Abstract
Upon emerging from the ribosome exiting tunnel, polypeptide folding occurs immediately with the assistance of both ribosome-associated and free chaperones. While many chaperones known to date are dedicated folding catalysts, recent studies have revealed a novel chaperoning system that functions at the interface of protein biogenesis and quality control by using a special “holdase” activity in order to sort and channel client proteins to distinct destinations. The key component, Bag6/Bat3/Scythe, can effectively shield long hydrophobic segments exposed on the surface of a polypeptide, preventing aggregation or inappropriate interactions before a triaging decision is made. The biological consequences of Bag6-mediated chaperoning are divergent for different substrates, ranging from membrane integration to proteasome targeting and destruction. Accordingly, Bag6 can act in various cellular contexts in order to execute many essential cellular functions, while dysfunctions in the Bag6 system can cause severe cellular abnormalities that may be associated with some pathological conditions.
Keywords: Bag6/Bat3/Scythe, chaperone/holdase, ER-associated degradation/ERAD, proteasome, protein quality control, tail-anchored protein biogenesis, ubiquitin
Introduction
The HLA (human leukocyte antigen)-B-associated transcript 3 (Bat3) gene (also named Scythe) was originally identified as a member of a cluster of genes located in the human major histocompatibility complex (MHC) III locus on chromosome 6, a region containing many genes essential for immune functions [1, 2]. Sequence analysis showed that the major isoform of the human Bat3 gene encodes a 1,132 amino acid-long multidomain protein, consisting of an amino-terminal ubiquitin-like (UBL) domain, a proline-rich segment bearing short tracts of polyproline, polyglycine, and charged amino acids, and a zinc finger-like domain (Fig. 1A). In addition, the carboxyl terminus of Bag6 contains a conserved BAG domain that is found in a family of proteins. Therefore, Bat3/Scythe is also named as Bag6.
Figure 1.
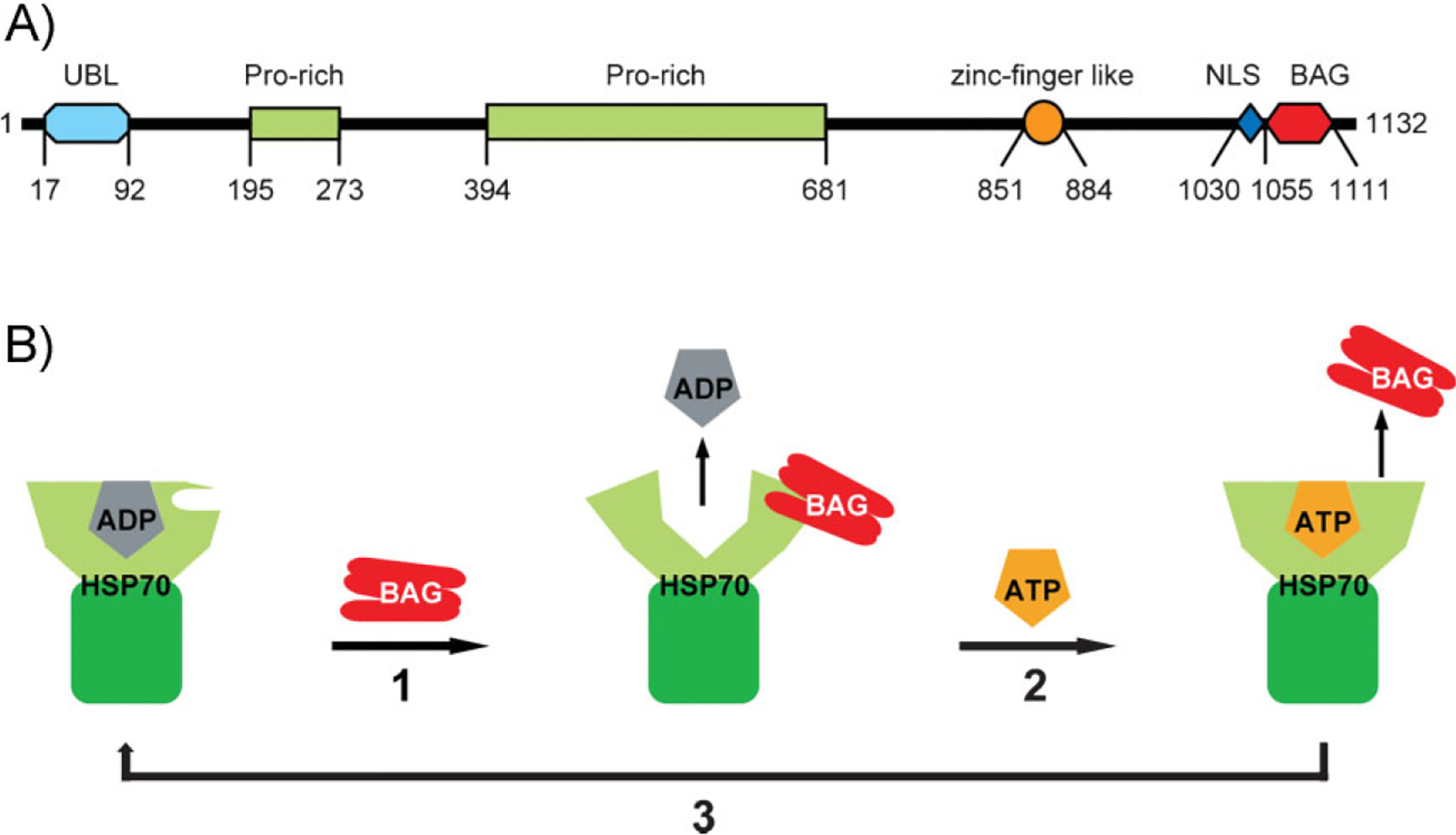
The BAG domain and BAG6. A: The domain structure of human Bag6. UBL, ubiquitin-like; Pro-rich, proline-rich domain; NLS, nuclear localization signal. B: A schematic model of the BAG domain and HSP70 interaction. Step 1: A BAG domain binds the nucleotide binding domain of HSP70 in ADP-bound state, promoting the exchange of ADP with ATP. Step 2: ATP binding causes a conformational change in HSP70, leading to the release of the BAG domain. Step 3: ATP is converted to ADP by HSP70. Whether this cycle can be leveraged by the BAG domain in Bag6 to engage HSP70 is unclear.
Bag6/Bat3/Scythe (hereafter referred to as Bag6) is an abundant cellular protein highly conserved in higher eukaryotes. It is ubiquitously expressed in multicellular organisms with high expression detected in rat testis, suggesting a potential function in meiosis and/or spermatogenesis [3–5]. Despite having a nuclear localization signal (NLS) [6], immunofluorescence studies showed that endogenous Bag6 is primarily localized to the cytoplasm with only a small fraction in the nucleus. This is at least in part due to the interaction of Bag6 with a cofactor named TRC35, which masks the NLS [7]. In addition, alternative splicing has been reported to generate a few Bag6 variants that lack the NLS and therefore are preferentially localized to the cytoplasm [8]. Interestingly, the nuclear Bag6 levels were found to be increased in osteosarcoma tissue [9], suggesting a potential role for nuclear Bag6 in tumorigenesis. Biochemical fractionation experiments also demonstrated the presence of a small pool of Bag6 that is associated with the intracellular membrane system including the endoplasmic reticulum (ER) [7].
The BAG domain in Bag6 is commonly found in a family of modulator proteins that interact with the family of HSP70 chaperones [10]. The BAG domains contain 110–124 amino acids that form three anti-parallel a-helices. The second and third helices are the sites to which the ATPase domain of HSC70/HSP70 binds. The interaction of the BAG domain with HSP70 is mediated by both electrostatic and hydrophobic interactions and is dependent on the nucleotide state of HSP70 (Fig. 1B). Accordingly, it was reported that Bag6 can bind the ATPase domain of a HSP70-like molecule named Stch [11]. Moreover, purified Bag6 has been shown to inhibit HSP70-mediated protein folding [12]. Another link between Bag6 and the HSP70-dependent folding machinery was established when a HSP70-interacting cofactor named SGTA was identified as a Bag6 interactor [13]. Based on these observations, Bag6 was proposed to function as a HSP70 co-chaperone. In addition to modulating the activity of HSP70, Bag6 also regulates its expression levels under heat shock conditions. In Bag6 deficient mouse embryonic fibroblast (MEF) cells, heat shock induced-HSP70 mRNA expression is reduced when compared to wild type MEF cells [14]. Moreover, Bag6 deficiency also leads to ablation of the testis-specific HSP70 member, HSP70-2/HSPA2 in male germ cells, although in this case its regulation appears to occur at a post-transcriptional level via ubiquitin-mediated degradation of HSP70-2 [5]. Thus, the interaction of Bag6 and HSP70 appears to be critical for stabilizing HSP70, at least during spermatogenesis. The above mentioned evidence establishes a functional link between Bag6 and HSP70/HSC70. However, the precise cellular function(s) executed by the interplays between these proteins is unknown.
Bag6 is a cell death regulator
The first cellular function attributed to Bag6 was as a regulator of apoptosis. Biochemical studies by Thress et al. [15, 16] showed that Bag6 binds the C-terminal 50 amino acids of Reaper, an apoptotic inducer in Drosophila. Bag6 also interacts with two other Drosophila apoptotic modulators, namely Grim and Hid, as well as with a mammalian apoptotic factor named Immediate Early gene X-1 (IEX-1) [17]. Using an in vitro cytochrome c releasing assay, Thress et al. [15] demonstrated that Reaper-induced cytochrome c release, the hallmark of mitochondria-mediated apoptosis, is dependent on Bag6. By contrast, depletion of Bag6 has no effect on Caspase-induced apoptosis, suggesting that it is a specific regulator for Reaperinduced apoptosis. Thress and colleagues named this Reaperinteracting protein Scythe. Subsequent studies revealed that a Bunyaviral nonstructural protein termed NSs bears similar apoptotic activities to Reaper [18]. Strikingly, both these factors interact with Bag6, which seems to cause the release of an apoptosis inducer normally sequestered by Bag6. These results suggest that Bag6 may be an anti-apoptotic regulator [16]. In addition to sequestration of pro-apoptotic factors, Bag6 may also protect cells from apoptosis by promoting the degradation of certain apoptotic factors [18, 19]. Importantly, genetic ablation of Bag6 in mice causes embryonic lethality as a result of developmental defects associated with increased apoptosis and aberrant cell proliferation, confirming the involvement of Bag6 in developmentally regulated apoptotic events in vivo [20].
Intriguingly, although Bag6 deficiency elevates the levels of apoptosis in numerous murine tissues, when compared to wild type cells, Bag6 deficient cortical neurons are actually more resistant to thapsigargin and menadione-induced apoptosis, chemicals that induce ER stress by affecting the calcium flux [20, 21]. Moreover, Bag6 itself contains a Caspase-3 cleavage site near the C-terminal BAG domain and Caspase-3-mediated cleavage may be critical for cell death induced by certain pathogenic bacteria [22, 23]. Thus, despite being an anti-apoptotic regulator that inhibits programmed cell death under normal conditions, Bag6 can also contribute to apoptosis when the cell death program has been initiated in cells under some stress conditions. The complex roles of Bag6 in apoptosis regulation may stem from the fact that as a special chaperone, Bag6 is capable of rendering diverse functional consequences to a large number of client proteins (see below).
Bag6 functions as a TMD specific chaperone in tail-anchored protein biogenesis
Despite extensive studies on Bag6 in the context of apoptosis, its precise cellular function(s) has remained elusive until recently when a series of biochemical studies on the biogenesis of a special class of ER-localized membrane proteins named tail-anchored (TA) proteins rediscovered Bag6.
TA proteins are inserted post-translationally into the ER membrane by a single C-terminal-localized transmembrane domain (TMD) in a manner that is independent of the Sec61 translocon. While transiting through the cytosol, the hydrophobic TMDs require constant chaperoning in order to prevent aggregation or inappropriate interactions. One chaperone that fulfils such function is TRC40 in mammals or Get3p in yeast [24–27]. Structural and biochemical studies showed that TRC40/Get3p forms a homo-dimeric complex, cycling between a nucleotide free “open” and an ATP-bound “close” state. This conformational change seems to occur while TRC40/Get3p is shuttling on-and-off the ER membrane [28–33]. In the cytosol, TRC40/Get3p transiently adopts the closed ATP bound state, allowing the capture of substrates released from the ribosome. Subsequently, the interaction of the TRC40/Get3p-substrate complex with its membrane receptors results in an ATPase-dependent release of the TA proteins from TRC40/Get3p for membrane insertion (Fig. 2) [25, 32–35]. However, numerous lines of evidence suggest that TRC40/Get3p alone cannot efficiently capture TA proteins released from the ribosome in a timely manner. Instead, it requires a cofactor(s) that helps upload the substrates. Using chemical crosslinking in an elegant in vitro TA protein targeting assay, Hegde and colleagues identified a three-protein complex that acts upstream of TRC40 to facilitate TA protein biogenesis [36]. The central component of this complex is Bag6, a TMD selective chaperone that was also independently identified by Leznicki et al. [37]. Bag6 appears to be recruited to the ribosome in a TA protein dependent manner, and the interaction of the Bag6 complex with TMDs in ribosome-associated nascent TA proteins effectively shields them from the aqueous cytosolic environment. Bag6 then communicates with TRC40 through two cofactors, Ubl4A and TRC35, leading to the transfer of the TA proteins to TRC40 for membrane targeting (Fig. 2) [36]. In addition to Bag6, another chaperone capable of recognizing TMDs to prevent aggregation in mammalian cells is SGTA. Like TRC40, SGTA also interacts with Bag6 through Ubl4A [38], and the interplays between these chaperones might have important roles in membrane targeting of certain classes of TA proteins in mammals [39]. The budding yeast does not have a Bag6 homolog, but it contains a SGTA homologue named Sgt2p, which serves as the sole TMD selective chaperone for capturing TA proteins emerging from the ribosome. Interestingly, despite sharing no sequence similarity, both Bag6 and Sgt2p can transiently interact with the ribosome, bind TMDs located at the C-terminus of the TA proteins, and transfer them to TRC40 (in mammals) or Get3p (in yeast) through two homologous cofactors (Ubl4A and TRC35 in mammals or Get5p and Get4p in yeast) (Fig. 2) [32, 36, 40]. These studies established a conserved chaperone relay that channels nascent TA proteins to facilitate their membrane targeting with Bag6 being a central component.
Figure 2.
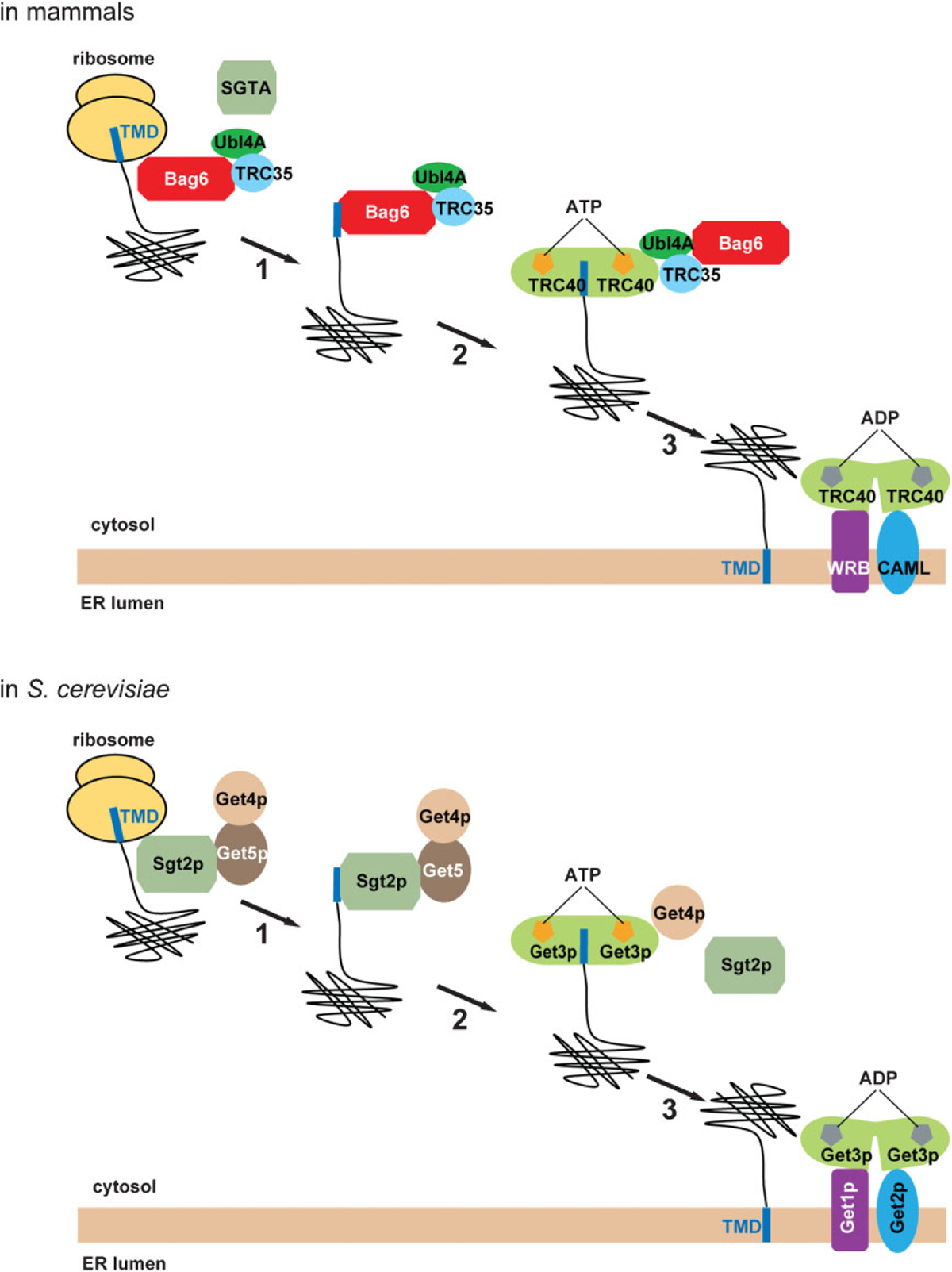
Roles of TMD selective chaperones in TA protein biogenesis. In mammals, Bag6 serves as a TMD selective chaperone that targets TA proteins to the ER membrane (upper panel). In step 1, Bag6 recognizes hydrophobic transmembrane domain (TMD) in a nascent TA protein exiting the ribosome translation tunnel. In step 2, the TA protein is transferred to the ATP-bound TRC40 dimer. This reaction requires the Bag6 cofactor TRC35 and Ubl4A, which serves as a major link between Bag6 and TRC40. In step 3, the TRC40-TA complex is recruited to the membrane by interacting with a membrane receptor complex consisting of WRB and CAML, which also promotes TA protein insertion into the membrane. In yeast, the process occurs in an analogous manner with the exception that Sgt2p serves as the initial TMD selective chaperone (lower panel) while Get1p and Get2p act as membrane receptors.
Bag6 is a master regulator of protein quality control
Degradation of defective cytosolic proteins requires Bag6
Although the first well-characterized biological function of Bag6 is to chaperone nascent TA proteins, several earlier pieces of evidence had hinted that Bag6 might also play pivotal roles in proteasome-dependent protein turnover. Firstly, sequence analysis identified a UBL domain present at the N-terminus of Bag6, a domain often implicated in protein turnover by the ubiquitin proteasome system [2]. Secondly, a high throughput yeast two hybrid study reported several Bag6 interactors, all of which are connected one way or the other to the ubiquitin proteasome system [41]. These include SGT1, a HSP70-associated factor that is also part of the Skp, Cullin, F-box containing (SCF) ubiquitin ligase complex [42, 43], a putative proteasome adaptor named UBL7 that contains both an ubiquitin-associated (UBA) and a UBL domain, and the really interesting gene (RING) finger ubiquitin ligase RNF126. Biochemical studies showed that Bag6 also associates with polyubiquitinated proteins, although it is unclear whether or not this interaction is direct [44]. Finally, it was found that Bag6 can also interact with a specific isoform of Rpn10 (Xrpn10c) in Xenopus [45]. Rpn10 contains an ubiquitin interacting motif (UIM) and is an integral component of the 26S proteasome with a proposed function in substrate recognition. The interaction of Bag6 with Xrpn10c is independent of the UIM domain and involves two distinct regions on Bag6, suggesting the existence of multivalent interactions between Bag6 and the proteasome. This interaction may also engage additional proteasomal subunits because mammals do not have this Rpn10 isoform, yet an interaction between Bag6 and the proteasome has been demonstrated by co-immunoprecipitation using human cells [44].
Minami et al. [44] first established a role for Bag6 in proteasomal degradation while investigating cellular factors associated with a model proteasome substrate (enhanced green fluorescent protein (EGFP)-CL1; Fig. 3A). The study identified Bag6 together with various other chaperones as the factors that interact with this proteasome-bound substrate carrying a C-terminally localized hydrophobic degradation signal (ACKNWFSSLSHFVIHL). shRNA-mediated depletion of Bag6 inhibits the turnover of EGFP-CL1, suggesting that Bag6 is functionally required for the degradation of this model substrate. In addition to EGFP-CL1, Bag6 also associates with a large number of defective nascent polypeptides synthesized by the ribosome (also named defective ribosome products or DRiPs) in order to facilitate their turnover, a process that is thought to generate the MHC class I-presenting antigenic peptides [46]. Accordingly, Bag6 depletion results in reduced cell surface presentation of antigen-loaded MHC class I molecules [44].
Figure 3.

Bag6 regulates protein turnover. A: Bag6 regulates the degradation of folding defective nascent polypeptides. Newly synthesized defective polypeptides are detected by Bag6 and ubiquitinated by a yet-to-be identified E3 ubiquitin ligase that interacts with Bag6. Bag6 may escort the substrates to the proteasome for degradation. B: Bag6 assists mislocalized proteins in reaching the proteasome. TA proteins bound by Bag6 can have different fates. Some may be transferred to the downstream ATPase TRC40 for membrane targeting, whereas others are ubiquitinated by Bag6-associated ubiquitin ligases for degradation by the proteasome. Bag6 may function as a timer, analogous to the one in the ER lumen that dictates the degradation of misfolded glycoproteins. For those TA proteins that fail to be efficiently targeted to the membrane, Bag6 would triage them to the degradation pathway. C: Bag6 escorts ERAD substrates to the proteasome in order to enhance degradation efficiency. The recruitment of the Bag6 system to the gp78- and p97-containing retro-translocation complex seems to mark Bag6 as the preferred chaperone that channels dislocated ERAD substrates to the proteasome. Bag6 may interact with the proteasome either directly or via an unknown adaptor indicated by the question mark.
Bag6 promotes ubiquitination and turnover of mislocalized proteins
Another quality control process that involves the Bag6 complex is the degradation of mislocalized membrane proteins (Fig. 3B). It is thought that membrane protein integration through either the canonical Sec61 pathway or the TRC40 system may not always proceed with 100% accuracy. Instead, certain membrane proteins may be integrated less efficiently than others, resulting in the accumulation of mislocalized proteins in the cytosol. This may be a particularly problematic issue when cells undergo ER stress, a condition that reduces the efficiency of membrane protein integration [47]. Mislocalized proteins often contain surface exposed hydrophobic TMD domains that are prone to aggregation. In an attempt to study how cells deal with such mislocalized polypeptides, Hegde and colleagues discovered that many mislocalized proteins are subject to ubiquitination by a cytosolic ubiquitin ligase in a manner dependent on the Bag6 complex. It appears that after the capture of newly synthesized TA proteins released from the ribosome by Bag6, a “sorting” decision is made, which ends up with either transferring the substrates to the TRC40 complex for membrane insertion or ubiquitination of the substrates by an unknown Bag6-associated ubiquitin ligase for proteasomal degradation [48]. The nature of this sorting step requires further investigation.
ER-associated Bag6 channels dislocated ERAD substrates to the proteasome
Finally, a connection between Bag6 and an ER quality control process named ER-associated protein degradation (ERAD) was recently established by our laboratory, while we investigated the function of the ER-associated ubiquitin ligase gp78 (Fig. 3C). ERAD is an evolutionarily conserved quality control system in eukaryotes that utilizes a sophisticated chaperoning network in the ER lumen to identify misfolded or unassembled proteins and subsequently target them for retrotranslocation into the cytosol and for elimination by the cytosolic proteasome [49, 50]. Upon emerging into the cytosol, these retrotranslocation products are subject to polyubiquitination by a few membrane-associated ubiquitin ligases, which include gp78 [51, 52]. Subsequently, ubiquitinated substrates are extracted from the membrane by a multiprotein complex consisting of the ATPase associated with diverse cellular activities (AAA), ATPase p97 and two cofactors Ufd1 and Npl4 [53–57]. The dislocated ERAD substrates are believed to be shuttled to the proteasome, either directly by p97 or with the assistance from some adaptor proteins, although the underlying mechanism is still obscure [58]. Using affinity chromatography, the Bag6-Ubl4A-TRC35 complex was identified as a new cytosolic component that is associated with the gp78- and p97-containing retrotranslocation complex [7]. Bag6 was also found to interact specifically with deglycosylated ERAD intermediates that have apparently undergone retrotranslocation [7, 59, 60], suggesting that it plays a role in ERAD downstream of p97-mediated retrotranslocation. Consistent with this notion, depletion of each component of the Bag6 complex stabilizes ERAD substrates, including both membrane and soluble proteins, and a fraction of the stabilized substrates still carry polyubiquitin conjugates that are generated during retrotranslocation. Importantly, a large amount of substrates accumulated in Bag6-depleted cells form protein aggregates that cannot be extracted by a non-ionic detergent, providing a plausible explanation on their resistance to proteasome-mediated turnover [7]. These observations suggest that the Bag6 chaperoning activity is required to maintain the solubility of dislocated ERAD substrates and facilitate their delivery to the proteasome for degradation.
A recent study also implicated the Bag6 partner SGTA in ERAD. SGTA contains a tetratricopeptide (TPR) domain that binds HSP70, although the biological significance of this interaction is unclear. SGTA also contains an N-terminal dimerization domain [61]. Upon dimerization, this domain forms an unusual UBL-binding motif, which interacts preferentially with the Ubl4A UBL, and only weakly with the Bag6 UBL, although the two UBL domains share significant sequence homology [38, 61]. On the other hand, the Bag6 UBL, but not Ubl4A UBL can bind specifically to the CUE (coupling of ubiquitin conjugation to ER degradation) domain in gp78, a motif previously implicated in ubiquitin recognition [62]. These observations suggest that the two UBLs in the Bag6 complex serve as distinct “joint bolts” that connect the chaperone complex to different ERAD components. SGTA appears to be an essential cofactor of Bag6 in ERAD as SGTA depletion also leads to accumulation of ERAD substrates in detergent insoluble aggregates. In addition, SGTA deficiency leads to reduced substrate binding by Bag6 [38]. These observations suggest that SGTA may serve as a co-chaperone that assists Bag6 in capturing substrates in ERAD. Interestingly, a recent report showed that purified SGTA can antagonize Bag6-mediated ubiquitination of mislocalized TA proteins in vitro, and also in cells overexpressing SGTA the expression of certain membrane proteins is increased [63]. If this phenotype is caused by inhibition of protein turnover as proposed, then it indicates that the functional interplays between SGTA and Bag6 can be variable in different cellular contexts. In TA protein biogenesis, some substrates might be constantly partitioned between SGTA and Bag6 in a dynamic and reversible way. The flow of substrates towards SGTA may favor membrane insertion whereas the flow of substrates in the opposing direction targets them for degradation. In ERAD, the substrates may be delivered in a unidirectional way from SGTA to Bag6 or another proteasome-interacting factor, which restricts the chaperone system to only function in favor of protein turnover. Sgt2p in Saccharomyces cerevisiae may also have a role in protein quality control given the observation that Sgt2p can interact with a yeast prion protein to modulate its propagation [64].
Is the Bag6 complex a direct mediator of protein degradation?
Since both the Bag6 complex and SGTA have been implicated in biogenesis of TA proteins, which have broad cellular functions including protein turnover, the question of whether the proteolysis defect associated with depletion of the Bag6 system is an indirect effect of abnormality in TA biogenesis should be addressed. Several pieces of evidence argue that Bag6 can serve a direct function in ERAD as well as other protein quality control processes. Firstly, as pointed out above, the Bag6 system is intimately connected with the ubiquitin proteasome system as demonstrated by its interaction with both ubiquitin ligases and the proteasome. Secondly, for proteasomal substrates that require Bag6 for turnover, an interaction between Bag6 and the substrates can often be established, suggesting a direct chaperone-client relationship [7, 44, 59, 60]. Most importantly, the involvement of the Bag6 system in proteasomal degradation has been established using several in vitro protein degradation assays. For instance, Minami et al. used an engineered luciferase transcript lacking stop codon to generate a model defective ribosome product. They found that in vitro ubiquitination and degradation of this substrate can be inhibited by an anti-Bag6 antibody [44]. Likewise, immunodepletion of Bag6 from the cytosol blocks in vitro ubiquitination of mislocalized membrane proteins [48]. Finally, in an in vitro retrotranslocation assay, depletion of the Bag6 complex results in reduced turnover of an ERAD substrate [7]. These in vitro studies showed that Bag6 acts upstream of ubiquitin ligases and is often required for substrate ubiquitination in the cytosolic quality control pathways, while in ERAD, Bag6 only captures substrates after they have been ubiquitinated. This is not entirely surprising given that ubiquitination is known to be required for retrotranslocation of misfolded ER proteins across the membrane and that retrotranslocation is an apparent prerequisite for Bag6 binding, at least for soluble ERAD substrates. Mechanistically, ubiquitin ligase-associated Bag6 may use different mechanisms to assemble ubiquitin chains in the ER and cytosol quality control pathways. The cytosolic E3s may use Bag6 as a “holdase” (see below) that prevents premature release of substrates prior to the completion of the polyubiquitination reaction, whereas the ERAD ubiquitin ligase gp78 is capable of transferring preassembled ubiquitin chains from its cognate E2 Ube2g2 to a substrate, thus alleviating the necessity of engaging a chaperone in ubiquitination [65, 66]. Collectively, these studies establish the Bag6 system as multifaceted cytosolic chaperone machinery, capable of acting at the interface of protein biogenesis and degradation.
The nuclear functions of Bag6
The presence of a NLS on Bag6 suggests that it must have a function(s) in the nucleus. Although the precise function of Bag6 in the nucleus is not entirely clear, a few recent studies have implicated it in gene transcription regulation/DNA damage response, possibly via modulating posttranslational modification of histones or other nuclear factors. The first nuclear function attributed to Bag6 was that of a regulator of the tumor suppressor p53 acetylation, which is essential for transactivation of some p53 target genes in response to genotoxic stress. Bag6 seems to enhance p53 acetylation by stabilizing an interaction between p53 and the acetyltransferase p300, which likely involves a direct interaction between Bag6 and p300 [67, 68]. More recently, Bag6 was shown to interact with two histone methyltransferases, SET1A, and DOT1L, which regulate histone H3K4 and H3K79 dimethylation, respectively [69, 70]. H3K4 dimethylation, associated with the recruitment of the Bag6-SET1A complex to DNA promoters, regulates the expression of certain pro-carcinogenic genes such as Myc and BRCA1 [69], whereas H3K79 dimethylation was proposed to mediate the recruitment of 53BP1 to the site of ionizing radiation-induced DNA damages, an event critical for DNA damage signaling and repair [70]. The nuclear function of Bag6 clearly requires its presence in the nucleus, thus excluding the possibility that TRC35 may also participate in these events. However, Ubl4A often accompanies Bag6 to the nucleus. Its potential involvement in Bag6-dependent DNA damage/transcriptional regulations awaits further elucidation.
A “holdase” activity rules all
How can Bag6 and its associated cofactors participate in so many different biological processes? Biochemical studies on both TA protein biogenesis and protein quality control have provided some important clues. Despite its association with HSP70, Bag6 itself contains a critical chaperoning activity distinct from that of HSP70. Initial studies show that Bag6 can directly interact with nascent TA proteins in a TMD dependent manner, suggesting that it may prefer binding polypeptides with exposed long hydrophobic segments [36]. This activity was similarly demonstrated for Sgt2p, the yeast homologue of human SGTA [40]. The precise mode of substrate recognition by Bag6 awaits structural characterization, but it is tempting to speculate that the interaction may to some extent resemble how TRC40/Get3p binds TA proteins. Structural studies show that Get3p cycles between an ATP bound closed conformation that contains a long hydrophobic groove, accommodating the TMDs in TA proteins, and a nucleotide free open conformation that likely represents a substrate free state [28, 29, 31, 71]. Bag6 may involve a conceptually similar conformational cycle coupled to substrate binding and release, but in this case, the trigger of substrate association and release must be controlled by Bag6-binding co-factors rather than the nucleotide. The binding of Bag6 to nascent TA proteins provides a sufficient time window that allows a sorting decision regarding the fate of the bound TA proteins to be made [39].
The chaperoning activity of Bag6 appears applicable to substrates without TMDs because in protein quality control processes, Bag6 can regulate the turnover of both soluble and membrane substrates [7, 44]. In addition, purified Bag6 is capable of binding heat-denatured luciferase that does not carry any TMDs. Intriguingly, unlike conventional chaperones such as HSP70, the binding of Bag6 to heat-denatured luciferase does not promote its refolding. Instead, Bag6 seems to hold its substrates tightly in an unfolded yet soluble state, a condition that favors their turnover by the proteasome [7]. The strong propensity of Bag6 to bind unfolded polypeptides raises the question of how Bag6-bound substrates are released.
The nuclear functions of Bag6 are likely to be mechanistically linked to this “holdase” activity. Bag6 may capture certain aggregation prone proteins in the nucleus to prevent promiscuous interactions/aggregation until these proteins meet their functional partners or have been delivered to the correct subcellular compartments. In support of this view, it has been demonstrated that under certain conditions, Bag6 can shuttle polypeptides from the cytosol into the nucleus, resulting in either the activation or inactivation of the bound polypeptides [37, 72–74].
Collectively, it seems that we can view Bag6 as a special transporter that uses its “holdase” activity to carry aggregation prone substrates, ensuring their safe transit through the aqueous cellular environment and proper integration into various functional systems. For those clients that fail to be properly targeted, Bag6 may use its connections with the ubiquitin proteasome system to turn them over. The final destinations of the Bag6-bound substrates may be determined by both the cofactors associated with Bag6 as well as by the precise interplays between substrates and the Bag6 system.
Bag6/Bat3/Scythe and human diseases
The diverse functions of the Bag6 system suggest that deficiency in Bag6 may be the cause of certain human diseases or developmental defects. Consistent with this view, a genetic study showed that Bag6 knockout mice die early during embryonic development [20]. In addition, human genetic studies have identified Bag6 as a potential candidate gene of which mutations may cause some diseases. For instance, haplotypes including a genetic variation in the Bag6 gene have been associated with the Kawasaki Syndrome, an autoimmune disease in which inflammation occurs in the medium-sized blood vessels throughout the body [75], as well as with rheumatoid arthritis [76]. Bi-allelic inactivating mutations were also found in the Bag6 gene in several colon cancer cell lines that showed microsatellite instability [77]. The association of Bag6 polymorphisms with rheumatoid arthritis suggests a potential function for Bag6 in immune regulation, particularly in inflammation control. This is consistent with the fact that Bag6 is genetically clustered with several other inflammation regulators such as TNF at the MHC class III region [78]. Bag6 likely influences the immune system by regulating a variety of substrate proteins essential to immune regulations. This is exemplified by the recently established association of Bag6 with Tim3, an inhibitory type I membrane receptor expressed on exhausted T cells during infection with human immunodeficiency virus (HIV)-1 and hepatitis C virus, which represses Tim3 function and thus protects helper type 1 (T(H)1) cells from galectin-9-mediated cell death [79]. Similar functional interplays between Bag6 and client proteins essential for neuronal functions have also been reported, which may play crucial roles in protecting neurons from apoptosis elicited under various disease conditions such as the Parkinson’s disease [80].
Finally, it is worth mentioning that the intracellular bacterial pathogen Legionella pneumophila can inject several F-box proteins into the host cells during infection. Several of these F-box proteins can join host factors to form multisubunit E3 ubiquitin ligase complexes that interact with Bag6. The precise function of these interactions in L. pneumophila infection is unclear, but it is likely that the Bag6 system is leveraged by the bacteria to ubiquitinate and/or shuttle certain host factors, which may promote the bacterial infection cycle [81].
Conclusions and perspectives
Recent studies have revealed a novel and important cellular chaperoning system with what appears to be an enormous functional capacity. A unique feature of the Bag6 system is that it interacts with both the ribosome and the proteasome, and therefore can function at the interface of protein biogenesis and turnover. The system uses a special “holdase” activity to “sort” client proteins and chaperone them to distinct destinations. Therefore, its functions may stem far beyond those described here. Thus, systematic identification of physiological substrates of Bag6 will certainly reveal its functional scope.
Another big mystery is how Bag6 can interact with so many different substrates with hydrophobic segments of different length and hydrophobicity. The answer to this question requires careful mapping of the substrate binding domain followed by structural characterization of the Bag6-substrate interactions. In addition, since Bag6 binds to its substrate with high affinity, it is unclear how the bound substrates are released after they reach the final destination. How Bag6 cooperates with other cofactors such as SGTA and whether HSP70 functions in various Bag6-mediated processes are also intriguing questions that await further studies. Whether or not the Bag6 system may be subject to regulation in the cell is also unknown. Finally, more genetic studies in animal models particularly with conditional deletion of Bag6 or its associated cofactors will contribute to unveil the physiological importance of this chaperoning system.
Acknowledgments
The authors’ research is supported by both the Intramural Research Program of the NIDDK and a grant from the NIH Intramural AIDS Targeted Antiviral Program (IATAP) to Y.Y.
Abbreviations:
- AAA
ATPase associated with diverse cellular activities
- Bat3
HLA (human leukocyte antigen)-B-associated transcript 3
- CUE
coupling of ubiquitin conjugation to ER degradation
- DRiPs
defective ribosome products
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- HIV
human immunodeficiency virus
- MEF
mouse embryonic fibroblast
- MHC
major histocompatibility complex
- NLS
nuclear localization signal
- RING
really interesting gene
- SCF
Skp, Cullin, F-box containing complex
- TA
tail-anchored
- TMD
transmembrane domain
- UBA
ubiquitin-associated
- UBL
ubiquitin-like
- UIM
ubiquitin interacting motif
Footnotes
The authors have declared no conflict of interest.
References
- 1.Gasser DL, Sternberg NL, Pierce JC, Goldner-Sauve A, et al. 1994. P1 and cosmid clones define the organization of 280 kb of the mouse H-2 complex containing the Cps-1 and Hsp70 loci. Immunogenetics 39: 48–55. [DOI] [PubMed] [Google Scholar]
- 2.Banerji J, Sands J, Strominger JL, Spies T. 1990. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc Natl Acad Sci USA 87: 2374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang R, Liew CC. 1994. The human BAT3 ortholog in rodents is predominantly and developmentally expressed in testis. Mol Cell Biochem 136: 49–57. [DOI] [PubMed] [Google Scholar]
- 4.Ozaki T, Hanaoka E, Naka M, Nakagawara A, et al. 1999. Cloning and characterization of rat BAT3 cDNA. DNA Cell Biol 18: 503–12. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki T, Marcon E, McQuire T, Arai Y, et al. 2008. Bat3 deficiency accelerates the degradation of Hsp70–2/HspA2 during spermatogenesis. J Cell Biol 182: 449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manchen ST, Hubberstey AV. 2001. Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem Biophys Res Commun 287: 1075–82. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Liu Y, Soetandyo N, Baek K, et al. 2011. A ubiquitin ligaseassociated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell 42: 758–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamper N, Kessler J, Temme S, Wegscheid C, et al. 2012. A novel BAT3 sequence generated by alternative RNA splicing of exon 11B displays cell type-specific expression and impacts on subcellular localization. PLoS ONE 7: e35972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukahara T, Kimura S, Ichimiya S, Torigoe T, et al. 2009. Scythe/BAT3 regulates apoptotic cell death induced by papillomavirus binding factor in human osteosarcoma. Cancer Sci 100: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doong H, Vrailas A, Kohn EC. 2002. What’s in the ‘BAG’? – a functional domain analysis of the BAG-family proteins. Cancer Lett 188: 25–32. [DOI] [PubMed] [Google Scholar]
- 11.Kaye FJ, Modi S, Ivanovska I, Koonin EV, et al. 2000. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett 467: 348–55. [DOI] [PubMed] [Google Scholar]
- 12.Thress K, Song J, Morimoto RI, Kornbluth S. 2001. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J 20: 1033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winnefeld M, Grewenig A, Schnolzer M, Spring H, et al. 2006. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res 312: 2500–14. [DOI] [PubMed] [Google Scholar]
- 14.Corduan A, Lecomte S, Martin C, Michel D, et al. 2009. Sequential interplay between BAG6 and HSP70 upon heat shock. Cell Mol Life Sci 66: 1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thress K, Henzel W, Shillinglaw W, Kornbluth S. 1998. Scythe: a novel reaper-binding apoptotic regulator. EMBO J 17: 6135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thress K, Evans EK, Kornbluth S. 1999. Reaper-induced dissociation of a Scythe sequestered cytochrome c-releasing activity. EMBO J 18: 5486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R, Lutz W, Frank E, Im HJ. 2004. Immediate early gene X-1 interacts with proteins that modulate apoptosis. Biochem Biophys Res Commun 323: 1293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colon-Ramos DA, Irusta PM, Gan EC, Olson MR, et al. 2003. Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol Biol Cell 14: 4162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minami R, Shimada M, Yokosawa H, Kawahara H. 2007. Scythe regulates apoptosis through modulating ubiquitin-mediated proteolysis of the Xenopus elongation factor XEF1AO. Biochem J 405: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desmots F, Russell HR, Lee Y, Boyd K, et al. 2005. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol Cell Biol 25: 10329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desmots F, Russell HR, Michel D, McKinnon PJ. 2008. Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J Biol Chem 283: 3264–71. [DOI] [PubMed] [Google Scholar]
- 22.Wu YH, Shih SF, Lin JY. 2004. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J Biol Chem 279: 19264–75. [DOI] [PubMed] [Google Scholar]
- 23.Grover A, Izzo AA. 2012. BAT3 regulates Mycobacterium tuberculosis protein ESAT-6-mediated apoptosis of macrophages. PLoS ONE 7: e40836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefanovic S, Hegde RS. 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128: 1147–59. [DOI] [PubMed] [Google Scholar]
- 25.Schuldiner M, Metz J, Schmid V, Denic V, et al. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134: 634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favaloro V, Vilardi F, Schlecht R, Mayer MP, et al. 2010. Asna1/TRC40 mediated membrane insertion of tail-anchored proteins. J Cell Sci 123: 1522–30. [DOI] [PubMed] [Google Scholar]
- 27.Leznicki P, Warwicker J, High S. 2011. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem J 436: 719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateja A, Szlachcic A, Downing ME, Dobosz M, et al. 2009. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461: 361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suloway CJ, Chartron JW, Zaslaver M, Clemons WM Jr. 2009. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA 106: 14849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagata A, Mimura H, Sato Y, Yamashita M, et al. 2010. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells 15: 29–41. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Li J, Qian X, Denic V, et al. 2009. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE 4: e8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Whynot A, Tung M, Denic V. 2011. The mechanism of tailanchored protein insertion into the ER membrane. Mol Cell 43: 738–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariappan M, Mateja A, Dobosz M, Bove E, et al. 2011. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477: 61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefer S, Reitz S, Wang F, Wild K, et al. 2011. Structural basis for tailanchored membrane protein biogenesis by the Get3-receptor complex. Science 333: 758–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Sakisaka T. 2012. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell 48: 387–97. [DOI] [PubMed] [Google Scholar]
- 36.Mariappan M, Li X, Stefanovic S, Sharma A, et al. 2010. A ribosomeassociating factor chaperones tail-anchored membrane proteins. Nature 466: 1120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leznicki P, Clancy A, Schwappach B, High S. 2010. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci 123: 2170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Cai M, Yang Y, Huang L, et al. 2012. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep 2: 1633–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde RS, Keenan RJ. 2012. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol 12: 787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Brown EC, Mak G, Zhuang J, et al. 2010. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehner B, Semple JI, Brown SE, Counsell D, et al. 2004. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83: 153–67. [DOI] [PubMed] [Google Scholar]
- 42.Tobaben S, Thakur P, Fernandez-Chacon R, Sudhof TC, et al. 2001. A trimeric protein complex functions as a synaptic chaperone machine. Neuron 31: 987–99. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa K, Skowyra D, Elledge SJ, Harper JW, et al. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33. [DOI] [PubMed] [Google Scholar]
- 44.Minami R, Hayakawa A, Kagawa H, Yanagi Y, et al. 2010. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol 190: 637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikukawa Y, Minami R, Shimada M, Kobayashi M, et al. 2005. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J 272: 6373–86. [DOI] [PubMed] [Google Scholar]
- 46.Schubert U, Anton LC, Gibbs J, Norbury CC, et al. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404: 770–4. [DOI] [PubMed] [Google Scholar]
- 47.Kang SW, Rane NS, Kim SJ, Garrison JL, et al. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127: 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hessa T, Sharma A, Mariappan M, Eshleman HD, et al. 2011. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475: 394–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum associated degradation. Nat Rev Mol Cell Biol 9: 944–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai B, Ye Y, Rapoport TA. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol 3: 246–55. [DOI] [PubMed] [Google Scholar]
- 51.Mehnert M, Sommer T, Jarosch E. 2011. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. BioEssays 32: 905–13. [DOI] [PubMed] [Google Scholar]
- 52.Fang S, Ferrone M, Yang C, Jensen JP, et al. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA 98: 14422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye Y, Meyer HH, Rapoport TA. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye Y, Meyer HH, Rapoport TA. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–6. [DOI] [PubMed] [Google Scholar]
- 55.Jarosch E, Taxis C, Volkwein C, Bordallo J, et al. 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4: 134–9. [DOI] [PubMed] [Google Scholar]
- 56.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, et al. 2001. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabinovich E, Kerem A, Frohlich KU, Diamant N, et al. 2002. AAAATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22: 626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Ye Y. 2011. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Res 21: 867–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ernst R, Claessen JH, Mueller B, Sanyal S, et al. 2011. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol 8: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Claessen JH, Ploegh HL. 2012. BAT3 guides misfolded glycoproteins out of the endoplasmic reticulum. PLoS ONE 6: e28542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chartron JW, VanderVelde DG, Clemons WM Jr. 2012. Structures of the Sgt2/SGTA dimerization domain with the Get5/Ubl4a UBL domain reveal a novel interaction that forms a conserved dynamic interface. Cell Rep 2: 1620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen B, Mariano J, Tsai YC, Chan AH, et al. 2006. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA 103: 341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leznicki P, High S. 2012. SGTA antagonizes BAG6-mediated protein triage. Proc Natl Acad Sci USA 109: 19214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiktev DA, Patterson JC, Muller S, Bariar B, et al. 2012. Regulation of chaperone effects on a yeast prion by cochaperone sgt2. Mol Cell Biol 32: 4960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Tu D, Li L, Wollert T, et al. 2009. Mechanistic insights into active site associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA 106: 3722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Tu D, Brunger AT, Ye Y. 2007. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446: 333–7. [DOI] [PubMed] [Google Scholar]
- 67.Sasaki T, Gan EC, Wakeham A, Kornbluth S, et al. 2007. HLA-B associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev 21: 848–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, et al. 2010. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest 120: 4569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen P, Bar-Sela G, Sun L, Bisht KS, et al. 2008. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol 28: 6720–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wakeman TP, Wang Q, Feng J, Wang XF. 2012. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cellcycle phases. EMBO J 31: 2169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, et al. 2009. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA 106: 21131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yong ST, Wang XF. 2012. A novel, non-apoptotic role for Scythe/BA T3: a functional switch between the proand anti-proliferative roles of p21 during the cell cycle. PLoS ONE 7: e38085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaczmarek K, Studencka M, Meinhardt A, Wieczerzak K, et al. 2011. Overexpression of peroxisomal testis-specific 1 protein induces germ cell apoptosis and leads to infertility in male mice. Mol Biol Cell 22: 1766–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamper N, Franken S, Temme S, Koch S, et al. 2012. Gamma-interferon-regulated chaperone governs human lymphocyte antigen class II expression. FASEB J 26: 104–16. [DOI] [PubMed] [Google Scholar]
- 75.Hsieh YY, Lin YJ, Chang CC, Chen DY, et al. 2010. Human lymphocyte antigen B-associated transcript 2, 3 and 5 polymorphisms and haplotypes are associated with susceptibility of Kawasaki disease and coronary artery aneurysm. J Clin Lab Anal 24: 262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harney SM, Vilarino-Guell C, Adamopoulos IE, Sims AM, et al. 2008. Fine mapping of the MHC Class III region demonstrates association of AIF1 and rheumatoid arthritis. Rheumatology (Oxford) 47: 1761–7. [DOI] [PubMed] [Google Scholar]
- 77.Ivanov I, Lo KC, Hawthorn L, Cowell JK, et al. 2007. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene 26: 2873–84. [DOI] [PubMed] [Google Scholar]
- 78.Deakin JE, Papenfuss AT, Belov K, Cross JG, et al. 2006. Evolution and comparative analysis of the MHC Class III inflammatory region. BMC Genomics 7: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rangachari M, Zhu C, Sakuishi K, Xiao S, et al. 2012. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat Med 18: 1394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long P, Samnakay P, Jenner P, Rose S. 2012. A yeast two-hybrid screen reveals that osteopontin associates with MAP1A and MAP1B in addition to other proteins linked to microtubule stability, apoptosis and protein degradation in the human brain. Eur J Neurosci 36: 2733–42. [DOI] [PubMed] [Google Scholar]
- 81.Ensminger AW, Isberg RR. 2010. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun 78: 3905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]


