Abstract
We are entering an era of medicine where increasingly sophisticated data will be obtained from patients to determine proper diagnosis, predict outcomes and direct therapies. We predict that the most valuable data will be produced by systems that are highly dynamic in both time and space. Three-dimensional (3D) organoids are poised to be such a highly valuable system for a variety of gastrointestinal (GI) diseases. In the lab, organoids have emerged as powerful systems to model molecular and cellular processes orchestrating natural and pathophysiological human tissue formation in remarkable detail. Preclinical studies have impressively demonstrated that these organs-in-a-dish can be used to model immunological, neoplastic, metabolic or infectious GI disorders by taking advantage of patient-derived material. Technological breakthroughs now allow to study cellular communication and molecular mechanisms of interorgan cross-talk in health and disease including communication along for example, the gut–brain axis or gut–liver axis. Despite considerable success in culturing classical 3D organoids from various parts of the GI tract, some challenges remain to develop these systems to best help patients. Novel platforms such as organ-on-a-chip, engineered biomimetic systems including engineered organoids, micromanufacturing, bioprinting and enhanced rigour and reproducibility will open improved avenues for tissue engineering, as well as regenerative and personalised medicine. This review will highlight some of the established methods and also some exciting novel perspectives on organoids in the fields of gastroenterology. At present, this field is poised to move forward and impact many currently intractable GI diseases in the form of novel diagnostics and therapeutics.
Keywords: stem cells, intestinal stem cell, gastrointestinal pathology, gastrointestinal physiology
Introduction
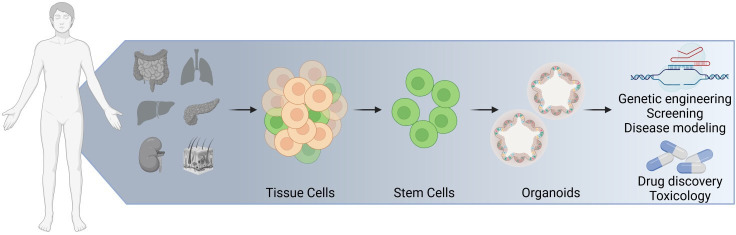
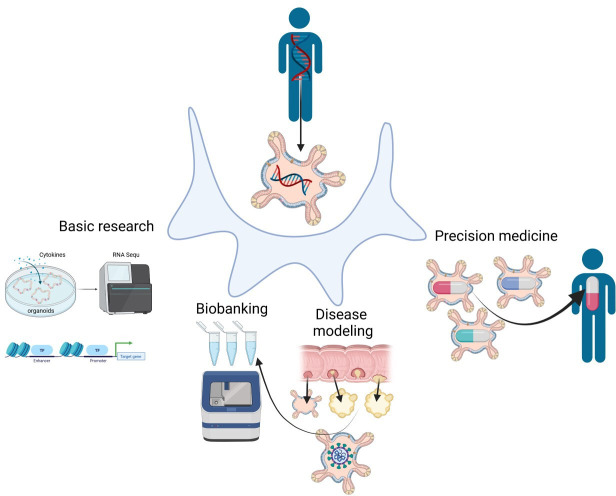
Organ cell cultures, also referred as organoids, are a powerful experimental system that have enabled important contributions to numerous disciplines of basic biology and led to many medical applications (figure 1). Examples abound in nearly every organ system including the central/enteric nervous systems, as well as respiratory, sensory, endocrine and reproductive systems.1–6 Researchers in these areas have tailored experimental settings to use these organoids to model numerous aspects of organ physiology including cell-to-cell communication, host–microbe interaction and interorgan cross-talk. Stemming from these basic research questions, organoids have been used to model human pathologies including aspects of inflammatory, infectious, metabolic and neoplastic diseases. These insights have opened new avenues in regenerative and personalised medicine. While the main area of today’s organoid technology is mainly based on initial landmark publications in the early 2000, the idea of stem-cell based organ cultures is not new and goes back to the 1970/1980 (box 1).
Figure 1.
Organoids, assembly of single cells. The term organoid generally applies to three-dimensional cellular cultures derived from stem cells that resemble components of a complex tissue or organ.
Box 1. History of stem cell cultures and organoids.
Preclinical model systems such as immortalised cell lines and animal models have strongly contributed to improve our understanding of cellular signalling pathways and to identify novel therapeutic targets for pathologies including GI inflammation, infection and cancer. However, mouse models, despite the genetic similarity to humans, display some restrictions such as their failure to accurately mimic various aspects of human disease phenotypes. Cell lines have often been transformed or genetically immortalised. Thus they fail to reflect for example important epithelial functions, such as polarisation, barrier formation and cell differentiation. However, beside these disadvantages, previous experimental work on organ development and physiology has opened new avenues for the evolution of more physiological and reliable preclinical in vitro systems. Accordingly, research on stem cell biology has uncovered improved methods on how to control stemness, including processes that determine self-renewal and differentiation along defined tissue lineages. Supported by this work, scientific achievements from the field of regenerative medicine have provided an understanding on how tissue damage can be repaired by dissociated stem cells that differentiate into one or more of the required mature cell types within the affected organ.212
The field of stem cell research has been inspired by the formation of primitive tissues that form organs naturally occurring during embryogenesis. These studies provided the crucial information and tools to understand patterning events and signalling during organogenesis. Investigators in other fields, such as skin keratinocytes, had identified factors required for in vitro culture decades ago.213 In the field of gastroenterology, this proved to be more challenging. For decades the critical factors for in vitro culture were elusive. It was in vivo work studying the role of R-spondin1 in 2005 that suggested this might be the missing factor in vitro.214 In deed it was the missing factor that Sato and Clevers established in 2009.215 They demonstrated that mouse small intestinal multipotent organ-specific adult stem cells (ASCs) have the intrinsic ability to self-organise into epithelial 3D structures, when cultured in an extracellular matrix covered by a medium supplemented with combinations of different mitogens, morphogens and cytokines that guide the self-renewal and organised differentiation of somatic stem cells (multipotent, can give rise to more than one cell type).215 216 Pioneering work from Thomson and colleagues established the derivation of embryonic stem cells (ESCs). They isolated and cultured human embryonic stem cells (hESC) by deriving them from blastocysts of donated embryos originally produced for in vitro fertilisation.217 At this stage the cells are no longer totipotent (able to give rise to a complete organism including the extraembryonic tissues), but pluripotent (able to give rise to basically all tissues in the body). Many of the hESC lines generated from these studies are still available in the stem cell community (https://grants.nih.gov/stem_cells/registry/current.htm). The first reprogrammed cells termed induced pluripotent stem cells (iPSC) were generated in the laboratory of Nobel prize winner Yamanaka by retroviral expression of four transcription factors Oct3/4, Sox2, Klf4 and c-myc (or alternatively Nanog) from adult fibroblasts, jump-starting their continuous expression.218 James Wells followed the work by Sasai and colleagues on hESC219 and showed that human-induced pluripotent stem cells could be differentiated towards hindgut endoderm and further developed to mature intestinal structures. Both ASC-derived and iPSC-derived 3D structures were able to resemble the native organ in terms of gene and protein expression, metabolic function, microscale tissue architecture, as well as cellular composition. We nowadays call these structures ‘organoids’, which defines 3D tissues or structures that resemble an organ or components of an organ. Organoids as such should match the following criteria: cultured cells should (I) retain the identity of the organ being modelled, (II) contain progenitor and terminal differentiated cell types that are present in the organ itself, (III) imitate key functions of the respective organ and (IV) self-organise according to the same intrinsic organising principles as in the organ itself. Interestingly, the emergence of submerged 2D cultures of ASC/ESC goes back to the 1970/1980.213 220 However, until the early 2000s these stem cells had to be cultured on growth-inactivated feeder cells.221 The groundbreaking work from 2009 now allows long-term, stem cell-based organotypic cultures, which have the potential to overcome previous limitations and thus represent a substantial advance over traditional in vitro cell culture systems.
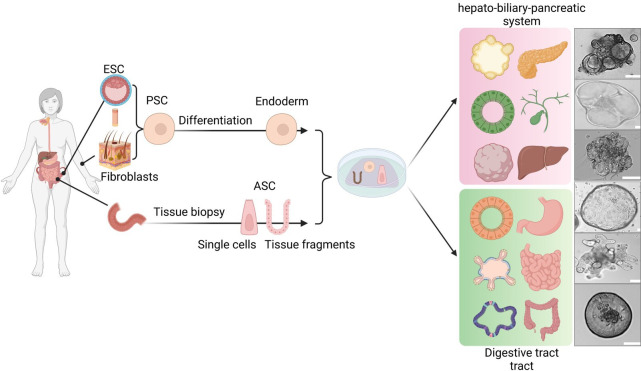
Organoid technology in nearly all fields of gastroenterology has shown substantial growth over the past decade. An increasing number of organoid culture methods have been developed, including protocols to culture human and murine adult stem cells (ASCs) as well as human pluripotent stem cells (PSC). Both of these systems can produce intestinal organoids resembling the oesophagus, stomach or small and large intestine (figure 2). Further modifications of the established luminal organ protocols enabled the expansion of organoid development to include components of the hepato-pancreato-biliary system: the liver (separated into organoids containing hepatocytes or biliary epithelial cells),7 8 gallbladder9 and pancreas.10 11 A common theme across all these tissues is that ASCs are composed only of epithelial cells and represent the tissue from which they were isolated and PSCs are composed of mixtures of epithelial and mesenchymal cells (MSCs) and are manipulated experimentally to undergo development and differentiation to mimic a particular area of the GI tract.
Figure 2.
The GI organoid culture system. GI organoids can be derived from PSCs and ASCs. PSC-derived organoids are established by directed differentiation of either ESCs or iPSCs (generated from fibroblast of skin biopsies) towards the endoderm. ASCs of epithelial origin can be isolated from tissue biopsies or surgical resections. They can be embedded in ECM and grown from either single cells or crypts that give rise to organoids that contain crypt-like structures. Adaption of the culture medium can foster the development of spheroids that are enriched for stem cells.222 Depending on the culture condition both PSCs and ASCs can give rise to three-dimensional organ-like structures of the hepatic-biliary-pancreatic tract and digestive tract containing gastric, small intestinal and colonic organoids. Representative pictures of organoid cultures are derived from the authors (unpublished, scale bars: 50 µm). ASCs, adult stem cells; ESCs, embryonic stem cells; ECM, extracellular matrices; PSCs, pluripotent stem cells.
In 2017, organoids were thus named ‘Method of the year’ by Nature (Organoids. Nat Methods 2017), demonstrating their enormous potential across all disciplines. Today, 3D organ cultures represent a central experimental tool among clinicians as well as medical scientists in the field of GI research, with the number of publications including organoids constantly increasing. In this review, we discuss GI organoids from experimental models to clinical translation with potential applications including state-of-the-art and current advances in the field, such as novel complex culture systems, and finally evaluate current challenges that have to be overcome.
GI organoids: a collection of different mucosal epithelial cell cluster
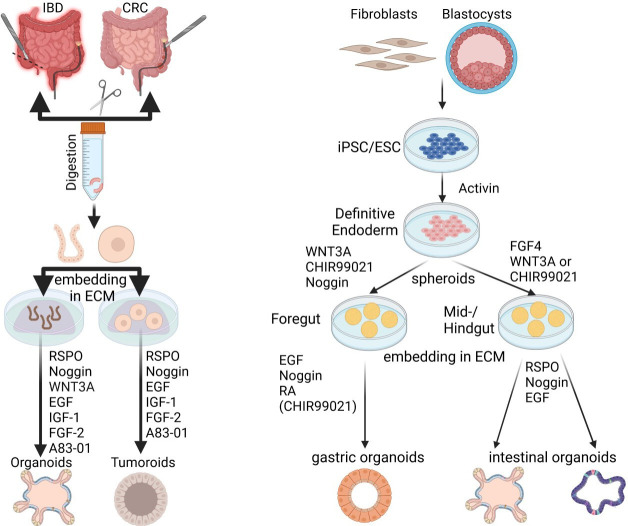
ASCs are the initial source for all primary GI organoid cultures. These stem cells are tissue resident and retain features of their site of origin in the GI tract as well as any genetic or epigenetic mutations that occur during their residence in the host. These stem cells can be isolated from hosts of any age including foetal stem cells.12–14 In contrast PSCs are either of embryonic origin (embryonic stem cell (ESC)) or are reprogrammed somatic cells to induce PSCs (iPSCs) (figure 3). To produce organoids from PSCs, developmental programmes of choice must be induced that in time will be permissive for differentiation into tissue-specific organoids. ASCs and PSCs, both display advantages and disadvantages depending on the scientific question as well as their preclinical/clinical application range. Beyond all doubt, the rapid rate and the genetic as well as epigenetic stability reflect the major advantages of ASC, their susceptibility to genetic manipulation, cellular composition and availability for organs without direct access to ASCs clearly highlight the benefits of PSC (box 2). Key limitations include the lack of cellular components of the microenvironment particularly in ASC-derived organoids, their heterogeneity driven by various protocols particularly for differentiation of PSC. Accordingly, researchers should consider this before starting to work with organoids.
Figure 3.
Overview of organoid derivation. GI organoids can be either derived from ASCs (left panel) or PSCs (right panel). Left panel: tissue-resident stem cells can be isolated from GI biopsies collected during colonoscopy, gastroscopy or surgery. Following an ethylenediaminetetraacetic acid (EDTA) or enzymatic digestion, either single cells or tissue fragments (crypt structures or ductal fragments) are embedded into an ECM and covered with a defined culture medium supplemented with a combination of different mitogens, morphogens and cytokines that guide the self-renewal and organised differentiation of somatic stem cells. Beside ASCs, organoids can be generated from cancer stem cells (isolated from tumour biopsies) and are referred to as tumouroids. Right panel: GI organoids can be also derived from PSCs (ESCs and iPSCs) in a stepwise differentiation process that recapitulates the signalling programme active during development. In an initial step, PSCs follow a guided differentiation towards an endodermal fate by exposure to Activin A. This is followed by the differentiation towards foregut, midgut/hindgut endoderm depending on the targeted tissue (foregut for gastric, hepatic, biliary or pancreatic organoids; midgut/hindgut for intestinal organoids). These precursor spheroids can be further instructed towards defined tissues and organs depending on different growth factors that allow proper terminal differentiation. ASCs, adult stem cells; CRC, colorectal cancer; ECM, extracellular matrices; ESCs, embryonic stem cells; IBD, inflammatory bowel disease; iPSCs, induced pluripotent stem cells.
Box 2. GI organoids as model system for human biology and medicine:
Key information and available techniques
Key factors
ASC-derived organoids: can be derived from endoscopic biopsies, surgical resection (gastric,223 jejunal,198 ileal,224 colonic,224 biliary37 186 225 and pancreatic186 226 organoids).
iPSC-derived organoids (can be derived from any somatic cell for example fibroblasts or blood cells) including patients with familial or complex genetic diseases on ethics consent. They can be differentiated towards intestinal,39 pancreatic duct-like,227 liver33 228 and gastric223 organoids).
ESC-derived organoids are available, if the country-specific regulations are followed (eg, Wicell Internations Stem Cell Bank).
ECM hydrogels: basement membrane secreted by Engelbreth-Holm-Swarm mouse sarcoma cells (various trade names) or other biological/synthetic matrices.
Culture medium: basal medium + growth factors (recombinant or cell lines (available for R-Spondin (family members 1–3), Wnt-3a and Noggin), alternatively ready to use products are available.
Translational benefits
Genetic and epigenetic signatures of derived tissue are stable in organoids (ASCs).28–32
Easy access and rapid set up of cultures (ASC).
Robustness: once established, expansion and long-term culturing allows large-scale genomic screening and drug screening.229–232
Several established cultures are available in biobanks (PSC/ASC).
ASC/PSC can be genetically modified using genome editing (eg, CRISPR/Cas9).37 186 233–240
Precision medicine (ASC, PSC).
Available techniques
Apical-out (polarity inversion, allows access to the apical but not basal site).114 241
Planar cultures: monolayer, cells adherent to plate (easy to perform, allows access to the apical but not basal site),241–243 air–liquid Interface (epithelial injury responses, host–microbe interaction, allows access to both, the apical and basal site).121 241 243
Microinjection (technique equipment is needed, host–microbe interaction).118 120 144
Gut-on-a-chip (more complex, comprise several specialised cell types, enable integration of mechano-physiological parameters and anaerobic bacteria).134 244–247
Orthotopic organoid transplantation.234 248–250
Microbe-organoid cocultures.97 118 242
Organoid-immune cell cocultures.5 48 49 251 252
Innervated organoids.63 253
Vascularised organoids.71 74
Organoids skewed to specific differentiation (enteroendocrine cells,254 goblet cells,255 wound associated epithelial cells,256 microfold cells257).
In the GI tract, epithelial ASCs are multipotent cells, modified during development to continuously and perpetually produce differentiated cells needed for the function of a particular location of the gut. The stem cells are a mixed population of active (ie, proliferative) and less-active cells that are interdependent and can interconvert.15 These stem cells are housed in specialised locations: at the base of intestinal crypts, the middle portion of gastric glands and in as yet undefined locations of the biliary tract as examples.16–19 Depending on the tissue origin and modifications of the defined culture medium, ASCs are capable of differentiating into both two-dimensional (2D) and 3D structures that contain polarised, specialised epithelial cell types highly similar to their counterparts from the tissue of isolation. Initial high-resolution single-cell transcriptomics of mouse intestinal organoids supported the claim that organoids reflect the in vivo epithelium and include less abundant cell populations such as enteroendocrine cells.20–23 Early studies using patient-derived organoids showed evidence of retention of their epithelial characteristics in vivo and these features appear to be stable over time.7 24 25 A more recent study supported this by demonstrating regional differences in the human biliary epithelium comparing intrahepatic, gallbladder, common bile duct, pancreatic duct sources; these regional differences were maintained in corresponding in vitro-derived organoids.26 This feature is similar to ASC-derived organoids along the intestine.27 Moreover, ASC-derived organoids can preserve epigenetic modifications associated with exposure to inflammation in vitro. Several studies have demonstrated that organoids established from the inflamed mucosal tissue of patients with IBD preserved differences in gene expression and DNA methylation patterns compared with organoids isolated from non-IBD subjects.28–30 Moreover genome-wide DNA methylation profiling of intestinal epithelial organoids derived either from foetal, paediatric or adult small and large bowel showed stable gut segment-specific profiles that closely mimicked the profiles from the corresponding primary gut epithelial source.31 In a similar fashion, promoter hyper-methylation patterns were observed in gastric intestinal metaplasia organoids similar to those observed in vivo.32 Going forward, these organoid systems will allow for functionally testing such genetic/epigenetic patterns and their impact on biology. In addition the use of intrapatient control organoids will be a powerful method to develop personalised medicine approaches for a variety of diseases.
Pluripotent stem cells (PSCs) include two different types, ESCs and iPSCs. Both are characterised by their plasticity and by their ability to self-renew. In general, PSC-derived organoid models require a complex, time-consuming and highly specific stepwise developmental protocol which starts with a guided differentiation towards endoderm. This is followed by the differentiation towards foregut, midgut or hindgut endoderm, which can be further instructed towards defined tissues and organs depending on the addition of permissive growth factors. The addition of defined mitogens, morphogens and cytokines at a precise dose and time point is absolutely crucial, not only to successfully generate 3D structures but also to strongly influence tissue specificity of the developing organoids33–39. Human PSCs are particularly interesting for modelling liver diseases that involve hepatocytes as well as MSCs. For example, Ouchi et al developed a reproducible method to generate multicellular human liver organoids including hepatocyte-like cells, Kupffer-like cells and stellate-like cells.40 These organoids were able to recapitulate progressive features of steatohepatitis, including steatosis, inflammation and fibrosis.40 Similarly patient-derived iPSCs cultures on a microwell chip successfully differentiated into the pancreatic ductal compartment including several mature duct-like cell populations and a few non-ductal cell types.41 This potential of PSCs to differentiate towards all lineages within a tissue is a major advantage of this system. However there remain challenges with PSC-derived organoids. They possess a limited application for clinical use due to genomic instability caused by the exposure of factors that induce reprogramming as well as a rather premature phenotype. However, despite this, a recent study identified that iPSC-derived human intestinal organoids display ASC properties similar to those of cultures obtained from human biopsies.42 Another study demonstrated that iPSCs-derived intestinal organoids derived from patients with ulcerative colitis recapitulate features of colitic activity.43 This is rather surprising given that these fibroblasts used to produce iPSCs undergo reprogramming which must affect the epigenetic makeup of these cells. The combination of human iPSC technology with recent developments in gene editing makes iPSC-based platforms a powerful novel tool for precision medicine.44 45
Multicellular organoids: modelling the tissue microenvironment
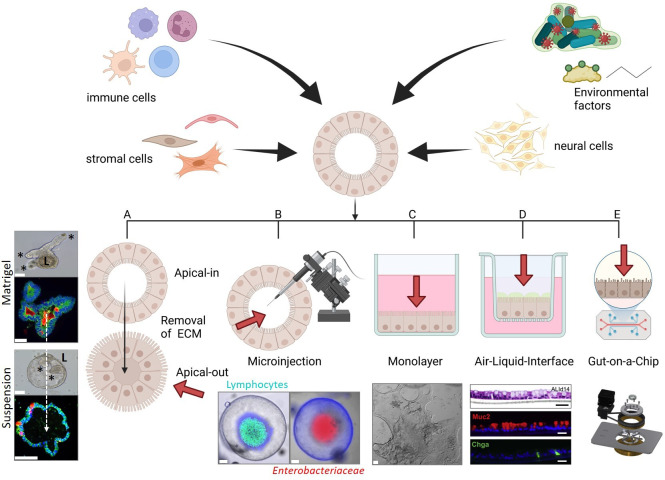
Epithelial surfaces of the GI mucosa are highly dynamic structures that process bidirectional signals from the lumen (including the microbiome and ingested substances) and underlying lamina propria (including the enteric nervous, immune and vascular systems). This bidirectional communication has profound effects on organ physiology and is involved, but not limited to the pathophysiology of intestinal and hepatic disorders. Chronic gut and liver diseases are typically multifactorial, including a dysfunction or dysregulated communication between different cell compartments, in addition to the gut microbiome.5 46 Thus the challenge for GI organoid models is incorporating these other factors in a fashion that will reveal new biology or new insights into a particular disease. For ASCs, organoids completely lack the in vivo microenvironment and PSCs contain MSCs but not luminal factors. However, key communication occurs between epithelial cells and fibroblasts (and their related lineages), a wide variety of innate and adaptive immune cells as well as neural-system, glial-system and vascular-/lymph-system cells. Moreover, exposure to gut microbiota-derived products and metabolites are absent in these ex vivo organoids. Accordingly, the field is beginning to make initial attempts to integrate these factors and cell types in PSC/ASC-derived epithelial organoids thus increasing the complexity of these cultures (figure 4). Currently iPSC-derived organoids allow addition of stromal cell types by using dedicated differentiation protocols and therefore enable the investigation of more complex relationships.47 We caution that this area remains a work in progress. For now, modelling binary interactions will be informative in many cases. Increased single cell analysis technical capacities (flow sorting for labelled cells for marker, protein and transcriptome) will be critical for proper analysis. We expect these experimental systems to increase in sophistication and the data from them will lead to new hypotheses that can be further tested using in vivo systems.
Figure 4.
Organoid coculture applications to increase the complexity of GI organoids, cellular and microbial components can be integrated into organoid cultures. This can be achieved by several protocols. (A) Reversal of the polarity in combination with removal of the ECM results in organoid cultures in which the apical/luminal surface faces towards the medium (Apical-out). (B) 3D-derived submerged monolayers can be grown as a thin monolayer to study the interaction of epithelial cells with environmental factors. (C) This monolayer technique has been expanded to air–liquid interface cultures in which monolayers are plated on transwells. This allows the investigation of the tissue microenvironment by adding defined cell populations (eg, immune cells, fibroblasts, endothelial cells) to the basal side of the epithelium and environmental factors to the apical surface. (D) Microinjections of luminal factors (eg, bacteria, viruses, dietary factors, metabolites) allow the investigation of the effect of environmental elements on epithelial homeostasis without changing architecture or polarity of organoids. (E) The organoid-on-a-chip concept is a more complex culture method in which self-organisation of stem cells can be extrinsically guided by a 3D microstructured scaffold. The advantage of this method is to integrate several cell types and mechano-physiological parameters. Asterisks indicate crypt region; L: lumen; red arrows indicate luminal side; air–liquid interface (C): pictures are derived from Wang et al., Cell 2019,121 Other representative pictures of organoid cultures are derived from the authors (unpublished, scale bars: 50 µm). 3D, three-dimensional; ECM, extracellular matrices.
Immune-epithelial cross-talk is an essential key factor for organ functionality, since a direct and functional interaction between these two cell compartments exists in vivo. This interaction is important for maturation of the adaptive immune system and activation of immune cells, while signals from the haematopoietic compartment influence epithelial function and turnover, particularly during the response to injury and infection. To identify novel therapeutic targets and to better understand the function of existing therapies directed against inflammation, we require a more detailed understanding of the molecular and cellular interactions of this interplay and their pathophysiological consequences when this communication goes awry. Several protocols have been established to coculture organoids with various types of immune cells such as T lymphocytes, innate lymphoid cells, macrophages, neutrophils, dendritic cells and intraepithelial lymphocytes.5 13 48–56 For translational approaches, researchers depend on autologous immune cells, which partially limits these cocultures as organoids require a certain time until cultures are established and expanded for experiments or preclinical validation. Cryopreservation of peripheral blood lymphocytes and further isolation and expansion of defined cell lineages such as T cells or macrophages is a widely used solution to overcome these limitations. While most protocols include 3D cocultures, additional improved culture methods including organoid-derived monolayer (taking advantage of an air–liquid interface53 57) have been developed. These monolayer cultures contain a luminal as well as basal side that is normally exposed to cells of the immune cell compartment. Thus specific scientific questions such as studying host responses to enteric pathogens or immune-epithelial communication ithelial communication can be addressed.57
Importantly, all cell types require specific culture media containing defined growth factors, which makes it often a challenge to find a combination of factors that allows survival and functionality of all populations in one experimental compartment. We expect these technical hurdles to be overcome and for a new wave of information to be generated.
Innervated organoids (epithelial–neural interaction)
The enteric nervous system (ENS) strongly influences mucosal immunity and epithelial function and is currently suggested as an important contributor to IBD development and progression.58 59 Moreover, disruption of ENS function may have bidirectional consequences for the gut and the central nervous system (CNS) as various neurotransmitters are released from ENS neurons that can also act on the intestinal epithelium (gut–brain axis).60 Although intestinal inflammation leads to ENS-associated alterations in function (altered peristalsis, abdominal pain, cramping and diarrhoea), the functional significance of the ENS for chronic intestinal inflammatory disorders and the mechanism for its impact on the intestinal epithelium remains poorly understood. The emerging field of epithelial–neural communication and its implication in GI disorders also fostered the development of coculture models including intestinal organoids and cells of the ENS.61 This has led to new insights into the innervation of enterochromaffin cells by sensory neurons in the ENS.62 Current models have also utilised the PSC system that can be differentiated towards intestinal organoids, as well as cells of the ENS such as neural crest cells (NCCs).63–65 NCCs that were cocultured with developing intestinal organoids migrated into the mesenchyme, differentiated into neurons and glial cells and showed neuronal activity.63 ENS-containing intestinal organoids formed neuroglial structures similar to a myenteric and submucosal plexus in vivo. Similarly, another study developed an organoid assembly approach including enteric neuroglial, mesenchymal and epithelial precursors, all differentiated from iPSCs.66 These organoids could be developed into a new section of intestine that showed innervation.63 iPSCs have also been used to successfully generate gastric tissue containing differentiated glands.66 These structures were surrounded by layers of smooth muscle composed of functional enteric neurons.
Vascularised epithelial organoids
The vascular endothelium also represents an important barrier in the GI system and varies in its permeability depending on the organ. It is organised in vascular units formed of endothelial cells, pericytes and enteric glial cells and regulates blood supply, nutrient transport, tissue fluid homeostasis and immune cell transmigration.67–69 Previous studies showed a pathophysiological role of the vasculature and lymphatics in the context of IBD.70 Moreover, transplantation of in vitro generated organoids is a promising approach towards regenerative medicine but requires blood supply within the organoid in order to replace functional and vascularised organs. Accordingly, in 2013, the group of Hideki Taniguchi addressed this emerging field and demonstrated the generation of a vascularised and functional human liver derived from iPSC-derived organ bud transplants.71 They uncovered that specific iPSC-derived liver precursors with a hepatic cell fate, self-organised into 3D organoids, by recapitulating organogenetic interactions between endogenous vascular endothelial cells (human umbilical vein endothelial cells; HUVECs) and MSCs. Interestingly, the vasculature within the liver organoids became functional after transplantation into mice. This proof-of-concept study demonstrated that organ-bud transplantation provides a promising new approach to study regenerative medicine. Using a similar approach, vascularisation of pancreatic organoids not only promoted functionalisation in culture but also substantially improved tissue survival after transplantation.72 In this study, the authors combined human isolated islets with HUVECs and MSCs. This resulted in a complex organoid culture fostered by ‘self-condensation’. In summary, these studies are particularly important in the context of therapeutic tissue transplantation as they clearly demonstrated that vascularisation prior to engraftment offers a promising strategy of enhancing efficacy. The opportunity to generate blood vessel organoids from iPSCs allows to efficiently combine iPSC-derived cell lineages towards vascularised organoids opening new avenues to work in an autologous setting.73 Indeed, several attempts have been made to introduce iPSC-derived endothelial cells into GI organoids.65 74 75 Interestingly, a recent study uncovered a population of endothelial cells which are present early in iPSC differentiation towards intestinal organoids that declines over time in culture. This study further developed a method to expand and maintain this endothelial cell popoulation within intestinal organoids as an alternative approach for classical vascularised organoid models.76
Organoids and the mesenchymal compartment
Intestinal stem and progenitor cells form within intestinal organoids, replicate and differentiate into distinct compartments, influenced by Wnt, Bmp and other cues, largely provided by MSCs in the lamina propria (canonical Wnts are produced by Paneth cells).77 78 Accordingly, central factors for ASC-derived mouse intestinal organoid culture medium include Wnt-3a and R-Spondin3, which are both essential for canonical Wnt signalling, as well as Noggin, a BMP agonist.79 Recently, some of the cellular sources of these essential signals in the mesenchyme were determined. Classical subepithelial myofibroblasts (SEMFs), also known as telocytes, were found to be an important sources of Wnt and R-Spondin ligands as well as diffusible BMP agonists/antagonists.80 81 Accordingly, cocultures of ASC-organoids with SEMFs permitted long-term culture of organoids in the absence of growth factors that normally are essential to ensure survival of organoids.82–85 Using coculture experiments, another group phenotypically and functionally examined fibroblast populations in the colon and found that CD90+ fibroblasts located in close proximity to the stem cells in vivo expressed crucial stem cell growth factors, such as Grem1, Wnt-2b and R-Spondin3.86 A recent study showed that fibroblasts could secrete extracellular vesicles with Wnt and EGF, thereby rescuing Wnt-deficient or Egf-deficient organoid growth.87 Going forward, as populations of fibroblasts and myofibroblasts are identified, their role in providing essential factors for intestinal epithelial stem cells can be in part assessed by coculture with ASC organoids.
Cancer-associated fibroblasts (CAFs) are mainly derived from normal fibroblasts and play a key role in the progression of cancer. However mechanistic understanding of cancer cell–CAF interactions are limited due to the lack of preclinical cellular models. Recently, an interesting study demonstrated the coculturing of primary liver tumour-derived organoids with CAFs.88 They found bidirectional signals via cell–cell contact and local secreted factors: CAFs promoted tumour organoid growth and cancer cells regulated CAF physiology. They further demonstrated that the response of tumour organoids to anticancer drugs was strongly dependent on CAFs. Similarly, a study by Öhlund et al established cocultures of human pancreatic ductal adenocarcinoma (PDAC) organoids and pancreatic stellate cells (PaSCs), myofibroblast-like cells that are located in exocrine regions of the pancreas. The authors recapitulated the desmoplastic reaction of PDAC with PaSCs converting from a resting quiescent state to activated, stroma-producing fibroblasts.89 They further demonstrated that PaSCs secrete factors that support growth of human PDAC organoids.89 90 Wnt was identified as the key element released by PaSCs that is essential to drive organoid growth in Wnt-non-producing PDAC subtypes. Using the same approach another study uncovered TGFβ and IL-1 as tumour-secreted ligands that promote myofibroblast heterogeneity.91 One step further Tsai et al established multicell type organotypic coculture models of the tumour microenvironment by coculturing human pancreatic cancer organoids with matching stromal and immune cells.52 Activation of CAFs and tumour-dependent lymphocyte infiltration were observed in these models. This 3D tumour microenvironment model system may be valuable for the assessment of immunotherapeutics such as checkpoint inhibitors in the context of T-cell infiltration.
The microbe–host interface
A highly important component of the mucosal microenvironment is the gut microbiota which can communicate with epithelial cells directly in the intestine (by direct attachment or infection), or indirectly with the gut and liver by secreting metabolites as well as proteins and nucleic acids contained in bacterial membrane vesicles.92–96 Accordingly, gut microbes as well as their derived factors have been implicated in human health and disease by regulating innate and adaptive immunity, metabolism and tissue homeostasis.
There are several excellent examples of direct infection of intestinal organoids which created essential models to determine the mechanism of infection and to develop new therapies. These examples include previously non-culturable viruses in the gut (ie, norovirus) but also formed a system to produce the entire life cycle of a parasite that could only be expanded experimentally in bovine calves.97 98 Both ASC-derived and iPSC-derived organoids have been cocultured with various commensal and pathogenic bacteria,99 parasites,100 as well as viruses including SARS-CoV-2.101–103 Host–microbe communication was analysed via several approaches including microinjection of microorganisms into the lumen of organoids, by taking advantage of monolayer or inverted organoids to mimic the physiological site of interaction. In 2020, Hans Clevers demonstrated how organoids can be used to better understand the role of the intestinal microbiota in colorectal cancer (CRC).104 By exposing human intestinal organoids to genotoxic pks+ Escherichia coli through repeated luminal injection over 5 months, they identified a distinct mutational signature that was also present in CRC. The organoid data generated within this study suggest that the underlying mutational process results directly from past exposure of the gut epithelium to bacteria carrying the pks pathogenicity island. Beside bacteria, viruses entering the host via the GI tract have been implicated in intestinal and extraintestinal inflammation. In this line, it is now well established that the GI route plays an essential role for SARS-CoV-2 infection. A study by Zhou et al established, for the first time, bat intestinal organoids to better understand the infection and transmission route of SARS-CoV-2.102 They could demonstrate that SARS-CoV-2 could efficiently infect not only bat organoids but also human intestinal organoids. Moreover, they could observe a robust SARS-CoV-2 replication in human intestinal organoids, demonstrating that the human intestinal tract might be a transmission route of SARS-CoV-2. In line with these results another study demonstrated efficient infection of foetal, paediatric and adult gastric organoids with SARS-CoV-2 further supporting faecal–oral transmission of this novel virus.105 These are only exemplified recent studies to visualise the power of microbe-organoid cocultures to better understand how gut microorganisms can cause intestinal inflammation. The broad spectrum on these cocultures would go beyond the scope of this review but is in depth described in specialised reviews.106
Fecal metabolite screens generated using untargeted metabolomics of intestinal contents from humans and in vivo model systems have been extensively performed and GI organoids do define cell types that can respond to these microbial products. A few recent examples (of many papers) include that succinate promotes tuff cell differentiation, dipeptide aldehydes from Clostridium sporogenes that can enhance endoplasmic reticulum stress, and secondary bile acids that affect intestinal epithelial stem cells to stimulate regeneration or negatively impact Paneth cell function.107–110 These screens have the potential to uncover many new avenues of basic biology. The challenge will be to determine which effects will translate to whole organisms in vivo.
Towards next generation of intestinal organoids
Epithelial organoids provide a highly attractive platform to study primary epithelial cells in settings that closely resemble the in vivo situation. Based on their experimental success and widespread distribution across the different preclinical and clinical disciplines over the last years, several novel culture systems have been developed. Accordingly, organoids may be integrated into microfluidic devices (organ-on-a-chip systems), microengineered systems that not only allow a more complex but also fluidic culture system. To study the interaction with environmental factors as well as the underlying mucosal immune system, several attempts have been made to change the architecture of these organoids, such as changing their morphology (inside-out) or maintaining them as planar 2D cultures.
Epithelial polarity and its downside: two sides of the same coin
The advantage of organoids to form normal apical junctional complexes and maintain this epithelial polarity also creates a disadvantage when it comes to specific scientific questions. Organoids embedded in Matrigel display an apical (luminal) surface facing inward enclosed within the organoids and a basolateral surface facing the outside of the organoid interacting with the extracellular matrix. Thus, factors that are added to the culture medium predominantly interact with the basolateral surface. However, many applications require access to the apical/luminal surface, as this represents the mucosal surface that is constantly exposed to environmental factors such as dietary nutrients, bacterial metabolites and intestinal microbes. This might be particularly relevant for nutritional studies, to investigate the sites of invasion and cell tropism for viral and bacterial pathogens and to conduct drug screening and toxicity assays.102 111–113 Several attempts have been made to face this problem including methods to reverse the polarity or to gain access to the luminal side (figure 4, box 2). Co et al developed a protocol (organoid polarity reversal method) to control organoid polarity in suspension culture, resulting in an organoid population in which the apical surface faces outwards to the medium (apical-out organoids).114 Apical-out organoids (figure 4A) can be used to better recapitulate host–microbe interaction (eg, infection with SARS-CoV-2 or effects of probiotics), but might be also useful for evaluating therapeutics as many therapeutics are taken orally and thus have to enter the human body via the apical surface of the epithelium.115–117 Besides changing the polarity, microinjections of microbes, microbial metabolites or other environmental factors into the lumen of organoids is a widely used experimental setting104 118 119 (figure 4B). In this experimental setting, organoids maintain a cystic morphology which allows bacterial species with low oxygen tolerance to survive but at the same time prevent bacterial overgrowth. While microinjection of microbes into the lumen of gastric/intestinal organoids allows the investigation of host-microbe interaction under the most physiological conditions, standardisation and scalability of this protocol is difficult to achieve. A recent work by Williamson et al developed an automated high-throughput computer-driven microinjection system for cargo delivery to the organoid lumen and high-content sampling.120 While this clearly represents a next-generation in vitro approach to investigate GI luminal physiology, it also requires complex technical equipment which is not widely available. Accordingly, several recent attempts have been made to investigate apical responses by generating 3D-derived monolayer grown initially on a thin Matrigel layer97 118 121 (figure 4C). These methods has been improved by growing monolayers on transwells to generate air–liquid interfaces (ALI; figure 4D). Exposure of the apical surface of 2D monolayer to air–liquid interface allows the establishment of long-term, self-organising 2D epithelial monolayers with highly differentiated epithelial cells121. A huge advantage of these monolayer protocols is the homogenous exposure of epithelial cells to environmental factors as well as an enhanced scalability. Of note, beside host–microbial communication, this model can re-enact the ‘homeostasis-injury-regeneration’ cycles of epithelial alterations that occur in vivo and thus is also feasible to investigate wound healing and mucosal healing.121 Additional alternatives to successfully address the problem of lumen accessibility are more complex and require extensive technical expertise and mainly include contribution of other disciplines.
Engineering organoids
Microengineering and tissue engineering is currently a highly fetching field to overcome key limitations of exciting organoid techniques and are associated with increased reproducibility, experimental control and higher throughput screens.122–126 These modifications will be of high value for clinical translation studies. Engineered biomimetic platforms will allow investigators to mimic features of in vivo cells that are not accessible by the established static culture methods. Engineering techniques include micromanufacturing, bioprinting and laser cutting. The insertion of artificial boundaries by microengineered scaffolds, may represent a substantial advantage over classical organoids cultures as it is well known that shape-guided morphogenesis is an essential step during development.127–129 In this case several attempts have been made to use fabricated stamps to pattern synthetic matrices or hydrogels resulting in highly organised tubular backbones onto which organoid-derived cells can be seeded.123 127 130 131
Importantly, these model systems allow exposure of the apical surface while at the same time remain a 3D structure and thus represent a highly attractive model for studying host–microbe interaction. Additional upcoming technique to control cell assembly and to overcome their stochastic development includes bioprinting methods that are developed to pre-establish their compatibility with microfluidic devices, often referred to as organ-on-chip132 (figure 4E). These microfluidic cell devices contain perfused chambers in which living cells are arranged in such a manner that they reflect tissue-level and organ-level physiology.133 Initially, they were developed to mimic gradients of signalling molecules but subsequently they have been applied to better control cell fate patterning during organoid development. Accordingly, when tissue engineered intestines are connected to an external pumping system, the resulting microfluidic mini-guts allow the removal of dead cells comparable to the in vivo situation. This is associated with prolonged survival by several weeks, and also enables the interaction with microorganisms for modelling host–microorganism interactions without microbial overgrowth.130 134 It may be possible, using control of the upper chamber oxygen concentration, to flow through anaerobic bacteria. We anticipate that intestines-on-chip will facilitate many preclinical and clinical studies in the future with regard to reproducibility, experimental control and high throughput read outs.
GI organoids for disease modelling, drug discovery and personalised medicine
As mentioned above, the organoid technologies, including the classical organoid models and recent improvements, hold great promise for use in medical research and the development of new treatments (figure 5). Within the last 10 years, a wide-range of organoid-based disease models have been developed. This includes models to reproduce inflammatory and metabolic diseases,135–137 infection and cancer.1 3 5 138–140
Figure 5.
Applications of organoids from patients can be used for basic research to better understand organ development and to dissect the relevant intracellular pathways that control homeostasis and might be altered during organ dysfunction. These patient-derived organoids can be further used to preclinically screen for novel therapies and to predict response to drugs as well as for regenerative medicine (precision medicine). Another preclinical application includes disease modelling to better understand the mechanisms underlying GI diseases (including inflammatory, infectious, metabolic, inheritable genetic and neoplastic disorders) as well as the biobanking of patient-derived-organoids and -tumouroids.
Disease modelling and drug discovery
In 2020, the group of Toshiro Sato utilised organoids to analyse somatic mutations in the ulcerative colitis epithelium to decipher how a chronic inflammatory microenvironment shapes the genetic landscape.141 They generated 76 clonal human colon organoids including 55 cultures derived from patients with ulcerative colitis, 26 of whom had colitis-associated neoplasia, and 16 non-IBD individuals. These organoid cohorts were subjected to whole-genome sequencing to capture mutations at the clonal level. They identified a unique pattern of somatic mutagenesis in the inflamed epithelium, including somatic mutations in multiple genes that are related to IL-17 signalling. The results of this study were supported by another study published in the same year, also in Nature,142 demonstrating that organoids are reliable disease models for genomic and functional studies of human GI diseases. The group of Gerald Schwank performed pooled-library CRISPR screens by capturing sgRNA integrations in single wild-type and APC (adenomatous-polyposis-coli)-mutant human intestinal organoids.143 By optimised conditions that enabled genome-wide CRISPR screening in organoids, they identified and validated tumour suppressor genes that mediate transforming growth factor beta (TGFβ) resistance. This proof-of-principle study highlighted the power of this system that can be applied to a variety of assays including different organoid types to facilitate biological discovery in primary 3D tissue models. Gastric organoids derived from human or mouse antral or corpus glands or PSCs have been established to not only shed light on processes involved in gastric development but also to study the pathogenesis of several human gastric disorders. Helicobacter pylori infection is associated with a 10%–20% increased risk for developing peptic ulcer diseases and a 1%–2% risk for gastric cancer. In contrast to previous in vitro culture systems, gastric organoids now allow to study cell type specific responses, including gastric epithelial cells such as mucus gland cells or parietal cells, that could not be cultured by classical protocols.144 145 This allowed for the first time to discriminate and compare gene signatures of individual cell populations following Helicobacter pylori infection. For example, Sina Bartfeld could demonstrate that human gastric organoids composed of mainly cell lineages of the gland, produce higher levels of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) target genes compared with organoids that contain predominantly cell lineages located in the pit.145 Beside this initial work by the Clevers lab, several additional studies could demonstrate that characteristic hallmarks of H. pylori infection such as attachment, colonisation, NF-κB activation, CagA-c–MET interaction and increased host cell proliferation can be recapitulated in gastric organoids.38 146–148 Another recent study by Lo and colleagues used CRISPR-based genome engineering to investigate how common mutations, with so far unknown oncogenic consequences, affects human cells.149 By taking advantage of CRISPR/Cas9-mediated gene knockout, they could demonstrate that lack of ARID1A, which ranks among the most common molecular aberrations in human cancer, is not tolerated in human wild-type gastric organoids, but in the context of TP53 loss. This study showed in a very clever experimental approach the power of organoids to better decipher the function of unknown genetic mutations. Organoids can be further used to unravel the molecular mechanisms underlying injury-associated epithelial regeneration.121 150 Organoid based studies not only helped to better capture injury-repair cycles, but further uncovered that small-molecule epigenetic modulators promote regeneration of the gut epithelium on damage in organoids comparable to in vivo. Another recent study identified that lack of epithelial expression of the gasdermin family member gasdermin B (GSDMB) in organoids is associated with disrupted epithelial restitution/repair.151 Gasdermins are mainly recognised for their key role in pyroptosis through their function during pore formation. The work by Rana et al now identified that in contrast to the other gasdermins, GSDMB, does not intrinsically engage in pyroptosis, but appears to plays a role in the restoration of epithelial barrier function and the resolution of inflammation during IBD. Beside these genetic studies, organoids could be used to gain insight into cell type specific responses following exposure to define therapeutic. As an example, preclinical ex vivo experiments uncovered that JAK inhibitors partially exert their anti-inflammatory function by inhibiting IFN-induced epithelial cell death.152 153 Beside this therapeutic approach, this study also demonstrated that corticosteroids enhance epithelial barrier function, which was supported by another translational study.152 154 Organoids can also be further used to screen for drugs targeting viral infection.155 For instance related to the current pandemic situation drugs for treating SARS-CoV-2 virus infections were recently pre clinically evaluated in PSC-derived intestinal organoids.156 Specifically, remdesivir effectively inhibited SARS-CoV-2 infection in organoids and restored organoid viability.157 Another recent study used lung and colon organoids and identified imatinib, mycophenolic acid and quinacrine dihydrochloride as efficient therapeutics to block entry of SARS-CoV-2.158 Beside preclinical evaluation and identification of drugs, there is increasing interest in using organoids as novel tools for preclinical testing of probiotics.159 160 Preincubation of organoids with the probiotic E. coli strain Nissle prevented toxicity of epithelial cells induced by pathogenic E. coli strains.160
Cancer research and precision medicine
Cancer is a major health problem and a leading cause of death worldwide. Novel therapeutic strategies such as targeted drugs offer improved opportunities to not only improve overall patient survival, but also offer new approaches for personalised cancer treatment. Precision medicine of course requires ex vivo model systems that can predict the response of individual patients. Cancer stem cells are a subpopulation of cells within tumours with capabilities of self-renewal, differentiation, and thus also can give rise to organoids, the so called patient-derived organoids (PDOs) or tumouroids. In 2015, Hans Clevers performed a proof-of-principle study using an organoid biobank of patients with CRC (including organoids derived from the tumour and from adjacent normal tissue) and screened for cancer treatments currently used in the clinic or in clinical trials.161 He could show that this technique is suitable to detect gene-drug associations that matched prior clinical observations and thus may fill the gap for personalised therapy design. In-depth OMIC analysis further demonstrated that PDOs not only recapitulated the somatic copy number alterations, but also the mutation spectra found in CRC including mutations in the typical suppressor genes as well as activating mutations. Personalised proteome profiles of healthy and tumour PDOs later confirmed these results on a post-translational level.162 One year later, the group of Toshiro Sato expanded this initial study and published a colorectal tumour organoid library including 55 different tumour organoid clones including rare subtypes and matched pairs of primary and metastatic organoids.163 He could demonstrate that niche-independent growth is predominantly associated with the adenoma-carcinoma transition. In a more recent study, authors established a biobank of PDOs from patients with metastatic GI cancer, who had previously been enrolled in phase I or II clinical trials.164 This allowed the authors to compare clinical responses of patients with organoid drug responses. Indeed, they could demonstrate examples of how drug responses of cancer organoids matched with those observed in the clinic, reinforcing their value as a platform for drug screening.165 Based on the success of these initial studies and scope of application in personalised medicine, several additional studies, focusing on various different tumour entities, soon followed. All studies impressively demonstrated that PDOs recapitulate the mutational landscape of their primary tumours and can be generated from the respective GI cancer entity including colorectal cancer,161 163 166–170 gastric cancer,32 164 171–175 oesophageal cancer,176–179 pancreatic cancer,10 90 180 181 gastroenteropancreatic neuroendocrine neoplasms,182 and liver cancer.183–185 Of particular interest for precision medicine, the molecular subtypes of cancers are maintained during PDO cultures, indicating that the culture conditions do not foster outgrowth of certain subtypes. Several clinical studies could further demonstrate that PDOs can be derived from needle biopsy, demonstrating that a few single cells are sufficient to allow extended ex vivo analyses.184 Finally, the opportunity to culture 3D multicellular structures from GI tissue, can be used to grow and compare side-by-side pre-cancerous and tumour cells from human patients.131 186
Previous studies have impressively demonstrated that PDOs derived from patient tumour material have not only extended our molecular understanding of human cancer but also hold great potential for personalised medicine.187 188 Accordingly the number of organoid biobanks is rapidly increasing including a variety of GI cancers (colorectal cancer, advanced rectal cancer, metastatic colorectal and oesophageal cancer, gastric cancer including oesophageal adenocarcinoma and oesophageal squamous cell carcinoma, PDAC, gastroenteropancreatic neuroendocrine neoplasms, and liver cancer including hepatocellular cancer, and cholangiocellular carcinoma).10 32 90 161 163–166 171 173 183 189–192 These biobanks include a detailed characterisation and molecular profiling of both the tissue of origin and the corresponding organoid line, a description of clinical phenotype and patient records. Since PDOs recapitulated patient- and disease-specific features, several drug-screening approaches on such biobanks have been reported.180 In a very recent study by the group of Gerald Schwank, they used PDOs from pancreatic cancer (PDAC) patients to demonstrate that organoids can be used for precision oncology, not only to allow drug screening, but also for the identification of novel candidates for drug repurposing via genome editing.180 In line with previous studies they observed a high diversity in PDOs phenotypes, which correlated strongly with in vivo morphology of primary PDAC tumours. To enable high-throughput compound screening in these organoid lines, they established a fully automated drug screening platform and compared different approaches that have been used in previous drug screening assays. Similar as previously observed they highlighted that Matrigel domes were not required to achieve drug sensitivity and finally used a Matrigel-supplemented media protocol. This liquid-overlay culture system is more scalable to screen a larger number of compounds and thus might be an easy associable and cost-effective alternative to classical Matrigel-dome cultures. With this protocol they conducted an automated drug-repurposing screen with 1,172 FDA-approved compounds and identified 26 compounds that effectively killed PDAC organoids, including 19 chemotherapy drugs currently approved for other cancer types. While this is only one example, several other studies have used PDO biobanks for drug screening approaches with slight variants in the experimental design.171 183 189
Regenerative medicine and organoid transplantation
Organoids are not only an emerging tool for preclinical drug screening, they further hold great potential as a source of transplantable tissues and organ therapy in regenerative medicine. Generation of autologous ex vivo tissue generated from patient-derived ASC or iPSCs offers advantages over conventional organ transplants, which are limited in availability and require immunosuppression. Recent advances in stem cell technology, including genetic engineering, now also offers the opportunity to transdifferentiate organoids into other cell types. For example, type 1 diabetes is an autoimmune disease characterised by destruction of insulin-producing pancreatic β-cells by the immune system and thus requires replacement of this cell type in particular. Interestingly, inhibition of the transcription factor Foxo1 in gut organoids promoted the generation of insulin-positive cells that express all markers of mature pancreatic β-cells, producing what could be a rich source of cells for transplantation studies and development.193 Similarly, creating complex structures in the liver, ultimately for transplantation, will require multiple epithelial cell types. One step here was the transdifferentiation of organoids containing hepatocytes in a cholangiocyte fate.14 These are only two GI-centric examples demonstrating the wide range of different organoid populations that can be used as organ/cell transplant. Already in 2012, ASC-derived colonic organoids were orthotopically transplanted into the mouse colon, where they were able to regenerate damaged epithelium.194 These transplanted donor cells formed functionally and histologically normal crypts that were able to self-renew. Similarly, foetal enterospheres contributed to epithelial regeneration in vivo.12 Later also human intestinal organoids were transplanted and repopulated the rectal epithelium. Interestingly, these cells retained self-renewal and multi differentiation capacities over months.195 In line with that, a study by the group of Toshiro Sato xenotransplanted human ileum organoids in the mouse colon and thus replaced the native colonic epithelium with small intestinal epithelial cells. As expected, based on previously developmental studies, these organoids maintained their regional identity and generated a functional small intestinalised colon.196 197 In a more translational approach, a proof-of-concept study constructed autologous jejunal mucosal grafts using biomaterials from paediatric patients.198 Seeding of small intestinal organoids onto decellularised human intestinal matrices (as biological scaffold) reliably reconstructed grafts. These engraftments displayed several aspects of physiological jejunal function and survived up to 2 weeks after transplantation into the kidney capsule. As an alternative, organoids were implanted into the mesentery.199
Beside intestinal organoids, several previous studies showed that human liver organoids functionally colonise the damaged mouse liver.7 14 A study by Sampaziotis and Vallier could further demonstrate that human bile duct organoids self-organise into bile duct-like tubes, expressing biliary markers, following transplantation under the kidney capsule of immunocompromised mice.200 When embedded within a collagen tube scaffold, they could stably bridge the surgical defect in the mouse biliary tract. In a translational approach, they further confirmed that these cholangiocyte organoids were able to repair bile ducts after transplantation in the human liver.201 In summary, these data provide proof-of-principle for the use of GI organoids for regenerative purposes, and offer a feasible strategy for treatment of various diseases such as short bowel syndrome196 or other intestinal failures.198
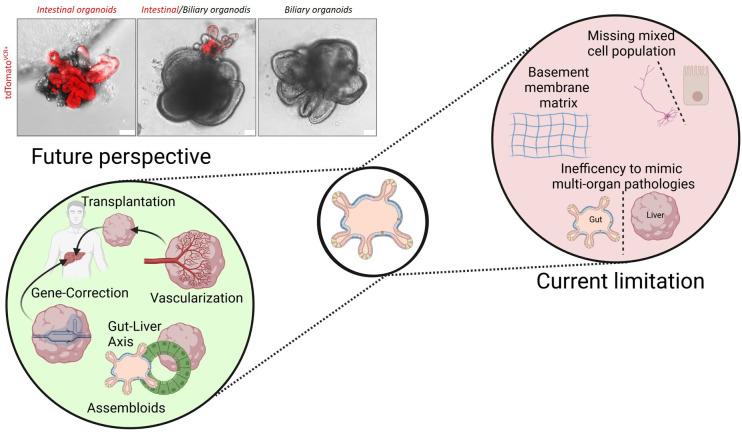
Current limitations and future direction
GI epithelial organoids and their related platforms have the capacity to mimic specific aspects of the three-dimensional architecture, cell type composition and functionality of different regions of the digestive tract, while at the same time maintaining the advantages of a simplified and easily accessible cell culture model. This makes organoids highly attractive for both basic tissue/organ biology, as well as disease modelling and clinical application. Despite the high level of enthusiasm and the seeming in exponential phase of growth for this field, organoids remain a potential revolutionising model system. Challenges remain with their future development (figure 6). One of the most important challenges, particularly for ASC-derived organoids, is the inability to easily model multiorgan pathologies. Organoids can impressively mimic key features of certain tissue or organ, but cannot replace the cellular diversity present in vivo and thus are unable to reflect the complexity of a human tissue organism. Accordingly, several attempts have been made to address these restrictions including coculturing and mixed cultures of iPSC.35 202 203 For example Takeishi et al biofabricated human livers using a mixture of iPSC-derived hepatocytes, biliary epithelial cells and vascular endothelial cells.204 They engineered human hepato-biliary-pancreatic organoids by fusing iPSC-derived anterior and posterior gut spheroids. This fostered the differentiation of the domains into segregated organs completed with developmentally relevant invagination and epithelial branching.33 35 These assembloids (organoids that have been generated by spatially organising multiple cell types) will potentially provide new insights into tissue function and will allow us to study the communication between the liver, pancreas and GI tract in a reductionist system. Again, engineering solutions here will be a critical component of these systems. The collaboration across diverse fields will likely be the key ingredient to overcome this hurdle. Another limitation is the undefined effect of the sometimes extremely complex extracellular matrices (ECM) that are purchased for organoid growth. The batch-to-batch variations as well as the differences between commercial available matrices need to be better controlled. Particularly for translational approaches it is unknown how and if these ECMs and variations in ECMs will influence preclinical drug or genetic screening and thus represent a major challenge for producing clinical-grade organoids. This is particularly relevant since the current gold standard for organoid culture is the basement membrane matrix (BMM) purified from Engelbreth-Holm-Swarm mouse sarcoma. Accordingly, diverse efforts have been made to replace this BMM for GI organoids. This includes natural ECMs derived from decellularised gut or liver tissue,205–207 hyaluronan elastin-like protein materials,208 collagen-nanocellulose hydrogel,209 alginate,210 as well as synthetic hydrogels for iPSC-derived organoids.211 For all these different systems, it will be extremely important to evaluate over time how they affect organoid cultures and to include ‘good manufacturing practice’ to allow translation into the clinic. Another source of variation is the media that is used for individual experiments. We have no method currently to rapidly and sensitively standardise the potency (ie, Wnt signalling) of media (either produced in the lab or purchased from a vendor) for each experiment. This lack of rigour will bring challenges in comparing experiments both within and from different labs. One can envision in the future, a rigorous approach similar to what was developed for cDNA microarrays to make them reproducible applied to the organoid system. Another interesting perspective for translation into the clinic includes gene-editing in iPSC. Tissue replacement using gene-corrected organoids would open a completely new avenue for precision medicine. Just recently, CRISPR-based adenine editors corrected nonsense mutations in a cystic fibrosis intestinal organoid biobank.190 Another study supported these results by demonstrating that restoration of a specific cystic fibrosis associated mutation in iPSCs restored intestinal organoid function.202 Widespread application of this technology is critical for the field to move forward.
Figure 6.
Current limitations and future perspective of GI organoids. Current limitations of GI organoids include their inability to model multiorgan pathologies and their limited cellular diversity. Assembloids (organoids derived from spatially organising multiple cell types) will partially address these limitations and allow us in the future to better understand cell-to-cell communication and organ-cross-talk. Finally the mechanistic impact of the EMC on organoids and the batch-to-batch variations are limiting factors for translational approaches. Several attempts have been made to replace the current ECM by natural or synthetic ECMs. Organoids were derived from the small intestine (intestinal organoids) or biliary system (biliary organoids) of Rosa26.tdTomatoxVillinCre reporter mice. Organoids were either cultures alone, or as assembloids (middle panel). Representative pictures of organoid cultures are derived from the authors (unpublished, scale bars: 50 µm). ECM, extracellular matrix.
Despite the remaining challenges, the rapid technical improvements in the field of human organoids will provide unprecedented avenues to improve human health. Within the past 10 years, organoids have evolved from basic 3D cell cluster systems with an expanded laboratory life cycle, to improved highly complex multidisciplinary translational model systems including recent advances such as forward genetic modelling, mechano-physiological stimulation by microfluidics as well as microengineered platforms to allow automated and continuous monitoring of physical and biological parameter. Thus, technologies and experimental protocols that were developed in other disciplines for different model systems can now be applied to human organoid systems, which will increase reproducibility, improve functional readouts and provide enhanced experimental control.
Acknowledgments
All figures were created with BioRender.com. We further thank Miriam Bittel, Heidrun Dorner, Iris Stolzer, Barbara Ruder and Laura Schickedanz for providing representative organoid pictures.
Footnotes
Twitter: @MIB_GuentherLab, @Winner_Lab@WinnerLab
Contributors: CG, TS, MFN and BW wrote the paper.
Funding: Support came from the Johannes und Frieda Marohn-Stiftung (CG, BW), the Bavarian Ministry of Science and the Arts in the framework of the ForInter network, the German Research Foundation, DFG WI 3567/2-1, 270949263/GRK2162 (BW), TRR241 (A02, CG), FOR 2886 (A02, CG), TRR305 (B08, CG) and DFG GU 1431/5-1. Further support was given by the Interdisciplinary Center for Clinical Research (IZKF) of the University Erlangen-Nürnberg (Jochen-Kalden funding program N5), 5U01 DK062413 U24 DK062429 R01 DK122790 R01 CA257523 (TS).
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1. Kim J, Koo B-K, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 2020;21:571–84. 10.1038/s41580-020-0259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kretzschmar K, Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev Cell 2016;38:590–600. 10.1016/j.devcel.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 3. Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet 2018;19:671–87. 10.1038/s41576-018-0051-9 [DOI] [PubMed] [Google Scholar]
- 4. Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut 2019;68:2228–37. 10.1136/gutjnl-2019-319256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol 2020;20:279–93. 10.1038/s41577-019-0248-y [DOI] [PubMed] [Google Scholar]
- 6. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125. 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- 7. Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299–312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huch M, Dorrell C, Boj SF, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013;494:247–50. 10.1038/nature11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lugli N, Kamileri I, Keogh A, et al. R-spondin 1 and noggin facilitate expansion of resident stem cells from non-damaged gallbladders. EMBO Rep 2016;17:769–79. 10.15252/embr.201642169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boj SF, Hwang C-I, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. 10.1016/j.cell.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. Embo J 2013;32:2708–21. 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fordham RP, Yui S, Hannan NRF, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 2013;13:734–44. 10.1016/j.stem.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schreurs RRCE, Baumdick ME, Sagebiel AF, et al. Human Fetal TNF-α-Cytokine-Producing CD4+ Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity 2019;50:462–76. 10.1016/j.immuni.2018.12.010 [DOI] [PubMed] [Google Scholar]
- 14. Hu H, Gehart H, Artegiani B, et al. Long-Term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018;175:1591–606. 10.1016/j.cell.2018.11.013 [DOI] [PubMed] [Google Scholar]
- 15. Takeda N, Jain R, LeBoeuf MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science 2011;334:1420–4. 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brittan M, Wright NA. Stem cell in gastrointestinal structure and neoplastic development. Gut 2004;53:899–910. 10.1136/gut.2003.025478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010;6:25–36. 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 18. van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol 2009;71:241–60. 10.1146/annurev.physiol.010908.163145 [DOI] [PubMed] [Google Scholar]
- 19. Manohar R, Komori J, Guzik L, et al. Identification and expansion of a unique stem cell population from adult mouse gallbladder. Hepatology 2011;54:1830–41. 10.1002/hep.24568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grün D, Lyubimova A, Kester L, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251–5. 10.1038/nature14966 [DOI] [PubMed] [Google Scholar]
- 21. Norkin M, Ordóñez-Morán P, Huelsken J. High-content, targeted RNA-seq screening in organoids for drug discovery in colorectal cancer. Cell Rep 2021;35:109026. 10.1016/j.celrep.2021.109026 [DOI] [PubMed] [Google Scholar]
- 22. Haber AL, Biton M, Rogel N, et al. A single-cell survey of the small intestinal epithelium. Nature 2017;551:333–9. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beumer J, Puschhof J, Bauzá-Martinez J, et al. High-Resolution mRNA and secretome atlas of human enteroendocrine cells. Cell 2020;181:1291–306. 10.1016/j.cell.2020.04.036 [DOI] [PubMed] [Google Scholar]
- 24. Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105–9. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011;141:1762–72. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 26. Rimland CA, Tilson SG, Morell CM, et al. Regional differences in human biliary tissues and corresponding in Vitro-Derived organoids. Hepatology 2021;73:247–67. 10.1002/hep.31252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015;64:911–20. 10.1136/gutjnl-2013-306651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut 2017;66:2069–79. 10.1136/gutjnl-2016-312609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howell KJ, Kraiczy J, Nayak KM, et al. DNA Methylation and Transcription Patterns in Intestinal Epithelial Cells From Pediatric Patients With Inflammatory Bowel Diseases Differentiate Disease Subtypes and Associate With Outcome. Gastroenterology 2018;154:585–98. 10.1053/j.gastro.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzuki K, Murano T, Shimizu H, et al. Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J Gastroenterol 2018;53:1035–47. 10.1007/s00535-018-1437-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kraiczy J, Nayak KM, Howell KJ, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut 2019;68:49–61. 10.1136/gutjnl-2017-314817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nanki K, Toshimitsu K, Takano A, et al. Divergent routes toward Wnt and R-spondin niche Independency during human gastric carcinogenesis. Cell 2018;174:856–69. 10.1016/j.cell.2018.07.027 [DOI] [PubMed] [Google Scholar]
- 33. Koike H, Iwasawa K, Ouchi R, et al. Engineering human hepato-biliary-pancreatic organoids from pluripotent stem cells. Nat Protoc 2021;16:919–36. 10.1038/s41596-020-00441-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Collin de l'Hortet A, Takeishi K, Guzman-Lepe J, et al. Generation of human fatty livers using Custom-Engineered induced pluripotent stem cells with modifiable SIRT1 metabolism. Cell Metab 2019;30:385–401. 10.1016/j.cmet.2019.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koike H, Iwasawa K, Ouchi R, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 2019;574:112–6. 10.1038/s41586-019-1598-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camp JG, Sekine K, Gerber T, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017;546:533–8. 10.1038/nature22796 [DOI] [PubMed] [Google Scholar]
- 37. Hendriks D, Artegiani B, Hu H, et al. Establishment of human fetal hepatocyte organoids and CRISPR-Cas9-based gene knockin and knockout in organoid cultures from human liver. Nat Protoc 2021;16:182–217. 10.1038/s41596-020-00411-2 [DOI] [PubMed] [Google Scholar]
- 38. McCracken KW, Catá EM, Crawford CM, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014;516:400–4. 10.1038/nature13863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCracken KW, Howell JC, Wells JM, et al. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc 2011;6:1920–8. 10.1038/nprot.2011.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ouchi R, Togo S, Kimura M, et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab 2019;30:374–84. 10.1016/j.cmet.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wiedenmann S, Breunig M, Merkle J, et al. Single-cell-resolved differentiation of human induced pluripotent stem cells into pancreatic duct-like organoids on a microwell chip. Nat Biomed Eng 2021;5:897–913. 10.1038/s41551-021-00757-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Forster R, Chiba K, Schaeffer L, et al. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Reports 2014;2:838–52. 10.1016/j.stemcr.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarvestani SK, Signs S, Hu B, et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat Commun 2021;12:262. 10.1038/s41467-020-20351-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi Y, Inoue H, Wu JC, et al. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 2017;16:115–30. 10.1038/nrd.2016.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet 2019;20:377–88. 10.1038/s41576-019-0100-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 2014;14:329–42. 10.1038/nri3661 [DOI] [PubMed] [Google Scholar]
- 47. McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 2017;144:958–62. 10.1242/dev.140731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schreurs RRCE, Baumdick ME, Drewniak A, et al. In vitro co-culture of human intestinal organoids and lamina propria-derived CD4+ T cells. STAR Protoc 2021;2:100519. 10.1016/j.xpro.2021.100519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cattaneo CM, Dijkstra KK, Fanchi LF, et al. Tumor organoid-T-cell coculture systems. Nat Protoc 2020;15:15–39. 10.1038/s41596-019-0232-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dijkstra KK, Cattaneo CM, Weeber F, et al. Generation of Tumor-Reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 2018;174:1586–98. 10.1016/j.cell.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nozaki K, Mochizuki W, Matsumoto Y, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol 2016;51:206–13. 10.1007/s00535-016-1170-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tsai S, McOlash L, Palen K, et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018;18:335. 10.1186/s12885-018-4238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neal JT, Li X, Zhu J, et al. Organoid modeling of the tumor immune microenvironment. Cell 2018;175:1972–88. 10.1016/j.cell.2018.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cook L, Stahl M, Han X, et al. Suppressive and Gut-Reparative functions of human type 1 T regulatory cells. Gastroenterology 2019;157:1584–98. 10.1053/j.gastro.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 55. Kong JCH, Guerra GR, Millen RM, et al. Tumor-Infiltrating lymphocyte function predicts response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. JCO Precis Oncol 2018;2:1–15. 10.1200/PO.18.00075 [DOI] [PubMed] [Google Scholar]
- 56. Schnalzger TE, de Groot MH, Zhang C, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. Embo J 2019;38. 10.15252/embj.2018100928. [Epub ahead of print: 17 06 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Noel G, Baetz NW, Staab JF, et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci Rep 2017;7:45270. 10.1038/srep45270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jarret A, Jackson R, Duizer C, et al. Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell 2020;180:e12:50–63. 10.1016/j.cell.2019.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Engel MA, Leffler A, Niedermirtl F, et al. TRPA1 and substance P mediate colitis in mice. Gastroenterology 2011;141:1346–58. 10.1053/j.gastro.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 60. Drokhlyansky E, Smillie CS, Van Wittenberghe N, et al. The human and mouse enteric nervous system at single-cell resolution. Cell 2020;182:e23:1606–22. 10.1016/j.cell.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takahashi T, Fujishima K, Kengaku M. Modeling intestinal stem cell function with organoids. Int J Mol Sci 2021;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bellono NW, Bayrer JR, Leitch DB, et al. Enterochromaffin cells are gut Chemosensors that couple to sensory neural pathways. Cell 2017;170:e16:185–98. 10.1016/j.cell.2017.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Workman MJ, Mahe MM, Trisno S, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017;23:49–59. 10.1038/nm.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Loffet E, Brossard L, Mahe MM. Pluripotent stem cell derived intestinal organoids with an enteric nervous system. Methods Cell Biol 2020;159:175–99. 10.1016/bs.mcb.2020.04.012 [DOI] [PubMed] [Google Scholar]
- 65. Park CS, Nguyen LP, Yong D. Development of colonic organoids containing enteric nerves or blood vessels from human embryonic stem cells. Cells 2020;9:2209. 10.3390/cells9102209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eicher AK, Kechele DO, Sundaram N, et al. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell 2022;29:36–51. 10.1016/j.stem.2021.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Spadoni I, Zagato E, Bertocchi A, et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 2015;350:830–4. 10.1126/science.aad0135 [DOI] [PubMed] [Google Scholar]
- 68. Rafii S, Butler JM, Ding B-S. Angiocrine functions of organ-specific endothelial cells. Nature 2016;529:316–25. 10.1038/nature17040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Carloni S, Bertocchi A, Mancinelli S, et al. Identification of a choroid plexus vascular barrier closing during intestinal inflammation. Science 2021;374:439–48. 10.1126/science.abc6108 [DOI] [PubMed] [Google Scholar]
- 70. Cromer WE, Mathis JM, Granger DN, et al. Role of the endothelium in inflammatory bowel diseases. World J Gastroenterol 2011;17:578–93. 10.3748/wjg.v17.i5.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481–4. 10.1038/nature12271 [DOI] [PubMed] [Google Scholar]
- 72. Takahashi Y, Sekine K, Kin T, et al. Self-Condensation culture enables vascularization of tissue fragments for efficient therapeutic transplantation. Cell Rep 2018;23:1620–9. 10.1016/j.celrep.2018.03.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wimmer RA, Leopoldi A, Aichinger M, et al. Generation of blood vessel organoids from human pluripotent stem cells. Nat Protoc 2019;14:3082–100. 10.1038/s41596-019-0213-z [DOI] [PubMed] [Google Scholar]
- 74. Takebe T, Enomura M, Yoshizawa E, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 2015;16:556–65. 10.1016/j.stem.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 75. Wörsdörfer P, Dalda N, Kern A, et al. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci Rep 2019;9:15663. 10.1038/s41598-019-52204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Holloway EM, Wu JH, Czerwinski M, et al. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Dev Cell 2020;54:516–28. 10.1016/j.devcel.2020.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McCarthy N, Kraiczy J, Shivdasani RA. Cellular and molecular architecture of the intestinal stem cell niche. Nat Cell Biol 2020;22:1033–41. 10.1038/s41556-020-0567-z [DOI] [PubMed] [Google Scholar]
- 78. Farin HF, Jordens I, Mosa MH, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 2016;530:340–3. 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- 79. McMahon JA, Takada S, Zimmerman LB, et al. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev 1998;12:1438–52. 10.1101/gad.12.10.1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McCarthy N, Manieri E, Storm EE, et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell 2020;26:391–402. 10.1016/j.stem.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Greicius G, Kabiri Z, Sigmundsson K, et al. PDGFRα + pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A 2018;115:E3173–81. 10.1073/pnas.1713510115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012;143:1518–29. 10.1053/j.gastro.2012.08.031 [DOI] [PubMed] [Google Scholar]
- 83. Lahar N, Lei NY, Wang J, et al. Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One 2011;6:e26898. 10.1371/journal.pone.0026898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lei NY, Jabaji Z, Wang J, et al. Intestinal subepithelial myofibroblasts support the growth of intestinal epithelial stem cells. PLoS One 2014;9:e84651. 10.1371/journal.pone.0084651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kabiri Z, Greicius G, Madan B, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 2014;141:2206–15. 10.1242/dev.104976 [DOI] [PubMed] [Google Scholar]
- 86. Karpus ON, Westendorp BF, Vermeulen JLM, et al. Colonic CD90+ crypt fibroblasts secrete semaphorins to support epithelial growth. Cell Rep 2019;26:3698–708. 10.1016/j.celrep.2019.02.101 [DOI] [PubMed] [Google Scholar]
- 87. Oszvald Ádám, Szvicsek Z, Sándor GO, et al. Extracellular vesicles transmit epithelial growth factor activity in the intestinal stem cell niche. Stem Cells 2020;38:291–300. 10.1002/stem.3113 [DOI] [PubMed] [Google Scholar]
- 88. Liu J, Li P, Wang L, et al. Cancer-Associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell Mol Gastroenterol Hepatol 2021;11:407–31. 10.1016/j.jcmgh.2020.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Öhlund D, Handly-Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017;214:579–96. 10.1084/jem.20162024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Seino T, Kawasaki S, Shimokawa M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 2018;22:454–67. 10.1016/j.stem.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 91. Biffi G, Oni TE, Spielman B, et al. Il1-Induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov 2019;9:282–301. 10.1158/2159-8290.CD-18-0710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bittel M, Reichert P, Sarfati I, et al. Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe-host-communication in vivo. J Extracell Vesicles 2021;10:e12159. 10.1002/jev2.12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hickey CA, Kuhn KA, Donermeyer DL, et al. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a Sulfatase-Dependent manner via outer membrane vesicles. Cell Host Microbe 2015;17:672–80. 10.1016/j.chom.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–52. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 2015;15:375–87. 10.1038/nri3837 [DOI] [PubMed] [Google Scholar]
- 96. Tilg H, Zmora N, Adolph TE, et al. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 2020;20:40–54. 10.1038/s41577-019-0198-4 [DOI] [PubMed] [Google Scholar]
- 97. Wilke G, Funkhouser-Jones LJ, Wang Y, et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell Host Microbe 2019;26:123–34. 10.1016/j.chom.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ettayebi K, Crawford SE, Murakami K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016;353:1387–93. 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Abuaita BH, Lawrence A-LE, Berger RP, et al. Comparative transcriptional profiling of the early host response to infection by typhoidal and non-typhoidal Salmonella serovars in human intestinal organoids. PLoS Pathog 2021;17:e1009987. 10.1371/journal.ppat.1009987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Holthaus D, Kraft MR, Krug SM, et al. Dissection of barrier dysfunction in Organoid-Derived human intestinal epithelia induced by Giardia duodenalis. Gastroenterology 2022;162:844–58. 10.1053/j.gastro.2021.11.022 [DOI] [PubMed] [Google Scholar]
- 101. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020;369:50–4. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou J, Li C, Liu X, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med 2020;26:1077–83. 10.1038/s41591-020-0912-6 [DOI] [PubMed] [Google Scholar]
- 103. Guo M, Tao W, Flavell RA, et al. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol 2021;18:269–83. 10.1038/s41575-021-00416-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020;580:269–73. 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Giobbe GG, Bonfante F, Jones BC, et al. SARS-CoV-2 infection and replication in human gastric organoids. Nat Commun 2021;12:6610. 10.1038/s41467-021-26762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hill DR, Spence JR. Gastrointestinal organoids: understanding the molecular basis of the host-microbe interface. Cell Mol Gastroenterol Hepatol 2017;3:138–49. 10.1016/j.jcmgh.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Liu T-C, Kern JT, Jain U, et al. Western diet induces Paneth cell defects through microbiome alterations and farnesoid X receptor and type I interferon activation. Cell Host Microbe 2021;29:988–1001. 10.1016/j.chom.2021.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sorrentino G, Perino A, Yildiz E, et al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 2020;159:956–68. 10.1053/j.gastro.2020.05.067 [DOI] [PubMed] [Google Scholar]
- 109. Ke X, You K, Pichaud M, et al. Gut bacterial metabolites modulate endoplasmic reticulum stress. Genome Biol 2021;22:292. 10.1186/s13059-021-02496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Banerjee A, Herring CA, Chen B, et al. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology 2020;159:2101–15. 10.1053/j.gastro.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yin Y-B, de Jonge HR, Wu X, et al. Enteroids for nutritional studies. Mol Nutr Food Res 2019;63:e1801143. 10.1002/mnfr.201801143 [DOI] [PubMed] [Google Scholar]
- 112. Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 2020;5. 10.1126/sciimmunol.abc3582. [Epub ahead of print: 13 05 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Heo I, Dutta D, Schaefer DA, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 2018;3:814–23. 10.1038/s41564-018-0177-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Co JY, Margalef-Català M, Monack DM, et al. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat Protoc 2021;16:5171–92. 10.1038/s41596-021-00607-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Co JY, Margalef-Català M, Li X, et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep 2019;26:2509–20. 10.1016/j.celrep.2019.01.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Salahudeen AA, Choi SS, Rustagi A, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 2020;588:670–5. 10.1038/s41586-020-3014-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Han X, Lee A, Huang S, et al. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes 2019;10:59–76. 10.1080/19490976.2018.1479625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Puschhof J, Pleguezuelos-Manzano C, Martinez-Silgado A, et al. Intestinal organoid cocultures with microbes. Nat Protoc 2021;16:4633–49. 10.1038/s41596-021-00589-z [DOI] [PubMed] [Google Scholar]
- 119. Holokai L, Chakrabarti J, Broda T, et al. Increased programmed Death-Ligand 1 is an early epithelial cell response to Helicobacter pylori infection. PLoS Pathog 2019;15:e1007468. 10.1371/journal.ppat.1007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Williamson IA, Arnold JW, Samsa LA, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol 2018;6:301–19. 10.1016/j.jcmgh.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wang Y, Chiang I-L, Ohara TE, et al. Long-Term culture captures Injury-Repair cycles of colonic stem cells. Cell 2019;179:e15:1144–59. 10.1016/j.cell.2019.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang Y, Kim R, Gunasekara DB, et al. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell Mol Gastroenterol Hepatol 2018;5:113–30. 10.1016/j.jcmgh.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang Y, Gunasekara DB, Reed MI, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017;128:44–55. 10.1016/j.biomaterials.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater 2021:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fu J, Warmflash A, Lutolf MP. Stem-cell-based embryo models for fundamental research and translation. Nat Mater 2021;20:132–44. 10.1038/s41563-020-00829-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Brassard JA, Lutolf MP. Engineering stem cell self-organization to build better organoids. Cell Stem Cell 2019;24:860–76. 10.1016/j.stem.2019.05.005 [DOI] [PubMed] [Google Scholar]
- 127. Nelson CM, Vanduijn MM, Inman JL, et al. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science 2006;314:298–300. 10.1126/science.1131000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Gjorevski N, Nikolaev M, Brown TE, et al. Tissue geometry drives deterministic organoid patterning. Science 2022;375:eaaw9021. 10.1126/science.aaw9021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Chrisnandy A, Blondel D, Rezakhani S, et al. Synthetic dynamic hydrogels promote degradation-independent in vitro organogenesis. Nat Mater 2022;21:479–87. 10.1038/s41563-021-01136-7 [DOI] [PubMed] [Google Scholar]
- 130. Nikolaev M, Mitrofanova O, Broguiere N, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020;585:574–8. 10.1038/s41586-020-2724-8 [DOI] [PubMed] [Google Scholar]
- 131. Khan S, Tiriac H. Synthetic scaffold for pancreatic organoids. Nat Mater 2022;21:9–11. 10.1038/s41563-021-01177-y [DOI] [PubMed] [Google Scholar]
- 132. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, et al. Development of a primary human small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep 2018;8:2871. 10.1038/s41598-018-21201-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Toda S, Blauch LR, Tang SKY, et al. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018;361:156–62. 10.1126/science.aat0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 2019;3:520–31. 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Schwärzler J, Mayr L, Vich Vila A, et al. PUFA-induced metabolic enteritis as a fuel for Crohn's disease. Gastroenterology 2022;162:1690–704. 10.1053/j.gastro.2022.01.004 [DOI] [PubMed] [Google Scholar]
- 136. Michiba K, Maeda K, Shimomura O, et al. Usefulness of human jejunal spheroid-derived differentiated intestinal epithelial cells for the prediction of intestinal drug absorption in humans. Drug Metab Dispos 2022;50:204-213. 10.1124/dmd.121.000796 [DOI] [PubMed] [Google Scholar]
- 137. Yan S, Conley JM, Reilly AM, et al. Intestinal Gpr17 deficiency improves glucose metabolism by promoting GLP-1 secretion. Cell Rep 2022;38:110179. 10.1016/j.celrep.2021.110179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Dedhia PH, Bertaux-Skeirik N, Zavros Y, et al. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology 2016;150:1098–112. 10.1053/j.gastro.2015.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Seidlitz T, Koo B-K, Stange DE. Gastric organoids-an in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ 2021;28:68–83. 10.1038/s41418-020-00662-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Fujii M, Sato T. Somatic cell-derived organoids as prototypes of human epithelial tissues and diseases. Nat Mater 2021;20:156–69. 10.1038/s41563-020-0754-0 [DOI] [PubMed] [Google Scholar]
- 141. Nanki K, Fujii M, Shimokawa M, et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 2020;577:254–9. 10.1038/s41586-019-1844-5 [DOI] [PubMed] [Google Scholar]
- 142. Kakiuchi N, Yoshida K, Uchino M, et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 2020;577:260–5. 10.1038/s41586-019-1856-1 [DOI] [PubMed] [Google Scholar]
- 143. Ringel T, Frey N, Ringnalda F, et al. Genome-Scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell 2020;26:431–40. 10.1016/j.stem.2020.02.007 [DOI] [PubMed] [Google Scholar]
- 144. Bartfeld S, Clevers H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J Vis Exp 2015. 10.3791/53359. [Epub ahead of print: 12 Nov 2015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bartfeld S, Bayram T, van de Wetering M, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015;148:126–36. 10.1053/j.gastro.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Schumacher MA, Feng R, Aihara E, et al. Helicobacter pylori-induced sonic hedgehog expression is regulated by NFκB pathway activation: the use of a novel in vitro model to study epithelial response to infection. Helicobacter 2015;20:19–28. 10.1111/hel.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Schlaermann P, Toelle B, Berger H, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 2016;65:202–13. 10.1136/gutjnl-2014-307949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wroblewski LE, Piazuelo MB, Chaturvedi R, et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 2015;64:720–30. 10.1136/gutjnl-2014-307650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Lo Y-H, Kolahi KS, Du Y, et al. A CRISPR/Cas9-Engineered ARID1A-Deficient Human Gastric Cancer Organoid Model Reveals Essential and Nonessential Modes of Oncogenic Transformation. Cancer Discov 2021;11:1562–81. 10.1158/2159-8290.CD-20-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Qu M, Xiong L, Lyu Y, et al. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res 2021;31:259–71. 10.1038/s41422-020-00453-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rana N, Privitera G, Kondolf HC, et al. GSDMB is increased in IBD and regulates epithelial restitution/repair independent of pyroptosis. Cell 2022;185:e17:283–98. 10.1016/j.cell.2021.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Günther C, Ruder B, Stolzer I, et al. Interferon lambda promotes Paneth cell death via STAT1 signaling in mice and is increased in inflamed ileal tissues of patients with Crohn's disease. Gastroenterology 2019;157:e13:1310–22. 10.1053/j.gastro.2019.07.031 [DOI] [PubMed] [Google Scholar]
- 153. Xu P, Becker H, Elizalde M, et al. Interleukin-28A induces epithelial barrier dysfunction in CD patient-derived intestinal organoids. Am J Physiol Gastrointest Liver Physiol 2021;320:G689–99. 10.1152/ajpgi.00064.2020 [DOI] [PubMed] [Google Scholar]
- 154. Xu P, Elizalde M, Masclee A, et al. Corticosteroid enhances epithelial barrier function in intestinal organoids derived from patients with Crohn's disease. J Mol Med 2021;99:805–15. 10.1007/s00109-021-02045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zhou T, Tan L, Cederquist GY, et al. High-Content screening in hPSC-Neural progenitors identifies drug candidates that inhibit Zika virus infection in Fetal-like organoids and adult brain. Cell Stem Cell 2017;21:274–83. 10.1016/j.stem.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Krüger J, Groß R, Conzelmann C, et al. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol Gastroenterol Hepatol 2021;11:935–48. 10.1016/j.jcmgh.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ebisudani T, Sugimoto S, Haga K, et al. Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening. Cell Rep 2021;35:109218. 10.1016/j.celrep.2021.109218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Han Y, Duan X, Yang L, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021;589:270–5. 10.1038/s41586-020-2901-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Son YS, Ki SJ, Thanavel R, et al. Maturation of human intestinal organoids in vitro facilitates colonization by commensal lactobacilli by reinforcing the mucus layer. FASEB J 2020;34:9899–910. 10.1096/fj.202000063R [DOI] [PubMed] [Google Scholar]
- 160. Pradhan S, Weiss AA. Probiotic properties of Escherichia coli Nissle in human intestinal organoids. mBio 2020;11. 10.1128/mBio.01470-20. [Epub ahead of print: 07 07 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Cristobal A, van den Toorn HWP, van de Wetering M, et al. Personalized proteome profiles of healthy and tumor human colon organoids reveal both individual diversity and basic features of colorectal cancer. Cell Rep 2017;18:263–74. 10.1016/j.celrep.2016.12.016 [DOI] [PubMed] [Google Scholar]
- 163. Fujii M, Shimokawa M, Date S, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 2016;18:827–38. 10.1016/j.stem.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 164. Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920–6. 10.1126/science.aao2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Yan HHN, Siu HC, Ho SL, et al. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut 2020;69:2165–79. 10.1136/gutjnl-2019-320019 [DOI] [PubMed] [Google Scholar]
- 166. Ganesh K, Wu C, O'Rourke KP, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med 2019;25:1607–14. 10.1038/s41591-019-0584-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yao Y, Xu X, Yang L, et al. Patient-Derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 2020;26:17–26. 10.1016/j.stem.2019.10.010 [DOI] [PubMed] [Google Scholar]
- 168. Knight JRP, Alexandrou C, Skalka GL, et al. MNK Inhibition Sensitizes KRAS-Mutant Colorectal Cancer to mTORC1 Inhibition by Reducing eIF4E Phosphorylation and c-MYC Expression. Cancer Discov 2021;11:1228–47. 10.1158/2159-8290.CD-20-0652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Buzzelli JN, Ouaret D, Brown G, et al. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res 2018;27:109–20. 10.1016/j.scr.2018.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ooft SN, Weeber F, Dijkstra KK, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 2019;11. 10.1126/scitranslmed.aay2574. [Epub ahead of print: 09 10 2019]. [DOI] [PubMed] [Google Scholar]
- 171. Yan HHN, Siu HC, Law S, et al. A comprehensive human gastric cancer organoid Biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 2018;23:e11:882–97. 10.1016/j.stem.2018.09.016 [DOI] [PubMed] [Google Scholar]
- 172. Steele NG, Chakrabarti J, Wang J, et al. An Organoid-Based preclinical model of human gastric cancer. Cell Mol Gastroenterol Hepatol 2019;7:161–84. 10.1016/j.jcmgh.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Seidlitz T, Merker SR, Rothe A, et al. Human gastric cancer modelling using organoids. Gut 2019;68:207–17. 10.1136/gutjnl-2017-314549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Togasaki K, Sugimoto S, Ohta Y, et al. Wnt signaling shapes the histologic variation in diffuse gastric cancer. Gastroenterology 2021;160:823–30. 10.1053/j.gastro.2020.10.047 [DOI] [PubMed] [Google Scholar]
- 175. Gao M, Lin M, Rao M, et al. Development of patient-derived gastric cancer organoids from endoscopic biopsies and surgical tissues. Ann Surg Oncol 2018;25:2767–75. 10.1245/s10434-018-6662-8 [DOI] [PubMed] [Google Scholar]
- 176. Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–75. 10.1038/nature20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Derouet MF, Allen J, Wilson GW, et al. Towards personalized induction therapy for esophageal adenocarcinoma: organoids derived from endoscopic biopsy recapitulate the pre-treatment tumor. Sci Rep 2020;10:14514. 10.1038/s41598-020-71589-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Li X, Francies HE, Secrier M, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun 2018;9:2983. 10.1038/s41467-018-05190-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zheng B, Ko K-P, Fang X, et al. A new murine esophageal organoid culture method and organoid-based model of esophageal squamous cell neoplasia. iScience 2021;24:103440. 10.1016/j.isci.2021.103440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Hirt CK, Booij TH, Grob L, et al. Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label treatment. Cell Genom 2022;2:100095. 10.1016/j.xgen.2022.100095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Driehuis E, van Hoeck A, Moore K, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A 2019. 10.1073/pnas.1911273116. [Epub ahead of print: 09 Dec 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kawasaki K, Toshimitsu K, Matano M, et al. An organoid Biobank of neuroendocrine neoplasms enables genotype-phenotype mapping. Cell 2020;183:1420–35. 10.1016/j.cell.2020.10.023 [DOI] [PubMed] [Google Scholar]
- 183. Broutier L, Mastrogiovanni G, Verstegen MM, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23:1424–35. 10.1038/nm.4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Nuciforo S, Fofana I, Matter MS, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep 2018;24:1363–76. 10.1016/j.celrep.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Li L, Knutsdottir H, Hui K, et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 2019;4. 10.1172/jci.insight.121490. [Epub ahead of print: 24 Jan 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Broutier L, Andersson-Rolf A, Hindley CJ, et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc 2016;11:1724–43. 10.1038/nprot.2016.097 [DOI] [PubMed] [Google Scholar]
- 187. Shi X, Yang J, Liu M, et al. Circular RNA ANAPC7 inhibits tumor growth and muscle wasting via PHLPP2-AKT-TGF-β signaling axis in pancreatic cancer. Gastroenterology 2022;162:2004–17. 10.1053/j.gastro.2022.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kato H, Tateishi K, Fujiwara H, et al. MNX1-HNF1B axis is indispensable for intraductal papillary mucinous neoplasm lineages. Gastroenterology 2022;162:1272–87. 10.1053/j.gastro.2021.12.254 [DOI] [PubMed] [Google Scholar]
- 189. Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 2018;8:1112–29. 10.1158/2159-8290.CD-18-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Geurts MH, de Poel E, Amatngalim GD, et al. CRISPR-Based adenine editors correct nonsense mutations in a cystic fibrosis organoid Biobank. Cell Stem Cell 2020;26:503–10. 10.1016/j.stem.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 191. Lee SH, Hu W, Matulay JT, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018;173:515–28. 10.1016/j.cell.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Li Z, Ge Y, Dong J, et al. BZW1 facilitates glycolysis and promotes tumor growth in pancreatic ductal adenocarcinoma through potentiating eIF2α phosphorylation. Gastroenterology 2022;162:1256–71. 10.1053/j.gastro.2021.12.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Bouchi R, Foo KS, Hua H, et al. FOXO1 inhibition yields functional insulin-producing cells in human gut organoid cultures. Nat Commun 2014;5:4242. 10.1038/ncomms5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Yui S, Nakamura T, Sato T, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5⁺ stem cell. Nat Med 2012;18:618–23. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- 195. Sugimoto S, Ohta Y, Fujii M, et al. Reconstruction of the Human Colon Epithelium In Vivo. Cell Stem Cell 2018;22:171–6. 10.1016/j.stem.2017.11.012 [DOI] [PubMed] [Google Scholar]
- 196. Sugimoto S, Kobayashi E, Fujii M, et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 2021;592:99–104. 10.1038/s41586-021-03247-2 [DOI] [PubMed] [Google Scholar]
- 197. Duluc I, Freund JN, Leberquier C, et al. Fetal endoderm primarily holds the temporal and positional information required for mammalian intestinal development. J Cell Biol 1994;126:211–21. 10.1083/jcb.126.1.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Meran L, Massie I, Campinoti S, et al. Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure. Nat Med 2020;26:1593–601. 10.1038/s41591-020-1024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Poling HM, Wu D, Brown N, et al. Mechanically induced development and maturation of human intestinal organoids in vivo. Nat Biomed Eng 2018;2:429–42. 10.1038/s41551-018-0243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Sampaziotis F, Justin AW, Tysoe OC, et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat Med 2017;23:954–63. 10.1038/nm.4360 [DOI] [PubMed] [Google Scholar]
- 201. Sampaziotis F, Muraro D, Tysoe OC, et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science 2021;371:839–46. 10.1126/science.aaz6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Fleischer A, Vallejo-Díez S, Martín-Fernández JM, et al. iPSC-Derived intestinal organoids from cystic fibrosis patients acquire CFTR activity upon TALEN-mediated repair of the p.F508del mutation. Mol Ther Methods Clin Dev 2020;17:858–70. 10.1016/j.omtm.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Miura Y, Li M-Y, Revah O, et al. Engineering brain assembloids to interrogate human neural circuits. Nat Protoc 2022;17:15–35. 10.1038/s41596-021-00632-z [DOI] [PubMed] [Google Scholar]
- 204. Takeishi K, Collin de l'Hortet A, Wang Y, et al. Assembly and function of a bioengineered human liver for transplantation generated solely from induced pluripotent stem cells. Cell Rep 2020;31:107711. 10.1016/j.celrep.2020.107711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Giobbe GG, Crowley C, Luni C, et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat Commun 2019;10:5658. 10.1038/s41467-019-13605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Giuffrida P, Curti M, Al-Akkad W, et al. Decellularized human gut as a natural 3D platform for research in intestinal fibrosis. Inflamm Bowel Dis 2019;25:1740–50. 10.1093/ibd/izz115 [DOI] [PubMed] [Google Scholar]
- 207. Gjorevski N, Sachs N, Manfrin A, et al. Designer matrices for intestinal stem cell and organoid culture. Nature 2016;539:560–4. 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- 208. Hunt DR, Klett KC, Mascharak S, et al. Engineered matrices enable the culture of human patient-derived intestinal organoids. Adv Sci 2021;8:2004705. 10.1002/advs.202004705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Curvello R, Alves D, Abud HE, et al. A thermo-responsive collagen-nanocellulose hydrogel for the growth of intestinal organoids. Mater Sci Eng C Mater Biol Appl 2021;124:112051. 10.1016/j.msec.2021.112051 [DOI] [PubMed] [Google Scholar]
- 210. Capeling MM, Czerwinski M, Huang S, et al. Nonadhesive alginate hydrogels support growth of pluripotent stem cell-derived intestinal organoids. Stem Cell Reports 2019;12:381–94. 10.1016/j.stemcr.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Cruz-Acuña R, Quirós M, Farkas AE, et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 2017;19:1326–35. 10.1038/ncb3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008;132:661–80. 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- 213. Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 1975;6:331–43. 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- 214. Kim K-A, Kakitani M, Zhao J, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 2005;309:1256–9. 10.1126/science.1112521 [DOI] [PubMed] [Google Scholar]
- 215. Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 216. Yokota J, Yamashita T, Inui T, et al. Comparison of culture media for human intestinal organoids from the viewpoint of pharmacokinetic studies. Biochem Biophys Res Commun 2021;566:115–22. 10.1016/j.bbrc.2021.06.007 [DOI] [PubMed] [Google Scholar]
- 217. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145–7. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 218. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–72. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 219. Eiraku M, Watanabe K, Matsuo-Takasaki M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008;3:519–32. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 220. Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981;292:154–6. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- 221. Todaro GJ, Green H. Successive transformations of an established cell line by polyoma virus and SV40. Science 1965;147:513–4. 10.1126/science.147.3657.513 [DOI] [PubMed] [Google Scholar]
- 222. Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 2013;8:2471–82. 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Broda TR, McCracken KW, Wells JM. Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat Protoc 2019;14:28–50. 10.1038/s41596-018-0080-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Pleguezuelos-Manzano C, Puschhof J, van den Brink S, et al. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr Protoc Immunol 2020;130:e106. 10.1002/cpim.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Tysoe OC, Justin AW, Brevini T, et al. Isolation and propagation of primary human cholangiocyte organoids for the generation of bioengineered biliary tissue. Nat Protoc 2019;14:1884–925. 10.1038/s41596-019-0168-0 [DOI] [PubMed] [Google Scholar]
- 226. Driehuis E, Gracanin A, Vries RGJ, et al. Establishment of pancreatic organoids from normal tissue and tumors. STAR Protoc 2020;1:100192. 10.1016/j.xpro.2020.100192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Breunig M, Merkle J, Melzer MK, et al. Differentiation of human pluripotent stem cells into pancreatic duct-like organoids. STAR Protoc 2021;2:100913. 10.1016/j.xpro.2021.100913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Sampaziotis F, de Brito MC, Geti I, et al. Directed differentiation of human induced pluripotent stem cells into functional cholangiocyte-like cells. Nat Protoc 2017;12:814–27. 10.1038/nprot.2017.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc 2020;15:3380–409. 10.1038/s41596-020-0379-4 [DOI] [PubMed] [Google Scholar]
- 230. Padmanaban V, Grasset EM, Neumann NM, et al. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nat Protoc 2020;15:2413–42. 10.1038/s41596-020-0335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Calandrini C, Drost J. Normal and tumor-derived organoids as a drug screening platform for tumor-specific drug vulnerabilities. STAR Protoc 2022;3:101079. 10.1016/j.xpro.2021.101079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Pascual-Sabater S, Raimondi G, Mato-Berciano A, et al. Preclinical testing of oncolytic adenovirus sensitivity in patient-derived tumor organoids. STAR Protoc 2021;2:101017. 10.1016/j.xpro.2021.101017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Fujii M, Matano M, Nanki K, et al. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc 2015;10:1474–85. 10.1038/nprot.2015.088 [DOI] [PubMed] [Google Scholar]
- 234. Roper J, Tammela T, Akkad A, et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat Protoc 2018;13:217–34. 10.1038/nprot.2017.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Lin S-C, Haga K, Zeng X-L, et al. Generation of CRISPR-Cas9-mediated genetic knockout human intestinal tissue-derived enteroid lines by lentivirus transduction and single-cell cloning. Nat Protoc 2022;17:1004–27. 10.1038/s41596-021-00669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Kaltenbacher T, Löprich J, Maresch R, et al. CRISPR somatic genome engineering and cancer modeling in the mouse pancreas and liver. Nat Protoc 2022;17:1142–88. 10.1038/s41596-021-00677-0 [DOI] [PubMed] [Google Scholar]
- 237. van Neerven SM, Ramadan R, van Driel MS, et al. Intestinal organoid co-culture protocol to study cell competition in vitro. STAR Protoc 2022;3:101050. 10.1016/j.xpro.2021.101050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Okamoto T, Natsume Y, Yamanaka H, et al. A protocol for efficient CRISPR-Cas9-mediated knock-in in colorectal cancer patient-derived organoids. STAR Protoc 2021;2:100780. 10.1016/j.xpro.2021.100780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Haga K, Ettayebi K, Tenge VR, et al. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for Secretor-Dependent human norovirus infection. mBio 2020;11. 10.1128/mBio.00251-20. [Epub ahead of print: 17 03 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Lin S-C, Qu L, Ettayebi K, et al. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci U S A 2020;117:23782–93. 10.1073/pnas.2010834117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Stroulios G, Stahl M, Elstone F, et al. Culture methods to study Apical-Specific interactions using intestinal organoid models. J Vis Exp 2021. 10.3791/62330. [Epub ahead of print: 23 03 2021]. [DOI] [PubMed] [Google Scholar]
- 242. Zhang J, Hernandez-Gordillo V, Trapecar M, et al. Coculture of primary human colon monolayer with human gut bacteria. Nat Protoc 2021;16:3874–900. 10.1038/s41596-021-00562-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Sayed IM, Tindle C, Fonseca AG, et al. Functional assays with human patient-derived enteroid monolayers to assess the human gut barrier. STAR Protoc 2021;2:100680. 10.1016/j.xpro.2021.100680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Lai BFL, Lu RXZ, Davenport Huyer L, et al. A well plate-based multiplexed platform for incorporation of organoids into an organ-on-a-chip system with a perfusable vasculature. Nat Protoc 2021;16:2158–89. 10.1038/s41596-020-00490-1 [DOI] [PubMed] [Google Scholar]
- 245. Shin W, Kim HJ. 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert. Nat Protoc 2022;17:910–39. 10.1038/s41596-021-00674-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Aleman J, Kilic T, Mille LS, et al. Microfluidic integration of regeneratable electrochemical affinity-based biosensors for continual monitoring of organ-on-a-chip devices. Nat Protoc 2021;16:2564–93. 10.1038/s41596-021-00511-7 [DOI] [PubMed] [Google Scholar]
- 247. Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol 2013;5:1130–40. 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 248. Watanabe S, Kobayashi S, Ogasawara N, et al. Transplantation of intestinal organoids into a mouse model of colitis. Nat Protoc 2022;17:649–71. 10.1038/s41596-021-00658-3 [DOI] [PubMed] [Google Scholar]
- 249. Fumagalli A, Suijkerbuijk SJE, Begthel H, et al. A surgical orthotopic organoid transplantation approach in mice to visualize and study colorectal cancer progression. Nat Protoc 2018;13:235–47. 10.1038/nprot.2017.137 [DOI] [PubMed] [Google Scholar]
- 250. Kasashima H, Duran A, Cid-Diaz T, et al. Mouse model of colorectal cancer: orthotopic co-implantation of tumor and stroma cells in cecum and rectum. STAR Protoc 2021;2:100297. 10.1016/j.xpro.2021.100297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Biton M, Haber AL, Rogel N, et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 2018;175:1307–20. 10.1016/j.cell.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Jowett GM, Norman MDA, Yu TTL, et al. ILC1 drive intestinal epithelial and matrix remodelling. Nat Mater 2021;20:250–9. 10.1038/s41563-020-0783-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Besikcioglu HE, Yurteri Ümmügülsüm, Munkhbaatar E, et al. Innervated mouse pancreas organoids as an ex vivo model to study pancreatic neuropathy in pancreatic cancer. STAR Protoc 2021;2:100935. 10.1016/j.xpro.2021.100935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), Canadian Interventional Radiology Association (CIRA), Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR), European Stroke Organization (ESO), Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR), Society of NeuroInterventional Surgery (SNIS), and World Stroke Organization (WSO), Sacks D, Baxter B, et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke 2018;13:612–32. 10.1177/1747493018778713 [DOI] [PubMed] [Google Scholar]
- 255. Patel KK, Miyoshi H, Beatty WL, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. Embo J 2013;32:3130–44. 10.1038/emboj.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Miyoshi H, VanDussen KL, Malvin NP, et al. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. Embo J 2017;36:5–24. 10.15252/embj.201694660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Kanaya T, Sakakibara S, Jinnohara T, et al. Development of intestinal M cells and follicle-associated epithelium is regulated by TRAF6-mediated NF-kappaB signaling. J Exp Med 2018;215:501–19. 10.1084/jem.20160659 [DOI] [PMC free article] [PubMed] [Google Scholar]