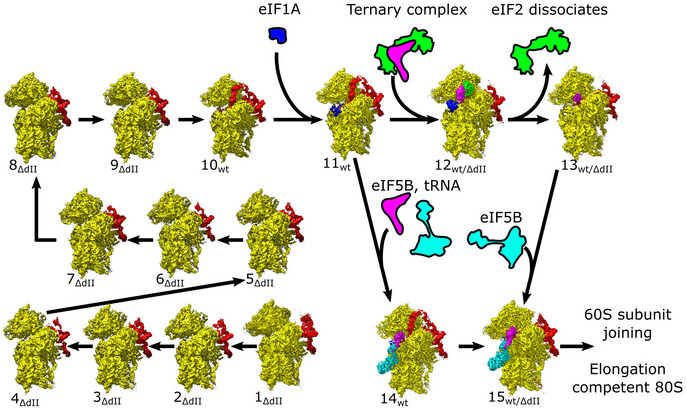
Ribosomal complexes organized in a putative IRES‐mediated initiation pathway. Maps are segmented showing the 40S subunit (yellow), IRES (red), eIF1A (blue), Met‐tRNAi Met (magenta), and eIF2 (green) or eIF5B (cyan).
Once the mRNA is loaded, initiation can proceed either along the canonical initiation pathway, in which eIF2 promotes attachment of Met‐tRNAi Met to form the 48S IC (structure 12wt/ΔdII) and then dissociates after GTP hydrolysis (likely structure 13wt/ΔdII) followed by binding of eIF5B (structure 15wt/ΔdII), or by a shortcut route, in which Met‐tRNAi Met is loaded directly by eIF5B, first forming the open‐state pre‐48S IC (structure 14wt) and then transitioning to the closed 48S IC (structure 15wt).
The eIF2‐containing 48S IC assembled on the IRES is structurally identical to the canonical 48S IC with respect to the conformation of the 40S subunit and the positions of Met‐tRNAi Met, eIF2, and eIF1A. The position of domain II of the IRES in 40S/IRESwt binary complexes is incompatible with the binding of eIF2 and the closed conformation of the 40S subunit, and consequently, in 48S ICs, domain II is oriented away from the subunit interface, toward the solvent side, occupying multiple conformations. In eIF5B‐containing pre‐48S ICs, the 40S subunit is in the open conformation and domain II is inserted into the E site. Upon 40S subunit closure in eIF5B‐containing 48S ICs, domain II becomes displaced from the E site and is again oriented away from the subunit interface like in eIF2‐containing 48S ICs. Importantly, compared with eIF2‐containing 48S ICs, in eIF5B‐containing pre‐48S ICs and 48S ICs, Met‐tRNAi Met is rotated by ∼14° and moved 15 Å from the head of the 40S subunit to a position that matches its orientation in 80S ribosomes. Thus, our data show how eIF5B repositions tRNA already on the 48S complex, preparing it for joining with the 60S subunit.
Materials and Methods
Plasmids
Vectors for expression of His6‐tagged eIF1A (Pestova et al, 1998a) and Escherichia coli methionyl tRNA synthetase (Lomakin et al, 2006) have been described. The plasmid HCV‐MSTN‐Stop (Hashem et al, 2013) containing HCV Type 1 nt 40–375 was used for the transcription of mRNA containing the wt HCV IRES. A derivative for transcription of HCV IRES lacking domain II (containing HCV nt. 125–375) was made by GenScript Corp. (Piscataway, NJ). The HCV plasmids were linearized by BamHI, and mRNAs were transcribed using T7 RNA polymerase (Thermo Scientific).
Purification of factors, ribosomal subunits, and aminoacylation of tRNA
Native mammalian 40S subunits, eIF2, eIF3, and eIF5B were purified from rabbit reticulocyte lysate (RRL; Green Hectares), as described (Pisarev et al, 2007). Recombinant eIF1A and E. coli methionyl tRNA synthetase were expressed and purified from E. coli as described (Pisarev et al, 2007).
For purification of native rabbit total tRNA, 200 ml RRL were centrifuged at 45,000 rpm (RCFmax = 244,000 x g) for 4.5 h in a Beckman 50.2 Ti rotor at 4°C in order to pellet polysomes. The supernatant was dialyzed overnight against buffer A (20 mM Tris [pH 7.5], 4 mM MgCl2, 250 mM KCl, 2 mM DTT) and applied to a DE52 (Whatman) column equilibrated with buffer A. The tRNA was eluted with buffer B (20 mM Tris [pH 7.5], 3 mM MgCl2, 700 mM NaCl, 2 mM DTT) and precipitated overnight with 2.5 volumes of ethanol at −80°C. The precipitate was centrifuged at 13,000 rpm (RCFmax = 20,000 x g) for 15 min and resuspended in 5 ml buffer C (100 mM Tris [pH 7.5], 5 mM MgCl2), phenol‐chloroform (pH 4.7) extracted and precipitated again with 0.3 M NaOAc and 2.5 volumes of ethanol. To isolate tRNA, the pellet was dissolved and subjected to gel filtration on a Superdex 75 column (Pestova & Hellen, 2001). Purified total tRNA was aminoacylated using E. coli methionyl tRNA synthetase (to obtain Met‐tRNAi Met) as described (Pisarev et al, 2007).
Assembly of ribosomal complexes
To form 48S initiation complexes, 7 pmol HCV IRES mRNA (wt or Δdomain ΙΙ mutant) were incubated with 3.5 pmol 40S subunits, 10 pmol eIF1A, 4.5 pmol eIF3, total native rabbit tRNA containing 3.5 pmol Met‐tRNAi Met, and 6 pmol eIF2 or 10 pmol eIF5B in 40 μl buffer D (20 mM Tris [pH 7.5], 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine, 1 mM ATP and 0.2 mM GTP) for 10 min at 37°C. The obtained complexes (containing 87.5 nM 40S subunits) were applied directly onto grids without dilution.
Grid preparation and electron microscopy
Gold foil grids were prepared from Quantifoil gold mesh grids (Passmore & Russo, 2016). Initially, Quantifoil R0.6/1.0300 mesh gold grids (Quantifoil Micro Tools GmbH) were visually inspected to check for uniformity and intactness of the Quantifoil layer and then placed into an Auto 306 Turbo Vacuum Coater (Edwards Vacuum) at a pressure of 103 Pa, and then, gold wire (Ted Pella, Inc) was evaporated for approximately 8 min to create a 500 Å layer. Deposition thickness was determined using the inbuilt film thickness monitor. To remove the underlying Quantifoil carbon foil layer the grids were then treated with plasma using a Gatan Solarus 950 (Gatan, Inc) operated at 25 W for 4 min with an argon/oxygen gas mixture.
To prepare hydrophilic grids, 30 min prior to sample application, grids were treated with plasma using a Gatan Solarus 950 (Gatan, Inc) operated at 25 W for 25 s with a hydrogen/oxygen gas mixture. These grids were then transferred to the environmental chamber of a Vitrobot Mark IV (Thermo Fisher Scientific) maintained at 4°C and 100% humidity. Here, 3 μl of sample was applied and then blotted for 4 s with blot force 3 before plunging into a cooled (77 K) ethane‐propane mixture (Tivol et al, 2008) and then transferred to liquid nitrogen. Selected grids were screened to confirm sample composition and ice thickness using a Tecnai F20 electron microscope (Thermo Fisher Scientific) equipped with a field emission gun (FEG) operating at 200 kV and a K2 Summit direct electron detector (Gatan, Inc).
After screening grids from each sample, data collection was performed on a Tecnai Polara F30 (Thermo Fisher Scientific) equipped with an FEG operating at 300 kV and a K3 direct electron detector (Gatan, Inc). Movies were collected at a nominal magnification of 52,000× and defocus range of −0.5 to −2.5 μm in counting mode with a pixel size of 0.95 Å using the automated collection software Leginon (Potter et al, 1999; Carragher et al, 2000; Suloway et al, 2005). Each movie consisted of 40 frames recorded over 4 s with a total dose of 70.9 e−/Å2. Due to sample conditions, the 40S ribosomal subunits were observed in a preferred orientation, and so, portions of the data were collected with a 30° stage tilt (Appendix Table S2). For the wt IRES eIF2‐containing sample 14,815 micrographs (14,815 at 30° stage tilt) were collected over two sessions, for the wt IRES eIF5B‐containing sample 27,263 micrographs (20,509 at 30° stage tilt) were collected over four sessions, for the ΔdII IRES eIF2‐containing sample 22,735 micrographs (17,695 at 30° stage tilt) were collected over three sessions, and for the ΔdII IRES eIF5B‐containing sample 13,809 micrographs (13,809 at 30° stage tilt) were collected over two sessions (Appendix Table S2). Optical groups of micrographs with similar beam tilt values were identified using k‐means clustering on the image shift values recorded by the microscope during data collection.
Image processing
Gain references for each session were produced by visually screening ∼1,000 micrographs to remove images that contained gold foil and then summing them using cisTEM sum_all_tifs (Grant et al, 2018). Movies were then aligned using MotionCor2 (Zheng et al, 2017) with dose weighting of 1.77 e−/Å2/frame and local patch correction with 8 × 5 patches. Initial CTF parameters were estimated using CTFFIND4 (Rohou & Grigorieff, 2015). Particle locations were identified using Topaz version 0.2.3 (Bepler et al, 2019) by initially downscaling all micrographs by 8× and then using the Topaz general model to identify particles. Particles with a confidence score below 0 were removed and the remaining positions rescaled for subsequent processing in Relion 3.1 (Scheres, 2012, 2016; Zivanov et al, 2018, 2019).
Initially, we identified 2,183,185 particles for the wt IRES eIF2‐containing sample (147 particles per micrograph), 1,133,335 particles for the wt IRES eIF5B‐containing sample (42 particles per micrograph), 2,213,826 particles for the ΔdII IRES eIF2‐containing sample (97 particles per micrograph), and 1,459,506 particles for the ΔdII IRES eIF5B‐containing sample (106 particles per micrograph). Particle locations were extracted from micrographs into downsampled boxes of 100 × 100 pixels (at 3.8 Å/pixel) to speed up initial classification. This corresponds to 400 × 400‐pixel boxes (at 0.95 Å/pixel) without downsampling. Twenty‐five iterations of 2D classification were performed to identify incorrectly‐picked particles, contamination, and other particles that were unable to be correctly aligned (e.g., due to poor signal‐to‐noise). Particles that were selected for removal were subjected to additional 2D classification to confirm that they did not contain clear 40S ribosome particles.
After the initial screening of the 2D classification data, the remaining particles for each sample were 1,201,923 particles for the wt IRES eIF2‐containing sample (81 particles per micrograph), 736,700 particles for the wt IRES eIF5B‐containing sample (36 particles per micrograph), 1,484,658 particles for the ΔdII IRES eIF2‐containing sample (84 particles per micrograph), and 1,119,610 particles for the ΔdII IRES eIF5B‐containing sample (81 particles per micrograph). For each sample, all particles were refined into a single model, which was used to estimate the defocus values on a per‐particle basis, followed by an additional refinement step, and then 3D classification without alignment into 10 classes for 25 iterations. This initial 3D classification was used to identify the major conformational states present in each sample, and further removal of poor‐quality particles. For the wt IRES eIF2‐containing sample, we identified 580,938 particles in the closed state, 176,793 particles in the open state, and removed 444,192 particles. For the wt IRES eIF5B‐containing sample, we identified 378,325 open state, 360,338 closed state, and removed 58,037 particles. For the ΔdII IRES eIF5B‐containing sample, we identified 883,893 particles in the closed state and removed 62,455 particles. For the ΔdII IRES eIF2‐containing sample, we identified 615,125 particles in the closed state, 198,920 particles in the intermediate‐open state, and removed 670,433 particles. All particles that were selected for removal were subjected to additional 2D classification and 3D refinement steps to confirm that they did not contain 40S ribosome complexes.
General processing pathway
All particles were re‐extracted at full‐size (400 × 400 pixel box, 0.95 Å/pixel) and underwent iterations of 3D refinement, followed by anisotropic magnification correction, defocus refinement, and beam tilt estimation. Multiple rounds of 3D classification (25 iterations, without alignment) were used to progressively remove poor‐quality particles. After each round of CTF refinement, each particle stack underwent 3D refinement and was checked for an increase in resolution and visual improvement of map density. Once CTF refinement no longer improved map quality, particle polishing using all frames was performed and then iterations of CTF refinement as outlined above were completed. Focused classification on consensus maps was performed to isolate desired conformational or compositional states. Full details of the processing pathway for each complex are included in the expanded information available online.
Model building and refinement
For all data, where applicable, we were able to unambiguously fit the head and body of the 40S (PDB: 6D9J; Pisareva et al, 2018), HCV IRES (PDB: 5FLX; Yamamoto et al, 2015), eIF1A (PDB: 4KZZ; Lomakin & Steitz, 2013), tRNA (PDB: 5K0Y; Simonetti et al, 2016) eIF2α subunit (PDB: 6O85; Kenner et al, 2019), and eIF5B (PDB: 4UJD; Yamamoto et al, 2014). Initial model fitting was performed using UCSF Chimera v1.14 (Pettersen et al, 2004) with additional modeling in Coot (Emsley & Cowtan, 2004). For regions of eIF5B that did not have available models (e.g., Switch 1), model building was performed independently and then cross‐checked for consistency. All models underwent one round of Phenix geometry minimization and multiple rounds of PHENIX real‐space refinement (Adams et al, 2010; Afonine et al, 2018).
Author contributions
Zuben P Brown: Conceptualization; data curation; software; formal analysis; investigation; writing – original draft; writing – review and editing. Irina S Abaeva: Investigation; methodology; writing – review and editing. Swastik De: Investigation; writing – review and editing. Christopher UT Hellen: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; methodology; writing – original draft; project administration; writing – review and editing. Tatyana V Pestova: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; methodology; writing – original draft; project administration; writing – review and editing. Joachim Frank: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Acknowledgements
This work was supported by grants from the National Institutes of Health (National Institute of General Medical Sciences), R01 GM55440 (to JF), R35 GM139453 (to JF), R35 GM122602 (to TVP), and R01 GM097014 (to CUTH), and from the National Institutes of Health (National Institute of Allergy and Infectious Diseases), R01 AI123406 (to CUTH).
Contributor Information
Tatyana V Pestova, Email: tatyana.pestova@downstate.edu.
Joachim Frank, Email: jf2192@cumc.columbia.edu.
Data availability
Primary models and maps (Appendix Table S3) reported in this study were deposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) under the following accession codes: structure 1ΔdII (PDB: 7SYG, https://www.rcsb.org/structure/7SYG; EMD‐25527, https://www.ebi.ac.uk/emdb/EMD‐25527), structure 2ΔdII (PDB: 7SYH, https://www.rcsb.org/structure/7SYH; EMD‐25528, https://www.ebi.ac.uk/emdb/EMD‐25528), structure 3ΔdII (PDB: 7SYI, https://www.rcsb.org/structure/7SYI; EMD‐25529, https://www.ebi.ac.uk/emdb/EMD‐25529), structure 4ΔdII (PDB: 7SYJ, https://www.rcsb.org/structure/7SYJ; EMD‐25530, https://www.ebi.ac.uk/emdb/EMD‐25530), structure 5ΔdII (PDB: 7SYK, https://www.rcsb.org/structure/7SYK; EMD‐25531, https://www.ebi.ac.uk/emdb/EMD‐25531), structure 6ΔdII (PDB: 7SYL, https://www.rcsb.org/structure/7SYL; EMD‐25532, https://www.ebi.ac.uk/emdb/EMD‐25532), structure 7ΔdII (PDB: 7SYM, https://www.rcsb.org/structure/7SYM; EMD‐25533, https://www.ebi.ac.uk/emdb/EMD‐25533), structure 8ΔdII (PDB: 7SYN, https://www.rcsb.org/structure/7SYN; EMD‐25534, https://www.ebi.ac.uk/emdb/EMD‐25534), structure 9ΔdII (PDB: 7SYO, https://www.rcsb.org/structure/7SYO; EMD‐25535, https://www.ebi.ac.uk/emdb/EMD‐25535), structure 10wt (PDB: 7SYP, https://www.rcsb.org/structure/7SYP; EMD‐25536, https://www.ebi.ac.uk/emdb/EMD‐25536), structure 11wt (PDB: 7SYQ, https://www.rcsb.org/structure/7SYQ; EMD‐25537, https://www.ebi.ac.uk/emdb/EMD‐25537), structure 12wt (PDB: 7SYR, https://www.rcsb.org/structure/7SYR; EMD‐25538, https://www.ebi.ac.uk/emdb/EMD‐25538), structure 12ΔdII (PDB: 7SYS, https://www.rcsb.org/structure/7SYS; EMD‐25539, https://www.ebi.ac.uk/emdb/EMD‐25539), structure 13wt (PDB: 7SYT, https://www.rcsb.org/structure/7SYT; EMD‐25540, https://www.ebi.ac.uk/emdb/EMD‐25540), structure 13ΔdII (PDB: 7SYU, https://www.rcsb.org/structure/7SYU; EMD‐25541, https://www.ebi.ac.uk/emdb/EMD‐25541), structure 14wt (PDB: 7SYV, https://www.rcsb.org/structure/7SYU; EMD‐25542, https://www.ebi.ac.uk/emdb/EMD‐25542), structure 15wt (PDB: 7SYW, https://www.rcsb.org/structure/7SYW; EMD‐25543, https://www.ebi.ac.uk/emdb/EMD‐25543), and structure 15ΔdII (PDB: 7SYX, https://www.rcsb.org/structure/7SYX; EMD‐25544, https://www.ebi.ac.uk/emdb/EMD‐25544). Additional maps (Appendix Table S5) showing the movement of HCV IRES domain II were deposited in the EMDB under the following accession codes: structure 16wt (EMD‐25545, https://www.ebi.ac.uk/emdb/EMD‐25545), structure 17wt (EMD‐25546, https://www.ebi.ac.uk/emdb/EMD‐25546), structure 18wt (EMD‐25547, https://www.ebi.ac.uk/emdb/EMD‐25547), structure 19wt (EMD‐25548, https://www.ebi.ac.uk/emdb/EMD‐25548), structure 20wt (EMD‐25549, https://www.ebi.ac.uk/emdb/EMD‐25549), structure 21wt (EMD‐25550, https://www.ebi.ac.uk/emdb/EMD‐25550), structure 22wt (EMD‐25551, https://www.ebi.ac.uk/emdb/EMD‐25551), structure 23wt (EMD‐25552, https://www.ebi.ac.uk/emdb/EMD‐25552), structure 24wt (EMD‐25553, https://www.ebi.ac.uk/emdb/EMD‐25553), structure 25wt (EMD‐25554, https://www.ebi.ac.uk/emdb/EMD‐25554), structure 26wt (EMD‐25555, https://www.ebi.ac.uk/emdb/EMD‐25555), structure 27wt (EMD‐25556, https://www.ebi.ac.uk/emdb/EMD‐25556), and structure 28wt (EMD‐25557, https://www.ebi.ac.uk/emdb/EMD‐25557). Maps obtained during data processing (Appendix Table S7)from particle stacks that were compositionally and conformationally identical and were later combined were deposited in the EMDB under the following accession codes: structure 29wt (EMD‐25588, https://www.ebi.ac.uk/emdb/EMD‐25588), structure 30wt (EMD‐25589, https://www.ebi.ac.uk/emdb/EMD‐25589), structure 31ΔdII (EMD‐25590, https://www.ebi.ac.uk/emdb/EMD‐25590), and structure 32ΔdII (EMD‐25591, https://www.ebi.ac.uk/emdb/EMD‐25591). Consensus maps (Appendix Table S7) that were used for focused classification were deposited in the EMDB under the following accession codes: structure 33ΔdII (EMD‐25592, https://www.ebi.ac.uk/emdb/EMD‐25592), structure 34ΔdII (EMD‐25593, https://www.ebi.ac.uk/emdb/EMD‐25593), structure 35wt (EMD‐25594, https://www.ebi.ac.uk/emdb/EMD‐25594), structure 36wt (EMD‐25595, https://www.ebi.ac.uk/emdb/EMD‐25595), structure 37ΔdII (EMD‐25596, https://www.ebi.ac.uk/emdb/EMD‐25596), structure 38wt (EMD‐25597, https://www.ebi.ac.uk/emdb/EMD‐25597), structure 39wt (EMD‐25598, https://www.ebi.ac.uk/emdb/EMD‐25598), and structure 40ΔdII (EMD‐25599, https://www.ebi.ac.uk/emdb/EMD‐25599). Other high‐resolution maps obtained during classification were deposited in the EMDB under the following accession codes: structure 41wt (EMD‐25600, https://www.ebi.ac.uk/emdb/EMD‐25600), structure 42wt (EMD‐25601, https://www.ebi.ac.uk/emdb/EMD‐25601), structure 43wt (EMD‐25602, https://www.ebi.ac.uk/emdb/EMD‐25602), structure 44wt (EMD‐25603, https://www.ebi.ac.uk/emdb/EMD‐25603), structure 45wt (EMD‐25604, https://www.ebi.ac.uk/emdb/EMD‐25604), and structure 46ΔdII (EMD‐25620, https://www.ebi.ac.uk/emdb/EMD‐25620). For each entry, the half maps, unsharpened map, mask used for postprocessing, and the Fourier correlation curve has been deposited as additional files. Primary map entries also have local resolution maps as additional files and consensus map entries have the focused classification masks included. Requests for materials and additional information can be directed to Dr. Tatyana Pestova (tatyana.pestova@downstate.edu) or Dr. Joachim Frank (jf2192@cumc.columbia.edu).
References
- Acker MG, Shin BS, Dever TE, Lorsch JR (2006) Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem 281: 8469–8475 [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD (2018) Real‐space refinement in PHENIX for cryo‐EM and crystallography. Acta Crystallogr D Biol Crystallogr 74: 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo J, Ulryck N, Deforges J, Chamond N, Lopez‐Lastra M, Masquida B, Sargueil B (2016) LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S‐HCV IRES complex. Nucleic Acids Res 44: 1309–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhab Y, Bulakhov AG, Pestova TV, Hellen CUT (2020) Dissemination of internal ribosomal entry sites (IRES) between viruses by horizontal gene transfer. Viruses 12: 612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhab Y, Miścicka A, Pestova TV, Hellen CUT (2022) Horizontal gene transfer as a mechanism for the promiscuous acquisition of distinct classes of IRES by avian caliciviruses. Nucleic Acids Res 22: 1052–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JL, Pestova TV, Hellen CU, Wagner G (2000) The eIF1A solution structure reveals a large RNA‐binding surface important for scanning function. Mol Cell 5: 109–119 [DOI] [PubMed] [Google Scholar]
- Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, Berger B (2019) Positive‐unlabeled convolutional neural networks for particle picking in cryo‐electron micrographs. Nat Methods 16: 1153–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck‐Lederer A, Lewis JD, Stark H (2005) Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Brito Querido J, Sokabe M, Kraatz S, Gordiyenko Y, Skehel JM, Fraser CS, Ramakrishnan V (2020) Structure of a human 48S translational initiation complex. Science 369: 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A (2000) Leginon: an automated system for acquisition of images from vitreous ice specimens. J Struct Biol 132: 33–45 [DOI] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Pestova TV, Hellen CU (2008) Factor requirements for translation initiation on the simian picornavirus internal ribosomal entry site. RNA 14: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y (2015) Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Imprima E, Floris D, Joppe M, Sánchez R, Grininger M, Kühlbrandt W (2019) Protein denaturation at the air‐water interface and how to prevent it. eLife 8: e42747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS (2011) HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA 17: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CU, Wagner G (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J 18: 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser RM (2021) Preparing better samples for cryo‐electron microscopy: biochemical challenges do not end with isolation and purification. Annu Rev Biochem 90: 451–474 [DOI] [PubMed] [Google Scholar]
- Grant T, Rohou A, Grigorieff N (2018) cisTEM, user‐friendly software for single‐particle image processing. eLife 7: e35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J (2013) Hepatitis‐C‐virus‐like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Ping LH, Rijnbrand RC, Amphlett E, Clarke B, Rowlands D, Lemon SM (1996) Structural requirements for initiation of translation by internal ribosome entry within genome‐length hepatitis C virus RNA. Virology 222: 31–42 [DOI] [PubMed] [Google Scholar]
- Huang BY, Fernández IS (2020) Long‐range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation. Proc Natl Acad Sci USA 117: 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin‐Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V (2014) Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Wimberly BT, Kieft JS, Ramakrishnan V (2016) Large‐scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Oguro A, Nakamura Y, Kieft JS (2016) Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. eLife 5: e21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA (2004) Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaledhonkar S, Fu Z, Caban K, Li W, Chen B, Sun M, Gonzalez RL, Frank J (2019) Late steps in bacterial translation initiation visualized using time‐resolved cryo‐EM. Nature 570: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner LR, Anand AA, Nguyen HC, Myasnikov AG, Klose CJ, McGeever LA, Tsai JC, Miller‐Vedam LE, Walter P, Frost A (2019) eIF2B‐catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science 364: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA (2001) Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU (2000) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhle B, Ficner R (2014) eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J 33: 1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V (2015) Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ (2007) HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA (2013) The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500: 307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Ma XX, Chang QY, Li LJ, Wang YN, Feng YP, Ma ZR, Zhou JH (2018) The effects of nucleotide usage in key nucleotide positions +4 and −3 flanking start codon on translation levels mediated by IRES of hepatitis C virus. Acta Virol 62: 441–446 [DOI] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR (2005) A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Malygin AA, Kossinova OA, Shatsky IN, Karpova GG (2013a) HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 41: 8706–8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Shatsky IN, Karpova GG (2013b) Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc) 78: 53–59 [DOI] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G (2003) Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc Natl Acad Sci USA 100: 1535–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Mauro VP (2014) Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES‐dependent translation initiation in vivo. Proc Natl Acad Sci USA 111: 15385–15389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag N, Lin KY, Edmonds KA, Yu J, Nadkarni D, Marintcheva B, Marintchev A (2016) eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res 44: 7441–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane R, Pisareva VP, Rodriguez CF, Pisarev AV, Fernández IS (2020) A complex IRES at the 5'‐UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. eLife 9: e54575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odreman‐Macchioli F, Baralle FE, Buratti E (2001) Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem 276: 41648–41655 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV IV, Tarry MJ, Ramakrishnan V (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Passmore LA, Russo CJ (2016) Specimen preparation for high‐resolution Cryo‐EM. Methods Enzymol 579: 51–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (2001) Preparation and activity of synthetic unmodified mammalian tRNAi(met) in initiation of translation in vitro. RNA 7: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU (1998a) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU (1998b) A prokaryotic‐like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU (2008) eIF2‐dependent and eIF2‐independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J 27: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV (2006) Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Fernández IS (2018) Dual tRNA mimicry in the cricket paralysis virus IRES uncovers an unexpected similarity with the hepatitis C virus IRES. eLife 7: e34062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A et al (1999) Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 77: 153–161 [DOI] [PubMed] [Google Scholar]
- Prince JB, Taylor BH, Rhurlow DL, Ofengand J, Zimmermann RA (1982) Covalent crosslinking of tRNA1 Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc Natl Acad Sci USA 79: 5450–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N (2015) Cryo‐EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9‐Å resolution. Nat Commun 6: 7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibarkh, M , Yamamoto, Y , Singh, C.R, del Rio, F , Fahmy, A , Lee, B , Luna, RE , Ii, M , Wagner, G , and Asano, K (2008) Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5‐binding faces important for multifactor assembly and AUG selection. J Biol Chem 283: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ (1996) Internal initiation of translation of hepatitis C virus RNA: The ribosome entry site is at the authentic initiation codon. RNA 2: 867–878 [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012) RELION: implementation of a Bayesian approach to cryo‐EM structure determination. J Struct Biol 180: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2016) Processing of structurally heterogeneous cryo‐EM data in RELION. Methods Enzymol 579: 125–157 [DOI] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera‐Martinez E, Renaud A, Kuhn L, Hashem Y (2016) eIF3 peripheral subunits rearrangement after mRNA binding and start‐codon recognition. Mol Cell 63: 206–217 [DOI] [PubMed] [Google Scholar]
- Simonetti A, Guca E, Bochler A, Kuhn L, Hashem Y (2020) Structural insights into the mammalian late‐stage initiation complexes. Cell Rep 31: 107497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU (1998) Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA‐induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151: 41–60 [DOI] [PubMed] [Google Scholar]
- Tang S, Collier AJ, Elliott RM (1999) Alterations to both primary and predicted secondary structure of stem‐loop IIIc of the hepatitis C virus 1b 5′ untranslated region (5'UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild‐type 5'UTR sequence. J Virol 73: 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN (2008) Eukaryotic translation initiation machinery can operate in a bacterial‐like mode without eIF2. Nat Struct Mol Biol 15: 836–841 [DOI] [PubMed] [Google Scholar]
- Thakur A, Gaikwad S, Vijjamarri AK, Hinnebusch AG (2020) eIF2α interactions with mRNA control accurate start codon selection by the translation preinitiation complex. Nucleic Acids Res 48: 810280–810296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ (2008) An improved cryogen for plunge freezing. Microsc Microanal 14: 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon‐anticodon base‐pairing and hydrolysis of eIF2‐bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang J, Shin BS, Kim JR, Dever TE, Puglisi JD, Fernández IS (2020) Structural basis for the transition from translation initiation to elongation by an 80S‐eIF5B complex. Nat Commun 11: 5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Schäfer T, Leibundgut M, Böhringer D, Aylett CHS, Ban N (2017) Structural and functional insights into human re‐initiation complexes. Mol Cell 67: 447–456 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Bürger J, Mielke T, Spahn CM (2014) Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV‐IRES RNA. Nat Struct Mol Biol 21: 721–727 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J et al (2015) Molecular architecture of the ribosome‐bound hepatitis C virus internal ribosomal entry site RNA. EMBO J 34: 3042–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Machida K, Iwasaki W, Shigeta T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M, Harada Y, Shigematsu H et al (2019) HCV IRES captures an actively translating 80S ribosome. Mol Cell 74: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Zheng A, Yu J, Yamamoto R, Ose T, Tanaka I, Yao M (2014) X‐ray structures of eIF5B and the eIF5B‐eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A. Acta Crystallogr D Biol Crystallogr 70: 3090–3098 [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsbery BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SHW (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. eLife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Scheres SHW (2019) A Bayesian approach to beam‐induced motion correction in cryo‐EM single‐particle analysis. IUCrJ 6: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]


