Abstract
Maltose metabolism was investigated in the hyperthermophilic archaeon Thermococcus litoralis. Maltose was degraded by the concerted action of 4-α-glucanotransferase and maltodextrin phosphorylase (MalP). The first enzyme produced glucose and a series of maltodextrins that could be acted upon by MalP when the chain length of glucose residues was equal or higher than four, to produce glucose-1-phosphate. Phosphoglucomutase activity was also detected in T. litoralis cell extracts. Glucose derived from the action of 4-α-glucanotransferase was subsequently metabolized via an Embden-Meyerhof pathway. The closely related organism Pyrococcus furiosus used a different metabolic strategy in which maltose was cleaved primarily by the action of an α-glucosidase, a p-nitrophenyl-α-d-glucopyranoside (PNPG)-hydrolyzing enzyme, producing glucose from maltose. A PNPG-hydrolyzing activity was also detected in T. litoralis, but maltose was not a substrate for this enzyme. The two key enzymes in the pathway for maltose catabolism in T. litoralis were purified to homogeneity and characterized; they were constitutively synthesized, although phosphorylase expression was twofold induced by maltodextrins or maltose. The gene encoding MalP was obtained by complementation in Escherichia coli and sequenced (calculated molecular mass, 96,622 Da). The enzyme purified from the organism had a specific activity for maltoheptaose, at the temperature for maximal activity (98°C), of 66 U/mg. A Km of 0.46 mM was determined with heptaose as the substrate at 60°C. The deduced amino acid sequence had a high degree of identity with that of the putative enzyme from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 (66%) and with sequences of the enzymes from the hyperthermophilic bacterium Thermotoga maritima (60%) and Mycobacterium tuberculosis (31%) but not with that of the enzyme from E. coli (13%). The consensus binding site for pyridoxal 5′-phosphate is conserved in the T. litoralis enzyme.
Thermococcus litoralis is a hyperthermophilic marine archaeon that grows optimally at 85°C (26). The order Thermococcales includes the species of the genera Thermococcus and Pyrococcus, and as expected, T. litoralis and Pyrococcus furiosus have many features of growth and metabolism in common. The species T. litoralis was initially described as growing on peptides and pyruvate but not on carbohydrates (26). However, later studies showed that maltose could stimulate growth on peptides (25, 47) and that T. litoralis produced as much extracellular amylolytic enzymes as P. furiosus in response to the presence of α-1,4-linked saccharides in the growth medium (6). The extracellular amylolytic enzymes degrade complex carbohydrates to disaccharides such as cellobiose and maltose. Maltose and trehalose are transported into T. litoralis by a recently characterized high-affinity transport system with a Km of about 20 nM (13, 47). The trehalose/maltose binding protein (TMBP) of T. litoralis was purified and characterized, and the gene encoding this protein, malE, was cloned, sequenced, and expressed in Escherichia coli (13). Cloning and sequencing of the T. litoralis malEFG gene cluster revealed a remarkable similarity between the organization of the respective operon and that of E. coli and other bacterial binding protein-dependent ABC transporters (13).
Like P. furiosus, which is the most studied of the hyperthermophilic archaea, T. litoralis utilizes a modified Embden-Meyerhof glycolytic pathway, involving two ADP-dependent kinases: hexokinase and phosphofructokinase (18, 38, 42). Carbohydrates are fermented mainly to acetate, alanine, CO2, and H2; when S0 is available, H2S is produced instead of hydrogen and only traces of alanine are formed.
It has been suggested that maltose metabolism in P. furiosus and T. litoralis is catalyzed by intracellular α-glucosidases that hydrolyze maltose to two glucose molecules (6, 7, 17). In fact, an α-glucosidase, induced by the presence of carbohydrates in the growth medium, has been purified from P. furiosus and characterized (7). A similar enzyme was found to exist in T. litoralis (17).
Following our study of the maltose transport system in T. litoralis (13, 47), we now investigate intracellular maltose metabolism in this organism. Maltose metabolism in the archetypal organism E. coli is well known (4, 40); it proceeds by the combined action of 4-α-glucanotransferase (amylomaltase) and maltodextrin phosphorylase (MalP). The former enzyme cleaves maltodextrins (releasing glucose or a short maltodextrin residue) and transfers the remaining portion onto the nonreducing end of an acceptor which may be glucose or a maltodextrin molecule, keeping the sum of glycosidic linkages constant. The action of 4-α-glucanotransferase on maltose (in the presence of maltodextrin primers) releases glucose and a series of longer maltodextrins that are then used as substrates for MalP, an enzyme catalyzing the phosphorolytic cleavage of maltodextrins with a minimal chain length of five glucose residues, to yield glucose 1-phosphate (40, 41, 46). (Maltose is not regarded as substrate in a strict sense; only in the presence of trace amounts of maltodextrins does it act as an acceptor [33]. However, for practical purposes of maltose degradation, this phenomenon is irrelevant. Even purified enzyme preparations can act on maltose, due to either maltodextrin impurities in maltose or maltodextrins bound to the enzyme [29].)
Here, a pathway for the catabolism of maltose in T. litoralis is proposed, based on the determination of the relevant enzymatic activities; in addition, two key enzymes in the pathway, 4-α-glucanotransferase and MalP, were purified and characterized, and growth conditions leading to their induction were investigated. While this work was in progress, a report on the sequencing, cloning, and expression in E. coli of the gene encoding T. litoralis 4-α-glucanotransferase, was published (15).
MATERIALS AND METHODS
Chemicals.
Peptone, tryptone, yeast extract, and dextran were obtained from Difco Laboratories. Maltose was obtained from Merck, and α-, β-, and γ-cyclodextrins were obtained from Wacker (Munich, Germany). Amylose from potato (type III), p-nitrophenyl-α-d-glucopyranoside (PNPG), and p-nitrophenol (PNP) were from Sigma Chemical Co. Amyloglucosidase was obtained from Roth (Karlsruhe, Germany). Chromatographic materials and molecular weight standards were from Pharmacia except for hydroxyapatite (Bio-Gel), which was purchased from Bio-Rad. Amylose resin was from New England Biolabs. 14C-labeled compounds were purchased from Amersham.
Organisms and growth conditions.
T. litoralis (DSM 5473) and P. furiosus (DSM 3638) were obtained from Deutsche Sammlung von Mikrooganismen und Zellkulturen GmbH (Braunschweig, Germany). T. litoralis was grown as previously described (47). Maltose (3 g/liter) dextran (3 g/liter), or yeast extract (1 g/liter) were used as the carbon source in a medium containing peptone (5 g/liter). Carbohydrate-free media containing peptone (5 g/liter) or tryptone (5 g/liter) as the sole carbon sources were also used for some cultures. P. furiosus was cultured in the medium described by Raven et al. (35) with peptone (2 g/liter) and maltose (3 g/liter) as carbon sources without the addition of sulfur. This organism was grown at 95°C in a 2-liter Braun fermentor with continuous bubbling of nitrogen gas and stirring at 100 rpm. Growth was monitored by measurements of the optical density at 600 nm (OD600). Cells were harvested by centrifugation (5,000 × g for 15 min at 27°C) and washed once with a solution of the same composition as that of the growth medium (pH 6.5 for T. litoralis and pH 6.8 for P. furiosus) but without the carbon sources. After cell harvesting, all manipulations were performed under aerobic conditions.
Purification of T. litoralis 4-α-glucanotransferase. (i) Preparation of cell extracts.
T. litoralis cells were collected during the late exponential phase (OD600 = 0.4), washed as described above, and frozen overnight at −20°C. Cells were broken by thawing and resuspension with equal volume of buffer (100 mM morpholinepropanesulfonic acid [MOPS; pH 7.0] containing 10 mM MgCl2 and 1 mM dithiothreitol). Leupeptin (2 μg per ml), antipain (2 μg per ml), and phenylmethylsulfonyl fluoride (85 μg per ml in methanol) were added as protease inhibitors. The extract was stirred with DNase I (30 μg per ml of extract), and the resulting suspension was centrifuged for 1 h at 100,000 × g; the supernatant typically contained around 20 mg of total protein per ml. The enzyme was purified, in most cases, from cell extracts derived from cultures grown on peptone and yeast extract. Measurements of enzyme activities of maltose metabolism were carried out with cells grown on maltose and peptone. For these studies, cell extracts were passed through a Sephadex G-25 column to remove residual maltose.
(ii) Affinity chromatography.
Two milliliters of freshly prepared cell extract (about 40 mg of protein) was heated for 5 min in an oven at 90°C and then loaded onto an amylose column at room temperature (4-ml bed volume). The column was preequilibrated with 100 mM MOPS (pH 7.0) containing 10 mM MgCl2 and 1 mM dithiothreitol. After the addition of the extract, the column was washed with 5 volumes of the same buffer. The enzyme was eluted with 20 mM maltose or maltotriose in the same buffer and appeared homogeneous on sodium dodecyl sulfate (SDS)-gels (Fig. 2). The pure enzyme was stored at −20°C and was stable for several months. When necessary, the purified protein and the cell extract were concentrated by ultrafiltration using a 10-kDa-cutoff membrane (YM10; Amicon, Lexington, Mass.).
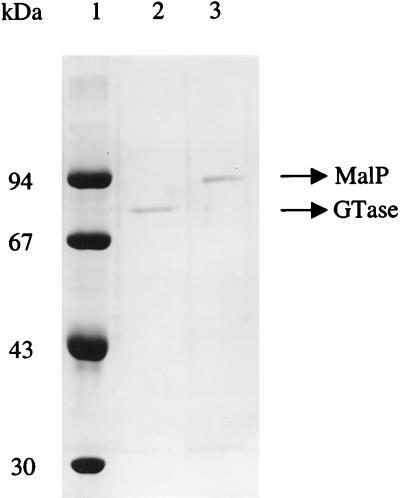
FIG. 2.
SDS-PAGE of purified T. litoralis 4-α-glucanotransferase and maltodextrin phosphorylase. Lane 1, molecular mass standards; lane 2, purified glucanotransferase (Gtase; 79 kDa); lane 3, purified MalP (94 kDa).
Purification of T. litoralis MalP. (i) Cell extract preparation.
T. litoralis cells were cultured in a 100-liter fermentor with maltose and peptone as carbon sources. Cells were harvested during the late exponential phase (OD600 = 0.4) and frozen at −20°C until used. Cells (65 g [wet mass]) were broken by thawing and resuspension in 65 ml of buffer A (20 mM Tris-HCl [pH 7.6]) containing 10 mM MgCl2. The extract was stirred with DNase I (30 μg per ml), the resulting suspension was centrifuged (100,000 × g for 1 h), and the supernatant was dialyzed overnight against buffer A.
(ii) Purification procedure.
All chromatographic steps were performed on a Hiload system (Pharmacia, Uppsala, Sweden) at 15°C, and the purification steps were monitored by activity measurements with the coupled assay described below. The cell extract was applied onto a Fast Flow DEAE column preequilibrated with buffer A. Elution was carried out with 15 bed volumes of a linear salt gradient (0 to 400 mM NaCl in buffer A). Phosphorylase activity eluted between 160 and 230 mM NaCl. These fractions were pooled, concentrated by ultrafiltration using a 30-kDa-cutoff membrane (YM30; Amicon), and applied onto a Superdex 200 column equilibrated with buffer A containing 150 mM NaCl. Fractions with phosphorylase activity were pooled and concentrated by ultrafiltration. The resulting sample was loaded onto a hydroxyapatite (Bio-Gel) column equilibrated with buffer A; after the buffer was changed to 1 mM potassium phosphate (pH 7.6), the adsorbed proteins were eluted with 15 bed volumes of a linear gradient of potassium phosphate, pH 7.6 (1 to 200 mM). Fractions with phosphorylase activity were pooled, and the buffer was exchanged with 10 mM potassium phosphate (pH 7.2) in 200 mM NaCl. The protein solution was then loaded onto a Red Sepharose CL-4B column equilibrated in the same buffer. The adsorbed proteins were eluted with 20 mM potassium phosphate (pH 8.0) containing 0.5 M NaCl. MalP activity was found in the fractions that did not bind to the column. These fractions were pooled, and the buffer was changed to buffer A; the sample was applied onto a Q Sepharose column and eluted with 15 bed volumes of a linear salt gradient (0 to 0.5 M NaCl in buffer A). Fractions with phosphorylase activity eluted between 270 and 300 mM NaCl. These fractions were pooled and after concentration were applied onto a Superdex 200 column equilibrated with buffer A containing 150 mM NaCl. After this step, MalP was judged pure by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 2).
Enzyme assays. (i) Assay for 4-α-glucanotransferase activity.
Routinely, enzyme activity was assessed by the production of maltodextrins and glucose from maltose or maltotriose; the products were identified by thin layer chromatography (TLC) as described below. The specific activity was determined from the amount of glucose released during incubation with maltose or maltotriose in a discontinuous assay. The reaction mixture, containing 100 mM MOPS (pH 7.0) and 10 mM maltose or maltotriose, was incubated at the desired temperature in a heating block. The reaction was initiated by the addition of the enzyme and stopped at different times (to allow determination of initial rates) by adding 400 μl of cold acetone to the reaction mixture (100 μl). The mixture was cooled in ice for at least 2 h, and precipitated material was removed in an Eppendorf centrifuge. Acetone was evaporated at 80°C, and the volume was adjusted to 500 μl with water. Glucose was quantified with a glucose enzymatic kit (Boehringer Mannheim). One unit of enzyme activity is defined as 1 μmol of glucose released per minute. The same assay was used to measure the rate of glucose release in cell extracts.
(ii) Assay for MalP activity.
Routinely, MalP activity was measured in a continuous assay at 60°C with maltoheptaose as the substrate. Glucose-1-phosphate was determined by monitoring the reduction of NADP, using phosphoglucomutase and glucose-6-phosphate dehydrogenase as auxiliary enzymes. The assay mixture contained 50 mM potassium phosphate buffer (pH 7.0), 10 mM MgCl2, 2 mM NADP, 5 mM maltoheptaose, phosphoglucomutase (rabbit muscle; 17 U/ml; Sigma), glucose-6-phosphate dehydrogenase (Torula yeast; 3 U/ml; Sigma), and cell extract or purified enzyme. The auxiliary enzymes were added shortly before initiation of the reaction, and it was confirmed that the enzymes were not rate limiting. To measure MalP activity at temperatures above 60°C, a discontinuous assay similar to that described for 4-α-glucanotransferase activity was used. The reaction mixture, containing 50 mM potassium phosphate (pH 7.0) and 5 mM maltoheptaose, was incubated at the desired temperature; the reaction was initiated by the addition of the extract or the enzyme solution. The final volume of the reaction mixture was 100 μl, and the reaction was stopped at different times by addition of 400 μl of water and cooling in ice. Glucose-1-phosphate was determined at room temperature with the coupled enzymatic assay described above, but in 100 mM triethanolamine buffer (pH 7.6). One unit of enzyme activity is defined as 1.0 μmol of glucose-1-phosphate formed per minute.
(iii) Assay for phosphoglucomutase.
The activity of phosphoglucomutase was measured at 85°C by a discontinuous assay. The reaction mixture contained 50 mM potassium phosphate (pH 7.0), 10 mM MgCl2, 5 μM glucose-1,6-bisphosphate, and 10 mM glucose-1-phosphate. One unit of enzyme activity is defined as 1.0 μmol of glucose-6-phosphate produced per minute.
(iv) Assay for PNPG hydrolysis.
The hydrolysis of PNPG, the typical artificial substrate for α-glucosidase, was measured at 80, 85, and 95°C by the formation of PNP (monitoring the absorbance at 405 nm). The reaction mixture contained 1.0 mM PNPG in 100 mM potassium phosphate buffer (pH 7.0). The reaction was started by the addition of the cell extract. Assays were performed in a double-beam spectrophotometer against a blank without the extract to account for chemical degradation of PNPG. The molar extinction coefficients of PNP in the buffer used were 13.6, 14.2, and 14.7 mM−1 · cm−1 at 80, 85, and 95°C, respectively.
TLC.
TLC was used to identify the products formed by incubation of the different maltodextrins with the cell extract or the pure enzymes (4-α-glucanotransferase and MalP). Reaction mixtures were applied on silica gel plates (type 60; Merck), and butanol-ethanol-water (5:3:2 by volume) was used for development. The plates were dipped into methanol containing 5% H2SO4. The carbohydrate spots were visualized by charring at 120°C for 15 min. When radioactively labeled substrates were used, the plates were dried and autoradiographed for 4 days.
Analysis of cyclodextrin production.
To verify whether cyclodextrins were produced by the action of 4-α-glucanotransferase on maltoheptaose, the reaction products were incubated with amyloglucosidase for 1 h at 30°C, and the final products were analyzed by TLC using α-, β-, and γ-cyclodextrins as standards.
Temperature dependence of 4-α-glucanotransferase and MalP activities.
Enzymatic activities were determined with the discontinuous assay described above, and the reaction was stopped by cooling in ice; for temperatures above 85°C, the reaction mixture was incubated in sealed glass capillaries. All assays were performed in triplicate.
Molecular mass determination.
Molecular mass was determined by SDS-PAGE (10% acrylamide) and gel filtration chromatography using a Superose 12 column (Pharmacia) at 25°C with the following proteins as molecular mass standards: cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), aldolase (158 kDa), and ferritin (440 kDa).
Isoelectric point determination.
The isoelectric point was determined by isoelectric focusing (Bio-Rad model 111 mini IEF cell) according to the method recommended by the manufacturer. A pH 3 to 10 isoelectric focusing gel and standards in a range of pI 4.5 to 9.6 were supplied by the manufacturer.
Cloning and sequencing of the gene encoding MalP (malP).
T. litoralis DNA was prepared (28) and partially digested with Sau3a. The digest was ligated into the BamHI site of plasmid pSU2718 (24) upstream of the DNA encoding the α-complement of β-galactosidase. E. coli TG1 (23) was transformed and plated for single-cell colonies on minimal glucose plates containing 0.2% dextrins. The plates were heated to 70°C overnight and stained with iodine vapor. Loss of blue iodine stain around the colony was taken as evidence for thermostable maltodextrin-degrading enzymatic activity. DNA from two heat-treated colonies was prepared (2) and used to transform strain TG1 once more. The two transformants were grown in liquid culture, and their cellular extracts were tested for activity toward maltodextrins. One contained MalP activity and the other contained 4-α-glucanotransferase activity at 85°C. The DNA of the phosphorylase-expressing plasmid was sequenced by GATC GmbH, Konstanz, Germany, and MWG-Biotech GmbH, Ebersbach, Germany. The plasmid did not contain the entire reading frame of malP.
To obtain the complete malP sequence, we proceeded in the following way. The original gene bank inserted in pSU2718 was amplified. Starting from about 80,000 transformants, the plasmid DNA was prepared. Two complementary 30-nucleotide-long DNA primers (primer 1 [5′-GGCAGAGAGGGAGATCAGTTTAACATGACCC-3′] and primer 2 [5′-GGGTCATGTTAAACTGATCTCCCTCTCTGCC-3′]) were chosen as representing the middle of malP. PCR was performed by using 1 μl of the plasmid preparation as the template and the following program: 4 min at 94°C; 25 cycles of 30 s at 69.5°C, 7 min at 74°C, and 30 s at 94°C; a final elongation step for 10 min at 74°C. The Turbo Pfu polymerase (Stratagene) was used. The remaining nonamplified plasmid DNA was digested with DpnI, a restriction enzyme that digests half-methylated and fully methylated DNA but not unmethylated DNA (43). After transformation, we obtained several clones covering the 3′-terminal portion of malP. To obtain the 5′-terminal portion, we had to repeat the procedure using a complementary pair of primers (primer 3 [5′-GCTTCTCCTCGATACTCCAGAGGAGAGATTAAAGG-3′] and primer 4 [5′-CCTTTAATCTCTCC-TCTGGAGTATCGAGGAGAAGC-3′]) that covered the 5′ end of the known sequence. The amplified plasmids were sequenced by GATC GmbH.
Protein induction by different carbohydrates.
To examine the induction by malto-oligosaccharides or trehalose, peptone medium (5 g/liter) was supplemented with maltose (3 g/liter), dextran (3 g/liter), or yeast extract (1 g/liter). The latter carbon source contains 113 mg of trehalose per g (47). As control conditions, peptone or tryptone (5 g/liter) was used as the only carbon source. Cells were harvested during different growth phases (early, mid, and late exponential phases and stationary phase). For enzymatic assays, cells derived from 200 ml of culture (OD600 = 0.2) were harvested and washed once as described above. After disruption of the cells by resuspending in 300 μl of water, 300 μl of 200 mM MOPS (pH 7.0) containing 20 mM MgCl2 and 20 μg of DNase I were added. The extract was centrifuged in an Eppendorf centrifuge, and the supernatant was used immediately to assess MalP activity, PNPG hydrolysis, and maltose and maltotriose degradation by TLC. Cell extracts for Western blots were prepared with cells derived from 60 ml of culture (OD600 = 0.1). Cells were resuspended in 300 μl of water containing 5 μg of DNase I and 300 μl of sample buffer for gel electrophoresis (20). After boiling for 5 min, samples were frozen at −20°C and used for Western blot analysis with antibodies raised against T. litoralis TMBP and 4-α-glucanotransferase.
Preparation of antibodies and Western blot analysis.
Antibodies against TMBP were prepared as previously described (13). For the preparation of antibodies against 4-α-glucanotransferase, a chicken was immunized five times with 80 μg of pure protein each. Fourteen days after the last immunization, antibodies were prepared from 10 eggs (34). Western blot analysis was done as described previously (11, 45), using the primary antibody (17 mg/ml) in a dilution of 1:10,000.
N-terminal amino acid sequencing.
The N-terminal amino acid sequences of pure 4-α-glucanotransferase and MalP were determined by the method of Edman and Begg (9), using an Applied Biosystem model 477A protein sequencer.
Protein determination and SDS-PAGE.
Protein quantification was performed as described by Bradford (5), with bovine serum albumin as the standard. SDS-PAGE was performed with 10% acrylamide, and gels were stained with Coomassie brilliant blue R-250 (20).
Fluorescence spectroscopy.
All spectra were run at room temperature with a SPEX Fluorolog 2002 spectrofluorometer. Spectra were recorded at an excitation wavelength of 345 nm and an emission scan from 340 to 650 nm.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to GenBank under accession no. AF115479.
RESULTS
Purification and properties of T. litoralis 4-α-glucanotransferase.
The presence of 4-α-glucanotransferase activity in T. litoralis cell extracts was revealed by the characteristic oligosaccharide pattern detected by TLC resulting from the incubation of a dialyzed cell extract with maltose (Fig. 1, lanes A) or other maltodextrins. Maltose and maltodextrins with chain length up to maltoheptaose produced glucose and maltodextrins of various lengths, some longer than the initially given substrate. The enzyme responsible for this activity was purified to homogeneity by affinity chromatography in a one-step procedure (Table 1). The molecular mass as determined by SDS-PAGE was 79 kDa (Fig. 2), but a value of 134 ± 30 kDa was found by gel filtration at 25°C. Thus, at this temperature, 4-α-glucanotransferase seems to be a dimeric protein. The isoelectric point was 3.5. The sequence of the first 35 amino acids in the N-terminal region was also determined and found to be identical to that deduced from the gene sequence recently published by Jeon et al. (15).
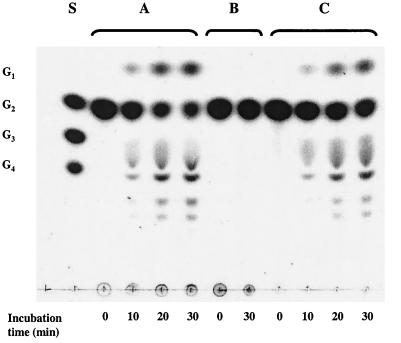
FIG. 1.
TLC of maltose reaction products obtained by a dialyzed T. litoralis cell extract (A), 4-α-glucanotransferase-free cell extract (B), and purified 4-α-glucanotransferase (C). Dialyzed cell extract (1.4 mg of protein per ml), glucanotransferase-free cell extract (4.6 mg of protein per ml), or purified glucanotransferase (11 μg of protein per ml) was incubated with 10 mM maltose in 100 mM MOPS (pH 7.0) at 85°C. Aliquots of the reaction mixture (10 μl) were applied to the TLC plate at different time intervals. S, standards; G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose.
TABLE 1.
Purification of 4-α-glucanotransferase from T. litoralis
| Purification step | Total protein (mg) | Total activity (U)a | Sp act (U/mg)a | Yield (%) |
|---|---|---|---|---|
| Cell extract | 40 | 6.4 | 0.16 | 100 |
| Amylose column | 0.059 | 1.5 | 25.7 | 23 |
4-α-Glucanotransferase activity was measured at 95°C with 10 mM maltotriose as the substrate.
The effect of temperature on 4-α-glucanotransferase activity was studied (Fig. 3). Incubation of the pure enzyme with [U-14C]glucose or [U-14C]maltose in the presence of unlabeled maltose led to the formation of 14C-maltodextrins longer than the substrates (Fig. 4). Therefore, maltose and glucose can be used as acceptors in this transfer reaction, and the enzyme catalyzes the transfer not only of 4-α-glucanosyl groups but apparently also of single glucosyl groups. The transfer of glucosyl groups is apparent from the formation of labeled maltose as a major product in the earliest stages of the reaction when [U-14C]glucose is used as an acceptor (Fig. 4A). The specific activities of 4-α-glucanotransferase at 95°C for 10 mM maltose and maltotriose are 13 and 26 U/mg of protein, respectively. These values were determined from the rates of glucose release upon incubation of the enzyme with the respective substrate. An immediate linear release of glucose was observed when maltotriose was provided, but when maltose was added, a linear release of glucose was observed only after a lag time, indicating that maltose is not an immediate substrate for the enzyme. In contrast to the report of Jeon et al. (15), cyclodextrins were not end products derived from the activity of T. litoralis 4-α-glucanotransferase. In fact, when the products of the reaction of 4-α-glucanotransferase with maltoheptaose were incubated with amyloglucosidase (an exoamylase), cyclodextrins were not detected on TLC plates, and glucose was the only final product (data not shown). We verified that α-, β-, and γ-cyclodextrins migrated in the TLC and that amyloglucosidase was unable to degrade these cyclodextrins. PNPG was not hydrolyzed by 4-α-glucanotransferase.
FIG. 3.
Temperature dependence of T. litoralis 4-α-glucanotransferase activity. The assay mixture, containing 7 μg of enzyme per ml in 100 mM MOPS (pH 7.0)–10 mM maltotriose, was incubated at different time points. Enzyme activity was assessed by the production of glucose as described in Materials and Methods.
FIG. 4.
TLC of 14C-labeled reaction products of purified 4-α-glucanotransferase with unlabeled maltose and [U-14C]glucose (A) or with unlabeled maltose and [U-14C]maltose (B). The reaction mixture (45 μl) contained 1.6 μg of enzyme, 10 mM unlabeled maltose in 100 mM MOPS (pH 7.0), and 0.33 nmol of [U-14C]glucose or 0.6 nmol of [U-14C]maltose. Aliquots of the reaction mixture (10 μl) were applied to the TLC plate at different time intervals. G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose.
Purification and properties of T. litoralis MalP.
Soluble extracts of T. litoralis contained MalP activity, and the specific activity leading to glucose-1-phosphate production from maltoheptaose was 0.82 U/mg at 85°C. This enzyme was purified by the procedure summarized in Table 2. Glutamate dehydrogenase, the most prominent contaminant of MalP-containing fractions (27), was removed by using a Red Sepharose resin. Purified MalP originated a single band in SDS-PAGE corresponding to a molecular mass of 94 kDa (Fig. 2), but a value of 270 ± 30 kDa was determined by gel filtration at 25°C, suggesting that the protein could be a trimer at this temperature. Most known phosphorylases have a subunit molecular mass of around 90 kDa and usually exist as homodimers or tetramers (31). A firm conclusion on the oligomerization state of the protein from T. litoralis demands a more reliable technique for determination of the molecular mass. The isoelectric point was 4.5. The sequence of the first 19 amino acids in the N-terminal region as determined by Edman degradation was Met-Glu-Thr-Val-Val-Asn-Gln-Ile-Lys-Ser-Lys-Leu-Pro-Glu-Asn-Leu-Glu-Gly-Leu.
TABLE 2.
Purification of MalP from T. litoralis
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg)a | Yield (%) |
|---|---|---|---|---|
| Cell extract | 2.9 × 103 | 203 | 0.07 | 100 |
| DEAE | 535 | 80 | 0.15 | 39 |
| Superdex 200 | 324 | 74 | 0.23 | 36 |
| Hydroxyapatite | 97 | 65 | 0.67 | 32 |
| Red Sepharose | NDb | ND | ND | ND |
| Q Sepharose | ND | ND | ND | ND |
| Superdex 200 | 0.215 | 2.5 | 11.5 | 1.2 |
MalP activity was measured at 60°C with 5 mM maltoheptaose as the substrate, using the assay with coupled enzymes described in Materials and Methods.
ND, not determined.
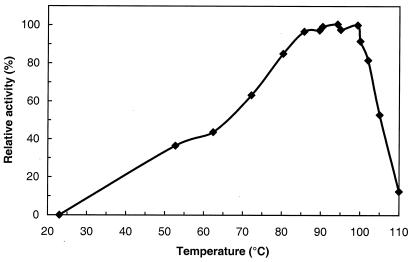
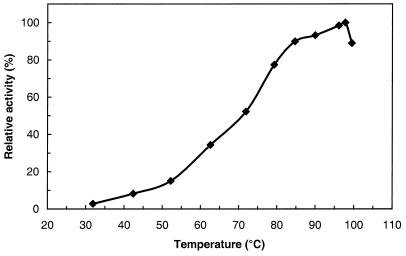
The temperature dependence of MalP activity is shown in Fig. 5. The specific activity of MalP using maltoheptaose as the substrate was 66 U/mg at the temperature for maximal activity of the enzyme (98°C). Phosphorylase activity was measured at 60°C for maltoheptaose concentrations of between 0.125 and 5 mM. A Km of 0.46 mM was determined from a Hanes plot. The substrate specificity of the pure enzyme was determined for a series of maltodextrins from maltose to maltoheptaose (substrate concentration, 4 mg/ml). Maximum activity (11.3 ± 1.7 U/mg at 60°C) was observed with heptaose, hexaose, or pentaose; with tetraose the enzyme activity decreased to 6 ± 2 U/mg, while triose and maltose were not used. It was verified that long maltodextrins (8 to 15 glucose units) as well as amylose and starch were not as good substrates as maltoheptaose. PNPG was not hydrolyzed by the MalP.
FIG. 5.
Temperature dependence of T. litoralis MalP activity. The assay mixture containing 2 μg of enzyme per ml in 50 mM potassium phosphate (pH 7.0)–5 mM maltoheptaose was incubated for 15 min. The enzyme activity was assessed by the production of glucose-1-phosphate as described in Materials and Methods. For all temperatures tested, glucose-1-phosphate production was linear with time.
The UV-visible spectrum of T. litoralis phosphorylase had an absorption band at 336.5 nm, indicating that this enzyme, like all known MalPs, contains pyridoxal 5′-phosphate as a cofactor. Upon excitation at 345 nm, the fluorescence emission spectrum of the enzyme showed a maximum at 460 nm.
Cloning and sequencing the malP gene.
The gene encoding MalP (malP) was isolated by functional screening in E. coli at 70°C after transformation with a gene bank of T. litoralis in pSU2718 (24). One clone expressing MalP activity contained an in-frame fusion to the α-complement of lacZ. The deduced amino acid sequence of the cloned fragment showed homology with a predicted phosphorylase of P. furiosus (Blast archaeal genome sequence at Center of Marine Biotechnology, University of Maryland). Even though the encoded fusion protein exhibited thermostable phosphorylase activity, it did not contain the complete open reading frame expected from its homology to the P. furiosus enzyme. Approximately 50 and 120 amino acids were missing from the N and C termini, respectively. The remaining part of the sequence was obtained by screening the amplified genomic library for plasmids containing parts of the coding sequence of malP. The start of the open reading frame of malP was corroborated by the sequence in the N terminus of the purified protein. Distal to the 3′ end of malP can be observed a series of T’s which could serve as a transcription terminator. Neither immediately upstream nor downstream of malP could we detect malQ, the gene encoding 4-α-glucanotransferase (15). Thus, in contrast to organization in E. coli, malP and malQ do not form an operon in T. litoralis. The calculated molecular mass for T. litoralis MalP is 96,622 Da, and the protein contains at position 584 the sequence EASGTSGMKAGLN, corresponding to the signature sequence for the pyridoxal 5′-phosphate binding site (EA[S/C]GX[G/S]XMKXX[L/M]N) (10). The protein exhibits over its entire length sequence identity of 66% to a putative phosphorylase from the hyperthermophilic archaeon Pyrococcus horikoshii (16) and 60% identity to the characterized phosphorylase from the hyperthermophilic bacterium Thermotoga maritima (1). The alignment of the T. litoralis protein to these two hyperthermophilic enzymes is shown in Fig. 6. Surprisingly MalP from T. litoralis is still 31% identical to the corresponding enzyme from the mesophilic bacterium Mycobacterium tuberculosis (Swiss-Prot accession no. Q10639) but only 13% to MalP from E. coli (32). The homology to the α-glucan phosphorylase of E. coli, encoded by glgP and supposedly involved in glycogen metabolism (48), was 12%.
FIG. 6.
Sequence alignment of the MalP from T. litoralis (Tli MalP) with phosphorylases from other thermophilic organisms. Pho PH1512, homologous sequence from hyperthermophilic archaeon P. horikoshii OT3 (16); Tma AgpA, MalP from the hyperthermophilic bacterium T. maritima (1). The pyridoxal 5′-phosphate binding consensus sequence begins at position 584 of the Tli MalP sequence.
Effect of carbon source on expression of proteins involved in maltose metabolism by T. litoralis.
Higher activities of MalP were found in extracts of cells grown in media containing maltose or maltodextrins than in extracts of cells grown in peptone or tryptone alone (Table 3). Furthermore, the phosphorylase activity did not increase in cells derived from growth in medium containing peptone plus yeast extract, indicating that this activity was not induced by trehalose present in the yeast extract. PNPG hydrolysis was constitutive under all growth conditions examined. The activities of both enzymes did not vary appreciably with the growth phase of the cultures (results not shown). The product patterns derived from maltose and maltotriose degradation by cell extracts in the absence of phosphate (conditions preventing the action of MalP) were identical to those obtained with purified 4-α-glucanotransferase (data not shown). Western blot analysis with antibodies against T. litoralis 4-α-glucanotransferase suggested that this enzyme was constitutive. On the other hand, the results of the Western blot analysis using antibodies against TMBP revealed that this component of the trehalose/maltose transport system was expressed in cells grown in the presence of maltose, maltodextrins, or trehalose (from yeast extract) but not in cells grown on peptone alone. In maltodextrin-containing medium, the expression of TMBP increased along with growth of cells, probably due to formation of maltose from maltodextrins by the action of extracellular hydrolytic enzymes. Under other growth conditions, expression of TMBP was independent of the growth phase (data not shown).
TABLE 3.
Specific enzyme activities in cell extracts from T. litoralis grown with different carbon sources
| Carbon source | Activity (mU/mg)a
|
|
|---|---|---|
| MalPb | PNPG hydrolysisc | |
| Peptone | 48 | 17 |
| Tryptone | 51 | 16 |
| Yeast extract + peptone | 57 ± 8 | 16 ± 1 |
| Maltose + peptone | 88 ± 10 | 15 ± 1 |
| Dextrins + peptone | 108 ± 9 | 21 ± 3 |
Values for activities measured in extracts of cells grown in peptone or tryptone correspond to the averages of triplicate determinations in a single experiment. The others are averages of activities measured at least in three independent extracts.
Measured at 60°C with 5 mM maltoheptaose as the substrate, using the assay with coupled enzymes described in Materials and Methods.
Measured at 80°C with 1 mM PNPG.
Contribution of 4-α-glucanotransferase and α-glucosidase to maltose metabolism.
A dialyzed cell extract (15 mg of protein), derived from T. litoralis grown on peptone and maltose, was passed twice through an amylose column for complete removal of 4-α-glucanotransferase. The initial cellular extract converted maltose to glucose at a rate of 213 nmol/min/mg of protein, but after these two chromatographic steps the rate for this conversion in the flowthrough decreased to 5 nmol/min/mg (assayed at 85°C). The efficient removal of maltose-degrading enzymes by the affinity columns is illustrated in Fig. 1 (compare lanes A and B). However, the specific activity of PNPG hydrolysis was identical in the initial cell extract (15.0 mU/mg) and in the flowthrough from the amylose column (14.8 mU/mg). Moreover, the protein fractions eluted from the amylose column contained the pure 4-α-glucanotransferase and degraded maltose with a product pattern similar to the cell extract (Fig. 1, lanes A and C). These results show that the PNPG-hydrolyzing enzyme did not bind to the amylose column and was unable to degrade maltose; furthermore, 4-α-glucanotransferase was the only maltose-degrading enzyme detected.
Combined action of 4-α-glucanotransferase and MalP.
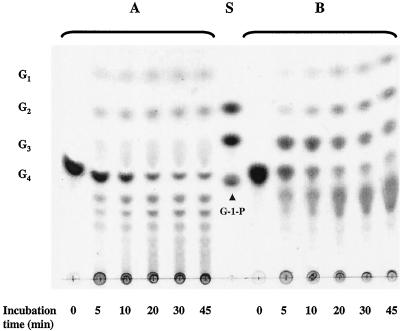
The dependence of MalP activity on inorganic phosphate allowed the visualization of the activity of 4-α-glucanotransferase alone or the combined action of the two enzymes on the degradation of maltotetraose in cell extracts of T. litoralis (Fig. 7). In the absence of phosphate, the products ranged from glucose to very long dextrins, whereas in the presence of phosphate, the longer dextrins were rapidly degraded by MalP and therefore were not detectable by TLC.
FIG. 7.
TLC of maltotetraose reaction products obtained with T. litoralis cell extracts in the presence and absence of phosphate. Reaction mixtures containing 5 mM maltotetraose and the cell extract (1 mg of protein per ml) in 100 mM MOPS (pH 7.0) (A) or in 50 mM potassium phosphate (pH 7.0) (B) were incubated at 85°C. Aliquots of the reaction mixture (10 μl) were applied to the TLC plate at different time intervals. S, standards; G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G-1-P, glucose-1-phosphate.
Comparison of maltose metabolism in T. litoralis and in P. furiosus.
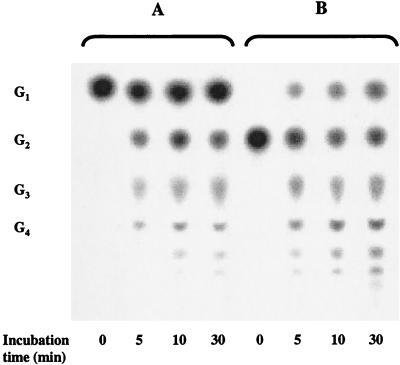
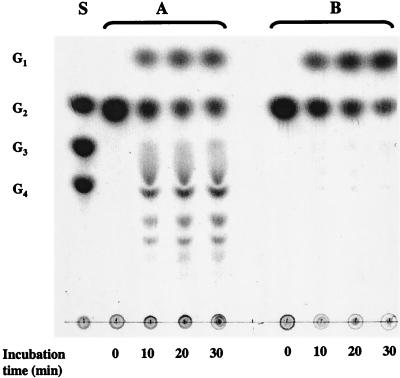
Extracts derived from T. litoralis and P. furiosus cells grown on the same carbon sources (maltose and peptone) were incubated with maltose in the absence of phosphate at the optimal growth temperature of each organism (85 and 95°C, respectively), and the products were analyzed by TLC. In T. litoralis, maltose is degraded to glucose and several maltodextrins, as expected by the action of 4-α-glucanotransferase. Glucose, on the other hand, is the only product detected from the metabolism of maltose in P. furiosus, as expected by the action of α-glucosidase (Fig. 8). The specific activities leading to glucose production from 10 mM maltose by T. litoralis and P. furiosus cell extracts are 168 mU/mg (at 85°C) and 266 mU/mg (at 95°C), respectively. The specific activity for PNPG hydrolysis is higher in P. furiosus (99 and 269 mU/mg at 85 and 95°C, respectively) than in T. litoralis (25 and 67 mU/mg at 85 and 95°C, respectively). Despite the difference in PNPG hydrolysis in both cell extracts, the rates of maltose consumption (or glucose production from maltose) are not so different. The incubation of maltotriose with P. furiosus cell extracts led to the formation of longer maltodextrins, similar to those observed in T. litoralis cell extracts, indicating the presence of 4-α-glucanotransferase with a different substrate specificity.
FIG. 8.
TLC of maltose reaction products obtained with T. litoralis and P. furiosus cell extracts. Reaction mixtures of T. litoralis (A) and P. furiosus (B) dialyzed cell extracts (5 mg of protein per ml) were incubated with 10 mM maltose in 100 mM MOPS (pH 7.0) at 85 and 95°C, respectively. Aliquots of the reaction mixture (10 μl) were applied to the TLC plate at different time intervals. Notation is as in Fig. 1.
Phosphoglucomutase activity in T. litoralis cell extracts.
Activity was measured at 85°C in extracts from cells grown with maltose and peptone. The specific activity was 61 mU/mg. The specific activities of several enzymes involved in maltose metabolism are summarized in Table 4.
TABLE 4.
Specific activities of enzymes involved in maltose metabolism in T. litoralis cell extracts
| Enzyme | Temp (°C) | Sp acta (U/mg) |
|---|---|---|
| 4-α-Glucanotransferase | 85 | 0.17 |
| MalP | 85 | 0.82 |
| Phosphoglucomutase | 85 | 0.06 |
| Hexokinase (ADP dependent)b | 50 | 0.37 |
| Phosphofructokinase (ADP dependent)b | 50 | 0.20 |
Average of results obtained with at least two independent cell batches. 4-α-Glucanotransferase activity corresponds to the rate of glucose release from maltose by cell extracts in the absence of phosphate. MalP activity was measured at 85°C with 5 mM maltoheptaose as substrate, using the discontinuous assay as described in Materials and Methods.
Data from Selig et al. (42).
DISCUSSION
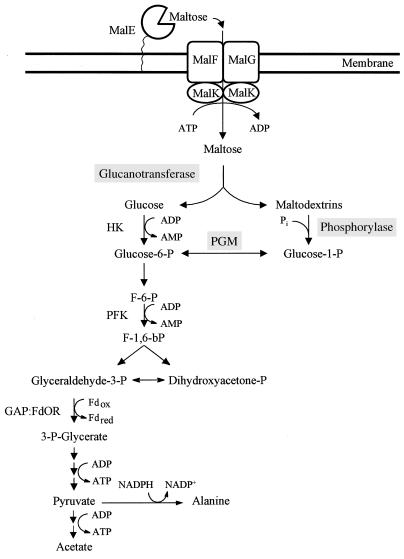
This study allows us to propose that maltose metabolism in T. litoralis proceeds according to the pathway presented in Fig. 9. After entering the cell via a very high affinity binding protein-dependent ABC transporter, maltose is degraded by the concerted action of 4-α-glucanotransferase and MalP. The first enzyme produces glucose and a series of maltodextrins with increasing chain lengths; maltodextrins with four or more glucose residues can be acted upon by MalP, with production of glucose-1-phosphate. The combined action of these two enzymes enables the cell to conserve the energy of the glycosidic bond in maltose. Glucose-1-phosphate is converted to glucose-6-phosphate by a phosphoglucomutase activity also detected in T. litoralis cell extracts. Glucose derived from the action of 4-α-glucanotransferase is phosphorylated by the activity of an ADP-dependent hexokinase, leading to the formation of glucose-6-phosphate that is subsequently catabolized via an Embden-Meyerhof-type glycolytic pathway (18, 42).
FIG. 9.
Proposed pathway for maltose metabolism in T. litoralis. MalE, maltose/trehalose binding protein; MalF, MalG, and MalK, components of the maltose/trehalose ABC transport system; PGM, phosphoglucomutase; HK, hexokinase; PFK, phosphofructokinase; GAP:FdOR, glyceraldehyde-3-phosphate:ferredoxin oxidoreductase; Fd, ferredoxin; F-6-P, fructose-6-P; F-1,6-bP, fructose-1,6-bisphosphate.
Given the close phylogenetic relationship between the genera Thermococcus and Pyrococcus, it is interesting that T. litoralis and P. furiosus use different enzymatic strategies to metabolize maltose. In the case of the former organism, the pattern of products was typical of a 4-α-glucanotransferase-catalyzed reaction, whereas with P. furiosus cell extracts, glucose was the only product, as expected from a reaction catalyzed by α-glucosidase (Fig. 8). In fact, it has been suggested that in P. furiosus, maltose is metabolized via an α-glucosidase (7, 17). Therefore, we looked for the existence of such an enzyme and its role in maltose metabolism in T. litoralis. Cell extracts could hydrolyze PNPG, the typical artificial substrate for α-glucosidase, an enzyme able to release d-glucose residues at the nonreducing end of a variety of substrates, such as maltose and other oligosaccharides. However, after removal of 4-α-glucanotransferase from a T. litoralis cell extract, the extent of maltose degradation decreased drastically (about 45-fold) even though the activity responsible for the hydrolysis of PNPG remained constant. In contrast, the purified α-glucosidase from P. furiosus hydrolyzes maltose rather efficiently (7). Thus, we conclude that the first step of maltose metabolism in T. litoralis and in P. furiosus is catalyzed by different enzymes, T. litoralis using a metabolic strategy similar to that used by E. coli. However, the presence of hydrolytic activity for PNPG in T. litoralis cell extracts remains intriguing. Maltotriose and maltotetraose could not be degraded in cell extracts depleted of 4-α-glucanotransferase (data not shown). Therefore, the observed PNPG hydrolytic activity is probably not due to the presence of α-glucosidase, or another enzyme able to hydrolize short maltodextrins. A likely candidate would be trehalose-6-phosphate hydrolase, since this enzyme from E. coli can hydrolyze PNPG (37). However, trehalose-6-phosphate hydrolase activity was not present in T. litoralis cell extracts (data not shown). Hence, the identity of the PNPG-hydrolyzing enzyme in T. litoralis is not known. In contrast to maltose metabolism, the patterns of products resulting from maltotriose degradation in P. furiosus and in T. litoralis cell extracts are similar. This is likely to result from the action of intracellular α-amylase earlier detected in P. furiosus (19) that could have some transferase activity, as previously suggested (44). This α-amylase has an amino acid sequence highly homologous to the sequences of 4-α-glucanotransferases from T. litoralis and Pyrococcus strain KOD1 (15, 44) and resembles these enzymes in some of its properties, such as the ability to produce maltose, glucose, and longer dextrins from maltotriose (19).
In this study, we purified and characterized two key enzymes involved in maltose degradation in T. litoralis: 4-α-glucanotransferase and MalP. The purification and characterization of the former enzyme have been recently reported by Jeon et al. (15); herein, we describe an efficient purification procedure, extend the characterization of the enzyme, and investigate its role in metabolism. Like the mesophilic counterpart from E. coli (amylomaltase), the T. litoralis 4-α-glucanotransferase can catalyze the transfer not only of 4-α-glucanosyl oligomers but apparently also of single glucosyl groups. Whether this results from a direct transfer of glucosyl units or from the cleavages of shorter oligosaccharides after the transfer of larger oligosaccharides to the acceptor remains unclear. It is clear, though, that maltose and glucose can both be used as acceptors. The 4-α-glucanotransferase from T. litoralis exhibits characteristics similar to those of its counterpart in Pyrococcus strain KOD1 (44) but different from those reported for 4-α-glucanotransferase from the hyperthermophilic bacterium T. maritima (12, 22), which is unable to disproportionate maltose or maltotriose. In this case, a maltosyl residue is the minimum unit transferred, and glucose cannot function as an acceptor, nor is it found as a reaction product.
The MalP described here is the most thermophilic phosphorylase known so far, with an optimum temperature for activity of 98°C. The α-glucan phosphorylases from other thermophilic sources, Thermus thermophilus and T. maritima, have optimum temperatures of 70 and 75 to 80°C, respectively (1, 3). These enzymes belong to a large group of highly homologous phosphorylases, comprising glycogen and MalPs from bacteria, eukaryotic unicellular organisms, plants, and mammals (31). To our knowledge, the T. litoralis MalP is the first enzyme of this kind to be purified and characterized from an organism belonging to the domain Archaea. It has many properties in common with the other known phosphorylases of the same group, despite low amino acid sequence homology. The E. coli enzyme, like most other known phosphorylases, is not able to degrade maltodextrins with chain lengths shorter than maltopentaose, so in this respect the T. litoralis phosphorylase resembles more the T. thermophilus α-glucan phosphorylase that also degrades maltotetraose (3). The affinity of the T. litoralis enzyme for maltoheptaose at 60°C (Km = 0.5 mM) was similar to that of the E. coli enzyme (8, 39) but higher than the affinity of the T. thermophilus enzyme, Km of 3.6 mM at 70°C (3).
Except for a few examples (14), all known phosphorylases contain tightly bound pyridoxal 5′-phosphate (30). This cofactor is also present in the T. litoralis enzyme, since the UV-visible spectrum exhibits a band at 336.5 nm, and the amino acid sequence contains the signature of a typical pyridoxal 5′-phosphate binding site. Most phosphorylases have a characteristic fluorescence maximum at 535 nm when excited at 345 nm. In the T. litoralis enzyme the fluorescence maximum is shifted to 460 nm, suggesting a more polar microenvironment for pyridoxal 5′-phosphate, resembling the recently characterized phosphorylase from the thermophilic bacterium T. thermophilus (3).
MalP and 4-α-glucanotransferase are constitutive in T. litoralis, whereas the trehalose/maltose transport system is induced by trehalose, maltose, and maltodextrins. Thus, the genes encoding the enzymes of maltose metabolism are not coregulated with those encoding the transport proteins, whereas in E. coli both are part of a regulon controlled by the transcriptional activator MalT (36). Furthermore, the genes for MalP and 4-α-glucanotransferase are apparently not part of an operon as in E. coli.
Although maltose and trehalose are transported with similar efficiencies by a single high-affinity transport system (47), our attempts to detect trehalose degradation in T. litoralis cell extracts were unsuccessful. In this organism, it is possible that trehalose has a very low metabolic turnover since it has been shown to be taken up from the medium to serve as a compatible solute (21).
In conclusion, the hyperthermophilic archaeon T. litoralis uses a biochemical strategy similar to that of the archetypal mesophile E. coli for the metabolism of maltose, despite the fact that these organisms belong to distinct phylogenetic domains and have optimal growth temperatures differing by about 50°C. Major differences are the remarkable thermophily of the key enzymes, the hyperaffinity of the transport system, and the independent regulation of transport and metabolism in T. litoralis. It appears as though major changes in metabolic strategies are not required to support life at high temperature.
ACKNOWLEDGMENTS
This work was supported by the European Community Biotech Programme (Extremophiles as Cell Factories, BIO4-CT96-0488), by PRAXIS XXI and FEDER, Portugal (PRAXIS/2/2.1/BIO/1109/95), and by the Deutsche Forschungsgemeinschaft. K. B. Xavier acknowledges a Ph.D. grant from PRAXIS XXI (BD/2760/94).
We thank Isabel Pacheco for valuable expertise on protein purification.
REFERENCES
- 1.Bibel M, Brettl C, Gosslar U, Kriegshäuser G, Liebl W. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol Lett. 1998;158:9–15. doi: 10.1111/j.1574-6968.1998.tb12793.x. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. Rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeck B, Schinzel R. Purification and characterisation of an α-glucan phosphorylase from the thermophilic bacterium Thermus thermophilus. Eur J Biochem. 1996;239:150–155. doi: 10.1111/j.1432-1033.1996.0150u.x. [DOI] [PubMed] [Google Scholar]
- 4.Boos W, Shuman H A. The maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown S H, Kelly R M. Characterization of amylolytic enzymes, having both α-1,4 and α-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl Environ Microbiol. 1993;59:2614–2621. doi: 10.1128/aem.59.8.2614-2621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costantino H R, Brown S H, Kelly R M. Purification and characterization of an α-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J Bacteriol. 1990;172:3654–3660. doi: 10.1128/jb.172.7.3654-3660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drueckes P, Boeck B, Palm D, Schinzel R. Mutational analysis of the oligosaccharide recognition site at the active site of Escherichia coli maltodextrin phosphorylase. Biochemistry. 1996;35:6727–6734. doi: 10.1021/bi951938l. [DOI] [PubMed] [Google Scholar]
- 9.Edman P, Begg G. A protein sequenator. Eur J Biochem. 1967;1:80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 10.Fukui T, Shimomura S, Nakano K. Potato and rabbit muscle phosphorylases: comparative studies on the structure, function and regulation of regulatory and nonregulatory enzymes. Mol Cell Biochem. 1982;42:129–144. doi: 10.1007/BF00238507. [DOI] [PubMed] [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 12.Heinrich P, Huber W, Liebl W. Expression in Escherichia coli and structure of the gene encoding 4-α-glucanotransferase from Thermotoga maritima. Classification of maltodextrin glycosyltransferases into two distantly related enzyme subfamilies. Syst Appl Microbiol. 1994;17:297–305. [Google Scholar]
- 13.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hüwel S, Haalck L, Conrath N, Spener F. Maltose phosphorylase from Lactobacillus brevis: purification, characterization, and application in a biosensor for ortho-phosphate. Enzyme Microb Technol. 1997;21:413–420. doi: 10.1016/s0141-0229(97)00014-8. [DOI] [PubMed] [Google Scholar]
- 15.Jeon B-S, Taguchi H, Sakai H, Ohshima T, Wakagi T, Matsuzawa H. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis. Enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli. Eur J Biochem. 1997;248:171–178. doi: 10.1111/j.1432-1033.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 16.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y T F, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 17.Kelly R M, Adams M W W. Metabolism in hyperthermophilic microorganisms. Antonie Leeuwenhoek. 1994;66:247–270. doi: 10.1007/BF00871643. [DOI] [PubMed] [Google Scholar]
- 18.Kengen S W M, De Bok F A M, van Loo N-D, Dijkema C, Stams A J M, De Vos W M. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–17541. [PubMed] [Google Scholar]
- 19.Laderman K A, Davis B R, Krutzsch H C, Lewis M S, Griko Y V, Privalov P L, Anfinsen C B. The purification and characterization of an extremely thermostable α-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem. 1993;268:24394–24401. [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lamosa P, Martins L O, da Costa M S, Santos H. Effect of temperature, salinity, and the medium composition on compatible solute accumulation by Thermococcus spp. Appl Environ Microbiol. 1998;64:3591–3598. doi: 10.1128/aem.64.10.3591-3598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liebl W, Feil R, Gabelsberger J, Kellermann J, Schleifer K-H. Purification and characterization of a novel thermostable 4-α-glucanotransferase of Thermotoga maritima cloned in Escherichia coli. Eur J Biochem. 1992;207:81–88. doi: 10.1111/j.1432-1033.1992.tb17023.x. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 24.Martinez E, Bartolome B, De La Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 25.Mukund S, Adams M W W. Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis. A role for tungsten in peptide catabolism. J Biol Chem. 1993;268:13592–13600. [PubMed] [Google Scholar]
- 26.Neuner A, Jannasch H W, Belkin S, Stetter K O. Thermococcus litoralis sp. nov.: a new species of extremely thermophilic marine archaebacteria. Arch Microbiol. 1990;153:205–207. [Google Scholar]
- 27.Ohshima T, Nishida N. Purification and characterization of extremely thermo-stable glutamate dehydrogenase from a hyperthermophilic archaeon, Thermococcus litoralis. Biocatalysis. 1994;11:117–129. [Google Scholar]
- 28.Owen R J, Borman P. A rapid biochemical method for purifying high molecular weight bacterial chromosomal DNA for restriction enzyme analysis. Nucleic Acids Res. 1987;8:3631. doi: 10.1093/nar/15.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pajatsch M, Böck A, Boos W. Enzymatic preparation of radiolabeled linear maltodextrins and cyclodextrins of high specific activity from [14C] maltose using amylomaltase, cyclodextrin glucosyltransferase and cyclodextrinase. Carbohydr Res. 1998;307:375–379. doi: 10.1016/s0008-6215(97)10113-6. [DOI] [PubMed] [Google Scholar]
- 30.Palm D, Klein H W, Schinzel R, Buehner M, Helmreich E J M. The role of pyridoxal 5′-phosphate in glycogen phosphorylase catalysis. Biochemistry. 1990;29:1099–1107. doi: 10.1021/bi00457a001. [DOI] [PubMed] [Google Scholar]
- 31.Palm D, Goerl R, Burger K J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985;313:500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- 32.Palm D, Goerl R, Burger K J, Buhner M, Schwartz M. DNA sequence of the malP gene and primary structure of Escherichia coli maltodextrin phosphorylase. Prog Clin Biol Res. 1984;144A:209–221. [PubMed] [Google Scholar]
- 33.Palmer T N, Ryman B E, Whelan W J. The action pattern of amylomaltase from Escherichia coli. Eur J Biochem. 1976;69:105–115. doi: 10.1111/j.1432-1033.1976.tb10863.x. [DOI] [PubMed] [Google Scholar]
- 34.Polson A, von Wechmar M B, van Regenmortel M H. Isolation of viral IgY antibodies from yolks of immunized hens. Immunol Commun. 1980;9:475–493. doi: 10.3109/08820138009066010. [DOI] [PubMed] [Google Scholar]
- 35.Raven N, Ladwa N, Cossar D, Sharp R. Continuous culture of the hyperthermophilic archaeum Pyrococcus furiosus. Appl Microbiol Biotechnol. 1992;38:263–267. [Google Scholar]
- 36.Richet E, Raibaud O. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 1989;8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rimmele M, Boos W. Trehalose-6-phosphate hydrolase of Escherichia coli. J Bacteriol. 1994;176:5654–5664. doi: 10.1128/jb.176.18.5654-5664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schäfer T, Xavier K B, Santos H, Schönheit P. Glucose fermentation to acetate and alanine in resting cell suspensions of Pyrococcus furiosus: proposal of a novel glycolytic pathway based on 13C labelling data and enzyme activities. FEMS Microbiol Lett. 1994;121:107–114. [Google Scholar]
- 39.Schinzel R, Palm D. Escherichia coli maltodextrin phosphorylase: contribution of active site residues glutamate-637 and tyrosine-538 to the phosphorolytic cleavage of α-glucans. Biochemistry. 1990;29:9956–9962. doi: 10.1021/bi00494a028. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz M. The maltose regulon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1482–1502. [Google Scholar]
- 41.Schwartz M, Hofnung M. La maltodextrine phosphorylase d’Escherichia coli. Eur J Biochem. 1967;2:132–145. doi: 10.1111/j.1432-1033.1967.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 42.Selig M, Xavier K B, Santos H, Schönheit P. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch Microbiol. 1997;167:217–232. doi: 10.1007/BF03356097. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan M, Folk W R. An improved procedure for measuring DNA replication with transient assays in eukaryotic cells. Gene Anal Tech. 1988;5:54–56. doi: 10.1016/0735-0651(88)90016-7. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana Y, Fujiwara S, Takagi M, Imanaka T. Cloning and expression of the 4-α-glucanotransferase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J Ferment Bioeng. 1997;83:540–548. [Google Scholar]
- 45.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson K A, Schinzel R, Palm D, Johnson L N. The crystal structure of Escherichia coli maltodextrin phosphorylase provides an explanation for the activity without control in this basic archetype of a phosphorylase. EMBO J. 1997;16:1–14. doi: 10.1093/emboj/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xavier K B, Martins L O, Peist R, Kossmann M, Boos W, Santos H. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1996;178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu F, Jen J, Takeuchi E, Inouye M, Nakayama H, Tagaya M, Fukui T. α-Glucan phosphorylase from Escherichia coli. Cloning of the gene and purification and characterization of the protein. J Biol Chem. 1988;263:13706–13711. [PubMed] [Google Scholar]