Abstract
Purpose:
Black women have a 40% increased risk of breast cancer-related mortality. These outcome disparities may reflect differences in tumor pathways and a lack of targetable therapies for specific subtypes that are more common in Black women. Hepatocyte Growth Factor (HGF) is a targetable pathway that promotes breast cancer tumorigenesis, is associated with basal-like breast cancer, and differentially expressed by race. This study assessed whether a 38-gene HGF expression signature is associated with recurrence and survival in Black and non-Black women.
Methods:
Study participants included 1,957 invasive breast cancer cases from the Carolina Breast Cancer Study. The HGF signature was evaluated in association with recurrence (n=1,251, 171 recurrences), overall, and breast-cancer specific mortality (n=706, 190/328 breast cancer/overall deaths) using Cox proportional hazard models.
Results:
Women with HGF positive tumors had higher recurrence rates [HR 1.88, 95%CI (1.19, 2.98)], breast cancer specific mortality [HR: 1.90, 95%CI (1.26, 2.85)], and overall mortality [HR: 1.69; 95%CI (1.17, 2.43)]. Among Black women, HGF positivity was significantly associated with higher 5-year rate of recurrence [HR: 1.73; 95%CI (1.01, 2.99)], but this association was not significant in non-Black women [HR 1.68; 95%CI (0.72, 3.90)]. Among Black women, HGF-positive tumors had elevated breast cancer-specific mortality [HR 1.80, 95%CI (1.05, 3.09)], which was not significant in non-Black women [HR:1.52; 95%CI (0.78, 2.99)].
Conclusion:
This multi-gene HGF signature is a poor-prognosis feature for breast cancer and may identify patients who could benefit from HGF-targeted treatments, an unmet need for Black and triple negative patients.
Keywords: Breast cancer, Hepatocyte Growth Factor
Introduction
In the United states, Black women experience earlier breast cancer recurrence, higher breast cancer specific mortality rates, and poorer overall survival compared to white women[1–6]. It is unclear why these disparities in breast cancer outcomes persist. One explanation is more prevalent aggressive tumor subtypes; triple-negative/basal-like breast cancer has been shown to be more than twice as common among Black women than other racial groups[1,7]. However, this tumor subtype is challenging to target because it lacks hormone receptors and HER2. While these tumors are positive for epidermal growth factor receptor [8], clinical trials targeting EGFR in triple negative breast cancer patients have had limited success[9,10]. Thus the current standard of care is to treat basal-like cancers with chemotherapy, and while many basal-like tumors are sensitive to chemotherapy, these tumors are more likely to recur and have poorer short-term survival[11]. Identifying novel, targetable approaches is therefore of high importance for addressing outcome disparities.
The hepatocyte growth factor (HGF) pathway is an important pathway regulating the tumor microenvironment and has been found to be associated with breast tumorigenesis [12–16]. Clinical and laboratory based studies have found that the HGF/c-MET axis may be an important feature of triple negative/basal-like tumors [14,17–19]. Charafe-Jauffret et al. molecularly characterized 31 breast cell lines for breast cancer subtype classification (luminal vs basal-like) and found that the gene for HGF receptor c-MET was one of 10 genes associated with basal-like cell line classification[17]. Clinical trials have targeted the HGF pathway in breast cancer patients; however these studies have lacked methods for identifying patients who are most likely to benefit; there is an ongoing need for an effective predictive biomarker for HGF expression[20,21].
Here we present a 38-gene HGF gene expression signature as a candidate biomarker for HGF pathway function in invasive breast tumors. We examined the association of the HGF signature with breast cancer recurrence and survival outcomes in the racially diverse Carolina Breast Cancer study.
Methods
Study population
The Carolina Breast Cancer Study (CBCS) is a North Carolina population-based study that has been described in detail previously[22,23]. Briefly, CBCS utilized rapid case ascertainment from the North Carolina Central Cancer Registry to identify new breast cancer cases. Inclusion criteria for all three study phases included North Carolina (NC) residency at diagnosis, English fluency, and age from 20–74 years old. Black women and women under the age of 50 were oversampled for participation, such that 50% of the population was Black and 50% is under age 50.
Phases 1 and 2 of CBCS were conducted in 24 central NC counties from 1993–2001. Overall and breast cancer specific survival were collected via linkage to the National Death Index through December 2018. Phase 3 of the Carolina Breast Cancer Study extended the original 24-county area to 44 counties. Phase 3 also collected recurrence information by medical record abstraction through December 2018 to calculate disease free survival. Phase 3 has not yet been linked to the National Death Index because patients are still being followed by medical-record; thus NDI-recorded deaths for Phase 3 participants are not yet available.
Formalin fixed paraffin embedded (FFPE) invasive breast cancer tumors were collected from all three phases of CBCS to assess RNA expression. Among the CBCS cases with gene expression data (n= 4,162), only women with invasive tumors and complete expression data for the 38-gene HGF expression signature were included in the current analysis (n=1,975). Among these, 706 women were from CBCS-1/2 and had breast cancer-specific and overall mortality data, and 1,251 women were from CBCS-3 and had recurrence data. Informed consent was obtained from each study participant under a protocol approved by the University of North Carolina at Chapel Hill- Office of Human Ethics and Institutional Review Board.
Clinical and patient demographics
All patient demographics (race, age at diagnosis, and family history of breast cancer) were self-reported and obtained from Carolina Breast Cancer Study questionnaires. Body mass index was recorded by the study nurse. Clinical factors including estrogen receptor (ER) status, tumor stage, and combined grade were obtained from medical records, and pathology reports. Stage 4 participants were removed from the survival analysis because treatment of metastatic patients follows very distinct clinical pathways [CBCS-1/2 (n=20) and in CBCS-3 (n= 47)]. Information on tumor grade was only available from CBCS Phase 1 & 3 and thus analyses regarding tumor grade excluded Phase 2 participants (n=454).
Gene expression data
RNA was isolated from Formalin fixed paraffin embedded (FFPE) invasive breast cancer tumor tissues using the Qiagen FFPE RNeasy isolation kit (Germantown, MD). RNA was quantified using Nanostring nCounter technology (Seattle, Washington), using a custom panel that included signatures for PAM50 (for classification of intrinsic breast cancer subtypes: luminal A, luminal B, HER2 overexpressing, basal-like- and normal-like) and the HGF 38-gene signature (classified as positive vs negative as described previously)[24,25]. Gene expression data were normalized using the RUVg function from the RUVSeq Bioconductor package as previously described by Bhattacharya et al.[26,27]. The HGF signature is a 38-gene weighted sum gene expression signature: TMEM45B, AKR7L, AQP5, C1QTNF3, C2ORF27A, C4ORF31, C9ORF98, CAPN13, CASKIN1, CMYA5, DTX3, EFHD1, F7, FMNL2, FUT8, GCNT2, HRC, INPP4B, ISLR2, KCNMA1, KCNN4, KIF3A, MAGI2, MARVELD2, NME5, PKIB, PRRG2, PRRT2, PVRL2, REEP6, RIMS4, SCUBE2, SHROOM3, SKAP1, SYBU, TFF3, and TMSB15B [25]. Tumors are characterized as HGF-positive if expression profiles match expression profiles of HGF protein treated breast cancer cells as described in Casbas-Hernandez et al[19].
Statistical methods
Descriptive analyses for demographic variables were calculated using frequency data for each clinical and patient characteristic. For survival analyses, proportional hazard assumptions were assessed using visual inspection of Kaplan Meier plots on the distribution of HGF gene expression signature and survival outcomes (disease-free, overall and breast cancer specific survival). Schoenfeld tests and residual plots were also used to test the proportional hazards assumption. HGF gene signature expression violated the proportional hazard assumption for overall/breast cancer-specific and disease-free survival. For this analysis 5-year risks/hazards are reported, as well as log-rank p-values over multiple time points (5-year & 10-year).
Overall survival is defined as time from study enrollment to death of any cause, and breast cancer specific survival is defined as time from breast cancer diagnosis to breast cancer-related death. In breast cancer specific survival analyses, death due to other causes is a censoring event. Disease free survival was defined as time from study enrollment to subsequent breast cancer recurrence. Hazard ratios and 95% confidence intervals for the association of HGF gene expression signature and survival outcomes were produced using Cox proportional hazard models. Effect measure modification in this study was assessed using likelihood ratio tests. Age (<50, 50+ years) and race (Black vs non-Black) were evaluated as effect measure modifiers for recurrence (p-values < 0.07) and mortality outcomes with statistical significance thresholds set at p < 0.10. To retain power in the study, race classifications of Black vs non-Black were used, although sensitivity analyses removed women who did not identify as Black or non-Hispanic white (n= 37 for recurrence, n= 8 for overall/breast cancer specific survival) and did not significantly change the study findings. Hazard ratios stratified by race are presented in the current analysis. To control for confounding, inverse probability of exposure weights were applied to both recurrence (CBCS-3) and mortality data (CBCS-1/2). For CBCS 3 stabilized weights included adjustment for grade, age and stage. Mortality data used stabilized weights to adjust for age and stage only because grade information was missing for CBCS 2 participants. All standardized hazard ratios and risks used robust variance estimation for calculation of confidence intervals. All statistical analysis were completed in Stata 15 SE. This analysis is in accordance with the criteria described in the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) and Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines[28–30].
Results
In Table 1 we present HGF status according to demographic characteristics and CBCS study phase. HGF positive tumors were more prevalent among Black women [CBCS 1/ 2: 64% vs 36%; CBCS3: 72% vs. 28%] than in non-Black women and in women under the age of 50 [CBCS 1/ 2: 65% vs 35%; CBCS 3: 60% vs 40%]. HGF positivity was strongly associated with PAM50 basal-like subtype. Women with HGF positivity tended to have higher tumor stage, grade, and BMI. Family history was not associated with HGF positivity. Figure 1 shows unadjusted survival curves for recurrence (1A), breast cancer specific mortality (1B) and all-cause mortality (1C) according to HGF-positivity. Median follow-up time for recurrence was 6.8 years (min: 0.4. years, max:10.7 years), 17.8 years for breast cancer -specific and overall survival (min: 0.17 years, max: 23.6 years). HGF-positivity was associated with early recurrence. HGF-positive and -negative curves were significantly different at 5 years of follow up (5- year log-rank p-value= 0.006), but this effect was attenuated over time and was no longer statistically significant at 10 years (10-year log-rank p-value= 0.07). Next, we assessed the association with HGF and breast cancer specific survival (Figure 1B). We found a pattern similar to that for recurrence, where HGF- positivity was related to early mortality (5-year log-rank p-value= 0.001), but the association attenuated with time (10- year log-rank p-value=0.45). Finally, the overall survival curves (Figure 1C) also showed HGF-positivity was a contributor to poorer survival compared to HGF negative tumors within the first 5 years of diagnosis (5-year log-rank p-value=0.006), but differences were not statistically significant at the 10- year mark (10-year log-rank p-value=0.37). HGF signature expression was not associated with overall mortality or breast cancer specific -mortality at longer periods of follow-up (? 10 years, p-value >0.05) (data not shown), however data were truncated at 10 years due to crossing hazards.
Table 1.
38-gene HGF status, patient, and clinical characteristics among Carolina Breast Cancer Study participants by study phase (CBCS-1/2, 1993–2001; CBCS-3, 2008–2013)
| CBCS 1 & 2 (N=706) | CBCS 3 (N=1251) | |||
|---|---|---|---|---|
| HGF Negative (N=428) |
HGF Positive (N= 278) |
HGF Negative (N= 901) |
HGF Positive (N=350) |
|
| Race | ||||
| Black | 171 (40%) | 179 (64%) | 432 (48%) | 251 (72%) |
| Non-Black | 257 (60%) | 99 (36%) | 469 (52%) | 99 (28%) |
| Age at diagnosis | ||||
| < 50 | 208 (49%) | 181 (65%) | 442 (49%) | 211 (60%) |
| 50+ | 220 (51%) | 97 (35%) | 459 (51%) | 139 (40%) |
| ER status | ||||
| Positive | 315 (74%) | 54 (20%) | 798 (90%) | 67 (21%) |
| Negative | 108 (26%) | 221 (80%) | 89 (10%) | 253 (79%) |
| PAM50 | ||||
| Luminal A | 283 (69%) | 20 (8%) | 535 (62%) | 11 (3%) |
| Luminal B | 74 (18%) | 6 (2%) | 214 (25%) | 13 (4%) |
| HER2- Enriched | 40 (10%) | 23(9%) | 82 (9%) | 35 (10%) |
| Basal | 13 (3%) | 216 (81%) | 32 (4%) | 277 (83%) |
| Stage** | ||||
| Stage I | 152 (38%) | 74 (28%) | 347 (40%) | 84 (25%) |
| Stage II | 208 (52%) | 164 (62%) | 397 (46%) | 185 (55%) |
| Stage III | 40 (10%) | 25 (10%) | 124 (14%) | 67 (20%) |
| Grade* | ||||
| I/II | 86 (62%) | 28 (26%) | 605 (68 %) | 43 (12%) |
| III | 52 (38%) | 81 (74%) | 285 (32%) | 303 (88%) |
| BMI | ||||
| Median BMI (IQR) | 27.10 (8.85) | 28.99 (9.50) | 29.66 (9.73) | 30.52 (9.30) |
| Family history of breast cancer | ||||
| Yes | 346 (83%) | 217 (82%) | 707 (81%) | 280 (82%) |
| No | 73 (17%) | 49 (18%) | 162 (19%) | 61 (18%) |
Tumor grade was not collected in CBCS-2 and therefore was missing from 454 participants in CBCS-2. Tumor grade was missing from less than 2% of patients in CBCS-1 & CBCS-3.
Stage 4 women were excluded (<3% for CBCS-1/2, <4% CBCS-3)
IQR: Interquartile range; ER: Estrogen receptor; BMI: Body Mass Index;
Fig 1. Kaplan Meier Plot of HGF association with survival outcomes in Carolina Breast Cancer Study.
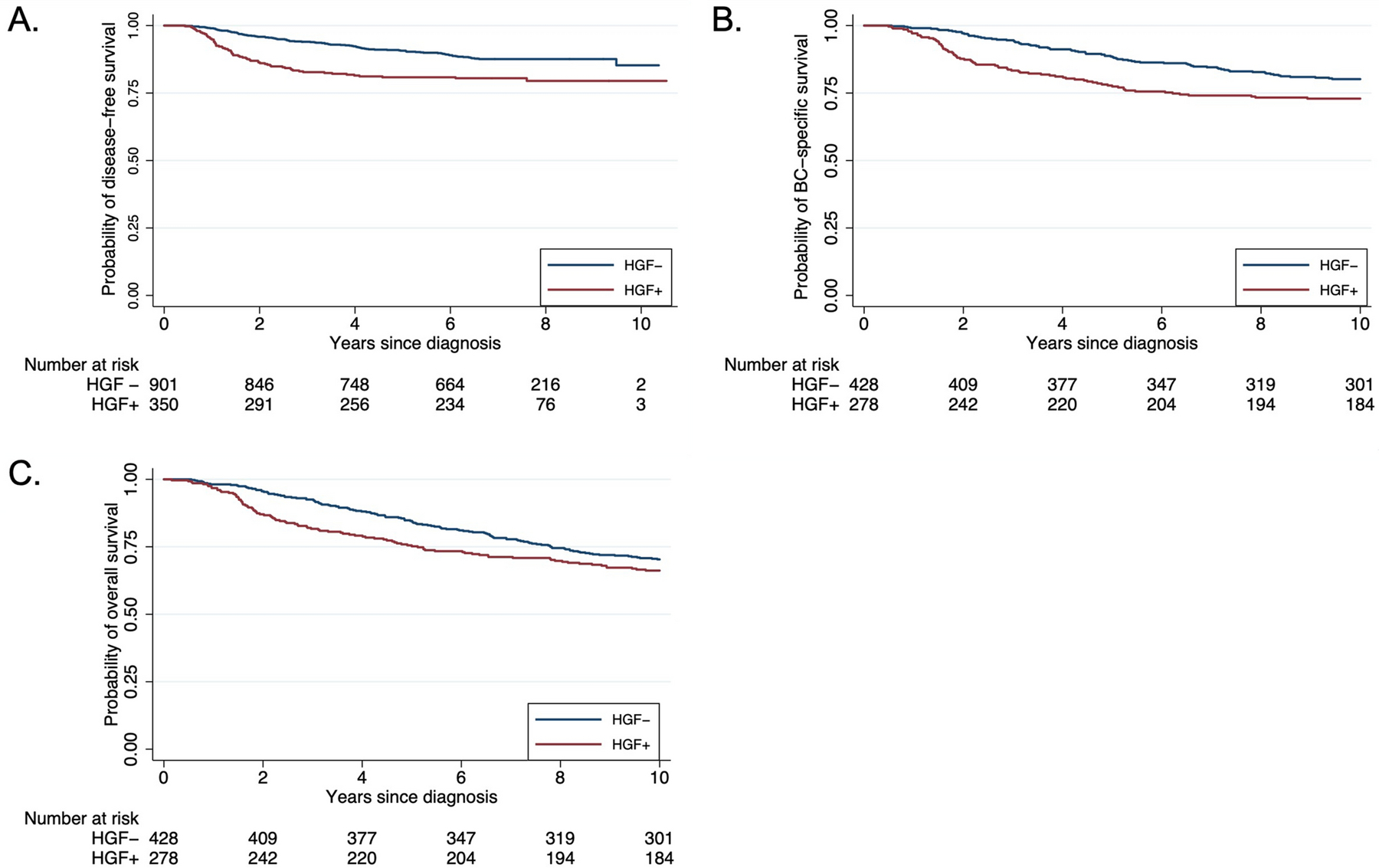
Panel A- disease-free survival (CBCS-3), Panel B- breast cancer specific survival (CBCS-1/2), Panel C- overall survival (CBCS-1/2).
HGF-positivity is associated with basal-like subtype and with higher proliferation rates, both of which may mediate effects of this pathway on outcomes[25]. Therefore, we did not adjust for molecular subtype in assessing the effects of HGF-positivity on outcomes. However, we were interested to know whether HGF-positivity was associated with outcomes independent of standard clinical features (stage and grade). Table 2 shows estimates of the magnitude of association between HGF-positivity and breast cancer recurrence, overall and stratified on age and race. Women with HGF positive tumors had higher recurrence than women with HGF negative tumors [HR: 1.88; 95% CI (1.19, 2.98)]. Standardized 5-year risk of recurrence for HGF positive tumors was 18% compared to HGF negative tumors with a 10% standardized risk of recurrence. This pattern was apparent in analyses restricted to Black women [HR 1.73; 95%CI (1.01, 2.99)], but not significant among non-Black women [HR 1.68; 95%CI (0.72, 3.90)]. HGF-positivity was less common (17%) among non-Black women compared to Black women (37%). Black women with HGF positive tumors had the highest 5-year risk of recurrence [20%; 95%CI (12%, 29%)]. Age did not modify the association between HGF-positivity and recurrence, with similar hazard ratio estimates for both age-defined strata [HR: 1.95; 95%CI (1.09, 3.50) for women <50 vs HR: 1.82; 95%CI (0.88, 3.75) for 50+]. Supplemental Table 1 shows 10-year risk of recurrence remains elevated for HGF-positive (vs. HGF-negative women) although differences were attenuated compared to at 5 years. Analyses stratified on basal-like subtype showed a slight, non-significant, increased risk of recurrence among Basal-like HGF positive tumors (HGF+ = 21.93% risk of recurrence, HGF- = 19.78%, data not shown).
Table 2.
Risk or recurrence and hazard ratios by HGF signature status, race and age in Carolina Breast Cancer Study phase 3
| HGF | N (recurrences) | Crude 5-year risk of recurrence | *Standardized 5-year risk of recurrence | *Standardized 5-year Hazard ratio HR (95%CI) | |
|---|---|---|---|---|---|
| HGF- | 901 (79) | 8.76% (6.92, 10.61) | 10.29% (8.01, 12,57) | Referent | |
| HGF+ | 350 (65) | 18.57%(14.49, 22.65) | 18.00% [11.62, 24.38) | 1.88 (1.19, 2.98) | |
|
| |||||
| Non-Black | HGF- | 469 (31) | 6.61% (4.35,8.86) | 7.90% (5.06, 10.74) | Referent |
| HGF+ | 99 (18) | 18.18% (10.55,25.81) | 12.61% (3.54, 21.69) | 1.68 (0.72, 3.90) | |
|
| |||||
| Black | HGF- | 432 (48) | 11.11% (0.81,14.08) | 12.76% (9.18,16.32) | Referent |
| HGF+ | 251 (47) | 18.72% (13.89,23.56) | 20.42% (12.21, 28.62) | 1.73 (1.01, 2.99) | |
|
| |||||
| < 50 | HGF- | 442 (43) | 9.72% (6.96,12.49) | 11.42% (8.00,14.86) | Referent |
| HGF+ | 211 (40) | 18.96% (13.65,24.25) | 20.22% (11.43, 29.01) | 1.95 (1.09, 3.50) | |
|
| |||||
| 50+ | HGF- | 459 (36) | 7.84% (5.38,10.31) | 9.06% (6.10, 12.03) | Referent |
| HGF+ | 139(25) | 17.99% (11.57,24.39) | 15.65% (6.47,24.84) | 1.82 (0.88, 3.75) | |
Recurrence was standardized using inverse probability of exposure weights for tumor grade, age, and stage.
Patterns for mortality were similar to those for recurrence and suggested an early impact of HGF status on outcomes (Table 3). HGF positive tumors had almost twice the rate of breast cancer specific mortality of HGF negative tumors [HR: 1.90; 95%CI (1.26, 2.85)], and again, the increase was statistically significant among Black [HR: 1.80; 95%CI (1.05, 3.09)] but not non-Black women [HR:1.52; 95%CI (0.78, 2.99)]. For 5-year breast cancer specific mortality, HGF-positivity was significantly associated with outcomes among women over the age of 50 [HR: 2.81; 95%CI (1.38, 5.70)], but not among women under 50 [HR: 1.53, 95%CI (0.94, 2.50)]. Similarly, 5-year overall mortality was associated with HGF positivity [HR: 1.69; 95%CI 1.17, 2.43)], but was only significant in women over 50 [HR: 2.08, 95%CI (1.19, 3.64) vs. HR:1.43, 95%CI (0.89, 2.29) for women <50]. HGF-positivity was not associated with 10-year breast cancer-specific and 10 year-overall mortality (Supplemental Table 1).
Table 3.
Risk of mortality and hazard ratios by HGF signature status, stratified by race and age in Carolina Breast Cancer study phase 1 and 2
| HGF | N (deaths) | Crude 5-year risk | *Standardized 5-year risk | *Standardized 5-year HR (95%CI) | |
|---|---|---|---|---|---|
|
Breast cancer specific mortality
| |||||
| HGF- | 428 (48) | 11.21% (8.22, 14.21) | 11.37% (8.12, 14.62) | Referent | |
| HGF+ | 278 (62) | 22.30% (17.40,27.20) | 20.09% (15.18, 24.99) | 1.90 (1.26,2.85) | |
|
| |||||
| Non-Black | HGF- | 257 (24) | 9.34% (5.78, 12.90) | 9.84% (5.91, 13.77) | Referent |
| HGF+ | 99 (17) | 17.17% (9.70, 24.64) | 14.45% (7.45, 21.46) | 1.52 (0.78,2.99) | |
|
| |||||
| Black | HGF- | 171(24) | 14.03% (8.81, 19.26) | 13.79% (8.14, 19.44) | Referent |
| HGF+ | 179 (45) | 25.13% (18.77,31.52) | 23.16% (16.66, 29.66) | 1.80 (1.05, 3.09) | |
|
| |||||
| <50 | HGF- | 208 (32) | 15.38% (10.47,20.29) | 15.00% (9.87, 20.12) | Referent |
| HGF+ | 181 (43) | 23.76% (17.53,29.97) | 21.51% (15.40, 27.63) | 1.53 (0.94,2.50) | |
|
| |||||
| 50+ | HGF- | 220 (16) | 7.27 (3.83, 10.71) | 6.95% (3.43, 10.47) | Referent |
| HGF+ | 97 (19) | 19.59% (11.64,27.52) | 18.34% (10.39, 26.29) | 2.81 (1.38,5.70) | |
|
| |||||
|
Overall mortality
| |||||
| HGF- | 428 (66) | 15.42% (11.99,18.85) | 14.82% (11.24, 18.40) | Referent | |
| HGF+ | 278 (69) | 24.82% (19.73,29.91) | 23.29% (18.06, 28.52) | 1.69 (1.17, 2.43) | |
|
| |||||
| Non-Black | HGF- | 257 (34) | 13.23% (9.07, 17.37) | 12.48% (8.19, 16.76) | Referent |
| HGF+ | 99 (18) | 18.18% (10.54,25.81) | 15.35% (8.18, 22.51) | 1.28 (0.68,2.39) | |
|
| |||||
| Black | HGF- | 171 (32) | 18.71% (12.85,24.57) | 18.53% (12.28, 24.78) | Referent |
| HGF+ | 179 (51) | 28.49% (21.86,35.12) | 27.63% (20.66, 34.59) | 1.60 (0.99,2.58) | |
|
| |||||
| <50 | HGF- | 208 (35) | 16.83% (11.73,21.92) | 16.54% (11.21, 21.86) | Referent |
| HGF+ | 181 (44) | 24.31% (18.04,30.58) | 22.09% (15.91, 28.27) | 1.43 (0.89,2.29) | |
|
| |||||
| 50+ | HGF- | 220 (31) | 14.09% (9.48, 18.70) | 12.73% (8.15, 17.32) | Referent |
| HGF+ | 97 (25) | 25.77% (17.02,34.52) | 24.75% (15.89, 33.61) | 2.08 (1.19,3.64) | |
Mortality outcomes were standardized using inverse probability of exposure weights for age, and stage.
Discussion
We found that HGF positive tumors have poorer 5-year recurrence and mortality, especially among Black women where HGF positivity is more prevalent. Associations with HGF positivity were attenuated at the 10-year mark, supporting a potential role for HGF as an early prognostic factor in breast cancer-related outcomes. This early recurrence pattern also aligns with prior literature for basal-like breast cancer, showing that more aggressive subtypes like basal-like tend to recur early compared to less aggressive subtypes (i.e. luminal)[11,25], underscoring HGF gene expression as one hallmark of basal-like breast cancer.
Our results are concordant with other studies that have assessed the prognostic value of HGF expression with breast cancer recurrence and survival. Raghav et al. measured HGF pathway expression via c-MET and phosphorylated-MET protein levels in 257 breast cancers and found that HGF overexpression was correlated with increased recurrence and poorer overall survival within 5 years[31]. Also two separate meta-analyses examining the prognostic value of c-MET overexpression by a variety of RNA and protein based detection methods concluded that c-MET overexpression is associated with both breast cancer recurrence and overall survival[32,33]. In contrast, a large Dutch male breast cancer cohort (n= 841) found that HGF protein expression (as measured by immunohistochemistry) was protective against overall survival[34], however, this population is quite distinct and male breast cancer is predominantly of luminal subtype, which we found to have lower prevalence of HGF expression[34,35]. Our results add important new data based on a large, diverse study population. Racial diversity in the previous published literature on HGF is lacking. The MET/HGF pathway has also been implicated in radioresistance, chemoresistance and targeted therapy resistance in several studies[36]. Also, some compounds in phase 3 clinical trials have not been able to sufficiently suppress HGF/MET signaling[36]. Identifying high risk populations that could benefit from HGF/MET targeted therapies in combination with traditional cancer treatment regimens and/or targeted therapies may improve breast cancer outcomes.
Our study has several strengths. One strength is that our HGF gene expression biomarker can be applied to formalin fixed paraffin embedded tumors. Furthermore, HGF is a soluble protein and c-MET is a receptor tyrosine kinase that can translocate to cell nuclei and both may be difficult to assay in a clinical setting [37]. Our multi-gene HGF expression signature captures that pathway as a whole and may be a candidate for clinical studies. Previous HGF-targeted trials have not identified predictive biomarkers for identifying participants that could benefit from HGF targeted therapies. Our study was also statistically powered to assess expression of this signature in relation to breast cancer outcomes in Black and young women, populations known to have the highest burden of adverse breast cancer outcomes, and who, in our study, had higher rates of HGF-positivity. Within our study we recognize race is a social construct. Race captures the interplay between social factors (e.g., discrimination or barriers to care) and biological factors (e.g., ancestry) that may contribute to breast cancer recurrence and mortality. Our findings support that there are racial differences in the prevalence of the HGF positive signature expression, and this could be the result of differences in genomic regulation of HGF as reported in clinical studies of HGF expression by race [38,39].
Our study also has some limitations. There was the potential for some selection bias in the tumors assayed, namely because some CBCS-1/2 tumor blocks had been depleted (13%). This would most likely bias the proportion of HGF-positive tumors upward, because tumors with residual blocks tended to have larger tumor size. However, we do not expect these missing data to distort the relationship between HGF positivity and survival outcomes. We also did not have the same outcomes on all participants (CBCS 1/ 2 had overall & breast-cancer specific mortality and CBCS 3 had recurrence data), however this allowed us to perform separate, independent time-to-event analyses in two similar populations. The concordance in direction of effect across these distinct datasets underscores the consistency of the associations we observed. Finally, we did not assess the effects of the HGF pathway independent of tumor subtype. This is because HGF is highly prevalent in basal-like breast cancers, verging on being a defining feature of this subtype, and we were interested in assessing whether it predicted outcomes, even if mediated by basal-like or proliferation-related gene expression.
Identification of pathways that can be targeted in triple negative/ basal-like tumors is important because of the poor prognosis and lack of available therapies for these subtypes. HGF is a stroma-derived targetable factor that may reflect a microenvironment-mediated pathway to aggressiveness in breast cancer[19,40–43]. Future studies should focus on evaluating the HGF gene expression signature to identify patients that may experience clinical benefit from HGF-targeted therapies. Predictive biomarkers that lead to targeted treatment of basal-like breast cancers could play an important role in reducing disparities in breast cancer outcomes.
Supplementary Material
Acknowledgements
We would like to acknowledge and thank all the patients and families of the Carolina Breast Cancer study for their contributions to this work. We are indebted for their participation in bringing this study into fruition.
Funding:
Gieira Jones was supported by the UNC Lineberger Cancer Control Education Program (T32CA057726). This research was supported by a grant from UNC Lineberger Comprehensive Cancer Center, which is funded by the University Cancer Research Fund of North Carolina, the Susan B Komen Foundation (OGUNC1202), the National Cancer Institute of the National Institutes of Health (P01CA151135), the National Cancer Institute (U54 CA156733), and the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/NCI P50-CA58223).
Footnotes
Conflict of Interest: The University of North Carolina is an interest owner in University Genomics, the patent holder of the PAM50 assay.
Data availability
The datasets generated during the current study are available from the corresponding author upon reasonable request. The code in this study is available from the corresponding author upon reasonable request.
REFERENCES
- 1.American Cancer Society. Breast Cancer Facts & Figures 2019–2020. Am Cancer Soc 2019;
- 2.Benefield HC, Allott EH, Reeder-Hayes KE, Perou CM, Carey LA, Geradts J, et al. Borderline Estrogen Receptor–Positive Breast Cancers in Black and White Women. JNCI J Natl Cancer Inst 2019; [DOI] [PMC free article] [PubMed]
- 3.Collin LJ, Yan M, Jiang R, Ward KC, Crawford B, Torres MA, et al. Oncotype DX recurrence score implications for disparities in chemotherapy and breast cancer mortality in Georgia. npj Breast Cancer 2019; [DOI] [PMC free article] [PubMed]
- 4.Pastoriza JM, Karagiannis GS, Lin J, Lanjewar S, Entenberg D, Condeelis JS, et al. Black race and distant recurrence after neoadjuvant or adjuvant chemotherapy in breast cancer. Clin Exp Metastasis 2018; [DOI] [PMC free article] [PubMed]
- 5.Kabat GC, Ginsberg M, Sparano JA, Rohan TE. Risk of Recurrence and Mortality in a Multi-Ethnic Breast Cancer Population. J Racial Ethn Heal Disparities 2017; [DOI] [PubMed]
- 6.Newman LA, Griffith KA, Jatoi I, Simon MS, Crowe JP, Colditz GA. Meta-analysis of survival in African American and white American patients with breast cancer: Ethnicity compared with socioeconomic status. J Clin Oncol 2006; [DOI] [PubMed]
- 7.Troester MA, Sun X, Allott EH, Geradts J, Cohen SM, Tse CK, et al. Racial Differences in PAM50 Subtypes in the Carolina Breast Cancer Study. J Natl Cancer Inst 2017;110(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Modern Pathology 2011. [DOI] [PubMed]
- 9.Costa R, Shah AN, Santa-Maria CA, Cruz MR, Mahalingam D, Carneiro BA, et al. Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: New discoveries and practical insights for drug development. Cancer Treatment Reviews 2017. [DOI] [PubMed]
- 10.Maennling AE, Tur MK, Niebert M, Klockenbring T, Zeppernick F, Gattenlöhner S, et al. Molecular targeting therapy against egfr family in breast cancer: Progress and future potentials. Cancers (Basel) 2019; [DOI] [PMC free article] [PubMed]
- 11.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 2007; [DOI] [PubMed]
- 12.Lengyel E, Prechtel D, Resau JH, Gauger K, Welk A, Lindemann K, et al. c-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer 2005; [DOI] [PubMed]
- 13.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, et al. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res 2003; [PubMed]
- 14.Graveel CR, DeGroot JD, Su Y, Koeman J, Dykema K, Leung S, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A 2009; [DOI] [PMC free article] [PubMed]
- 15.Ho-Yen CM, Green AR, Rakha EA, Brentnall AR, Ellis IO, Kermorgant S, et al. C-Met in invasive breast cancer: Is there a relationship with the basal-like subtype? Cancer 2014; [DOI] [PubMed]
- 16.Gastaldi S, Comoglio PM, Trusolino L. The Met oncogene and basal-like breast cancer: Another culprit to watch out for? Breast Cancer Research 2010. [DOI] [PMC free article] [PubMed]
- 17.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adélaïde J, Cervera N, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006; [DOI] [PubMed]
- 18.Garcia S, Dalès JP, Charafe-Jauffret E, Carpentier-Meunier S, Andrac-Meyer L, Jacquemier J, et al. Poor prognosis in breast carcinomas correlates with increased expression of targetable CD146 and c-Met and with proteomic basal-like phenotype. Hum Pathol 2007; [DOI] [PubMed]
- 19.Casbas-Hernandez P, Troester M a., Perez ER, Sandhu R, Kirk E, D’arcy M, et al. Role of HGF in epithelial-stromal cell interactions during progression from benign breast disease to ductal carcinoma in situ. Cancer Res 2012;72(5):LB-501-LB-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveres H, Pineda E, Maurel J. MET inhibitors in cancer: pitfalls and challenges. Expert Opinion on Investigational Drugs 2020. [DOI] [PubMed]
- 21.Parikh RA, Wang P, Beumer JH, Chu E, Appleman LJ. The potential roles of hepatocyte growth factor (HGF)-MET pathway inhibitors in cancer treatment. OncoTargets and Therapy 2014. [DOI] [PMC free article] [PubMed]
- 22.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich TE, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat 1995;35(1):51–60. [DOI] [PubMed] [Google Scholar]
- 23.Emerson MA, Golightly YM, Tan X, Aiello AE, Reeder-Hayes KE, Olshan AF, et al. Integrating access to care and tumor patterns by race and age in the Carolina Breast Cancer Study, 2008–2013. Cancer Causes Control 2020; [DOI] [PMC free article] [PubMed]
- 24.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol 2009; [DOI] [PubMed]
- 25.Jones GS, Hoadley KA, Olsson LT, Hamilton AM, Bhattacharya A, Kirk EL, et al. Hepatocyte growth factor pathway expression in breast cancer by race and subtype. Breast Cancer Res 2021;23(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya A, Hamilton AM, Furberg H, Pietzak E, Purdue MP, Troester MA, et al. An approach for normalization and quality control for NanoString RNA expression data. bioRxiv 2020; [DOI] [PMC free article] [PubMed]
- 27.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014; [DOI] [PMC free article] [PubMed]
- 28.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 2005. Aug 22;93(4):387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 2012/05/29. 2012;9(5):e1001216–e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandenbroucke JP, Elm E von, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and Elaboration. Ann Intern Med 2007. Oct 16;147(8):W-163–W-194. [DOI] [PubMed] [Google Scholar]
- 31.Raghav KP, Wang W, Liu S, Chavez-MacGregor M, Meng X, Hortobagyi GN, et al. cMET and phospho-cMET protein levels in breast cancers and survival outcomes. Clin Cancer Res 2012; [DOI] [PMC free article] [PubMed]
- 32.Yan S, Jiao X, Zou H, Li K. Prognostic significance of c-Met in breast cancer: A meta-analysis of 6010 cases. Diagn Pathol 2015; [DOI] [PMC free article] [PubMed]
- 33.Zhao X, Qu J, Hui Y, Zhang H, Sun Y, Liu X, et al. Clinicopathological and prognostic significance of c-Met overexpression in breast cancer. Oncotarget 2017; [DOI] [PMC free article] [PubMed]
- 34.Qiu SQ, van Rooijen J, Nienhuis HH, van der Vegt B, Timmer-Bosscha H, van Leeuwen-Stok E, et al. High hepatocyte growth factor expression in primary tumor predicts better overall survival in male breast cancer. Breast Cancer Res 2020; [DOI] [PMC free article] [PubMed]
- 35.Gucalp A, Traina TA, Eisner JR, Parker JS, Selitsky SR, Park BH, et al. Male breast cancer: a disease distinct from female breast cancer. Breast Cancer Research and Treatment 2019. [DOI] [PMC free article] [PubMed]
- 36.Huang X, Li E, Shen H, Wang X, Tang T, Zhang X, et al. Targeting the HGF/MET Axis in Cancer Therapy: Challenges in Resistance and Opportunities for Improvement. Vol. 8, Frontiers in Cell and Developmental Biology 2020. p. 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dua R, Zhang J, Parry G, Penuel E. Detection of hepatocyte growth factor (HGF) ligand-c-MET receptor activation in formalin-fixed paraffin embedded specimens by a novel proximity assay. PLoS One 2011; [DOI] [PMC free article] [PubMed]
- 38.Ma J, DeFrances MC, Zou C, Johnson C, Ferrell R, Zarnegar R. Somatic mutation and functional polymorphism of a novel regulatory element in the HGF gene promoter causes its aberrant expression in human breast cancer. J Clin Invest 2009;119(3):478–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of Breast Cancer Molecular Features and Survival by African and European Ancestry in The Cancer Genome Atlas. JAMA Oncol 2017. Dec 1;3(12):1654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parr C, Watkins G, Mansel RE, Jiang WG. The Hepatocyte Growth Factor Regulatory Factors in Human Breast Cancer. Clin Cancer Res 2004; [DOI] [PubMed]
- 41.Haslam SZ, Woodward TL. Host microenvironment in breast cancer development: Epithelial-cell–stromal-cell interactions and steroid hormone action in normal and cancerous mammary gland. Breast Cancer Res 2003; [DOI] [PMC free article] [PubMed]
- 42.Owusu BY, Galemmo R, Janetka J, Klampfer L. Hepatocyte growth factor, a key tumor-promoting factor in the tumor microenvironment. Cancers 2017. [DOI] [PMC free article] [PubMed]
- 43.Spina A, De Pasquale V, Cerulo G, Cocchiaro P, Della Morte R, Avallone L, et al. HGF/c-MET Axis in Tumor Microenvironment and Metastasis Formation. Biomedicines 2015;3(1):71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author upon reasonable request. The code in this study is available from the corresponding author upon reasonable request.


