ABSTRACT
Nucleoid-associated proteins (NAPs) help structure bacterial genomes and function in an array of DNA transactions, including transcription, recombination, and repair. In most bacteria, NAPs are nonessential in part due to functional redundancy. In contrast, in Bacillus subtilis the HU homolog HBsu is essential for cell viability. HBsu helps compact the B. subtilis chromosome and participates in homologous recombination and DNA repair. However, none of these activities explain HBsu’s essentiality. Here, using two complementary conditional HBsu alleles, we investigated the terminal phenotype of the mutants. Our analysis revealed that cells without functional HBsu fail to initiate DNA replication. Importantly, when the chromosomal replication origin (oriC) was replaced with a plasmid origin (oriN) whose replication does not require the initiator DnaA, cells without HBsu initiated DNA replication normally. However, HBsu was still essential in this oriN-containing strain. We conclude that HBsu plays an essential role in the initiation of DNA replication, likely acting to promote origin melting by DnaA, but also has a second essential function that remains to be discovered.
IMPORTANCE While it is common for a bacterial species to express multiple nucleoid-associated proteins (NAPs), NAPs are seldomly essential for cell survival. In B. subtilis, HBsu is a NAP essential for cell viability. Here, using conditional alleles to rapidly remove or inactivate HBsu, we show that the absence of HBsu abolishes the initiation of DNA replication in vivo. Understanding HBsu’s function can provide new insights into the regulation of DNA replication initiation in bacteria.
KEYWORDS: replication initiation, HU, HBsu, nucleoid-associated proteins, NAPs, Bacillus subtilis
INTRODUCTION
All living organisms are challenged with organizing their DNA within the confines of their cells. In eukaryotes, histones are a major player for packaging the DNA in the nucleus. Bacteria do not have histones, but they contain histone-like proteins called nucleoid-associated proteins, or NAPs (1). The vast majority of bacterial species contain many well-known NAPs, including HU, H-NS, IHF, and Fis (2). Generally, these proteins bind to DNA and introduce constraints like bending, wrapping, and bridging, which compact the DNA and structure the nucleoid. In addition to these structuring roles, many NAPs play other functions, such as global regulation of gene expression, controlling DNA replication initiation, and facilitating DNA recombination (3 to 9).
One of the most well-studied NAPs is HU. In Escherichia coli, the heterodimer of HU is among the most abundant proteins during exponential growth (10). In vitro, HU binds to DNA nonspecifically and induces or stabilizes DNA bending (10, 11), consistent with the role of facilitating nucleoid compaction and long-range DNA contacts in live cells (12). Using a plasmid that contains the chromosomal replication origin, oriC, in vitro work showed that HU interacts with the replication initiator protein, DnaA, and stimulates the unwinding of oriC at replication initiation (13–15). In vivo studies have also shown that HU influences DNA replication initiation. In E. coli, HU mutants exhibit asynchronous replication (16), cell division defects, and anucleate cell formation (17, 18). In Mycobacterium smegmatis, mutations in the HU homolog caused a delay in DNA replication initiation as well as lowered the frequency for reinitiation of replication (19). Despite these important roles, HU is not essential in E. coli or M. smegmatis (17, 19).
The HU homolog in the Gram-positive bacterium Bacillus subtilis is called HBsu, which is encoded by the hbs gene. HBsu is the only prominent NAP in B. subtilis and has been shown to be essential for viability (20–22). Like HU, HBsu binds nonspecifically to DNA in vitro and compacts DNA (23–25). In vivo, HBsu localizes to the entire nucleoid and is implicated in constraining DNA supercoiling (24, 26) and facilitating DNA repair and recombination (27). However, none of these functions explain why HBsu is essential for cell growth and survival.
Here, we investigate the function of HBsu in B. subtilis. We constructed conditional mutants either to remove the protein by combined depletion and degradation or to inactivate the protein with a temperature-sensitive allele. Using fluorescence microscopy and whole-genome sequencing, we found that when HBsu was absent or inactivated, DNA replication initiation was blocked. Interestingly, this effect is specific to the chromosomal replication origin, oriC, and not when replication is initiated from a plasmid replication origin, oriN. Thus, HBsu is required for replication initiation in B. subtilis in vivo and is likely acting through oriC or its initiator, DnaA.
RESULTS
Construction of conditional mutants of HBsu.
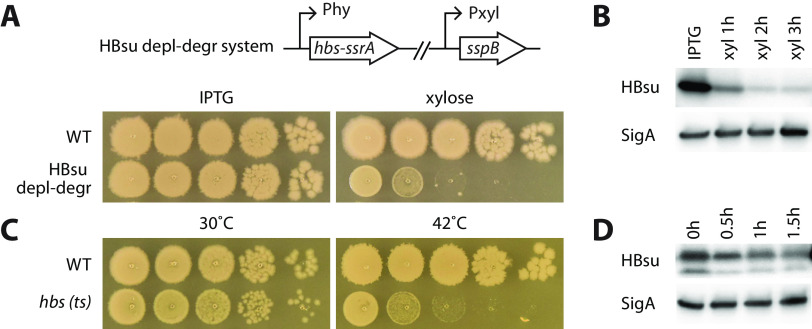
To investigate how the HBsu protein impacts chromosome organization and cell physiology, we sought to remove the protein by depletion or degradation. For depletion, at the hbs endogenous gene locus, we replaced the hbs promoter with an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (Phy) and grew the cells in the absence of inducer. For degradation, we replaced the hbs gene with hbs-ssrA fusion (28) and, at a separate genetic locus (lacA), expressed a xylose-inducible (E. coli) sspB gene, which encodes an adaptor protein for the degradation of HBsu-SsrA through the ClpXP proteosome (28). We found that either depleting or degrading HBsu was not efficient at removing the protein. Therefore, we combined these two approaches and constructed a system that could simultaneously deplete and degrade HBsu (Fig. 1A). We first grew the cells in the presence of IPTG to produce HBsu-SsrA. We then washed off the IPTG and resuspended the cells in fresh medium containing 0.5% xylose, so that transcription of hbs-ssrA was halted because of the absence of IPTG and the existing HBsu-SsrA proteins were degraded because of the xylose-induced SspB protein. In the presence of IPTG, cells grew similarly to the wild type (WT). In the absence of IPTG and presence of xylose, cell growth was severely retarded (Fig. 1A). Based on the immunoblot analysis and quantification, the HBsu-SsrA protein levels dropped to 21%, 15%, and 13% after depletion-degradation for 1 h, 2 h, and 3 h, respectively (Fig. 1B).
FIG 1.
HBsu is required for the growth of Bacillus subtilis. (A) An HBsu depletion-degradation (depl-degr) system was constructed. The gene fusion hbs-ssrA was expressed from an IPTG-inducible promoter (Phy) at the endogenous locus of hbs. E. coli sspB adaptor protein was expressed from a xylose-inducible promoter (Pxyl). The HBsu depletion-degradation strain (BWX1336) was grown in liquid medium containing 0.5 mM IPTG. Cultures were washed three times using medium without the inducer and then normalized to an OD600 of 2. The cultures were serially diluted and spotted on agar plates containing 0.5 mM IPTG (HBsu+) or 0.5% xylose (depletion-degradation). Wild-type B. subtilis (PY79) was used as a control. The plates were incubated at 37°C for 12 h. (B) Immunoblot analysis of the HBsu depletion-degradation system. The HBsu depletion-degradation strain (BWX1336) was grown in CH medium in the presence of 0.5 mM IPTG. Cells were washed three times using CH medium and then resuspended into CH medium containing 0.5% xylose (xyl). Samples were taken 1 h, 2 h, and 3 h after resuspension. When OD600 reached 0.5, the culture was diluted 1:5 in warm CH containing 0.5% xylose to maintain exponential growth. The same growth medium and condition was used for microscopy and whole-genome sequencing experiments throughout the study. Samples were blotted using HBsu polyclonal antibodies. SigA controls for loading. (C) Serial dilutions of WT and hbs(ts) (BWX4331) strains grown at 30°C and 42°C. The two strains were grown at 30°C, normalized to an OD of 2, serially diluted, and spotted on LB plates, which were incubated at indicated temperatures for 14 h. (D) Immunoblot analysis of hbs(ts) (BWX4331) strain. Cells were grown in CH medium at 30°C and shifted to 42°C for indicated times. When OD600 reached 0.5, the culture was diluted 1:5 in warm CH to maintain exponential growth. The same growth condition was used for microscopy and whole-genome sequencing experiments in this study. The membranes were blotted using HBsu polyclonal antibodies. SigA controls for loading.
As an alternative approach, we isolated temperature-sensitive (ts) alleles of hbs using a method described before (29). Briefly, the hbs gene was amplified by error-prone PCR and assembled with a spectinomycin resistance gene as well as a 2.5-kb region upstream and a 2.2-kb region downstream of the hbs gene using isothermal assembly (30). The product was transformed directly into B. subtilis, replacing the wild-type gene and selecting for spectinomycin resistance at 30°C. Transformants were arrayed and then screened for colony formation at permissive (30°C) and restrictive (42°C) temperatures. Approximately 6,900 transformants were screened. Mutants with growth rates and nucleoid morphologies most similar to those of the wild type at 30°C were characterized further. We selected the mutant that had the strongest growth defect at 42°C for our analysis and named it the hbs(ts) mutant (Fig. 1C), which contains two amino acid substitutions, D40G and P77S. Immunoblot analysis revealed an additional HBsu species that was smaller than the intact protein, indicating that HBsu(ts) is not as stable as the WT (Fig. 1D). After temperature shift, there was still a substantial fraction of the protein remaining, indicating that HBsu(ts) was inactivated at 42°C largely due to changes in protein conformation.
Removing or inactivating HBsu generated anucleate cells and larger gaps between nucleoids.
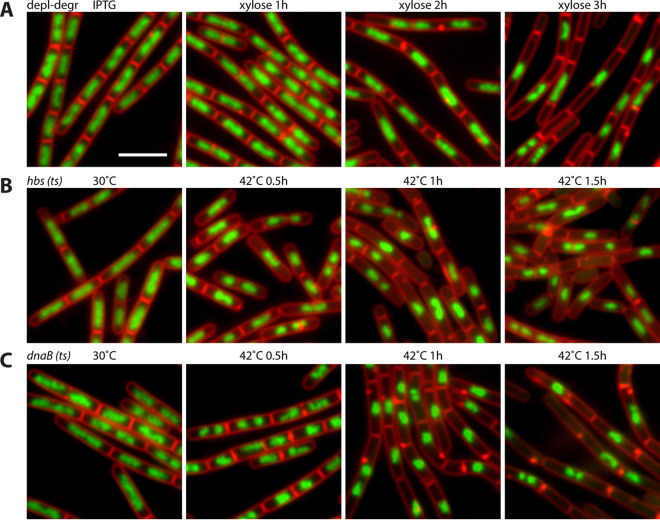
To investigate the cytological phenotype upon depletion-degradation of HBsu-SsrA, we visualized the nucleoid morphology using fluorescence microscopy. Cells were grown in the presence of 0.5 mM IPTG to mid-exponential phase. The cultures were washed three times and resuspended using prewarmed medium containing 0.5% xylose. Cultures were diluted as necessary so that the optical density at 600 nm (OD600) was below 0.6 at all time points. In the presence of IPTG, cells grew similarly to the wild type. Upon HBsu-SsrA depletion-degradation, we observed bigger gaps between nucleoids (Fig. 2A) and a dramatic increase of anucleate cells or minicells. Two hours after depletion-degradation, 10% of cells (n = 1,656) were anucleate; after another hour, the proportion of anucleate cells became 40% (n = 2,069) (Fig. 3D). A similar phenotype was observed for hbs(ts) mutant when HBsu was inactivated by shifting the temperature to 42°C (Fig. 2B). This phenotype was reminiscent of cells arrested for replication initiation, such as in dnaB(ts) (31) (Fig. 2C).
FIG 2.
Gaps between nucleoids increased upon HBsu removal or inactivation. (A) HBsu depletion-degradation strain (BWX1340) was grown in the presence of 0.5 mM IPTG and then washed three times and resuspended in warmed medium containing 0.5% xylose for indicated times. FM4-64-stained membrane (red) and DAPI-stained nucleoids (green) are shown. Bar, 4 μm. (B) hbs(ts) (BWX4414) strain was grown at 30°C, shifted to 42°C, and imaged at the indicated time points. (C) dnaB(ts) (BWX2533) strain was grown at 30°C, shifted to 42°C, and imaged at the indicated time points.
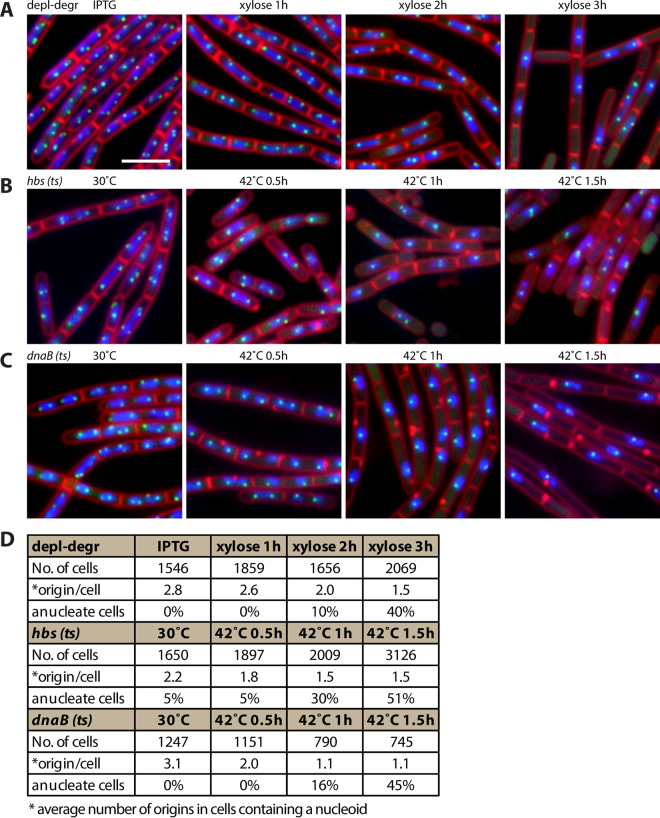
FIG 3.
Number of replication origins was reduced upon HBsu removal or inactivation. (A to C) Representative images of DAPI-stained nucleoids (blue), FM4-64-stained membrane (red), and replication origin (tetO/TetR-CFP, green) of cells growing under indicated conditions in CH medium. (A) HBsu depletion-degradation strain (BWX1340) was grown in the presence of 0.5 mM IPTG, washed three times, and resuspended in warmed medium containing 0.5% xylose. Bar, 4 μm. (B) hbs(ts) strain (BWX4414) was grown at 30°C, shifted to 42°C, and imaged at the indicated time points. (C) dnaB(ts) strain (BWX2533) was grown at 30°C, shifted to 42°C, and imaged at the indicated time points. (D) Analysis of number of origins per cell that contained nucleoids and the percentage of anucleate cells.
Removing or inactivating HBsu decreased the number of replication origins per nucleoid.
To test the effect of HBsu on DNA replication initiation, we analyzed the number of replication origins upon HBsu depletion-degradation. Replication origins were visualized using an array of 48 tetO genes bound by TetR-CFP (32). In the presence of IPTG, there were 2 to 4 origins per cell (Fig. 3A), with an average of 2.8 origins per cell (n = 1,546) (Fig. 3D). After depletion-degradation of HBsu-SsrA, the number of origins decreased to an average of 1.5 origins per cell (n = 2,069) (Fig. 3A and D). A similar effect was observed for hbs(ts) mutant when HBsu was inactivated at 42°C (Fig. 3B and D). The removal or inactivation of HBsu phenocopied the dnaB(ts) mutant, which is known to arrest DNA replication initiation (31) (Fig. 3C and D). These results indicate that HBsu is required for replication initiation in B. subtilis.
Removing or inactivating HBsu decreased ori-to-ter ratio.
To quantify the relative copy number of chromosome loci across the entire genome, we performed whole-genome sequencing (WGS). Cells were grown as described in previous sections, and samples were taken before and after HBsu depletion-degradation. The genomic DNA was extracted, processed, and sequenced using Illumina NextSeq. The samples were normalized for the total number of reads and mapped to the wild-type genome (PY79). Before HBsu-SsrA depletion-degradation, the WGS result showed the typical profile for exponential growth, with an ori-to-ter ratio of 2.8 (Fig. 4A). Upon depletion-degradation, the WGS profile became flat, and the ori-to-ter ratio dropped to 1.3 and 1.0 at 2 h and 3 h (Fig. 4A). A similar phenomenon was observed when HBsu was inactivated in hbs(ts) mutant (Fig. 4B). Again, removing or inactivating HBsu phenocopied the dnaB(ts) mutant, in which the ori-to-ter ratio dropped from 2.6 to 1.2 after temperature shift from 30°C to 42°C (Fig. 4C). These data further corroborate the conclusion that HBsu is required for replication initiation. We note that WGS measures the ori-to-ter ratio, which is lower than the origin/cell ratio measured by microscopy (Fig. 3D).
FIG 4.
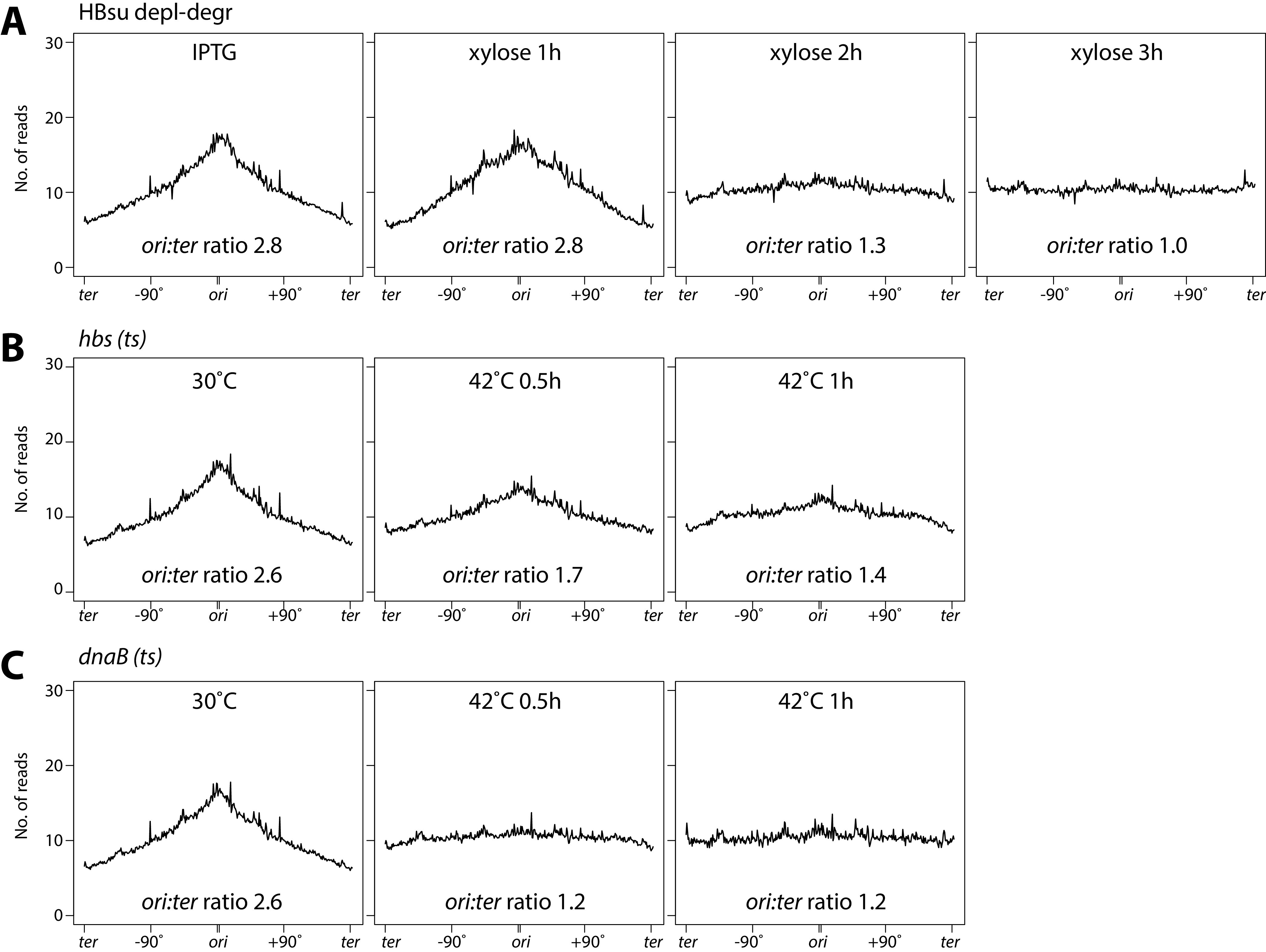
Ratio of ori to ter was reduced upon HBsu removal or inactivation. Whole-genome sequencing analysis of cells growing under indicated conditions (see Materials and Methods). The x axis represents genome location. The y axis represents the normalized number of reads. Data are plotted in 10-kb bins. (A) HBsu depletion-degradation strain (BWX1336) was grown in the presence of 0.5 mM IPTG, washed three times, and resuspended in warmed medium containing 0.5% xylose. (B) The hbs(ts) strain (BWX4331) was grown at 30°C, shifted to 42°C, and collected at indicated time points. (C) The dnaB(ts) strain (BWX2299) was grown at 30°C, shifted to 42°C, and collected at indicated time points.
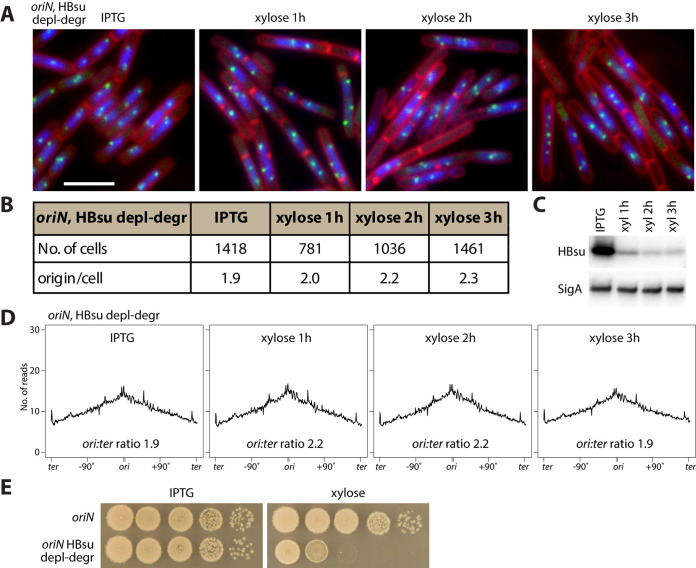
HBsu was not required for replication initiation when oriC was replaced by a plasmid replication origin, oriN.
To test whether the requirement of HBsu for replication initiation is specific to B. subtilis oriC, we introduced the HBsu depletion-degradation system to a strain containing the plasmid replication origin, oriN, as the only replication origin on the chromosome (33, 34). This strain does not require oriC or DnaA for replication. Upon HBsu depletion-degradation, the oriN strain had HBsu protein levels drop to levels similar to that of the oriC (WT) strain (compare Fig. 5C with 1B). However, in contrast to the oriC strain in which we saw a decrease in origin number per nucleoid (Fig. 3B), the average number of origin foci per cell in oriN strain did not decrease (Fig. 5A and B). In parallel, we performed whole-genome sequencing on samples collected before and after HBsu depletion-degradation in the oriN strain. We found that the replication profiles of the oriN strain remained largely unchanged upon HBsu depletion-degradation (Fig. 5D), consistent with the microscopy data (Fig. 5A and B). From these results, we concluded that HBsu is not required for replication initiation at oriN but is only required for replication initiation at oriC. Thus, the requirement of HBsu for replication initiation is likely acting through DnaA. Interestingly, in the absence of HBsu, although the oriN strain could replicate normally, it could not survive (Fig. 5E). Thus, HBsu has one or more other essential functions that are independent of its role in replication initiation.
FIG 5.
HBsu is not required for replication initiation at oriN. (A) Visualizing replication origin in oriN strain containing HBsu depletion-degradation system. BWX4530 was grown in the presence of 0.5 mM IPTG, washed three times, and resuspended in warmed medium containing 0.5% xylose. DAPI-stained nucleoids (blue), FM4-64-stained membrane (red), and replication origin (tetO/TetR-CFP, green) are shown. Bar, 4 μm. (B) Analysis of number of origins per cell that contained nucleoids and the percentage of anucleate cells. (C) Immunoblot analysis of the oriN strain containing the HBsu depletion-degradation system (BWX4397). Cells were grown in CH medium in the presence of 0.5 mM IPTG. Cells were washed three times using CH medium and then resuspended into CH medium containing 0.5% xylose. Samples were taken 1 h, 2 h, and 3 h after resuspension. When OD600 reaches 0.5, the cultures were diluted 1:5 in warm CH containing 0.5% xylose to maintain exponential growth. SigA controls for loading. The same growth condition was used for panels A, B, and D. (D) Whole-genome sequencing analysis of cells growing under indicated conditions. The x axis represents genome location. The y axis represents the normalized number of reads. Data are plotted in 10-kb bins. (E) Serial dilutions of cells containing oriN as the only replication origin with (bottom, BWX4397) or without (top, BWX4349) the HBsu depletion-degradation system. The agar plates contain 0.5 mM IPTG (HBsu+, left) or 0.5% xylose (depletion-degradation, right). The plates were incubated at 37°C for 18 h.
DISCUSSION
Although HU homologs have been shown to be required for in vitro replication experiments (13, 15), in vivo, HU is generally not essential for cell survival. The absence of HU only causes mild defects in DNA replication, such as asynchrony or delay in initiation (16, 19). In contrast, B. subtilis HBsu is essential for cell survival. Using conditional alleles to remove or inactivate HBsu, here we show that HBsu is essential for DNA replication initiation in vivo. We postulate that this essentiality is likely due to the lack of redundancy: in E. coli, two other NAPs, IHF and Fis, have been implicated in replication initiation (5), while B. subtilis lacks IHF and Fis. Consistent with this idea, although E. coli could survive the absence of one or two of HU, IHF, and HN-S, it could not survive the absence of all three (35).
We found that HBsu’s requirement for replication initiation is specific for the chromosomal origin, oriC; when oriC was changed to a plasmid origin, oriN, which uses a different initiator protein (33, 34), initiation was unchanged without functional HBsu. In E. coli, HU was shown to interact with DnaA, stabilize DnaA oligomers that are assembled on oriC, and facilitate strand opening (13). We speculate that HBsu plays a similar role in B. subtilis. Alternatively, HBsu may act on oriC DNA first, like bending the DNA, to facilitate DnaA acting on oriC. Biochemical and structural work has provided much understanding of DnaA interactions with oriC DNA sequence (36) and with initiation regulators Soj and SirA (37–39). Similar approaches aimed at identifying how HBsu interacts with DnaA or oriC will lead to better understanding of DNA replication initiation.
Here, we show that one reason HBsu is essential is its being required for replication initiation, but simply restoring replication was not sufficient to rescue cell growth. Specifically, although oriN permitted DNA replication in the absence of HBsu, it did not rescue cell viability (Fig. 5E). We infer that HBsu has other roles that are likely unrelated to DNA replication. Since HBsu is shown to be a nucleoid-associated protein and binds to the entire nucleoid (24), it is possible that structuring the nucleoid is important for various aspects of cell physiology, including but not limited to cell division and/or chromosome segregation. Moreover, pleiotropy may explain why the HU homolog is essential in B. subtilis but not in other organisms. In the future, isolation of separation-of-function mutants could identify HBsu’s essential functions and reveal the contribution of HBsu to different aspects of cell biology.
MATERIALS AND METHODS
General methods.
Bacillus subtilis strains were derived from the prototrophic strain PY79 (40). oriN strains were derived from JH642 (34). Strains used in this study are listed in Table 1; oligonucleotides and next-generation sequencing samples used in this study can be found in Tables S1 and S2 in the supplemental material. Cells were grown in defined rich casein hydrolysate (CH) medium (41). When oriN strains were grown, CH medium was supplemented with 20 μg/mL tryptophan and 50 μg/mL phenylalanine. For depletion and degradation experiments, cells were grown in the presence of 0.5 mM IPTG, washed three times with warm medium, and resuspended in medium containing 0.5% xylose. Cells were grown at 37°C, 30°C, or 42°C as indicated. Cultures were diluted in warm medium so that the cells were in mid-exponential-phase growth with an OD600 below 0.6 at all time points.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source | Figure |
|---|---|---|---|
| PY79 | Wild type | 40 | 1A–D |
| BWX1336 | Phy hbs-ssrA (spec), lacA::PxylA (Ec) sspB (loxP-erm-loxP) | This study | 1A and B, 2A, 4A |
| BWX4331 | hbs(ts) (loxP-spec-loxP) | This study | 1C and D, 2B, 4B |
| BWX2299 | dnaB134(ts), zhb83::Tn917 (erm) | This study | 2C, 4C |
| BWX1340 | yycR (−7˚)::tetO48 (cat), ycgO::PftsW tetR-cfp (phleo), lacA::PxylA (Ec) sspB loxP (no antibiotic marker) Phy hbs-ssrA (spec) | This study | 3A and D |
| BWX4414 | yycR (−7˚)::tetO48 (cat), ycgO::PftsW tetR-cfp (phleo), hbs(ts) (loxP-spec-loxP) | This study | 3B and D |
| BWX2533 | yycR (−7˚)::tetO48 (cat), ycgO::PftsW tetR-cfp (phleo), dnaX-yfp (spec), dnaB134(ts), zhb83::Tn917 (erm) | 43 | 3C and D |
| BWX4530 | trpC2, pheA1, 359˚::oriN (kan), ΔoriC-S, oriN-based replication, Phy hbs-ssrA (spec), lacA::PxylA (Ec) sspB (loxP-erm-loxP), yycR (−7˚)::tetO48 (cat), ycgO::PftsW tetR-cfp (phleo) | This study | 5A and B |
| BWX4397 | trpC2, pheA1, 359˚::oriN (kan), ΔoriC-S, oriN-based replication, Phy hbs-ssrA (spec), lacA::PxylA (Ec) sspB (loxP-erm-loxP) | This study | 5C, D, and E |
| BWX4349 (MMB208) | trpC2, pheA1, 359˚::oriN (kan), ΔoriC-S, oriN-based replication | 34 | 5E |
| KPL69 | trpC2, pheA1, dnaB134 (ts) - zhb83::Tn917 (erm) | 31 | |
| MMB208 | trpC2, pheA1, 359˚::oriN (kan), ΔoriC-S, oriN-based replication | 34 | |
| BWX1214 | hbs-ssrA (kan) | This study | |
| BWX1332 | Phy hbs-ssrA (spec) | This study |
Fluorescence microscopy.
Fluorescence microscopy was performed using a Nikon Eclipse Ti2 microscope equipped with a Plan Apo 100×/1.45-numeric-aperture phase contrast oil objective and a scientific complementary metal-oxide semiconductor (sCMOS) camera. Membranes were stained with N-(3-triethylammoniumpropyl)-4-(6-(4-[diethylamino] phenyl) hexatrienyl) pyridinium dibromide (FM4-64; Molecular Probes) at 3 μg/mL. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes) at 2 μg/mL. Images were cropped and adjusted using MetaMorph software. Final figure preparation was performed in Adobe Illustrator.
Construction of Phy hbsu-ssrA (spec) strain.
Phy hbsu-ssrA (spec) strain (BWX1332) was built in two steps (see below). First, a degradation-only strain, hbs-ssrA (kan) (BWX1214), was built. Secondly, hbs-ssrA (kan) strain was used to build Phy hbsu-ssrA (spec) strain.
hbs-ssrA (kan) strain (BWX1214) was built by direct transformation of an isothermal assembly product that contained three PCR fragments: (i) a 0.6-kb fragment containing the upstream region and open reading frame of the hbs gene (amplified from wild-type genomic DNA using oligonucleotides oWX442 and oWX443); (ii) the ssrA kan fragment from pWX499 (1.7 kb) (amplified using oligonucleotides oWX444 and oWX445); and (iii) a 0.6-kb fragment downstream of the hbs gene (amplified from wild-type genomic DNA using oligonucleotides oWX446 and oWX447). pWX499 is a cloning vector that contains the ssrA tag and the kanamycin resistance gene. After transformation into wild-type B. subtilis, the hbs gene region of the resulting strain was amplified and sequenced using oWX448 and 449.
Phy hbsu-ssrA (spec) strain (BWX1332) was built by direct transformation of an isothermal assembly product that contained five PCR fragments: (i) a 1.3-kb fragment containing the upstream region of the hbs gene (amplified from wild-type genomic DNA using oligonucleotides oWX475 and oWX476); (ii) a spectinomycin resistance gene and Phy promoter from pDR111 (1.6 kb) (amplified using oligonucleotides oWX477 and oWX478); (iii) the hbs-ssrA region (amplified from BWX1214 genomic DNA using oligonucleotides oWX479 and oWX480); (iv) a lacI fragment from pDR111 (amplified using oligonucleotides oWX481 and oWX482); and (v) a 1.2-kb fragment downstream of the hbs gene (amplified from wild-type genomic DNA using oligonucleotides oWX483 and oWX484). pDR111 is a cloning vector that contains the Phy promoter, a spectinomycin resistance gene, and the lacI gene. After transforming the isothermal assembly product to wild-type B. subtilis, the entire Phy hbsu-ssrA (spec) region was amplified using oWX475 and oWX484 and sequenced using oWX485-oWX489.
Generation of hbs mutant libraries.
We constructed an hbs mutant library by direct transformation of an isothermal assembly product that contained four PCR fragments: (i) a 2.5-kb fragment containing the upstream region of the hbs gene (amplified from wild-type genomic DNA using Phusion polymerase and oligonucleotides oWX853 and oWX854); (ii) the hbs gene (352 bp) generated by error-prone PCR (GeneMorph II random mutagenesis kit; Stratagene) and oligonucleotides oWX855 and oWX856, under conditions in which each amplicon had, on average, 1 mutation per 300 bp; (iii) a spectinomycin resistance cassette (amplified from pWX466 using oligonucleotides oWX438 and oWX439); and (iv) a 2.2-kb fragment downstream of the hbs gene (amplified from wild-type genomic DNA using Phusion polymerase and oligonucleotides oWX857 and oWX858). The transformation was plated on LB agar plates supplemented with spectinomycin (100 μg/mL) at 30°C. More than 40,000 colonies were pooled, aliquoted, and frozen in LB medium containing 14% glycerol.
Screening for hbs(ts) mutants.
Frozen aliquots of the mutant library were thawed, diluted, and plated onto rectangular plates (OmniTray; Thermo Scientific) containing LB agar supplemented with spectinomycin at a density of ~200 colonies per plate. The plates were incubated 16 to 18 h at 30°C. A colony-picking robot (BioMatrix; S&P Robotics) picked and arrayed independent transformants onto rectangular LB agar plates, which were incubated overnight at 30°C. The rearrayed colonies were replica plated onto two LB agar plates. One was placed at 30°C and the other at 42°C. Mutants that grew well at 30°C and grew poorly at 42°C were streaked for single colonies and retested to make sure the phenotype is reproducible. Genomic DNA from these mutants were used to transform the wild type to confirm linkage. The mutagenized region was then sequenced. Approximately 6,900 transformants were screened. Fifteen candidate clones were retested for streaking. Four mutants had growth close to that of the wild type at 30°C. These 4 mutants were characterized further for nucleoid morphologies. The mutant that was the most similar to the wild type for growth rate and nucleoid morphology and had the strongest defect at 42°C was named the hbs(ts) mutant and used for our analysis. The hbs region was amplified using oWX447 and oWX448 and sequenced using oWX442. Two amino acid substitutions were present in hbs(ts) (D40G and P77S). Note that the hbs(ts) mutant has a lower growth rate than the wild type at 30°C (Fig. 1C).
Whole-genome sequencing for DNA replication profile.
Cells were grown in CH medium containing desired supplements (such as 0.5 mM IPTG or 0.5% xylose) at 37°C, 30°C, or 42°C as indicated. Samples were collected at the desired time points. Genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit (69504; Qiagen). DNA was sonicated using a Qsonica Q800L sonicator for 12 min at 20% amplitude to achieve an average fragment size of 250 bp. DNA library was prepared using NEBNext Ultra II kit (E7645; NEB) and sequenced using Illumina NextSeq. Sequencing reads were mapped to the B. subtilis PY79 genome (NCBI reference sequence NC_022898.1) using CLC Genomics Workbench (Qiagen). The mapped reads were normalized to the total number of reads for that sample and plotted in R.
Immunoblot analysis.
Cells were collected at appropriate time points and resuspended in lysis buffer (20 mM Tris, pH 7.0, 1 mM EDTA, 10 mM MgCl2, 1 mg/mL lysozyme, 10 μg/mL DNase I, 100 μg/mL RNase A, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1% proteinase inhibitor cocktail (P-8340; Sigma) to a final OD600 of 10 for equivalent loading. The cell resuspensions were incubated at 37°C for 10 min for lysozyme treatment, followed by the addition of an equal volume of 2× Laemmli sample buffer (1610737; Bio-Rad) containing 10% β-mercaptoethanol. Samples were heated for 5 min at 80°C prior to loading. Proteins were separated by precast 4 to 20% polyacrylamide gradient gels (Bio-Rad 4561096) and electroblotted onto mini-polyvinylidene difluoride membranes using Bio-Rad Transblot Turbo system and reagents (1704156; Bio-Rad). The membranes were blocked in 5% nonfat milk in phosphate-buffered saline (PBS) with 0.5% Tween 20 and then probed with anti-HBsu (1:5,000) (gift from David Rudner) or anti-SigA (1:10,000) (42) diluted into 3% bovine serum albumin in 1× PBS–0.05% Tween 20. Primary antibodies were detected using Immun-Star horseradish peroxidase-conjugated goat anti-rabbit antibodies (1705046; Bio-Rad) and Western Lightning Plus ECL chemiluminescence reagents as described by the manufacturer (NEL1034001; Perkin Elmer). The signal was captured using ProteinSimple FluorChem system. The intensity of the bands was quantified using ProteinSimple AlphaView software.
Data availability.
Whole-genome sequencing data were deposited in the NCBI Sequence Read Archive (SRA) (accession no. PRJNA817244).
ACKNOWLEDGMENTS
We thank all members of the Wang laboratory for stimulating discussions. We thank David Rudner for HBsu antibodies, Alan Grossman for oriN strains, Olive Tang for help with hbs(ts) mutant screen, and Dan Kearns for reading our manuscript. This work was conceived when X.W. was in David Rudner’s lab at Harvard Medical School.
Support for this work comes from the NIH grants R01GM141242 and R01GM143182 (X.W.).
Footnotes
Supplemental material is available online only.
Contributor Information
Xindan Wang, Email: xindan@indiana.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Dame RT, Kalmykowa OJ, Grainger DC. 2011. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet 7:e1002123. 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 3.Dame RT, Rashid FM, Grainger DC. 2020. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat Rev Genet 21:227–242. 10.1038/s41576-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 4.Dame RT, Tark-Dame M. 2016. Bacterial chromatin: converging views at different scales. Curr Opin Cell Biol 40:60–65. 10.1016/j.ceb.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Leonard AC, Grimwade JE. 2005. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol Microbiol 55:978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haniford DB. 2006. Transpososome dynamics and regulation in Tn10 transposition. Crit Rev Biochem Mol Biol 41:407–424. 10.1080/10409230600987415. [DOI] [PubMed] [Google Scholar]
- 7.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13:773–780. 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Dorman CJ, Kane KA. 2009. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev 33:587–592. 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- 9.Rimsky S, Travers A. 2011. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol 14:136–141. 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Stavans J, Oppenheim A. 2006. DNA-protein interactions and bacterial chromosome architecture. Phys Biol 3:R1–R10. 10.1088/1478-3975/3/4/R01. [DOI] [PubMed] [Google Scholar]
- 11.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. 2006. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol 156:262–272. 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, Boccard F, Koszul R. 2018. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172:771–783. 10.1016/j.cell.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. 2008. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol Microbiol 67:781–792. [DOI] [PubMed] [Google Scholar]
- 14.Hwang DS, Kornberg A. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem 267:23083–23086. 10.1016/S0021-9258(18)50059-4. [DOI] [PubMed] [Google Scholar]
- 15.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. 2002. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol Microbiol 46:113–124. 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 16.Bahloul A, Boubrik F, Rouviere-Yaniv J. 2001. Roles of Escherichia coli histone-like protein HU in DNA replication: HU-beta suppresses the thermosensitivity of dnaA46ts. Biochimie 83:219–229. 10.1016/S0300-9084(01)01246-9. [DOI] [PubMed] [Google Scholar]
- 17.Huisman O, Faelen M, Girard D, Jaffe A, Toussaint A, Rouviere-Yaniv J. 1989. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol 171:3704–3712. 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada M, Kano Y, Ogawa T, Okazaki T, Imamoto F. 1988. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol 204:581–591. 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- 19.Hołówka J, Trojanowski D, Janczak M, Jakimowicz D, Zakrzewska-Czerwińska J. 2018. The origin of chromosomal replication is asymmetrically positioned on the mycobacterial nucleoid, and the timing of its firing depends on HupB. J Bacteriol 200:e00044-18. 10.1128/JB.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein W, Marahiel MA. 2002. Structure-function relationship and regulation of two Bacillus subtilis DNA-binding proteins, HBsu and AbrB. J Mol Microbiol Biotechnol 4:323–329. [PubMed] [Google Scholar]
- 21.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 22.Micka B, Groch N, Heinemann U, Marahiel MA. 1991. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J Bacteriol 173:3191–3198. 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groch N, Schindelin H, Scholtz AS, Hahn U, Heinemann U. 1992. Determination of DNA-binding parameters for the Bacillus subtilis histone-like HBsu protein through introduction of fluorophores by site-directed mutagenesis of a synthetic gene. Eur J Biochem 207:677–685. 10.1111/j.1432-1033.1992.tb17095.x. [DOI] [PubMed] [Google Scholar]
- 24.Kohler P, Marahiel MA. 1997. Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J Bacteriol 179:2060–2064. 10.1128/jb.179.6.2060-2064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham TG, Wang X, Song D, Etson CM, van Oijen AM, Rudner DZ, Loparo JJ. 2014. ParB spreading requires DNA bridging. Genes Dev 28:1228–1238. 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Hegarat F, Salti-Montesanto V, Hauck Y, Hirschbein L. 1993. Purification and characterization of the HU-like protein HPB9 from the Bacillus subtilis nucleoid. Biochim Biophys Acta 1172:101–107. 10.1016/0167-4781(93)90275-I. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez S, Rojo F, Alonso JC. 1997. The Bacillus subtilis chromatin-associated protein Hbsu is involved in DNA repair and recombination. Mol Microbiol 23:1169–1179. 10.1046/j.1365-2958.1997.3061670.x. [DOI] [PubMed] [Google Scholar]
- 28.Griffith KL, Grossman AD. 2008. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol 70:1012–1025. 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Tang OW, Riley EP, Rudner DZ. 2014. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol 24:287–292. 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 31.Rokop ME, Auchtung JM, Grossman AD. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol 52:1757–1767. 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Montero Llopis P, Rudner DZ. 2014. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA 111:12877–12882. 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan AK, Moriya S, Ogura M, Tanaka T, Kawamura F, Ogasawara N. 1997. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J Bacteriol 179:2494–2502. 10.1128/jb.179.8.2494-2502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkmen MB, Grossman AD. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol Microbiol 63:150–165. 10.1111/j.1365-2958.2006.05505.x. [DOI] [PubMed] [Google Scholar]
- 35.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. 1992. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene 122:9–15. [DOI] [PubMed] [Google Scholar]
- 36.Richardson TT, Harran O, Murray H. 2016. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534:412–416. 10.1038/nature17962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jameson KH, Rostami N, Fogg MJ, Turkenburg JP, Grahl A, Murray H, Wilkinson AJ. 2014. Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA. Mol Microbiol 93:975–991. 10.1111/mmi.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard TA, Butler PJ, Lowe J. 2005. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J 24:270–282. 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahn-Lee L, Merrikh H, Grossman AD, Losick R. 2011. The sporulation protein SirA inhibits the binding of DnaA to the origin of replication by contacting a patch of clustered amino acids. J Bacteriol 193:1302–1307. 10.1128/JB.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci USA 80:2305–2309. [6300908]. 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, New York, NY. [Google Scholar]
- 42.Fujita M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5:79–88. 10.1046/j.1365-2443.2000.00307.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. 2015. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev 29:1661–1675. 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S2. Download jb.00119-22-s0001.pdf, PDF file, 0.4 MB (47.2KB, pdf)
Data Availability Statement
Whole-genome sequencing data were deposited in the NCBI Sequence Read Archive (SRA) (accession no. PRJNA817244).