Abstract
COVID-19 is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus affecting the world population. Early detection has become one of the most successful strategies to alleviate the epidemic and pandemic of this contagious coronavirus. Surveillance testing programs have been initiated in many countries worldwide to prevent the outbreak of COVID-19. In this study, we demonstrated that our previously established clustered regularly interspaced short palindromic repeats (CRISPR)-Cas12a-based assay could detect variants of concern during 2021 in Thailand, including Alpha, Beta, and Delta strains as well as Omicron strain in early 2022. In combination with the newly designed saliva collection funnel, we established a safe, simple, economical, and efficient self-collection protocol for the COVID-19 screening process. We successfully utilized the assay in an active case finding with a total number of 578 asymptomatic participants to detect the SARS-CoV-2 in saliva samples. We finally demonstrated that the validation and evaluation in a large-scale setting could provide valuable information and elaborate the practicality of the test in real-world settings. Our optimized protocol yielded effective results with high sensitivity, specificity, and diagnostic accuracy (96.86%). In addition, this study demonstrates COVID-19 active case findings in low-resource settings, which would be feasible and attractive for surveillance and outbreak prevention in the future.
Keywords: COVID-19, active case findings, saliva samples, CRISPR-Cas12a, RPA, qRT-PCR
Impact Statement
Active case finding programs have been initiated in many countries worldwide to control the outbreak of COVID-19. In this study, we demonstrated that our previously established clustered regularly interspaced short palindromic repeats (CRISPR)-Cas12a-based assay could detect several variants of concern including Alpha, Beta, Delta, and Omicron strains of SARS-CoV-2. Moreover, saliva self-collection combined with the CRISPR-based assay was a simple, field-deployable, and cost-effective approach, which would be practical and attractive for COVID-19 surveillance programs in low-resource settings.
Introduction
COVID-19 is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As of February 7, 2022, there were more than 394 million infected cases and 5.7 million deaths worldwide. The outbreak of this disease caused the World Health Organization (WHO) to declare the COVID-19 pandemic on March 11, 2020 (https://covid19.who.int/). A key approach to prevent the spread of disease is to have routine testing of this virus.
A standard technique for COVID-19 testing is the detection of SARS-CoV-2 RNA by quantitative reverse transcription real-time PCR (qRT-PCR) from the nasopharyngeal swab. This method has high specificity and sensitivity. However, it takes at least a few hours and requires a real-time PCR machine, making it difficult for mobile testing or laboratory settings in low- or middle-income countries. Apart from inadequate availability, the costly machines must be operated and analyzed by professionally trained technicians.1–3 These limitations, together with the requirement of authorized healthcare personnel to collect nasopharyngeal swabs from patients and other time-consuming steps, the deadly disease has not been under control in many countries. Another convenient approach is based on the detection of SARS-CoV-2 antigen by lateral flow assay. This approach is fast (15–30 min), but it has a high false-negative rate due to the late onset of the antigen. 4
Meanwhile, the use of the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas system in molecular diagnostics has increased exponentially.2,5,6 Several CRISPR/Cas-based assays have been developed because the system offers highly specific nucleic acid detection. In our previous studies, we reported the detection of SARS-CoV-2 RNA based on CRISPR-Cas12a. Our assay acquired the limit of detection of 10 copies/µL. 7 The highest detectable Ct value was approximately 35. 8 We showed that this approach could detect SARS-CoV-2 RNA from nasopharyngeal swabs with high specificity (100%), high accuracy (⩾ 95%) and with less than 2 h turnaround time.7,8 However, the SARS-CoV-2 virus constantly mutates. During the previous study, only wild-type and Alpha strains were prevalent. Subsequently, there were new emerging strains such as Beta, Delta, and Omicron in 2021 (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). Hence, we tested the diagnostic performance of our CRISPR-Cas12a-based assay with new variants of concern (VOC), especially those reported in Thai patients.
In addition to developing the assay for a non-invasive active case findings purpose, we also designed a saliva collection funnel and tested the diagnostic performance of the CRISPR-Cas12a-based detection using saliva samples. We would like to improve and extend the usage of our assay for large-scale active case findings and make it accessible for the low-resource settings. Here, we show that our protocol consisting of a saliva self-collection step followed by CRISPR-Cas12a-based assay could provide highly sensitive and specific results suitable for applying to the surveillance programs in local areas. The assay has also been confirmed to detect all VOC, including the Omicron strain.
Materials and methods
Sample collection
Participants registered and signed the informed consent for the COVID-19 active case findings program during April–August 2021. A saliva collection kit was provided to participants for self-saliva collection with the instructions. The saliva collection kit consists of a funnel and 2.0 mL screw-cap tube containing 0.5 mL of 2X DNA/RNA shield (Zymo Research, USA), which lysed the viral particle and preserved nucleic acid at ambient temperature. The saliva samples (approximately 0.5 mL) were collected and sent to the express analysis mobile unit within 24 h after saliva collection.
Nucleic acid extraction
Saliva samples were mixed by vortexing, and were heated at 95°C for 10 min. Then, the 200 µL of each sample was used for viral nucleic acid extraction using Nextractor® NX-48 S Viral NA kit (Genolusion, South Korea) according to the manufacturer’s recommendations. The extracted nucleic acids (approximately 50 µL) were used for SARS-CoV-2 detection or kept at −80°C until used.
Reverse transcription, recombinase polymerase amplification, and CRISPR-Cas12a-based assay
To detect the SARS-CoV-2 gene, the RT, recombinase polymerase amplification (RPA), and CRISPR assay were performed following the previously reported protocol 7 with minor modification. Briefly, the RT mixtures consisting of 1X reaction buffer, 1 mM dNTPs, 5 µM random hexamer, 20 U RiboLock RNase inhibitor (Thermo Scientific, USA) and 100 U RevertAid RT (Thermo Scientific, USA) were mixed with 11.5 µL of extracted RNA. Then, the reaction was incubated at 39°C for 30 min, and then at 70°C for 10 min. The RPA reaction consisting of 0.48 µM forward primer, 0.48 µM reverse primer, 14 mM MgOAc, 29.5 µL rehydration buffer, and lyophilized RPA enzyme (TwistAmp®, UK) was prepared and mixed with 5 µL of cDNA. The RPA reaction was incubated at 39°C for 30 min followed by heat inactivation at 75°C for 5 min. The CRISPR assay mixture consisted of 1X NEBuffer 2.0 (NEB, USA), 30 mM crRNA-S1, 30 mM crRNA-S2, 33 nM EnGen® Lba Cas12a (NEB, USA), 200 nM fluorescence reporter, and 1 µL of RPA product. The reactions were incubated at 39°C for 15 min, and then the fluorescent signal was visualized using BluPAD Dual LED Blue/White Light Transilluminator (BIO-HELIX, Taiwan). Two of three concordant results from three interpreters were used as a final result. For the reactions performed using undiluted RNA from specimens compared to their 1:10 and 1:100 dilutions, the purified RNA samples were 10-fold serially diluted with diethylpyrocarbonate (DEPC) treated water before the RT-RPA-Cas steps.
qRT-PCR reaction
The SARS-CoV-2 detection was performed using primers and the probe for the CDC-N1 gene from the previous report. 9 The reaction mixture consisted of 1X Luna® Universal Probe qPCR Master Mix (New England Biolabs, USA), 7.5 U WarmStart® RTx Reverse Transcriptase (New England Biolabs, USA), 0.5 µM of each primer, 0.125 µM TaqMan probe, and 2.5 µL of extracted RNA in the final volume of 10 µL. All samples were replicated at least twice. The qRT-PCR reaction was carried out in the CFX96™ Real-Time PCR instrument (Bio-Rad, USA) using the thermal profile as follows: RT at 55°C for 10 min, initial denaturation at 95°C for 1 min, followed by 45 cycles of 95°C for 10 s, and 55°C for 1 min. The samples obtaining the mean Ct ⩽ 35 were interpreted as positive for SARS-CoV-2.
Clinical performance of CRISPR-Cas12a assay using saliva samples
To evaluate the clinical performance of the CRISPR-Cas12a assay tested with saliva samples, a comparison between the results obtained from CRISPR-Cas12a and standard qRT-PCR (CDC-N1 gene) was analyzed. The diagnostic test evaluation calculator (https://www.medcalc.org/calc/diagnostic_test.php) was used to calculate sensitivity, specificity, positive predictive values (PPVs), negative predictive values (NPVs), and diagnostic accuracy.
DNA sequencing for identification of SARS-CoV-2 variants
Samples with false-negative results obtained from the CRISPR-Cas12a assay were subjected to confirmation by PCR and direct nucleotide sequencing. Briefly, the cDNAs were amplified by PCR using primers specific to the S gene of SARS-CoV-2, as described previously. 10 The PCR products were separated by 1% agarose gel electrophoresis and purified by QIAquick® gel extraction kit (QIAGEN, Germany). The Sanger sequencing was performed by the nucleotide sequencing service company (BIONIC, South Korea). The nucleotide sequences were aligned with reference sequences using BioEdit and identified by nucleotide BLAST analysis.
Results
Detection of various variants of SARS-CoV-2 by CRISPR-Cas12a-based assay
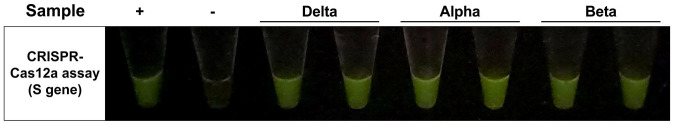
To become an effective and practical screening assay, the approach must successfully detect all VOC suspected to be contagious in the area during the testing period. We investigated the performance of our CRISPR-Cas12a-based assay in detecting three VOC reported to be found in Thailand in April–August 2021, including Alpha, Beta, and Delta strains in nasopharyngeal (NP) swab samples, as shown in Figure 1. The results showed that the assay could efficiently detect those VOC.
Figure 1.
The representative result obtained from CRISPR-Cas12a assay targeting S gene 7 for three SARS-CoV-2 variants of concern detection. The assay was tested against the RNA extracted from patients infected with different strains of SARS-CoV-2 (verified by Sanger sequencing). The Ct values of tested samples range between 16 and 30. The positive control is the RNA of the SARS-CoV-2 wild-type strain.
Saliva collection funnel design
To accomplish the goal of the COVID-19 active case findings pipeline with the most effective method, we decided that the sample collection step should be simple, inexpensive, and minimize the exposure risks between health-care workers and the participants. Recently, saliva has become a more attractive and suitable option for surveillance testing because the saliva test has shown comparable performance to the NP swabs, while sample collection is non-invasive.11–15
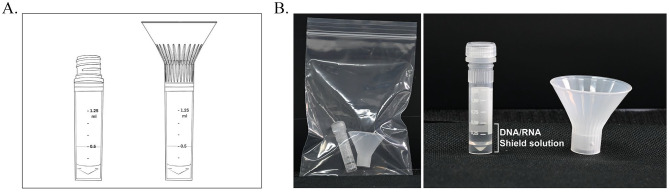
Since we proposed to develop the screening procedure for saliva samples which were aimed to be self-collected individually to reduce the risks of the personnel contacting with infectious clinical specimens, we designed a saliva collection funnel to be attached on top of the uncapped sample collection vial as shown in Figure 2. The funnel perfectly fits the vial, and it was tested to ensure there would be no leakage during the saliva collection step. Inside the vials, 0.5 mL of the 2XDNA/RNA Shield solution (Zymo research, USA) was added before saliva collection to lyse the viral particles and preserve the viral RNA for the extraction step. More importantly, to ensure that all infectious agents were completely inactivated, the saliva collection vials were incubated in the heat block at 95°C for 5 min before RNA extraction. Our sample collecting tool helped provide an easy-to-manage and safe procedure for both testing participants and health-care professionals.
Figure 2.
Saliva collection vial and funnel. (A) Design of the assembled device and (B) self-collected saliva collection package.
Active case finding program and sample collection
Next, we set up a COVID-19 active case finding program and enrolled 578 participants in Bangkok, Thailand, during April–August 2021. We wished to determine the performance of our screening procedure using CRISPR-Cas12a assay tested with RNA extracted from saliva samples. All volunteering individuals were asymptomatic or presymptomatic during sample collection.
To proceed the surveillance screening with self-collected samples, the package, including a vial and a saliva collection funnel packed in a zipped lock plastic bag (Figure 2), was provided to each participant. Each participant collected the specimen individually by following the instructions. The saliva collection tubes were disinfected with 70% ethanol, carefully packed in double zipped lock bags, and then sent back to the express analysis mobile unit by the following day. All the experimental steps are depicted in Figure 3.
Figure 3.
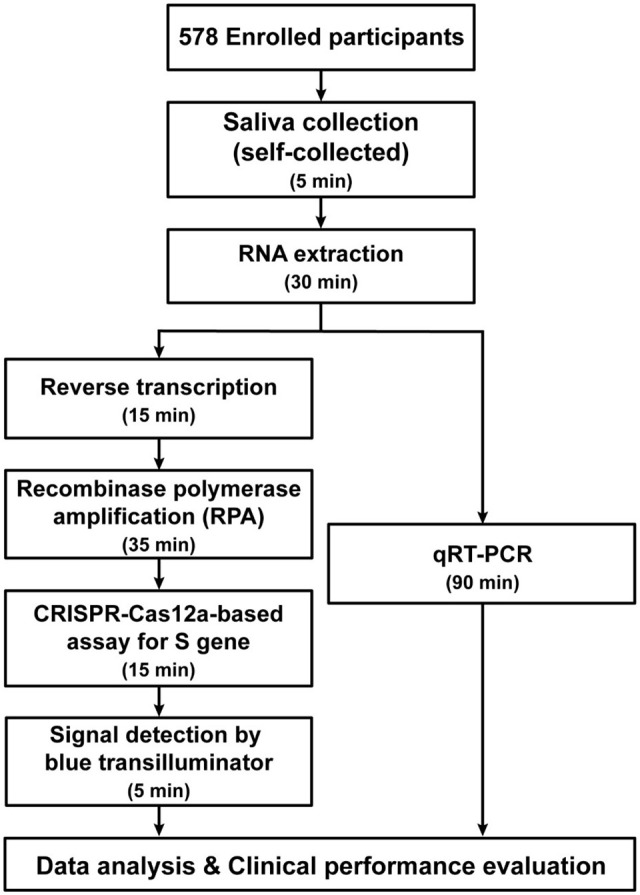
Flow chart demonstrating the experimental plan.
SARS-CoV-2 detection in saliva samples by CRISPR-Cas12a-based assay
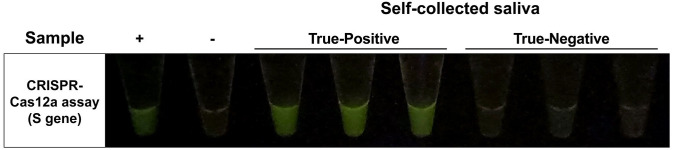
After the samples were transferred to the mobile analysis unit, the vials were disinfected with 70% ethanol and heated at 95°C for 10 min to inactivate viruses and to ensure no infectious agent was left outside or inside the vials. Then, viral nucleic acids were purified by magnetic-based automated extraction. The protocol for the CRISPR-Cas12a-based assay used in this study was slightly modified from our previously published work. 7 The RT and RPA steps were separated to maximize the detection efficiency of samples with low viral loads because all enrolled participants had no symptoms. The representative of positive and negative results from our screening is shown in Figure 4. There were 69 positive and 509 negative cases based on CRISPR-Cas12a, compared to 67 positive and 511 negative cases using qRT-PCR. The results from CRISPR-Cas12a were compared in parallel with qRT-PCR to evaluate the sensitivity, specificity, and diagnostic accuracy of the assay tested with RNA extracted from saliva specimens (Table 1). The results indicated that our CRISPR-Cas12a protocol achieved a sensitivity, specificity, and diagnostic accuracy of 88.06%, 98.04%, and 96.89%, respectively.
Figure 4.
The representative of positive and negative results from CRISPR-Cas12a-based assay targeting SARS-CoV-2 S gene tested with RNA extracted from saliva specimens. The Ct values of positive representative samples range between 23 and 29.
Table 1.
Comparative performance of the CRISPR-Cas12a-based assay for SARS-CoV-2 detection between self-collected saliva and nasopharyngeal swab.
| Parameters | CRISPR-Cas12a-based assay for SARS-CoV-2 detection | |
|---|---|---|
| Self-collected saliva (this study) | NP swab 6 | |
| Total samples tested | 578 | 164 |
| True-positive | 59 | 51 |
| True-negative | 501 | 111 |
| False-positive | 10 | 0 |
| False-negative | 8 | 2 |
| Sensitivity | 88.06% | 96.23% |
| Specificity | 98.04% | 100.00% |
| Positive predictive value (PPV) | 85.51% | 100.00% |
| Negative predictive value (NPV) | 98.43% | 98.23% |
| Diagnostic accuracy | 96.89% | 98.78% |
CRISPR: clustered regularly interspaced short palindromic repeats; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; NP: nasopharyngeal.
Delta strain detection using CRISPR-Cas12a-based assay
During the period of our surveillance program (April–August 2021), there was an increasing number of COVID-19 cases infected with the SARS-CoV-2 Delta variant. We suspected that the mutated virus might cause the false-negative results due to reducing the binding affinities of primers in the amplification step or crRNA in the CRISPR-Cas12a detection step. Therefore, we investigated whether the false-negative samples contained Delta strain virus. We performed Sanger sequencing of the amplified S gene and found that the sequences of those false-negative samples belonged to the Delta variant of SARS-CoV-2. When aligning the nucleotide sequence with the original primers and crRNA, the results showed no major mismatches within the region critical for the bindings. 7 We hypothesized that the variants of SARS-CoV-2 might not significantly affect the assay’s efficacy, but the quality of some saliva samples and the very low viral titers of asymptomatic participants would. To prove this hypothesis, we further modified the primer set by changing the S1 RPA primer to perfectly match the Delta strain sequence. We investigated whether the performance of the CRISPR-Cas12a-based assay would be improved by the new set of primers. We tested on the samples identified as Delta strain of SARS-CoV-2 at different dilutions. However, the original and new primer sets revealed similar results without significantly different fluorescence levels (Supplemental material Figure S1).
Omicron strain detection using CRISPR-Cas12a-based assay
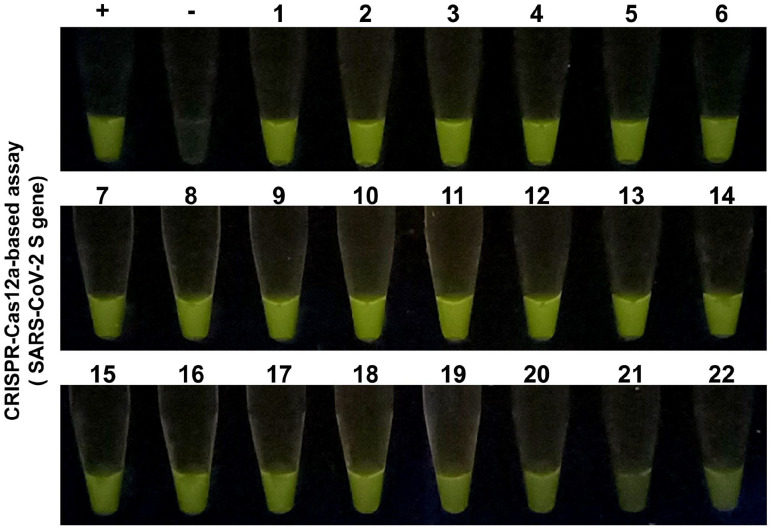
In November 2021, the WHO announced the emergence of the newest VOC. The strain was named Omicron (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). We also have acquired several clinical specimens from patients confirmed to have COVID-19 Omicron strain. Therefore, we investigated how our assay would detect the novel VOC of COVID-19. As shown in Figure 5, the results indicated that the assay could successfully deliver strong fluorescence signals from the reactions.
Figure 5.
The results of CRISPR-Cas12a-based assay tested with a total of 22 samples (labeled 1 to 22) of SARS-CoV-2 (Omicron variant) positive nasopharyngeal swab samples. The representative samples contain different mean Ct values ranging from 20 to 28 based on qRT-PCR analysis. (A color version of this figure is available in the online journal.)
Discussion
To increase the surveillance effectiveness and decrease the COVID-19 transmission rate, an appropriate procedure for a fast, safe, and effective alternative workflow is required to accelerate the speed of the test. The gold standard protocol such as qRT-PCR requires multiple critically controlled steps, expensive instruments, and trained professionals. Although the use of antigen test kits (ATKs) from many developers has been playing an important role in promoting a hygienic environment in the communities, there still are various concerns regarding the sensitivity and specificity of these rapid assays.4,16,17 Moreover, when new VOC carrying many mutations become dominant strains, the diagnostic accuracy of the rapid test kits might not be as high as previously validated. Therefore, the molecular diagnostic strategy with more precise and accurate detection is still in need for specific circumstances. Considering these challenges, we have validated that our previously published workflow could potentially detect major VOC in this study.
CRISPR-Cas-based diagnostic assay workflows have been in the spotlight during the pandemic of COVID-19 because of their ability to offer the users more precisely targeted tools. In combination with the isothermal amplification technique, several research groups have reported the advancement of diagnostic tool designs originating from the CRISPR-Cas system, and have developed various assays based on Cas12 and Cas13 to detect SARS-CoV-2.2,5,6 Saliva samples were used as an attractive choice owing to their non-invasive, repeatable, and simple collection procedure. Moreover, during the first week of infection, it has been reported that more viral particles could be retrieved from saliva compared to NP swab samples. 13 Saliva-based CRISPR-Cas detecting tools have been introduced by many groups and all of them were aimed to promote fast, easy, and accurate protocols or devices.18–22
Thus, we have put our efforts into optimizing the protocol to be employed with self-collected saliva samples using custom-made, user-friendly collection funnels. We validated the analytical sensitivity of the specimens by performing qRT-PCR in parallel. The results clearly indicated that our modified CRISPR-Cas12a-based protocol to detect SARS-CoV-2 could potentially become an alternative assay for active case findings with high diagnostic accuracy at 96.89% without an expensive real-time PCR machine. Although the performance might be comparatively lower than our previous report, we speculated that it was mainly because our enrolled participants were entirely asymptomatic with very low viral titer (Ct values > 28). In addition, the use of the self-collection protocol may affect the quality of some collected specimens, affecting the assay performance. Nonetheless, studies have shown that the patients with high Ct values (> 33) can almost be non-contagious with very low infectivity.23–25
Although many RNA-extraction free protocols have been suggested by several groups,19,21,22 and we, therefore, also considered skipping this step. We have tried multiple protocols to detect SARS-CoV-2 RNA from crude saliva; however, we subsequently found that the diagnostic capabilities could possibly become significantly decreased and affected the efficiency of the whole workflow. Therefore, our final decision was to maintain the extraction step in the protocol to achieve high efficacy and have an equivalent limit of detection compared to our published protocol. 7 Also, there are several options for the extraction kits, which are simple yet efficient.
This study investigated the feasibility of using the CRISPR-Cas12a-mediated diagnostic assay and applying it with saliva samples. Overall results suggest that saliva is suitable for screening purposes and worked well in comparison with our established CRISPR-Cas12a-based assay. The whole process can be accelerated, using less well-trained health-care professionals, and without the risk of close contact between infected patients and medical staff during sample collection. Moreover, this study might be considered as a showcase prior to larger-scale surveillance for COVID-19 in the future. One beneficial point is that the overall cost per sample (~US$20) is relatively cheaper than the regular qRT-PCR service price (~US$100). We also provide the total cost breakdown of the whole process to perform our CRISPR-Cas12-based assay, as shown in Supplemental material Table S1.
Besides the visual inspection of fluorescence signals by the blue light transilluminator, our group has been developing the machine learning–driven smartphone-based portable fluorescence detector, 26 which could supplement the process in the future. Based on our current protocol, we might also consider applying it to other detection platforms such as lateral flow-based assay or automated machines.
More recently, in early 2022, there were many cases infected with the Omicron variant of SARS-CoV-2 in Thailand. The Omicron outbreak has rapidly expanded to many countries within less than 3 months. From our data, we are confident that our modified established protocol will also work efficiently for active case findings with the Omicron strain. In summary, the self-saliva collection combined with the CRISPR-Cas12a assay is a simple, field-deployable, and cost-effective approach, which would be practical and attractive for COVID-19 active case findings in low-resource settings.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221090181 for COVID-19 active case findings based on self-collected saliva samples with CRISPR-Cas12a detection by Naphat Chantaravisoot, Pornchai Kaewsapsak, Oraphan Mayuramart, Pattaraporn Nimsamer, Suwanan Mankhong, Nantinee Chomta, Rungnapa Bootsri, Isara Alee, Piriya Wongkongkathep, Sombat Treeprasertsuk and Sunchai Payungporn in Experimental Biology and Medicine
Acknowledgments
We would like to thank Prof. Yong Poovorawan and Dr. Jiratchaya Puenpa, Center of Excellence in Clinical Virology, Faculty of Medicine, Chulalongkorn University, for providing RNA extracted from nasopharyngeal swabs of patients infected with SARS-CoV-2 (Omicron strain). We also would like to express our gratitude to Asst. Prof. Juthamas Ratanavaraporn, Faculty of Engineering, Chulalongkorn University and the Department of Disease Control, Ministry of Public Health for providing the express analysis mobile unit.
Footnotes
Authors’ Contributions: NC, PK, PW, and SP participated in the study design and data analysis. OM, PN, SM, NC, RB, and IA performed the experiments. ST and SP provided suggestions on the study design and discussion. NC, PK, OM, PN, and SP wrote the article. All authors participated in the execution of the active case-finding activity. All authors read and approved the final article.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: The study protocols were approved by the Institutional Review Board, Faculty of Medicine, Chulalongkorn University (IRB No. 302/63).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research is supported by the Health & Aging Platform [764002-HE08], Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University, Research Grants for Talented Mid-Career Researchers, The National Research Council of Thailand (NRCT) [C17F640216], Administration and Capital Management Unit for Enhancing the Competitiveness of The Country (AEC) [N41A640077], and The Innovation Fund to fight against COVID-19 [Taejai].
ORCID iD: Sunchai Payungporn  https://orcid.org/0000-0003-2668-110X
https://orcid.org/0000-0003-2668-110X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Benda A, Zerajic L, Ankita A, Cleary E, Park Y, Pandey S. COVID-19 testing and diagnostics: a review of commercialized technologies for cost, convenience and quality of tests. Sensors (Basel) 2021;21:6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mardian Y, Kosasih H, Karyana M, Neal A, Lau CY. Review of current COVID-19 diagnostics and opportunities for further development. Front Med (Lausanne) 2021;8:615099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Safiabadi Tali Seyed H, LeBlanc Jason J, Sadiq Z, Oyewunmi Oyejide D, Camargo C, Nikpour B, Armanfard N, Sagan Selena M, Jahanshahi-Anbuhi S. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev 2021;34:e00228–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayer J, Kasapic D, Zemmrich C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: a systematic meta-analysis of available data as of November 20, 2020. Int J Infect Dis 2021;108:592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan KG, Ang GY, Yu CY, Yean CY. Harnessing CRISPR-Cas to combat COVID-19: from diagnostics to therapeutics. Life (Basel) 2021;11:1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Falzone L, Gattuso G, Tsatsakis A, Spandidos DA, Libra M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int J Mol Med 2021;47:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayuramart O, Nimsamer P, Rattanaburi S, Chantaravisoot N, Khongnomnan K, Chansaenroj J, Puenpa J, Suntronwong N, Vichaiwattana P, Poovorawan Y, Payungporn S. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp Biol Med (Maywood) 2021;246:400–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nimsamer P, Mayuramart O, Rattanaburi S, Chantaravisoot N, Saengchoowong S, Puenpa J, Poovorawan Y, Payungporn S. Comparative performance of CRISPR-Cas12a assays for SARS-CoV-2 detection tested with RNA extracted from clinical specimens. J Virol Methods 2021;290:114092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Puenpa J, Suwannakarn K, Chansaenroj J, Nilyanimit P, Yorsaeng R, Auphimai C, Kitphati R, Mungaomklang A, Kongklieng A, Chirathaworn C, Wanlapakorn N, Poovorawan Y. Molecular epidemiology of the first wave of severe acute respiratory syndrome coronavirus 2 infection in Thailand in 2020. Sci Rep 2020;10:16602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ehrenberg AJ, Moehle EA, Brook CE, Doudna Cate AH, Witkowsky LB, Sachdeva R, Hirsh A, Barry K, Hamilton JR, Lin-Shiao E, McDevitt S, Valentin-Alvarado L, Letourneau KN, Hunter L, Keller A, Pestal K, Frankino PA, Murley A, Nandakumar D, Stahl EC, Tsuchida CA, Gildea HK, Murdock AG, Hochstrasser ML, O’Brien E, Ciling A, Tsitsiklis A, Worden K, Dugast-Darzacq C, Hays SG, Barber CC, McGarrigle R, Lam EK, Ensminger DC, Bardet L, Sherry C, Harte A, Nicolette G, Giannikopoulos P, Hockemeyer D, Petersen M, Urnov FD, Ringeisen BR, Boots M, Doudna JA, IGI SARS-CoV-2 Testing Consortium. Launching a saliva-based SARS-CoV-2 surveillance testing program on a university campus. PLoS ONE 2021;16:e0251296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams E, Bond K, Zhang B, Putland M, Williamson Deborah A, McAdam Alexander J. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020;58:e00776–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman O-E, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 2020;383:1283–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marais G, Hsiao N-y, Iranzadeh A, Doolabh D, Enoch A, Chu C-y, Williamson C, Brink A, Hardie D. Saliva swabs are the preferred sample for Omicron detection. medRxiv 2021. DOI: 10.1101/2021.12.22.21268246 [DOI] [Google Scholar]
- 15. Lai J, German J, Hong F, Tai SHS, McPhaul KM, Milton DK, for the University of Maryland StopCOVID Research Group. Comparison of saliva and mid-turbinate swabs for detection of COVID-19. medRxiv 2021. DOI: 10.1101/2021.12.01.21267147 [DOI] [Google Scholar]
- 16. Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Domen J, Dretzke J, Ferrante di, Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, McInnes MD, Spijker R, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 2021;3:CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jegerlehner S, Suter-Riniker F, Jent P, Bittel P, Nagler M. Diagnostic accuracy of a SARS-CoV-2 rapid antigen test in real-life clinical settings. Int J Infect Dis 2021;109:118–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arizti-Sanz J, Freije CA, Stanton AC, Petros BA, Boehm CK, Siddiqui S, Shaw BM, Adams G, Kosoko-Thoroddsen TF, Kemball ME, Uwanibe JN, Ajogbasile FV, Eromon PE, Gross R, Wronka L, Caviness K, Hensley LE, Bergman NH, MacInnis BL, Happi CT, Lemieux JE, Sabeti PC, Myhrvold C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun 2020;11:5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Azmi I, Faizan MI, Kumar R, Raj Yadav S, Chaudhary N, Kumar Singh D, Butola R, Ganotra A, Datt Joshi G, Deep Jhingan G, Iqbal J, Joshi MC, Ahmad T. A saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Front Cell Infect Microbiol 2021;11:632646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Puig H, Lee RA, Najjar D, Tan X, Soeknsen LR, Angenent-Mari NM, Donghia NM, Weckman NE, Ory A, Ng CF, Nguyen PQ, Mao AS, Ferrante TC, Lansberry G, Sallum H, Niemi J, Collins JJ. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv 2021;7:eabh2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding X, Yin K, Li Z, Sfeir MM, Liu C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens Bioelectron 2021;184:113218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ning B, Yu T, Zhang S, Huang Z, Tian D, Lin Z, Niu A, Golden N, Hensley K, Threeton B, Lyon Yin XM, Roy CJ, Saba NS, Rappaport J, Wei Q, Hu TY. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci Adv 2021;7:eabe3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hiroi S, Kubota-Koketsu R, Sasaki T, Morikawa S, Motomura K, Nakayama EE, Okuno Y, Shioda T. Infectivity assay for detection of SARS-CoV-2 in samples from patients with COVID-19. J Med Virol 2021;93:5917–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Platten M, Hoffmann D, Grosser R, Wisplinghoff F, Wisplinghoff H, Wiesmuller G, Schildgen O, Schildgen V. SARS-CoV-2, CT-values, and infectivity-conclusions to be drawn from side observations. Viruses 2021;13:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rabaan AA, Tirupathi R, Sule AA, Aldali J, Mutair AA, Alhumaid S, Muzaheed Gupta N, Koritala T, Adhikari R, Bilal M, Dhawan M, Tiwari R, Mitra S, Emran TB, Dhama K. Viral dynamics and real-time RT-PCR Ct values correlation with disease severity in COVID-19. Diagnostics (Basel) 2021;11:1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Samacoits A, Nimsamer P, Mayuramart O, Chantaravisoot N, Sitthi-Amorn P, Nakhakes C, Luangkamchorn L, Tongcham P, Zahm U, Suphanpayak S, Padungwattanachoke N, Leelarthaphin N, Huayhongthong H, Pisitkun T, Payungporn S, Hannanta-Anan P. Machine learning-driven and smartphone-based fluorescence detection for CRISPR diagnostic of SARS-CoV-2. ACS Omega 2021;6:2727–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221090181 for COVID-19 active case findings based on self-collected saliva samples with CRISPR-Cas12a detection by Naphat Chantaravisoot, Pornchai Kaewsapsak, Oraphan Mayuramart, Pattaraporn Nimsamer, Suwanan Mankhong, Nantinee Chomta, Rungnapa Bootsri, Isara Alee, Piriya Wongkongkathep, Sombat Treeprasertsuk and Sunchai Payungporn in Experimental Biology and Medicine