Abstract
DNA replication and differentiation are closely coupled during the Caulobacter crescentus cell cycle. We have previously shown that DNA topoisomerase IV (topo IV), which is encoded by the parE and parC genes, is required for chromosomal partitioning, cell division, and differentiation in this bacterium (D. Ward and A. Newton, Mol. Microbiol. 26:897–910, 1997). We have examined the cell cycle regulation of parE and parC and report here that transcription of these topo IV genes is induced during the swarmer-to-stalked-cell transition when cells prepare for initiation of DNA synthesis. The regulation of parE and parC expression is not strictly coordinated, however. The rate of parE transcription increases ca. 20-fold during the G1-to-S-phase transition and in this respect, its pattern of regulation is similar to those of several other genes required for chromosome duplication. Transcription from the parC promoter, by contrast, is induced only two- to threefold during this cell cycle period. Steady-state ParE levels are also regulated, increasing ca. twofold from low levels in swarmer cells to a maximum immediately prior to cell division, while differences in ParC levels during the cell cycle could not be detected. These results suggest that topo IV activity may be regulated primarily through parE expression. The presumptive promoters of the topo IV genes display striking similarities to, as well as differences from, the consensus promoter recognized by the major Caulobacter sigma factor ς73. We also present evidence that a conserved 8-mer sequence motif located in the spacers between the −10 and −35 elements of the parE and parC promoters is required for maximum levels of parE transcription, which raises the possibility that it may function as a positive regulatory element. The pattern of parE transcription and the parE and parC promoter architecture suggest that the topo IV genes belong to a specialized subset of cell cycle-regulated genes required for chromosome replication.
Differentiation in Caulobacter crescentus results from asymmetric cell division, which produces two distinct cell types: a motile swarmer cell with a set of differentiated structures, including a polar flagellum, bacteriophage receptors, and pili and a nonmotile stalked cell. Formation of the new swarmer cell results from a series of discrete morphogenic events that are closely coordinated with cell cycle progression and occur at the stalk-distal pole of the dividing cell (reviewed in references 3 and 24). This developmental sequence is dependent on completion of successive cell cycle checkpoints, and there is now evidence that the regulation of cell division and developmental events is mediated by two-component signal transduction pathways (6, 27, 44).
The progeny swarmer and stalked cells differ not only in morphology and motility but also in their capacities to initiate DNA replication. The stalked cell initiates chromosome replication (S phase) immediately upon division, and the period of replication is followed by a postsynthetic gap (G2 phase). The swarmer cell, by contrast, undergoes a presynthetic gap (G1 phase) and differentiates into a stalked cell before it initiates chromosome DNA replication (5). DNA replication is regulated in these cell types, at least in part, by the response regulator protein CtrA, which binds to the origin of replication in the swarmer cell and represses DNA initiation (34).
Several genes encoding proteins required for DNA replication or repair are also cell cycle regulated in C. crescentus. Their transcription is induced at or near the time of the swarmer-to-stalked-cell transition, which immediately precedes initiation of DNA replication. The first of these genes to be described is dnaC, which was originally identified genetically as a gene required for DNA chain elongation (26). dnaC is now known to encode a HolB homologue, a component of the DNA replication complex (28). Other genes with similar patterns of cell-cycle-regulated transcription include dnaA (45), which encodes a replication initiation protein; dnaN and dnaX (36, 42), which encode subunits of DNA polymerase; and gyrB (35, 36), which encodes a subunit of DNA gyrase. The dnaC gene has not been fully characterized, but the promoters of the other replication genes share two conserved sequence elements, an 8-mer motif (36) and a 13-mer motif, which appears to act as a negative regulatory element (42). The 8-mer motif (GnnTTTCG) is located at various positions in the vicinity of the −10 and −35 sequences (from −45 in dnaNpprox to +15 in dnaKp), but no functional analysis of this sequence had been reported prior to this study.
We have recently identified two other genes required for chromosome replication and segregation in C. crescentus, parE and parC (41). These genes encode subunits of DNA topoisomerase IV (topo IV), which is the enzyme responsible for the decatenation of replicated daughter chromosomes in bacteria (reviewed in reference 17). Conditional C. crescentus parE or parC mutants do not accurately segregate chromosomal DNA (41), but they differ from topo IV mutants in other bacteria that have been described (11, 14, 15, 18, 37). They fail to complete cell division, do not display asymmetrically located nucleoids, and do not give rise to anucleate cells (41). In addition to disrupting a late stage in cell division, C. crescentus topo IV mutants also fail to synthesize polar pili (40).
In this work, we have examined the expression of parE and parC in synchronous cell cultures and demonstrate that, like DNA replication genes examined previously in C. crescentus, their transcription is cell cycle regulated. Although activation of the parE and parC promoters coincides with the swarmer-cell-to-stalked-cell transition, when cells prepare for initiation of DNA replication, parE is the more strongly regulated of the two genes. These results and analysis of steady-state ParE and ParC protein levels suggest that topo IV activity may be regulated at the level of parE expression. Analysis of the 5′ regulatory regions of parE and parC suggests that these genes contain promoters that are similar in some respects to the consensus promoter recognized by the major Caulobacter sigma factor ς73 (19). The parE and parC promoters also contain a conserved 8-mer sequence in the spacer sequence between the −10 and −35 elements that is required for maximum levels of parE transcription and may function as a positive regulatory element. We discuss the possibility that parE and parC belong to a specialized class or subclass of developmentally regulated genes involved in DNA synthesis whose expression may be dependent on a secondary transcription factor(s).
MATERIALS AND METHODS
Strains and culture conditions.
C. crescentus strains used were all derived from strain CB15 (ATCC 19089). Strains were grown in PYE (32) medium or M2 minimal salts medium containing 0.2% glucose (12) supplemented with tetracycline (2 μg/ml) as indicated. Temperature-sensitive (Ts) alleles of parE (divC307 and divD308) and parC (divF310) have been characterized previously (41). Plasmids were introduced into C. crescentus by conjugation (30). Synchronous cultures were prepared on a density gradient of colloidal silica (no. 7631-86-9; Dupont) (7).
Radioimmune precipitation assays.
Synchronous cell preparations were diluted to an optical density at 660 nm of 0.1 in media prewarmed to 30°C. After a 10-min equilibration period, 2-ml samples were pulse-labeled for 10 min with 20 μCi of [35S]methionine (Amersham Corporation)/ml. After cell lysis, samples were divided. The rate of β-galactosidase or FlgE synthesis was monitored by radioimmune precipitation with either anti-β-galactosidase monoclonal antibody (Promega) or anti-FlgE antiserum (16). Immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by using a Molecular Dynamics PhosphorImaging system.
Western analysis of ParE and ParC proteins.
Carboxy-terminal peptides of ParC and ParE were used to raise rabbit polyclonal antibodies for use in Western analyses. The C-terminal BamHI-EcoRI fragment of the parC clone pDW131 (41) was cloned into pRSETC (Invitrogen) to create an N-terminally six-histidine (6×His)-tagged parC-derived peptide. Similarly, the C-terminal BamHI-XhoI fragment of pDW001 (41) was cloned into pRSETC to create an N-terminally 6×His-tagged parE-derived peptide. Each 6×His-tagged peptide was purified under denaturing conditions over Ni-nitrilotriacetic acid resin (QIAGEN Inc.) as described in reference 32a. The peptides were dialyzed overnight against 1,000× volumes of phosphate-buffered saline–0.1% Triton X-100 and used directly to raise polyclonal antibodies. Sera obtained were used at dilutions of 1:4,000 in the Western analyses discussed below.
Synchronous swarmer cells of strain CB15F were allowed to proceed through division. At each time point, an aliquot was removed, cells were pelleted by centrifugation, and the pellet was lysed by the addition of SDS-PAGE loading buffer. Equivalent volumes of culture were loaded in each sample and subjected to electrophoresis on a 7.5% polyacrylamide gel, transferred to Immobilon-P transfer membrane (Millipore), and blotted with either ParE or ParC antiserum.
In one set of experiments, alkaline phosphatase (AP) conjugated to anti-rabbit immunoglobulin G (IgG) (no. 1814206; Boehringer Mannheim) was used as the secondary antibody. In a second set of experiments, [125I]IgG (Amersham) was used as the secondary antibody to detect antibody binding, as described previously (31). For quantification, immunoblots using AP-conjugated antibody were scanned as TIF files by using Photolook (AGFA). Immunoblots using 125I-labeled IgGs were quantified by phosphorimaging (Molecular Dynamics), which gives a linear response to a wide range of signals (13). Quantification was performed with ImageQuant software (Molecular Dynamics).
Nuclease S1 protection assays.
To prepare a probe for determination of the start site of transcription for parE, a 493-bp fragment was amplified by PCR from pDW001 (41). The oligonucleotides used, DWPARE39 (5′CGATGTACATGCGCGGCCGCTTGCGCACCG3′) and DWPARE41 (5′CACCTGCAGCTGAAGGCCCGCTACGCGCCG3′), introduce mutations (underlined) which create NotI and PstI sites, respectively. The PCR product was cloned as a PstI-NotI fragment into pBluescript (Stratagene) to create pDW166, and the fragment was used as a probe in nuclease S1 protection assays. The 5′ end of the sequencing primer, DWPARE40 (5′GGCCGCTTGCGCACCGGCT3′), for the parE reference sequence ladder corresponds to the 5′ overhang generated by digestion of pDW166 with NotI. A 594-bp PstI-ApaLI fragment from pDW112, a pBluescript derivative of pDW148 (41) was used as a probe for determination of the transcriptional start site of parC. The 5′ end of the sequencing primer, DWPARC50 (5′TGCACGGGCTTCAAGCCATCGCG3′), for the parC reference sequence ladder corresponds to the 5′ overhang generated by ApaLI digestion.
Restriction fragments were 5′-end labeled by using T4 polynucleotide kinase (New England Biolabs). RNA was isolated from CB15 as previously reported (25). S1 assays were performed as previously described (2). First, 5′-end-labeled restriction fragments were hybridized to 100 μg of total cellular RNA at 65 and 60°C for parE and parC, respectively. After treatment with S1 nuclease, the resistant DNA fragments were electrophoresed through a polyacrylamide gel and visualized by autoradiography. DNA sequencing was performed on pDW112 and pDW001 double-strand template by using a Sequenase 7-deaza-dGTP sequencing kit (United States Biochemical) and [35S]dATP (Amersham Corp.).
Construction of lacZ promoter fusions.
To construct the parE or parC promoter fusions for determining cell cycle regulation of transcription, a SalI-PstI fragment of pSUPZ1 containing the promoterless lacZ gene (28) was cloned into pBluescript to create pDW100. To create the parE promoter fusion to lacZ, the 5′ XhoI-BamHI fragment of pDW001 (41) was cloned into pBluescript. An Asp718-SalI fragment was isolated from this construct and cloned into pDW100. The entire fusion was transferred as an Asp718-NotI fragment into pGH500 to create pDW104. To create the parC promoter fusion to lacZ, the 5′ PstI-XhoI fragment of pDW112 was transferred to pRSETA (Invitrogen) and reisolated as an Asp718-XhoI fragment. This fragment was cloned into pDW100, and the entire fusion was moved as an Asp718-NotI fragment into nonreplicative plasmid pGH500 (10) to create pDW138. Both promoter-reporter fusion vectors, pDW104 and pDW138, are nonreplicative plasmids in C. crescentus and must integrate by homologous recombination to confer tetracycline resistance to the host strain. Plasmids pDW104 and pDW138 were mated into synchronizable strain CB15F to create strains PC4828(parEp-lacZ) and PC4480(parCp-lacZ), respectively. An Asp718-XbaI fragment of pDW100 was cloned into plasmid pRK290 derivative pRK2L1 (23) to create pDW109(promoterless lacZ).
Mutations were introduced in the parE and parC gene promoters by sequential PCR as described previously (1) except that cloned Pfu polymerase (Stratagene) was employed. Reactions contained 20 mM (NH4)2SO4, 75 mM Tris (pH 9.0 at 25°C), 0.01% Tween 20, 2 mM MgSO4, and 10% dimethyl sulfoxide. Deoxyoligonucleotide primers used in PCR amplification were supplied by GIBCO BRL Custom Primers. Mutagenized parC constructs were amplified by using primers DWPARC71 (5′GTGTGGTACCTGCTGCAGACCATCC3′) and DWPARC72 (5′CTTGGGCTCGAGGACCAGACG3′). DWPARC71 contains the native PstI site and an Asp718 site (underlined [see Fig. 5B]) to facilitate cloning. DWPARC72 contains the native XhoI site (underlined [see Fig. 5B]). PCR products amplified by using DWPARC71 and DWPARC72 were cloned into pBluescript as Asp718-XhoI fragments, and the mutations were confirmed by DNA sequencing. The 5′ parC promoter deletions were generated by PCR using different 5′ deoxyoligonucleotides (see Fig. 5B). Mutagenized parE constructs were amplified by using primers DWPARE51 (5′CGCGCTCGAGGTCGGCAAGCT3′) and DWPARE86 (5′TATAAAGCTTGGGCATGCCGCGAC3′). The constructs contain the native XhoI site and an introduced HindIII site, respectively (underlined [see Fig. 5A]). The HindIII restriction site replaces a native SalI site in the parE gene. PCR products amplified by using DWPARE51 and DWPARE86 were cloned into pBluescript as XhoI-HindIII fragments, and the mutations were confirmed by sequencing. In addition, 5′ parE promoter deletions were generated (see Fig. 5A).
FIG. 5.
Sequence of parE and parC promoter regions and fusion junctions to the lacZ reporter gene. (A) Nucleotide sequence of the parE gene promoter region. (B) Nucleotide sequence of the parC gene promoter region. The 5′ ends of promoter deletion constructs are indicated by arrows followed, in parentheses, by their position relative to +1, and the promoter activity relative to the activity of the full-length fragment (100 U) (see text and Materials and Methods). An asterisk indicates transcription initiation sites. Restriction sites, −35 and −10 sequences, translation stop codons and transcription initiation sites are underlined. Transcription and translation initiation sequences are in boldface. The translated ParE and ParC sequences are given above the nucleotide sequence. The “…” represents a gap in the presented sequence, and is followed by the sequence of the transcriptional fusion junction to the lacZ reporter construct.
All pBluescript derivatives were cloned as Asp718-HindIII fragments into pRKlac290 (8). The pRKlac290 derivatives were mated into C. crescentus wild-type strain CB15 and assayed for β-galactosidase activity. (The sequences of the fusion junctions for both parE and parC are presented in Fig. 5.) The full-length parE and parC promoter fragments in the pRKlac290 derivatives correspond to those used in constructs pDW104 and pDW138 for examination of cell cycle regulation (see above).
β-Galactosidase activity assays.
Promoter activity of reporter constructs pDW104(parEp-lacZ) and pDW138(parCp-lacZ) in strains PC4828 and PC4480, respectively, and of control construct pDW109(promoterless lacZ) in strain CB15 was assayed as described previously (22). Expression from parEp-lacZ and parCp-lacZ yielded 101 and 98 U of activity, respectively. Expression from the control plasmid, pDW109, yielded 4 U of activity.
β-Galactosidase activity of the mutant promoter-lacZ fusion constructs (see Table 1 and Fig. 5) was assayed as described previously (39) except that overnight cultures were grown in PYE medium supplemented with 2 μg of tetracycline/ml and diluted 1:5 into fresh medium and incubated for 4 to 5 h. The activity of each construct was determined from an average of five to seven independent experiments. In each experiment, values for β-galactosidase activity were normalized; the wild-type fusions were assigned a value of 100, and the control plasmid pRKlac290 was assigned a value of 0. The normalized values from each experiment were then averaged to produce the data presented in Results (see Table 1).
TABLE 1.
Effect of promoter mutations on parCp-lacZ or parEp-lacZ expression
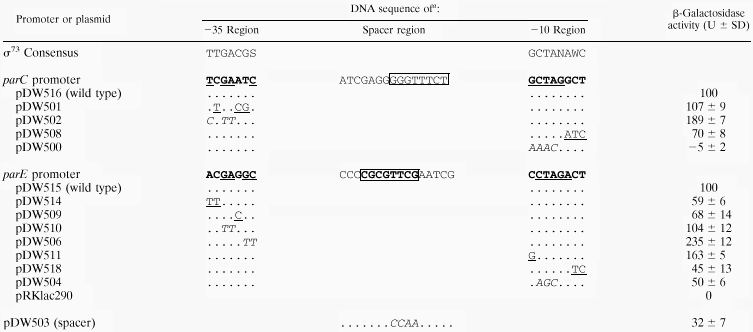 |
Underlining indicates either residues conserved in the −10 and −35 consensus sequence or changes made toward consensus. Italics indicate changes away from consensus. Periods indicate bases that were not changed in the mutant promoters. Boxes around sequences in the spacer region indicate the 8-mer sequences. Sequence elements subjected to mutagenesis are in boldface type. N = any base; S = G/C; W = A/T.
RESULTS
The parE and parC promoters are cell cycle regulated.
To examine topo IV regulation during the C. crescentus cell cycle, we determined the rates of parE and parC transcription in synchronous cultures. The transcription of other DNA topoisomerases, such as the Escherichia coli topA, gyrA, and gyrB genes, are known to respond to the superhelical state of DNA at their promoters (4, 20, 21). A similar effect has not been reported for parE or parC, but to minimize nonphysiological effects of transcription of the genes from multicopy plasmids, we examined expression from the parE and parC promoters by using transcription fusions to the lacZ reporter gene integrated in the chromosome (see Materials and Methods).
The parEp and parCp fusions were transferred into the synchronizable strain CB15F to construct the strains PC4828 (parEp-lacZ) and PC4480 (parCp-lacZ; see Materials and Methods). Swarmer cells were isolated from each strain and allowed to proceed through a synchronous round of division, and progeny swarmer and stalked cells were isolated after the completion of cell division. The rates of transcription from the parE and parC promoters were then determined in the synchronous swarmer and stalked cell cultures by radioimmune assays on cells pulse-labeled with [35S]methionine. The rate of flagellar hook protein (FlgE) synthesis, which is expressed late in the cell cycle at the S-to-G2-phase transition (38), was determined as an internal control.
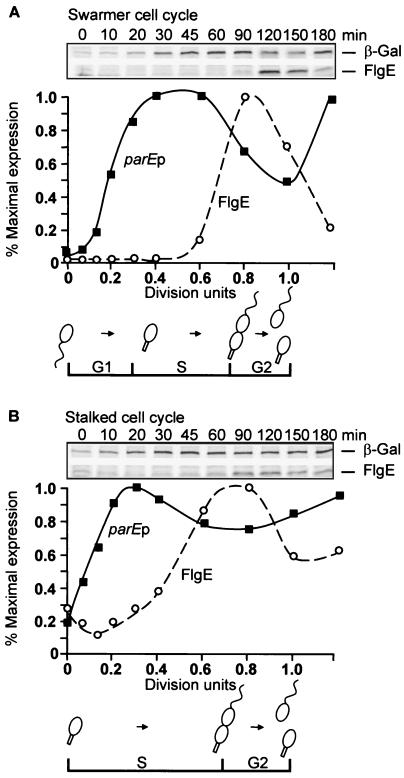
Transcription from the parE promoter (Fig. 1A) occurred at an extremely low rate in early G1-phase swarmer cells. After a lag of ca. 20 min, the rate of transcription increased ca. 20-fold in the cell cycle to maximum levels at 0.4 division unit, which corresponds to that of early S-phase cells that had just undergone stalk formation (as monitored by microscopic observation). Transcription continued at a high rate during the stalked cell portion of the cell cycle and decreased just before division as cells became highly pinched. Transcription began to increase as the cells entered the next cell cycle.
FIG. 1.
Transcriptional regulation of parEp in synchronous swarmer and stalked cell cultures. (A) Transcriptional activity of a parEp-lacZ fusion (PC4828) during the C. crescentus swarmer cell cycle as assayed by radioimmune precipitation of β-galactosidase. Division occurred at 150 min. (B) Transcriptional activity of a parEp-lacZ fusion (PC4828) during the C. crescentus stalked cell cycle as assayed by radioimmune precipitation of β-galactosidase. Division occurred at 120 min. Activity is plotted as a percentage of maximal expression. Note that 1.0 division unit for the stalked cell cycle corresponds approximately to the period of 0.4 to 1.0 division unit of the swarmer cell cycle. Expression of pulse-labeled FlgE protein served as an internal control for these synchrony experiments and those shown in Fig. 2. The times of swarmer to stalked cell differentiation and completion of cell division (1.0 division unit) are indicated at the bottoms of panels A and B along with the corresponding G1, S, and G2 phases.
Transcription from the parE promoter was also low in the isolated stalked cell population. However, it began to increase immediately and reached a maximum level of ca. fivefold of the initial rate between 0.2 and 0.4 division unit (Fig. 1B). Transcription decreased only slightly during the remainder of the cell cycle. These results demonstrated that the parE promoter is differentially regulated in the swarmer and stalked cell cycles. The rate of transcription was highest during S-phase in both swarmer and stalked cell synchronies, indicating that parE expression is coordinated in some way with DNA replication. In contrast to parE expression, FlgE synthesis reached a maximum both in the swarmer and stalked cell cycles at ca. 0.8 cell division unit, the time at which parE transcription was decreasing (Fig. 1A and B).
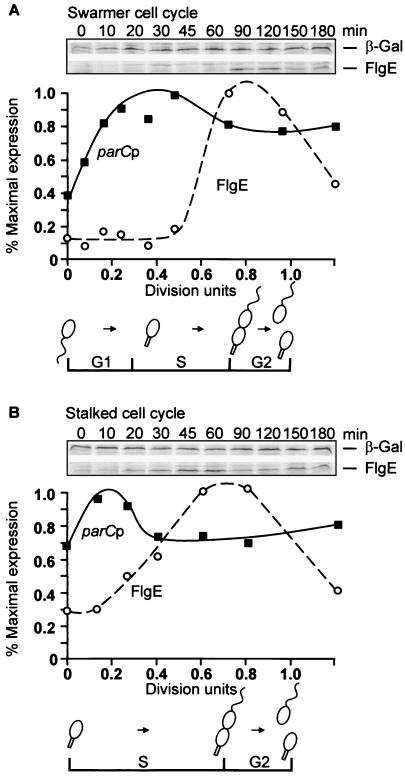
Transcription from the parC promoter in swarmer cells followed a similar but less pronounced pattern of cell cycle regulation than that displayed by the parE promoter (Fig. 2A). Levels of parCp activity displayed more than a twofold increase during the G1-to-S-phase transition in synchronous swarmer cells (Fig. 2A) and fell only slightly, later in the cell cycle just before division. Transcription from the parC promoter in synchronous stalked cell cultures increased only ca. 50% (Fig. 2B), which may be an artifact of the procedure used for cell synchrony (see below).
FIG. 2.
Transcriptional regulation of parCp in synchronous swarmer and stalked cell cultures. (A) Transcriptional activity of a parCp-lacZ fusion (PC4480) during the swarmer cell cycle as assayed by radioimmune precipitation of β-galactosidase. Division occurred at 120 min. (B) Transcriptional activity of a parCp-lacZ fusion (PC4480) during the stalked cell cycle as assayed by radioimmune precipitation of β-galactosidase. Division occurred at 90 min, which corresponds to 1.0 division unit. Activity is plotted as a percentage of maximal expression. The cell cycle periods are as described in the legend for Fig. 1 and the text.
We assessed the effects of the synchrony procedure on transcription by using the parEp-lacZ fusion construct. Mock synchronies in which cells were subjected to density centrifugation at 4°C were performed (see Materials and Methods). The resulting gradient was mixed to reconstitute an asynchronous culture. These cells were then incubated at 30°C, and the pattern of parEp activity was examined in a “mock” synchrony. A small, transient increase in transcription of ca. 1.5-fold was observed between 0 and 0.1 division unit of the cell cycle (data not shown). The increase is similar to the increase in transcription from the dnaN promoter reported previously in mock-synchronized cells (36). This transient increase in transcription presumably reflects the temperature shift to 30°C or some other aspect of the synchronization procedure. The increase occurred over a shorter period of the cell cycle than that observed for transcription from the parE promoter in swarmer and stalked cells or the parC promoter in swarmer cells (0.1 versus 0.4 division unit; Fig. 1A and B and 2A). Thus, the effects of the synchronization protocol should not contribute significantly to the pattern of parE or parC regulation seen in these three experiments where the amplitudes of changes in transcriptional activities were large. It could, however, account for the smaller change in transcription observed from the parC promoter in the stalked cell cycle (Fig. 2B).
Steady-state levels of ParE and ParC proteins during the cell cycle.
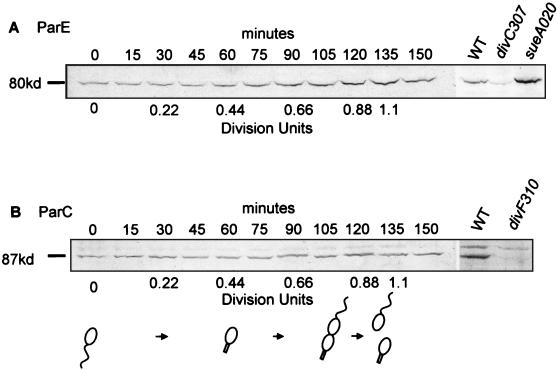
To determine the contribution of parE and parC transcription to the cellular levels of the topo IV subunits during the cell cycle, we examined the levels of the ParE and ParC proteins. The steady-state levels of ParE and ParC were assayed in synchronous swarmer cells of strain CB15F by Western blotting using rabbit polyclonal antibodies raised against C-terminal fragments of the two proteins and AP-conjugated anti-rabbit IgG as the secondary antibody (see Materials and Methods). Culture samples were removed at intervals throughout the cell cycle, the cells were collected by centrifugation and lysed, and the lysates were subjected to analysis by SDS-PAGE (Fig. 3; see Materials and Methods).
FIG. 3.
Levels of ParE and ParC in synchronous cell cultures. (A) The 80-kDa ParE protein was detected by Western analysis of samples from a synchronous culture of strain CB15F at the times indicated. The last three lanes contain lysates of wild-type strain CB15, strain PC8830[divC307(Ts)], and strain PC4885[divC307(Ts), sueA020(Cs)], a spontaneous revertant of the divC307(Ts) strain grown at the restrictive temperature of 37°C. The level of the ParE protein was reduced in the divC307(Ts) strain and increased in the sueA020(Cs) strain relative to wild-type levels. Levels of ParE and ParC were quantified as described in Materials and Methods, and the level in swarmer cells at 0 min was normalized to 1.0 U. These measurements showed that the level of ParE increased from 1.0 U at 0 min to 2.1 U immediately before cell division at 0.8 to 0.9 division unit and then decreased after cell division to 1.6 U at 1.2 division units. Similar results were obtained when 125I-labeled secondary antibodies were used; ParE levels determined in this assay (see Materials and Methods) increased from 1.0 U at 0 min to 1.9 U before division at 0.8 to 0.9 division unit and then decreased to 1.3 U after division at 1.2 division units. (B) The 87-kDa ParC protein was detected by Western analysis of lysates from a synchronous cell culture of strain CB15F. Assays of lysates of wild-type strains CB15 and PC8861[divF310(Ts)] grown at the restrictive temperature of 37°C are shown in the last two lanes. The level of the ParC protein was diminished in strain PC8861. Bands were quantified as described in Materials and Methods, but reproducible changes in ParC levels during the cell cycle were not detected.
We detected a discrete band at ca. 80 kDa by using the anti-ParE polyclonal antiserum (Fig. 3A). Although the ParE peptide is predicted to contain 667 residues with a calculated size of 73.3 kDa, we confirmed that the band detected corresponds to the parE gene product by comparing wild-type strain CB15 to a strain containing the Ts parE allele divC307(Ts) (29, 41) that had been grown at the nonpermissive temperature. The level of the 80-kDa protein was specifically reduced in the parE mutant strain. A spontaneous revertant of the divC307(Ts) mutant, sueA020(Cs) (strain PC4885), which can complete cell division at 37°C, showed an increase of the 80-kDa product under identical conditions of growth (Fig. 3A).
The amount of ParE protein was lower at the beginning of a synchronous swarmer cell cycle and increased ca. twofold during mid to late S phase. ParE levels peaked at the time of cell division and decreased immediately after division. The increase in ParE protein, although not as dramatic as the 20-fold increase observed for parE transcription, occurs in the cell cycle soon after the maximum rate of parE transcription is reached and transcription from the parE promoter has begun to decrease (Fig. 1A). Maximal ParE protein levels are, therefore, reached somewhat later in the cell cycle than maximal parE transcription.
Although the twofold increase in ParE observed using AP-conjugated anti-rabbit IgG as the secondary antibody was reproducible, we confirmed this result by using quantitative Western blots in which 125I-labeled IgG was used as the secondary antibody (see Materials and Methods and reference 31). Again, a twofold increase in ParE levels was observed during the synchrony with the maximum reached immediately before cell division, followed by a slight decrease immediately after cell division (see legend to Fig. 3 for results).
The anti-ParC polyclonal antibody detected a protein with an estimated size of 87 kDa relative to molecular mass markers (Fig. 3). The predicted peptide is 759 residues with a calculated size of 83 kDa. Confirming the identity of this protein as the parC gene product is its sharply decreased level in a strain containing the Ts parC allele divF310 (29) grown at the restrictive temperature (Fig. 3B). Changes in the steady-state levels of ParC could not be detected during the synchronous swarmer cell cycle (Fig. 3B). This result is consistent with the small changes in the rates of parC transcription observed during the cell cycle (Fig. 2A).
Mapping of parE and parC transcription start sites.
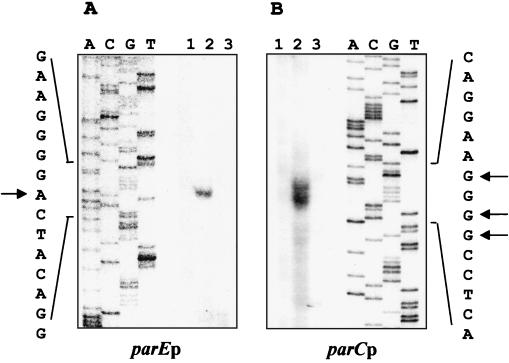
To locate the parE and parC promoters, we determined the transcription start sites by nuclease S1 protection assays using 5′-end-labeled probes (see Materials and Methods). A single protected fragment was observed for the parE gene and the start site mapped to an A residue on the template strand (Fig. 4A, lane 2). Multiple protected fragments observed for the parC gene correspond to potential start sites on three G residues on the template strand (Fig. 4B, lane 2). No protected fragments were observed in the control reactions with the labeled parE (Fig. 4A, lane 3) or parC probe (Fig. 4B, lane 3) when only tRNA was added. The proposed parE and parC start sites are indicated on the respective DNA sequences in Fig. 5, along with the −10 and −35 promoter sequences and the position of the predicted translation start sites.
FIG. 4.
Identification of transcription start sites for the parE and parC genes. (A) S1 nuclease protection of the parE transcript. Lanes: 1, probe plus 100 μg of CB15 RNA; 2, probe plus 100 μg of CB15 RNA plus S1 nuclease; 3, probe plus 100 μg of yeast tRNA plus S1 nuclease. The transcription start site is indicated. (B) S1 nuclease protection of the parC transcript. Lanes: 1, probe plus 100 μg of CB15 RNA; 2, probe plus 100 μg of CB15 RNA plus S1 nuclease; 3, probe plus 100 μg of yeast tRNA plus S1 nuclease. The transcription start sites are indicated.
To determine the extent of the functional parE and parC promoters, we constructed a series of deletions 5′ to the respective transcriptional start sites, as diagrammed in Fig. 5, and assayed their effect on promoter activity by using transcription fusions to the lacZ reporter gene (see Materials and Methods). The three deletions constructed for each parE and parC resulted in little or no reduction of β-galactosidase activity compared to the full-length promoter fragments starting at the 5′ XhoI (parE) and PstI (parC) sites (Fig. 5). These results indicate that all sequence elements required for transcriptional regulation lie within 141 bp 5′ of the parE transcriptional start and within 118 bp of the parC transcriptional start site.
parE and parC promoter analysis and effect of site-directed mutations.
As shown in Fig. 6, we aligned the parE and parC promoters with those of other genes that are required for DNA replication in C. crescentus and display a pattern of cell cycle-regulated transcription similar to these topo IV genes. The −10 and −35 sequences of the seven promoters display some similarity to the C. crescentus ς73-dependent promoters (Fig. 6) (19).
FIG. 6.
Alignment of parE and parC gene promoters with other C. crescentus gene promoters and to the ς73 consensus recognition sequence (Fig. 6) (19). Bases matching the consensus sequence are underlined. N = any base; S = C/G; W = A/T; Y = C/T. The references for promoter sequences and transcriptional start sites are as follows: dnaA (45), dnaN and dnaX (36, 42), dnaK (1a), and gyrB (35, 36).
We also examined the parE and parC promoters for two conserved sequence elements previously reported in C. crescentus replication genes. One of these, a 13-mer sequence with the consensus of ynCnCTCCGCnCs, is located most frequently in the −10, −35 spacer region (42). The second, an 8-mer motif with a consensus of GnnTTTCG, is found at various locations within these promoters (36). Although we were unable to identify a sequence corresponding to the 13-mer (Fig. 6), the −10, −35 spacers of parE and parC contain a sequence corresponding to the 8-mer consensus (Fig. 6). Because parE is under strong cell cycle regulation, we examined the contribution of this 8-mer sequence to the transcriptional activity of the parE promoter in vivo. Site-directed mutagenesis was used to change four conserved residues (TTCG) in this sequence without changing the −10, −35 spacing (pDW503; Table 1). The activity of the mutated promoter was decreased more than threefold, which suggests that the sequence mutated, and presumably the 8-mer sequence, is involved in the positive regulation of parE promoter activity.
To identify residues within the −10 and −35 elements of the parE and parC promoters responsible for determining transcriptional activity in vivo, we introduced mutations designed to make each promoter either more or less like the proposed ς73 promoter consensus sequence (Table 1). The −10 sequence elements conform more closely to the ς73 consensus than the −35 elements, with the strongest conservation in the first five or six residues of the −10 element (Fig. 6) (19). Site-directed mutations that changed any of the first five bases of the −10 element in either the parE (pDW511 and pDW504) or the parC (pDW500) promoter resulted in changes in transcriptional activity predicted from the −10 consensus (Table 1). Transcription was decreased substantially in constructs pDW500(parC) and pDW504(parE), which change residues away from the −10 consensus sequence (Table 1). Significantly, the activity of the parC construct pDW500 was reduced to levels equivalent to those of the promoterless control construct (pRKlac290), effectively eliminating transcriptional activity. The activity of parE construct pDW504 was reduced to 50% of wild-type levels. The one mutation in this set of three constructs that was toward consensus (pDW511; parE) resulted in an increased rate of transcription (Table 1). The transcriptional activity of the two site-directed mutations altering the last two bases of the −10 element, which is not strongly conserved at these positions, had either a small effect (pDW508; parC) or was not consistent with the predicted effect (pDW518; parE).
Mutations in the less-well-conserved −35 sequence elements of parE and parC generally resulted either in small effects on transcription or changes that did not conform to those expected from the ς73 consensus sequence. In the parE promoter, two changes toward consensus (pDW514 and pDW509) resulted in somewhat reduced transcription, while changes away from consensus resulted in either little change (pDW510) or in increased transcription (pDW506; Table 1). Similar results were obtained for mutations in the parC −35 sequence element, in which one mutation away from consensus (pDW502) increased transcription and a second mutation toward consensus (pDW501) produced little change (Table 1).
DISCUSSION
We have examined the cell cycle regulation of the parE and parC genes, which encode the subunits of the Caulobacter DNA topo IV. Our results show that transcription of the two genes is induced during the swarmer-to-stalked-cell transition when cells prepare for initiation of DNA synthesis. The expression of parE and parC is not strictly coordinated, however. The rate of parE transcription (Fig. 1) is much more strongly regulated in the cell cycle than that of parC (Fig. 2). The pattern of parE promoter activation in the G1-to-S phase of the cell cycle is similar to that of several other genes involved in DNA replication and repair (summarized in reference 36). However, the promoter architecture of parE and parC diverges in some respects from this group of replication-related genes. The topo IV genes do not contain the conserved 13-mer sequence, which has been proposed to function as a negative regulatory element in the dnaX promoter (42), but they do contain a conserved 8-mer sequence. We provide genetic evidence that this 8-mer sequence, whose function has not been previously reported, is required for maximum transcription from the parE promoter. We speculate that this sequence element may function as a positive regulatory element involved in the cell cycle regulation of this specialized set of replication genes.
parE transcription is induced more than 20-fold during the swarmer-to-stalked-cell transition (Fig. 1A), compared to a two- to threefold increase in parC transcription (Fig. 2A). The parE gene was also regulated in synchronous stalked cells, where its transcription was induced ca. fivefold very early in the cell cycle. We attribute the small increase in parC transcription in the stalked cell cycle to the effect of the synchronization protocol (compare Fig. 2B and 1B). These results suggest that topo IV activity may be regulated in part through parE expression. Consistent with this possibility is the regulation of ParE protein levels during the swarmer cell cycle. ParE increased ca. twofold after the swarmer-to-stalked-cell transition (Fig. 3A), while no change in the levels of ParC during the cell cycle could be detected (Fig. 3B).
Recent localization experiments in Bacillus subtilis show that ParC protein displays a bipolar localization pattern consistent with the suggestion that the topo IV subunits may function as part of a membrane-associated apparatus involved in chromosome segregation (11). Although ParE protein appears to be distributed throughout the B. subtilis cytoplasm, the polar localization of ParC depends on ParE function. In the absence of ParE, polar localization of ParC is abolished and ParC instead colocalizes with the nucleoid. These results may reflect a dynamic pattern of ParC localization or perhaps the fact that only a subpopulation of the ParC and ParE complex is involved in polar localization (11). Our results with Caulobacter do not address the question of subcellular localization of ParC and ParE; however, the results of Huang et al. (11) are consistent with the possibility that topo IV activity in Caulobacter may be regulated by the availability or activity of ParE during the cell cycle.
Many cell cycle and developmental events in Caulobacter are regulated at the level of transcription initiation. In eubacteria, the specificity of promoter recognition by RNA polymerase can be conferred by specialized sigma factor binding to the core RNA polymerase to reprogram RNA polymerase specificity (reviewed in reference 9). Thus, in the Caulobacter flagellar gene hierarchy, which contains four classes of genes (I to IV; reviewed in reference 43), transcription of the late class III and IV flagellar genes requires the specialized sigma factor ς54 and the transcriptional activator FlbD, which are encoded by class II genes. By contrast, the early class II flagellar genes, which contain noncanonical ς73-dependent promoters (44), depend on the transcriptional regulator CtrA for activation in vivo (33) and in vitro (44).
Alignment of the putative topo IV gene promoters with those of dnaA (45), dnaN and dnaX (42), gyrB (36), and dnaK (1a), reveal some similarity to the ς73 consensus. Mutagenesis of the −10 sequence elements of parE and parC generally yielded results expected of promoters recognized by ς73 (Table 1). By contrast, mutational analysis of the less-well-conserved −35 sequence elements of parE and parC did not yield expected results (Table 1). One explanation for the latter results is that the parE and parC promoters are recognized by an alternative sigma factor with a −10 recognition sequence similar to that of ς73. An alternative explanation, which we favor, is that the poorly conserved −35 elements reflect the requirement of an auxiliary regulatory factor(s) necessary for efficient transcription and cell cycle regulation of these ς73-dependent promoters. Since these promoters display a pattern of cell cycle-regulated transcription that is markedly different from CtrA-dependent genes and the promoters do not contain sequences conforming to the TTAAC direct repeat of the CtrA binding site (33), another transcription factor would presumably be required. As discussed below, the analysis of the 8-mer sequence in the parE promoter is consistent with this possibility.
Among the promoters aligned in Fig. 6, two features distinguish the parE and parC sequences. One is the spacing between the −10, −35 elements, which is 15 to 16 bp compared to the 10- to 14-bp spacing typical of ς73-dependent housekeeping promoters (19). The second is the absence of an identifiable 13-mer motif. This sequence, which is present in the other cell cycle-regulated promoters, has been suggested as a possible repressor binding site in dnaX (42). All of the promoters contain the previously described 8-mer motif (Fig. 6) (36) in various positions, however. The 8-mer motif appears to be required for efficient transcription from the parE promoter (Table 1), a result that is consistent with a positive regulatory role for this sequence. It remains to be determined if the 8-mer and 13-mer motifs are binding sites for auxiliary regulatory proteins involved in the cell cycle regulation of genes containing these sequences.
ACKNOWLEDGMENTS
We are grateful to Noriko Ohta for advice and encouragement, to Heping Jiang for valuable assistance in the preparation of media used in these experiments, and to Teresa Slover for her indispensable help in editing and assembling this manuscript.
This work was supported in part by Public Health Service grant GM22299 from the National Institutes of Health to A.N. and by National Institutes of Health Cellular and Molecular Biology Pre-doctoral Training grant 5-T326M07312 to D.V.W.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R, Moore D, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 1a.Avedissian M, Lessing D, Gober J W, Shapiro L, Gomes S L. Regulation of the Caulobacter crescentus dnaKJ operon. J Bacteriol. 1995;177:3479–3484. doi: 10.1128/jb.177.12.3479-3484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berk J A, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 3.Brun Y V, Marczynski G, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 4.Ching Y, Dinh T, Beran R K. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J Mol Biol. 1988;202:735–742. doi: 10.1016/0022-2836(88)90554-2. [DOI] [PubMed] [Google Scholar]
- 5.Degnen S T, Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 6.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 7.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gober J W, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht G B, Lane T, Ohta N, Sommer J N, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W M, Libbey J L, van der Hoeven P, Yu S X. Biopolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc Natl Acad Sci USA. 1998;95:4652–4657. doi: 10.1073/pnas.95.8.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R C, Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston R F, Pickett S C, Barker D L. Autoradiography using storage phosphor technology. Electrophoresis. 1990;11:355–360. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- 14.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 15.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornacker M, Newton A. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid residue sequence near the N-terminus. Mol Microbiol. 1994;14:73–85. doi: 10.1111/j.1365-2958.1994.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 17.Luttinger A. The twisted ‘life’ of DNA in the cell: bacterial topoisomerases. Mol Microbiol. 1995;15:601–606. doi: 10.1111/j.1365-2958.1995.tb02369.x. [DOI] [PubMed] [Google Scholar]
- 18.Luttinger A L, Springer A L, Schmid M B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 19.Malakooti J, Wang S P, Ely B. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthesis and housekeeping function. J Bacteriol. 1995;177:4372–4376. doi: 10.1128/jb.177.15.4372-4376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menzel R, Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Mullin D A, Newton A. Ntr-like promoters and upstream regulatory sequence ftr are required for transcription of a developmentally regulated Caulobacter crescentus flagellar gene. J Bacteriol. 1989;171:3218–3227. doi: 10.1128/jb.171.6.3218-3227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton A, Ohta N. Regulation of the cell division cycle and differentiation in bacteria. Annu Rev Microbiol. 1990;44:689–719. doi: 10.1146/annurev.mi.44.100190.003353. [DOI] [PubMed] [Google Scholar]
- 25.Ohta N, Chen L S, Swanson E, Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985;186:107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- 26.Ohta N, Masurekar M, Newton A. Cloning and cell cycle-dependent expression of DNA replication gene dnaC from Caulobacter crescentus. J Bacteriol. 1990;172:7027–7034. doi: 10.1128/jb.172.12.7027-7034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta N, Newton A. Signal transduction in the cell cycle regulation of Caulobacter differentiation. Trends Microbiol. 1996;4:326–332. doi: 10.1016/0966-842x(96)10050-0. [DOI] [PubMed] [Google Scholar]
- 28.Ohta, N., and A. Newton. Unpublished results.
- 29.Ohta N, Ninfa A J, Allaire A D, Kulick L, Newton A. Identification, characterization and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J Bacteriol. 1997;179:2169–2180. doi: 10.1128/jb.179.7.2169-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta N, Swanson E, Ely B, Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984;158:897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul K S, Bogan A A, Waters G M. Phosphatidylinositol transfer protein (PITPα) stimulates in vitro intra-Golgi transport. FEBS Lett. 1998;431:91–96. doi: 10.1016/s0014-5793(98)00722-4. [DOI] [PubMed] [Google Scholar]
- 32.Poindexter J S. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.QIAGEN Incorporated. The QIAexpressionist. 2nd ed. Valencia, Calif: QIAGEN Inc.; 1996. [Google Scholar]
- 33.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 34.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizzo M F, Shapiro L, Gober J. Asymmetric expression of the gyrase B gene from the replication-competent chromosome in the Caulobacter crescentus predivisional cell. J Bacteriol. 1993;175:6970–6981. doi: 10.1128/jb.175.21.6970-6981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts R C, Shapiro L. Transcription of genes encoding DNA replication proteins is coincident with cell cycle control of DNA replication in Caulobacter crescentus. J Bacteriol. 1997;179:2319–2330. doi: 10.1128/jb.179.7.2319-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid M B. A locus affecting nucleoid segregation in Salmonella typhimurium. J Bacteriol. 1990;172:5416–5424. doi: 10.1128/jb.172.9.5416-5424.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheffery M, Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981;24:49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- 39.Slauch J M, Silhavy T J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sommer J M, Newton A. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J Bacteriol. 1988;170:409–415. doi: 10.1128/jb.170.1.409-415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- 42.Winzeler E, Shapiro L. A novel promoter motif for Caulobacter cell cycle-controlled DNA replication genes. J Mol Biol. 1996;264:412–425. doi: 10.1006/jmbi.1996.0650. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–240. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zweiger G, Shapiro L. Expression of Caulobacter dnaA as a function of the cell cycle. J Bacteriol. 1994;176:401–408. doi: 10.1128/jb.176.2.401-408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]