ABSTRACT
The bacterial flagellar type III secretion system (fT3SS) is a suite of membrane-embedded and cytoplasmic proteins responsible for building the flagellar motility machinery. Homologous nonflagellar (NF-T3SS) proteins form the injectisome machinery that bacteria use to deliver effector proteins into eukaryotic cells, and other family members were recently reported to be involved in the formation of membrane nanotubes. Here, we describe a novel, evolutionarily widespread, hat-shaped structure embedded in the inner membranes of bacteria, of yet-unidentified function, that is present in species containing fT3SS. Mutant analysis suggests a relationship between this novel structure and the fT3SS, but not the NF-T3SS. While the function of this novel structure remains unknown, we hypothesize that either some of the fT3SS proteins assemble within the hat-like structure, perhaps including the fT3SS core complex, or that fT3SS components regulate other proteins that form part of this novel structure.
IMPORTANCE The type III secretion system (T3SS) is a fascinating suite of proteins involved in building diverse macromolecular systems, including the bacterial flagellar motility machine, the injectisome machinery that bacteria use to inject effector proteins into host cells, and probably membrane nanotubes which connect bacterial cells. Here, we accidentally discovered a novel inner membrane-associated complex related to the flagellar T3SS. Examining our lab database, which is comprised of more than 40,000 cryo-tomograms of dozens of species, we discovered that this novel structure is both ubiquitous and ancient, being present in highly divergent classes of bacteria. Discovering a novel, widespread structure related to what are among the best-studied molecular machines in bacteria will open new venues for research aiming at understanding the function and evolution of T3SS proteins.
KEYWORDS: bacteria, cryo-ET, flagella, secretion systems
INTRODUCTION
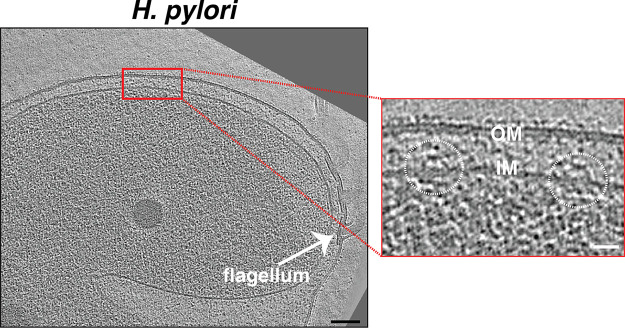
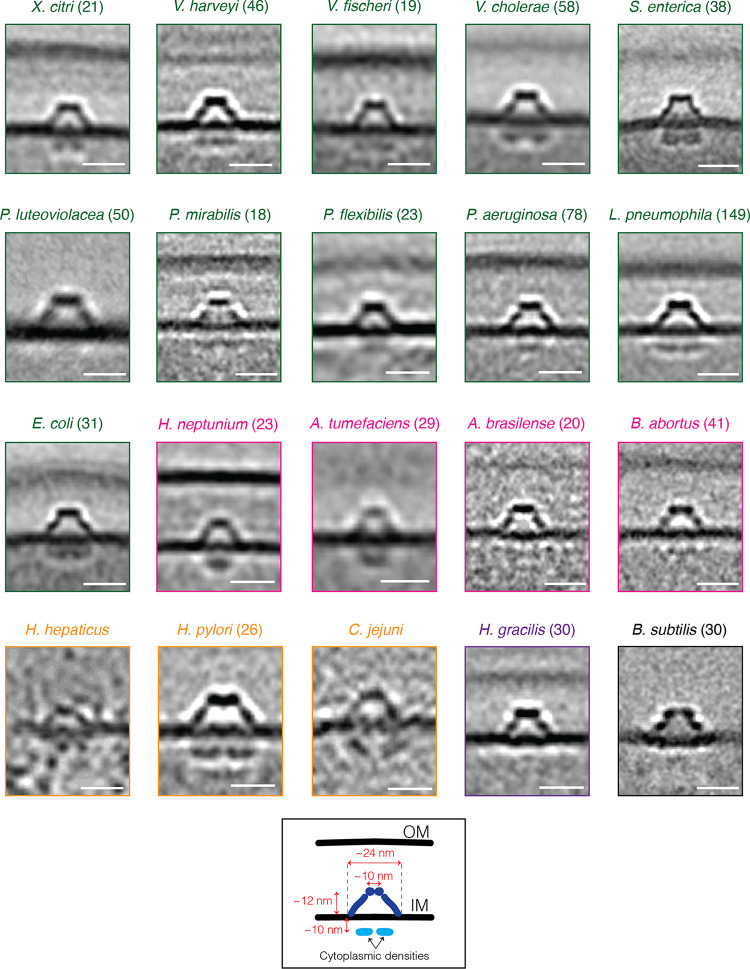
Cryogenic electron tomography (cryo-ET) images intact cells in 3D in a native, frozen-hydrated state, occasionally revealing unexpected structures (1–4). Examining a database of tens of thousands of electron cryo-tomograms of phylogenetically diverse bacterial species that our lab has imaged over the past 15 years (5, 6), we found an inner membrane (IM)-associated, hat-shaped structure in diverse Gram-negative and Gram-positive bacteria (Fig. 1 and S1). In many cases, we observed multiple such structures, up to 10, distributed around the cell (see Movie S1 for an example from an Escherichia coli cell that was partially lysed, enhancing visibility of periplasmic structures). Subtomogram averages of the structure from different species revealed conserved characteristics: a hat-shaped part in the periplasm (of diderms) or extracellular space (of monoderm Bacillus subtilis) and two cytoplasmic densities beneath, located ~10 nm below the IM (Fig. 2). In general, the periplasmic portion had a diameter of ~25 nm at its widest point at the outer leaflet of the IM, a diameter of ~10 nm at its top, and a height of ~12 nm.
FIG 1.
Identification of a novel hat-like structure in H. pylori. A slice through an electron cryo-tomogram of an H. pylori cell, showing two hat-like structures (white circles in the enlargement). Scale bars represent 100 nm in the main panel and 25 nm in the enlargement.
FIG 2.
The hat-like complex is a widespread bacterial structure. A gallery of the hat-like structure in various bacterial species (Xanthomonas citri, Vibrio harveyi, V. fischeri, V. cholerae, Salmonella enterica, Pseudoalteromonas luteoviolacea, Proteus mirabilis, Pseudomonas flexibilis, P. aeruginosa, Legionella pneumophila, Escherichia coli, Hyphomonas neptunium, Agrobacterium tumefaciens, Azospirillum brasilense, Brucella abortus, Helicobacter hepaticus, H. pylori, Campylobacter jejuni, Hylemonella gracilis, and Bacillus subtilis). Subtomogram averages are shown for all species except C. jejuni and H. hepaticus, for which insufficient data were available for averaging. For these two species, single tomographic slices are shown. Numbers in parentheses indicate how many particles were used to produce the subtomogram average. Color coding indicates taxonomic class: green, Gammaproteobacteria; pink, Alphaproteobacteria; light orange, Epsilonproteobacteria; purple, Betaproteobacteria; and black, Bacilli. Scale bars represent 20 nm.
The cytoplasmic densities were absent in the averages from three species: Pseudoalteromonas luteoviolacea, Hylemonella gracilis, and Bacillus subtilis. In P. luteoviolacea and B. subtilis, this could be due to the fact that the tomograms from these species, which were collected for other projects, were of lysed, not intact, cells. While lysing cells decreases their thickness, thereby increasing the quality of cryo-tomograms, it can also affect the integrity of macromolecular complexes, e.g., the flagellar motor (7). However, it is unclear why the cytoplasmic densities were also missing in H. gracilis, since these cryo-tomograms were of intact cells. In the averages from several other species, the cytoplasmic density did not resolve into two distinct sections (Fig. 2).
MUTANT ANALYSIS
We identified the same novel structure in an H. pylori strain which contains a naturally occurring point mutation that disrupts the function of FliP (8), the platform on which other flagellar type III secretion system (fT3SS) proteins assemble (Fig. 3). We imaged this strain (henceforth referred to as H. pylori fliP*) in the course of a study of flagellar assembly (9). Curiously, the diameter of the hat-like density in H. pylori fliP* was reduced to only ~20 nm at its widest part, and the two cytoplasmic densities were missing (Fig. 4A and C). These changes reminded us of the changes that we observed in flagellar intermediates in H. pylori fliP*. The flagellar membrane/supramembrane-ring complex (MS-complex) of H. pylori fliP* similarly lacks its two cytoplasmic densities, and the MS-ring (FliF) adopts a smaller diameter (~20 nm) compared to that of motile H. pylori (~25 nm) (Fig. 4B and D). In the MS-complex, the two cytoplasmic densities, which also appear ~10 nm below the IM, are known to be formed by the C-terminal domain of the fT3SS protein FlhA (FlhAC), which forms a nonameric ring (Fig. 3 and 4B) (10). The similar dimensions of the hat-like structure and the MS-complex as well as the comparable changes that occur in H. pylori fliP*suggested to us that the hat-like structure is related to the fT3SS and that its cytoplasmic densities could also be formed by FlhAC.
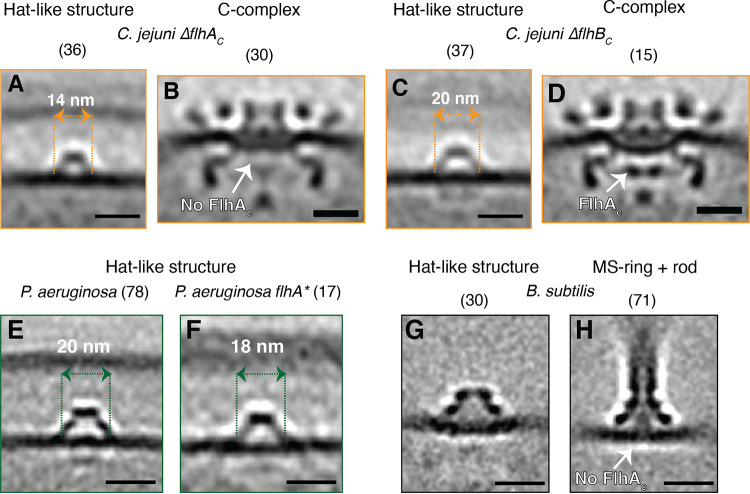
FIG 3.
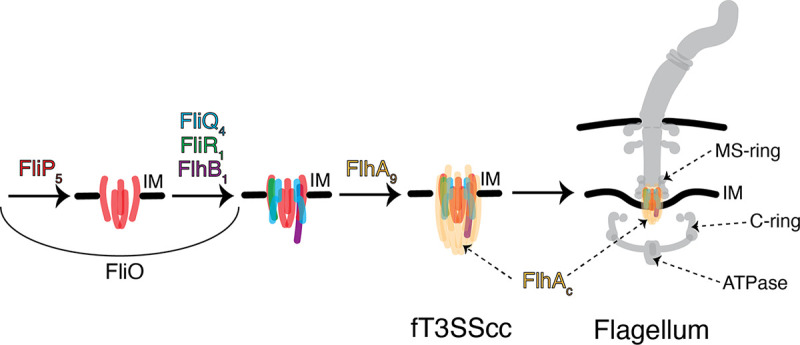
Schematic representation of the fT3SS assembly. The process starts with assembly of the fT3SScc proteins in the inner membrane (IM) and continues with the addition of the other components of the flagellum.
FIG 4.
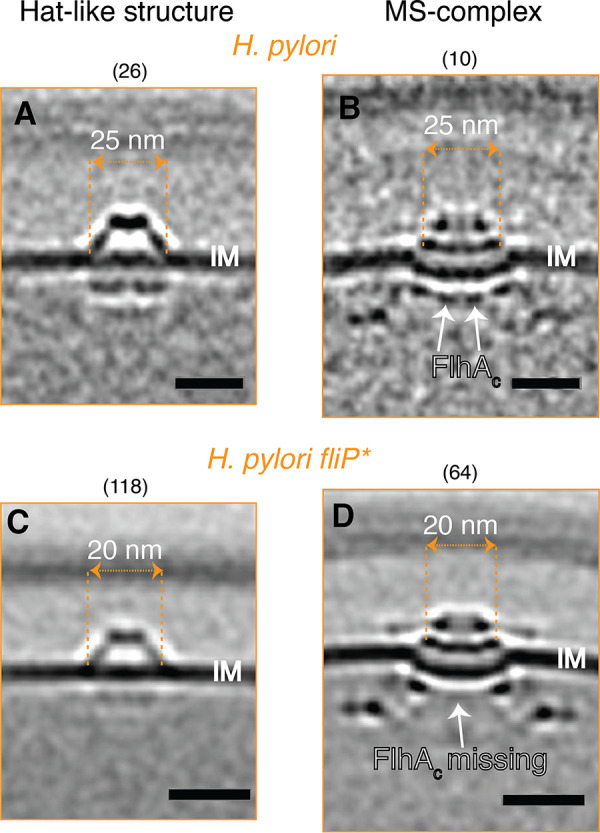
Comparison of the hat-like structure and MS-complexes in H. pylori strains. Central slices through subtomogram averages of the hat-like structures (A, C) and MS-complexes (B, D) of motile wild-type H. pylori (A, B) and nonmotile H. pylori fliP* (C, D). The diameters of the widest parts of the hat-like structures and the MS-rings are indicated. Numbers in parentheses indicate how many particles were used to produce the subtomogram average. (B, D) Adapted from (9). Scale bars represent 20 nm.
The fT3SS consists of a cytoplasmic part containing an ATPase and an IM-embedded part known as the core complex (fT3SScc). The fT3SScc is comprised of five proteins (FliP, FliQ, FliR, FlhB, and FlhA), with another protein, FliO, being required for assembly but not forming part of the complex (11, 12). Initially, FliP forms a pentameric platform on which FliQ, FliR, and FlhB assemble to create a FliP5FliQ4FliR1FlhB1 subcomplex upon which an FlhA ring is built (13) (Fig. 3). To explore our hypotheses that the hat-like structure is related to the fT3SS and that its cytoplasmic densities are formed by FlhAC, we further mined our database for mutants lacking other fT3SScc proteins in species in which we had identified the hat-like structure.
First, we examined Campylobacter jejuni mutants lacking the C-terminal domains of FlhA (ΔflhAc) and FlhB (ΔflhBc) (10). In the ΔflhAc cells, compared to wild-type cells, the periplasmic hat-like part was again smaller in diameter, and the two cytoplasmic densities again disappeared, supporting our hypothesis that they are formed by FlhAC (Fig. 5A). Of course, it is possible that FlhAC does not directly constitute the cytoplasmic densities but rather regulates the localization (or expression) of another protein or group of proteins that does. Such a regulatory role has been identified for another fT3SScc protein, FliO, which is responsible for the optimal expression of other flagellar genes (14). The ΔflhAc mutant arrests flagellar assembly at the C-complex (constituting the MS-ring, the fT3SS, the cytoplasmic [C-] ring, and other associated periplasmic components [9]), and we again observed that this intermediate lacked the cytoplasmic densities of FlhAC (Fig. 5B). In contrast, in ΔflhBc cells, the hat-like structure was indistinguishable from the wild-type complex, both in diameter and in the presence of the associated cytoplasmic densities (Fig. 5C), and the cytoplasmic FlhAC densities were also present in the flagellar C-complex (Fig. 4D). This lack of a visible difference was not surprising, since, unlike the large pentameric FliP ring or the nonameric FlhA ring, FlhB is a small protein present in a monomeric form in the fT3SScc. To confirm the generality of the relationship between the fT3SScc and the hat-like complex, we imaged an flhA mutant of P. aeruginosa (flhA*, obtained from a transposon insertion mutant library). Here, also, the hat-like structure appeared smaller in size (18 nm versus 20 nm) and lacked clear cytoplasmic densities compared to that of the wild-type cells (Fig. 5E and F), although the number of identified structures in the flhA* mutant was fewer than that of wild-type cells and produced a poorer-quality subtomogram average.
FIG 5.
Comparison of the hat-like structure and flagellar subcomplexes in fT3SScc mutants and lysed cells. (A–D) Central slices through subtomogram averages of the hat-like structure (A, C) and flagellar C-complex (B, D) in C. jejuni ΔflhAc (A, B) and ΔflhBc (C, D) cells. (E, F) Central slices through subtomogram averages of the hat-like structure in P. aeruginosa wild-type (E) and flhA* (F) cells. (G, H) Central slices through subtomogram averages of the hat-like structure (G) and a flagellar subcomplex constituting the MS-ring and rod (H) from lysed B. subtilis cells. (H) Adapted from (7). Numbers in parentheses indicate how many particles were used to produce the subtomogram average. Scale bars represent 20 nm.
Another similarity between the hat-like structure and the flagellar system was the absence of the cytoplasmic densities in both structures in the cryo-tomograms of lysed cells. Recently, we showed that cell lysis can lead to the loss of the C-ring and FlhAC densities in flagellar motors (7). In cryo-tomograms of lysed Bacillus subtilis, in which the hat-like structure lacked its two cytoplasmic densities, we identified a flagellar subcomplex consisting of the MS-ring and rod but lacking the cytoplasmic FlhAC densities (Fig. 5G and H). Moreover, the MS-ring in this flagellar subcomplex looked morphologically similar (at the resolution of our subtomogram averages) to the hat-like structure (Fig. 5G and H).
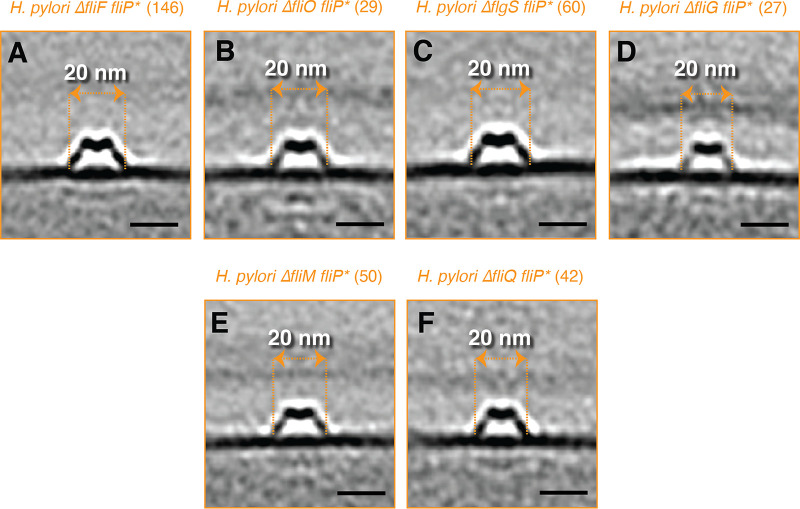
These observations strengthened our hypothesis that the hat-like structure is related to the fT3SS, and we speculated that the periplasmic hat may be formed by the flagellar MS-ring protein, FliF, which assembles on the fT3SScc during flagellar assembly (Fig. 3) in a different, more closed conformation than that observed in the flagellar motor. We therefore generated and imaged a ΔfliF mutant in the H. pylori fliP* background. To our surprise, while we could not identify any MS-complexes in this mutant, the hat-like complex was still present, though again with a smaller diameter and lacking clear cytoplasmic densities (Fig. 6A). The periplasmic hat, therefore, cannot be FliF. Subsequently, we speculated that it could be FliO, which is believed to have a large periplasmic domain in H. pylori (14) and is required for fT3SScc formation. We produced and imaged a ΔfliO fliP* H. pylori mutant but still observed hat-like structures (again lacking the cytoplasmic densities and with a reduced width of ~20 nm at the base), ruling out FliO, as well (Fig. 6B).
FIG 6.
The effect of various flagellar-related mutations on the hat-like structure in H. pylori fliP*. (A–F) Central slices through subtomogram averages of the hat-like structures in the indicated mutants of H. pylori. Numbers in parentheses indicate how many particles were used to produce the subtomogram average. Scale bars represent 20 nm.
From our study of flagellar assembly, we had cryo-tomograms of several flagellar-related mutants of H. pylori: ΔflgS fliP*, ΔfliM fliP*, ΔfliG fliP*, and ΔfliQ fliP*. These mutants remove an additional fT3SScc protein (ΔfliQ), the C-ring proteins (ΔfliM and ΔfliG), or the tyrosine kinase responsible for the expression of the class II flagellar genes (ΔflgS) (15), and they were all constructed in the fliP* background to capture early flagellar intermediates (9). In all of these mutants, we observed hat-like structures, again with reduced diameter (~20 nm at the widest part), with missing or less well-resolved cytoplasmic densities (Fig. 6C–F). These differences were not due to decreased resolution, since in each case, more particles were averaged than from wild-type cells (see numbers above, subtomogram averages). We also investigated whether any of the H. pylori mutants affected the abundance of the hat-like structure but found no obvious correlation between the strain and the number of hat-like structures per cell (Fig. S2). While H. pylori fliP* cells had more structures per cell than did motile cells, other mutants, namely, ΔfliM fliP*, ΔfliO fliP*, and ΔfliQ fliP*, had fewer. Interestingly, the hat-like structure was still present in two mutants that did not contain visible flagellar intermediates, ΔfliG fliP* and ΔfliF fliP* (Fig. S2). Taken together, our observations suggest that while the cytoplasmic densities of the novel complex are likely FlhAC, the periplasmic density is not formed by FliF or other fT3SScc proteins.
CORRELATION WITH FT3SS But Not With NF-T3SS
Next, we examined whether the hat-like structure is also related to the nonflagellar (NF-) T3SS. Bioinformatics analysis has shown that the NF-T3SS evolved from the flagellar system through a series of gene deletions, initially producing a machine thought to be involved in secreting proteins across the IM, an extant example of which is present in Myxococcales (16). Subsequently, the acquisition of an outer membrane-associated secretin resulted in the contact-dependent NF-T3SS (the injectisome) (16). First, we examined Myxococcus xanthus, which has NF-T3SS genes but no gene encoding a secretin, but found no hat-like structures in 95 cryo-tomograms (Fig. S3). Next, we examined Prosthecobacter vanneervenii, which has genes for the NF-T3SS and a secretin, but found no hat-like structures in 91 cryo-tomograms (Fig. S3). Enteropathogenic Escherichia coli (EPEC) has both the NF-T3SS injectisome and the fT3SS (17). In our database, we have cryo-tomograms of an EPEC strain that has the core complex proteins of the NF-T3SS (escR, escS, escT, escU, and escV) but lacks the other components of the injectisome and the fT3SScc (see Materials and Methods). We identified no hat-like structures in 200 cryo-tomograms of this strain (Fig. S3). Finally, as a negative-control, we investigated bacterial species that do not have any T3SS-related genes (neither fT3SS nor NF-T3SS). We examined Flavobacterium johnsoniae (206 cryo-tomograms), Coxiella burnetii (149 cryo-tomograms), and Amoebophilus asiaticus (9 cryo-tomograms) but did not identify any hat-like structures in any of these species (Fig. S3). Together, these results suggest that the hat-like structure is specifically related to the fT3SS (Table 1).
TABLE 1.
Summary of the species investigated in this study, indicating the presence (+) or absence (−) of fT3SS genes, NF-T3SS genes, and hat-like structures in each
| Species name | fT3SS genes | NF-T3SS genes | Hat-like structures |
|---|---|---|---|
| Helicobacter pylori | + | − | Yes |
| Xanthomonas citri | + | − | Yes |
| Vibrio harveyi | + | − | Yes |
| Vibrio fischeri | + | − | Yes |
| Vibrio cholerae | + | − | Yes |
| Pseudoalteromonas luteoviolacea | + | − | Yes |
| Pseudomonas flexibilis | + | − | Yes |
| Legionella pneumophila | + | − | Yes |
| Escherichia coli | + | − | Yes |
| Hyphomonas neptunium | + | − | Yes |
| Agrobacterium tumefaciens | + | − | Yes |
| Azospirillum brasilense | + | − | Yes |
| Brucella abortus | + | − | Yes |
| Helicobacter hepaticus | + | − | Yes |
| Campylobacter jejuni | + | − | Yes |
| Hylemonella gracilis | + | − | Yes |
| Salmonella enterica | + | + | Yes |
| Proteus mirabilis | + | + | Yes |
| Pseudomonas aeruginosa | + | + | Yes |
| Bacillus subtilis | + | + | Yes |
| Prosthecobacter vanneervenii | − | + | No |
| EPEC a | − | + | No |
| Myxococcus xanthus | − | + | No |
| Flavobacterium johnsoniae | − | − | No |
| Coxiella burnetii | − | − | No |
| Amoebophilus asiaticus | − | − | No |
This EPEC mutant lacks all the NF-T3SS genes except the core complex ones and has all the flagellar genes except the fT3SScc ones.
DISCUSSION
What could the function of the hat-like structure be? We do not know; we can only speculate. One possibility, for example, is that it plays a role in the assembly of early flagellar components. The fT3SScc is the first to assemble during flagellar biogenesis, followed by the MS- and C-rings (11, 12). Then, the process proceeds in a cooperative way in which the absence of a certain component prevents subsequent parts from assembling (18–20). However, we recently found that in an H. pylori fliP* strain, the MS-ring, which depends on Sec for secretion through the IM (21), can still assemble in the absence of the fT3SScc (9). One hypothesis for the function of the novel structure described here is that it is somehow involved in this process of early flagellar biogenesis. A physical association with the fT3SScc would explain the disappearance of the cytoplasmic densities and the smaller diameter of the periplasmic portion in the fliP* and flhA mutants. While bioinformatics analysis in the future might help to identify candidates for this structure, the fact that flagellar genes do not always cluster in the same way in all species (22) makes this challenging.
Another possibility is that this apparently ancient structure may have diverged to serve a function independent of flagellar assembly. Supporting an independent role, the hat-like structures did not show a preferred spatial distribution in the cell, even in species with polar flagella. It is also possible that the structure serves different functions in different species. This could explain the structural variations we observed, such as the absence of the cytoplasmic densities in H. gracilis. Whatever the function of the hat-like complex, it joins the already rich repertoire of the (f)T3SS, which has roles in flagellar motility and protein translocation and was recently implicated in membrane nanotube formation (23).
MATERIALS AND METHODS
Strains and growth conditions.
E. coli cells were grown as described (24). X. citri cells were grown in 2xTY medium for 14 h to stationary phase. V. cholerae, V. harveyi, and V. fischeri were grown as described (25). P. luteoviolacea were grown as described (4, 26). P. mirabilis were grown as described (27). P. aeruginosa were grown in LB medium at 37°C overnight. The P. aeruginosa flhA* mutant was obtained from a transposon library (mutant number 3296 from the nonredundant library http://pa14.mgh.harvard.edu/cgi-bin/pa14/downloads.cgi) from Dianne Newman’s lab at Caltech. L. pneumophila were grown as described (28). S. enterica were grown as described (29). P. flexibilis were grown in lactose growth medium. H. neptunium were grown to exponential phase in PYE medium. A. tumefaciens wild-type cells with a plasmid-borne VirC1-GFP translational fusion under control of the VirB promoter were grown in AB medium with 150 to 300 μg/mL of kanamycin. A. brasilense and B. abortus were grown as described (3). H. hepaticus ATCC 51449 and H. gracilis were grown as described (24, 30). C. jejuni and its mutants were grown as described (10, 29, 31). B. subtilis protoplasts and lysate were prepared with lysozyme, following protocol (32). A motile revertant H. pylori 26695 isolate was selected by serial passage in Brucella broth supplemented with 10% heat-inactivated fetal bovine serum at 37°C, 5% CO2 for 4 days until cultures reached an OD600 of ~0.4. Nonmotile H. pylori fliP* mutants were propagated on TSAII blood agar plates (BD Biosciences) at 37°C, 5% CO2 for either 24 or 48 h prior to collection with a sterile cotton swab for grid preparation. H. pylori mutants (ΔfliM fliP*, ΔfliO fliP*, ΔflgS fliP*, ΔfliG fliP*, ΔfliQ fliP*, and ΔfliF fliP*) were grown directly from glycerol stocks on sheep blood agar at 37°C with 5% CO2 for 48 h. Cells were either collected from the plate using a cotton swab, resuspended in PBS, then spun down and plunge-frozen directly, or passaged onto a new plate and allowed to grow for 24 h under the same conditions prior to collection and plunge-freezing. No difference could be discerned between the two samples by cryo-ET.
M. xanthus DK1622 cells were grown for 48 h in CTT medium at 30°C without antibiotics to OD600=0.7. F. johnsoniae strain CJ2618 was grown as described (4). P. vanneervenii cells were grown as described (33). A. asiaticus strain 452471 was grown as described (34). C. burnetii Nine Mile phase II clone 4 strain was grown in the laboratory of Robert Heinzen at the National Institute of Allergy and Infectious Diseases. EPEC cells strain 8612 (Δrorf1-escL Δrorf3-mpc ΔescN-espF ΔflhBA ΔfliOPQR ΔminCD) from a glycerol stock were grown in 2mL LB at 37°C in static conditions overnight. The next day, 1 mL of cells were spun down at 4,000 rpm for six minutes and concentrated 10 times. 3 μL of cells were spotted onto glow-discharged grids placed on DMEM high-glucose agar plates (1.5% agar) for 3 h. Then, the grids were washed twice in PBS and transferred to starvation-medium (DMEM without phenol red, vitamins, and amino acids) agar plates and incubated for 1 h at 37°C. Subsequently, the samples were washed with PBS and plunge-frozen.
H. pylori mutagenesis.
Flagellar mutants were generated in the nonmotile H. pylori 26695 background as previously described (9, 35).
Electron cryo-tomography sample preparation and imaging.
Sample preparation for cryo-ET imaging was done as described (20, 25, 36). All data were collected by the Jensen Lab at Caltech. For the total cumulative electron dose used for each tilt-series in each species, see Table 2.
TABLE 2.
Total electron dose used for each tilt-series in each species
| Species name | Class | Cumulative electron dose (e-/Å2) |
|---|---|---|
| Xanthomonas citri | Gammaproteobacteria | 120 |
| Vibrio harveyi | Gammaproteobacteria | 160 |
| Vibrio fischeri | Gammaproteobacteria | 150 |
| Vibrio cholerae | Gammaproteobacteria | 160 |
| Salmonella enterica | Gammaproteobacteria | 200 |
| Pseudoalteromonas luteoviolacea | Gammaproteobacteria | 180 |
| Proteus mirabilis | Gammaproteobacteria | 160 |
| Pseudomonas flexibilis | Gammaproteobacteria | 100 |
| Pseudomonas aeruginosa | Gammaproteobacteria | 170 |
| Legionella pneumophila | Gammaproteobacteria | 100 |
| Escherichia coli | Gammaproteobacteria | 130 |
| Hyphomonas neptunium | Alphaproteobacteria | 180 |
| Agrobacterium tumefaciens | Alphaproteobacteria | 200 |
| Azospirillum brasilense | Alphaproteobacteria | 200 |
| Brucella abortus | Alphaproteobacteria | 160 |
| Helicobacter hepaticus | Epsilonproteobacteria | 200 |
| Helicobacter pylori | Epsilonproteobacteria | 120 to 130 |
| Campylobacter jejuni | Epsilonproteobacteria | 200 |
| Hylemonella gracilis | Betaproteobacteria | 75 |
| Bacillus subtilis | Bacilli | 160 |
| Myxococcus xanthus | Deltaproteobacteria | 200 |
| Prosthecobacter vanneervenii | Verrucomicrobiae | 180 |
| Flavobacterium johnsoniae | Flavobacteria | 100 |
| Coxiella burnetii | Gammaproteobacteria | 130 |
| Amoebophilus asiaticus | Cytophagales | 200 |
| EPEC | Gammaproteobacteria | 120 |
Image processing and subtomogram averaging.
The three-dimensional reconstruction of the tilt-series was performed either automatically through the RAPTOR pipeline used in the Jensen Lab (5) or with the IMOD software package (37). Subtomogram averaging was done using the PEET program (38) with a 2-fold symmetrization applied along the particle y-axis. The numbers of particles included in each subtomogram average are indicated on the figures.
ACKNOWLEDGMENTS
This project was funded by the NIH (grant R01 AI127401 to G.J.J., and grant P20 GM130456 to C.L.S.) and the European Research Council (ERC) Synergy grant (no. 810186 awarded to S.B.Y. and I.R.). M.K. acknowledges a Baxter postdoctoral fellowship from Caltech. Cryo-ET work was done in the Beckman Institute Resource Center for Transmission Electron Microscopy at the California Institute of Technology. We are grateful to Marc Erhardt (Humboldt-Universität zu Berlin) for critically reading an initial version of this work. We thank Elitza I. Tocheva for collecting the A. tumefaciens data, Jian Shi for collecting the H. neptunium data, and Martin Pilhofer for collecting the P. luteoviolacea data. We thank Dianne Newman’s lab for providing the P. aeruginosa transposon mutant. We are grateful to the lab of Robert Heinzen at the National Institute of Allergy and Infectious Diseases in Montana for growing the Coxiella burnetii cells.
Footnotes
Supplemental material is available online only.
Contributor Information
Grant J. Jensen, Email: grant_jensen@byu.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Kaplan M, Nicolas WJ, Zhao W, Carter SD, Metskas LA, Chreifi G, et al. In situ imaging and structure determination of biomolecular complexes using electron cryo-tomography. In: Gonen T, Nannenga BL, editors. CryoEM: Methods and Protocols. New York, NY: Springer US; 2021. P 83–111. 10.1007/978-1-0716-0966-8_4. [DOI] [PubMed] [Google Scholar]
- 2.Ghosal D, Kaplan M, Chang Y-W, Jensen GJ. In situ imaging and structure determination of bacterial toxin delivery systems using electron cryotomography. In: Buchrieser C, Hilbi H, editors. Legionella. New York, NY: Springer New York; 2019. P 249–265. 10.1007/978-1-4939-9048-1_16. [DOI] [PubMed] [Google Scholar]
- 3.Dobro MJ, Oikonomou CM, Piper A, Cohen J, Guo K, Jensen T, Tadayon J, Donermeyer J, Park Y, Solis BA, Kjær A, Jewett AI, McDowall AW, Chen S, Chang Y-W, Shi J, Subramanian P, Iancu CV, Li Z, Briegel A, Tocheva EI, Pilhofer M, Jensen GJ. 2017. Uncharacterized bacterial structures revealed by electron cryotomography. Silhavy TJ, editor. J Bacteriol 199. 10.1128/JB.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan M, Chreifi G, Metskas LA, Liedtke J, Wood CR, Oikonomou CM, Nicolas WJ, Subramanian P, Zacharoff LA, Wang Y, Chang Y-W, Beeby M, Dobro MJ, Zhu Y, McBride MJ, Briegel A, Shaffer CL, Jensen GJ. 2021. In situ imaging of bacterial outer membrane projections and associated protein complexes using electron cryo-tomography. Elife 10:e73099. 10.7554/eLife.73099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding HJ, Oikonomou CM, Jensen GJ. 2015. The Caltech Tomography Database and automatic processing pipeline. J Struct Biol 192:279–286. 10.1016/j.jsb.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortega DR, Oikonomou CM, Ding HJ, Rees-Lee P, Jensen GJ, Alexandria . 2019. ETDB-Caltech: a blockchain-based distributed public database for electron tomography. Promponas VJ, editor. PLoS One 14:e0215531. 10.1371/journal.pone.0215531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan M, Tocheva EI, Briegel A, Dobro MJ, Chang Y-W, Subramanian P, McDowall AW, Beeby M, Jensen GJ. 2021. Loss of the bacterial flagellar motor switch complex upon cell lysis. mBio 12:e0029821. 10.1128/mBio.00298-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y-W, Shaffer CL, Rettberg LA, Ghosal D, Jensen GJ. 2018. In vivo structures of the Helicobacter pylori cag type IV secretion system. Cell Rep 23:673–681. 10.1016/j.celrep.2018.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan M, Oikonomou CM, Wood CR, Chreifi G, Subramanian P, Ortega DR, Chang Y‐W, Beeby M, Shaffer CL, Jensen GJ. 2022. Novel transient cytoplasmic rings stabilize assembling bacterial flagellar motors. EMBO J 41. 10.15252/embj.2021109523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabiani FD, Renault TT, Peters B, Dietsche T, Gálvez EJC, Guse A, Freier K, Charpentier E, Strowig T, Franz-Wachtel M, Macek B, Wagner S, Hensel M, Erhardt M. 2017. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. Stock A, editor. PLoS Biol 15:e2002267. 10.1371/journal.pbio.2002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukumura T, Makino F, Dietsche T, Kinoshita M, Kato T, Wagner S, Namba K, Imada K, Minamino T. 2017. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. Stock A, editor. PLoS Biol 15:e2002281. 10.1371/journal.pbio.2002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne‐Davies B, Wimmi S, Diepold A. 2021. Adaptivity and dynamics in type III secretion systems. Mol Microbiol 115:395–411. 10.1111/mmi.14658. [DOI] [PubMed] [Google Scholar]
- 14.Tsang J, Hoover TR. 2014. Requirement of the flagellar protein export apparatus component FliO for optimal expression of flagellar genes in Helicobacter pylori. J Bacteriol 196:2709–2717. 10.1128/JB.01332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abby SS, Rocha EPC. 2012. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. Achtman M, editor. PLoS Genet 8:e1002983. 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mare AD, Ciurea CN, Man A, Tudor B, Moldovan V, Decean L, Toma F. 2021. Enteropathogenic Escherichia coli—a summary of the literature. Gastroenterology Insights 12:28–40. 10.3390/gastroent12010004. [DOI] [Google Scholar]
- 18.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol 226:433–446. 10.1016/0022-2836(92)90958-M. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli: flagellar motor assembly. Mol Microbiol 80:886–899. 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan M, Subramanian P, Ghosal D, Oikonomou CM, Pirbadian S, Starwalt‐Lee R, Mageswaran SK, Ortega DR, Gralnick JA, El‐Naggar MY, Jensen GJ. 2019. In situ imaging of the bacterial flagellar motor disassembly and assembly processes. EMBO J 38:e100957. 10.15252/embj.2018100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, Ochman H. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol 189:7098–7104. 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Baidya AK, Pal RR, Mamou G, Gatt YE, Margalit H, Rosenshine I, Ben-Yehuda S. 2019. A ubiquitous platform for bacterial nanotube biogenesis. Cell Rep 27:334–342.e10. 10.1016/j.celrep.2019.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors: structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan M, Sweredoski MJ, Rodrigues JPGLM, Tocheva EI, Chang Y-W, Ortega DR, Beeby M, Jensen GJ. 2020. Bacterial flagellar motor PL-ring disassembly subcomplexes are widespread and ancient. Proc Natl Acad Sci USA 117:8941–8947. 10.1073/pnas.1916935117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. 2014. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343:529–533. 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Q, Jewett AI, Chang Y-W, Oikonomou CM, Beeby M, Iancu CV, Briegel A, Ghosal D, Jensen GJ. 2017. Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J 36:1577–1589. 10.15252/embj.201696235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosal D, Kim KW, Zheng H, Kaplan M, Truchan HK, Lopez AE, McIntire IE, Vogel JP, Cianciotto NP, Jensen GJ. 2019. In vivo structure of the Legionella type II secretion system by electron cryotomography. Nat Microbiol 4:2101–2108. 10.1038/s41564-019-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci USA 113:E1917–E1926. 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA 106:17181–17186. 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller A, Beeby M, McDowall AW, Chow J, Jensen GJ, Clemons WM. 2014. Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. MicrobiologyOpen 3:702–710. 10.1002/mbo3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeCastro-Costa MR, Landman OE. 1977. Inhibitory protein controls the reversion of protoplasts and L forms of Bacillus subtilis to the walled state. J Bacteriol 129:678–689. 10.1128/jb.129.2.678-689.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. 2011. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. Amos L, editor. PLoS Biol 9:e1001213. 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böck D, Medeiros JM, Tsao H-F, Penz T, Weiss GL, Aistleitner K, Horn M, Pilhofer M. 2017. In situ architecture, function, and evolution of a contractile injection system. Science 357:713–717. 10.1126/science.aan7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. Salama N, editor. PLoS Pathog 7:e1002237. 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan M, Ghosal D, Subramanian P, Oikonomou CM, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ. 2019. The presence and absence of periplasmic rings in bacterial flagellar motors correlates with stator type. Elife 8. 10.7554/eLife.43487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 38.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–948. 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3 and caption for Movie S1. Download jb.00144-22-s0001.pdf, PDF file, 6.4 MB (6.4MB, pdf)
Movie S1. Download jb.00144-22-s0002.mov, MOV file, 11.7 MB (11.7MB, mov)