Abstract
We report an incidence of natural infection of SARS-CoV-2 in free-ranging Indian leopard (Panthera pardus fusca). The case was detected during routine screening. Post-mortem and laboratory examination suggested virus-induced interstitial pneumonia. Viral genome could be detected in various organs including brain, lung, spleen, and lymph nodes by real-time PCR. Whole-genome sequence analysis confirmed infection of Pango lineage B.1.617.2 of SARS-CoV-2. Till now, only Asiatic lions have been reported to be infected by SARS-CoV-2 in India. Infections in animals were detected during peak phase of pandemic and all the cases were captive with close contacts with humans, whereas the present case was observed when human cases were significantly low. No tangible evidence linked to widespread infection in the wild population and the incidence seems to be isolated case. High nucleotide sequence homology with prevailing viruses in humans suggested spillover infection to the animal. This report underlines the need for intensive screening of wild animals for keeping track of the virus evolution and development of carrier status of SARS-CoV-2 among wildlife species.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10344-022-01608-4.
Keywords: SARS-CoV-2, COVID-19, Delta variant, Leopard, Wild animals
Introduction
There are increasing reports of spillover infection by severe acute respiratory syndrome coronavirus (SARS-CoV-2) in wide range of wild and companion animals (Hobbs and Reid 2021; Munir et al. 2020). Both in silico (Piplani et al. 2021) and experimental infections (Bosco-Lauth et al. 2020) of SARS-CoV-2 have been studied in a number of species that detailed out the immunogenicity and pathogenesis of the virus in these species. However, there are only isolated incidences of spillover infections which did not spread widely among the animals except in a few cases, for example, spread of the virus in white tailed deer (Odocoileus virginianus) population (Hale et al. 2022; Cool et al. 2022). In order to understand the potential role of animals in maintenance and transmission of virus in humans, large-scale screening of wide range of animals was required. We also initiated screening the animals for SARS-CoV-2 by real-time PCR, since the beginning of the pandemic. However, despite screening more than 500 samples of various species from various parts of the country, we could detect only two incidences in Asiatic Lions (Panthera leo persica) (Karikalan et al. 2021) whereas other two incidences of SARS-CoV-2 infections in Asiatic lions in India were reported by others (Mishra et al. 2021; Karikalan et al. 2021).
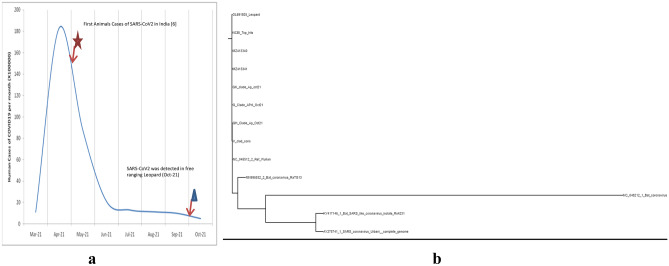
India experienced a huge second wave with a surge in cases of COVID-19 in humans from the month of March 2021 with peak reaching in the month of April 2021 with > 18 million cases reported in a single month (Fig. 1a; https://www.mohfw.gov.in/). Surge in the cases were attributed to spread of highly infectious Delta variant (Pango lineage B.1.617.2) of SARS-CoV-2 and lack of herd immunity in the human population (Novelli et al. 2021). All the incidences of animal infection of SARS-CoV-2 were observed during the second peak of COVID-19 in humans. With surge of infection and vaccination coverage, herd immunity in the population improved and infection rate declined.
Fig. 1.
a Daily number of COVID-19 cases recorded in India. First cases, where 12 Asiatic lions were found positive, were recorded during the peak of the pandemic, whereas incidence in leopard cub was recorded when the incidences in humans significantly subsided. b The leopard sample clustered into common SARS-CoV-2 G-clade (GISAID classification) or Delta variant (B.1.617.2 Pango lineage). Consensus sequences of each clade were generated by aligning using ClustalW tool in MEGA X version 10.1 followed by consensus generation in EMBOSS server. Phylogeny was constructed using NGphylogeny server (https://ngphylogeny.fr)
Detection of SARS-CoV-2
We continue to screen the animal samples collected either as preventive measure or submitted from dead animals for laboratory investigation. One animal was detected positive by real-time PCR with convincingly Ct values (Supplementary Table 1). In order to rule out the laboratory contamination, rectal and nasopharyngeal swabs along with tissue samples were again processed with all precautions and re-confirmed to be positive (Supplementary Table 1). SARS-CoV-2 was detected by RT-PCR kit (COVISure Viral detection kit, Genetixbiotech, India) as described previously (Karikalan et al. 2021). RT-PCR results were confirmed by the determination of partial spike protein gene sequence using Sanger’s method as described previously (Karikalan et al. 2021). Whereas the tissue samples (brain, liver, lungs, spleen, kidney, intestine, and lymph nodes) were negative for common feline pathogens such as haemoprotozoan or rickettsial parasites, canine distemper virus, feline panleukopenia virus, feline herpes-virus and feline calicivirus, and highly pathogenic avian influenza (H5 and H7). The animal was detected positive in the second week of October 2021, when human cases of COVID-19 dropped to less than 10,000/day in India (Fig. 1a).
The animal carcass of Indian leopard cub (Panthera pardus fusca) of approximately 1 year was recovered during the routine combing in the agricultural field of Mojipur village of Social Forestry, Bijnor range of Uttar Pradesh State (29.372442° N and 78.135849° E). The forest is located approximately 160 km from the national capital of New Delhi, India. Bijnor Social Forestry Division has approximately 14,400 hectare of reserve forest located in the foothills of the Himalaya. About 70% of the inhabited land is under sugarcane cultivation which favors the habitation of wild herbivores and carnivores in this area.
Post-mortem and histopathological examination
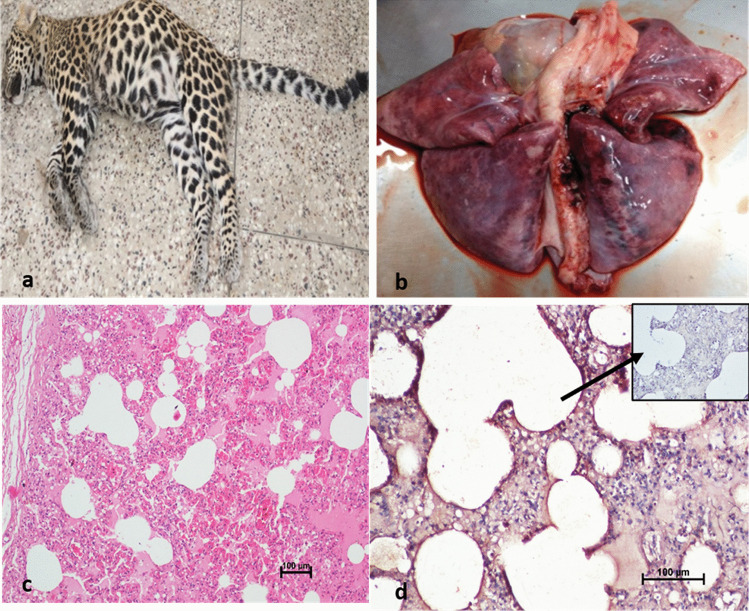
Necropsy examination revealed piercing wounds on both the sides of the ventral neck region (teeth marks of predator animal), subcutaneous contusions, and hemorrhages on neck and cranium suggesting case of infighting. Both the lungs were consolidated and severe vascular changes like congestion and hemorrhages in the visceral organs were observed (Fig. 2). The lung sections revealed diffuse areas of consolidation, hemorrhages, pneumocyte hyperplasia, septal thickening, and perivascular infiltration of mononuclear cells in histopathological analysis (Fig. 2). Viral antigen was demonstrated in the sections of lungs by immunohistochemistry using human anti-SARS-CoV-2 hyperimmune sera and goat-antihuman IgG-HRPO antibodies (Real Gene, Germany) as per previously described protocol (Sun et al. 2021; Karikalan et al. 2021). Abundant viral RNA (Ct value = 27.6 ± 0.4) was observed in lung sections. Similarly, viral antigen was noticed in the septal lining cells, alveolar macrophages, endothelial cells of pulmonary vessels, and bronchiolar epithelial cells (Fig. 2). Several natural infections in minks and humans and experimental infection of SARS-CoV-2 in laboratory animals showed similar lung histopathological lesions as observed in the present case (Molenaar et al. 2020; Caramaschi et al. 2021; Mohandas et al. 2021). Severe vascular changes were also noticed in the heart, brain, liver, and kidneys. The spleen and lymph nodes showed mild depletion of lymphoid follicles. Viral RNA was detected in these organs (Supplementary Table 1).
Fig. 2.
Lung from necropsied leopard showing severe inflammatory changes. a Carcass of leopard cub. b Representative macroscopic image of affected lung of animal. c Representative microscopic image (objective × 20) of section of lungs fixed in 10% formalin and stained with HE, showing diffuse inflammatory changes like hemorrhages and congestion and thickening of alveolar septum. d Representative microscopic image showing strong immune reactivity to SARS-COV-2 in the alveolar septal cells and inflammatory cells. IHC DAB × 200. Inset: antibody control, IHC DAB × 20
Isolation and characterization of SARS-CoV-2
SARS-CoV-2-positive nasopharyngeal swab was subjected to virus isolation in Vero E6 cells as described previously and blindly passaged 7 times. The obvious cytopathic effects (CPE) were observed after P-7 and presence of virus in the infected cells was confirmed by RT-PCR and immuno-staining with serum collected from SARS-CoV-2-infected and recovered human followed by detection by FITC-labeled anti-human secondary antibodies (Real Gene, Germany).
We generated whole-genome sequence directly from the nasal swab specimen. VTM containing nasal swabs were submitted to Eurofins India as for generation of whole-genome sequence following targeted sequencing method of viral genome enrichment and sequencing. The generated reads were cleaned and aligned with the reference sequence. Aligned sequence was submitted to NCBI with GenBank accession ID OL691925. The SRA project was submitted with accession number PRJNA786974. BLAST tool indicated > 99.9% identity with SARS-CoV-2 sequence (OK189615-OK189617) collected to Kerala state of India during the month of September 2021. Compared to the Wuhan-Hu-1 sequence (NC-045512), 40 amino acid substitutions were observed of which 3 were present in the 5’UTR and 2 were in 3’UTR (Supplementary Table 2). Six amino acid substations, viz. Thr19Arg, Asn99Lys, Leu452Arg, Thr478Lys, Asp614Gly, and Pro681Arg, were observed in the spike protein. Spike protein sequence of virus from leopard showed resemblance with that of sequences of Delta variant and with the sequences generated in previous study from infected Asiatic lions from Jaipur and Etawah in India (Karikalan et al. 2021). However, two amino acid substitutions (Thr77Lys and Asp142Gly) were observed when compared to genome sequences from infected Asiatic lions of Tamil Nadu (Mishra et al. 2021). Although very few animals have been found infected by SARS-CoV-2, high resemblance of spike protein sequences with that of human Delta variant suggest possible spillover from humans with no major genetic changes in the spike protein.
To elucidate the phylogenetic and temporal analysis, we downloaded representative sequences from GISAID database of each clade of SARS-CoV-2 samples collected from India during the period of March to Oct 2021. Consensus sequences of each clade were generated by aligning using ClustalW tool in MEGA X version 10.1 followed by consensus generation in EMBOSS server. Phylogeny was constructed using NGphylogeny server as described previously (Karikalan et al. 2021). The leopard sample clustered into common SARS-CoV-2 G-clade (GISAID classification) or Delta variant (B.1.617.2 Pango lineage) (Fig. 1b). SARS-CoV-2 nucleotide sequence from leopard was closely matching with the prevailing Delta mutant in the area suggesting possible spillover infection in the animal.
Discussion
Though the necropsy findings suggested traumatic injuries as the immediate cause of death, detection of virus in various tissues indicates systemic SARS-CoV-2 infection of the animal prior to sudden death due to trauma inflicted by another carnivore. Presence of virus in the brain section suggests infection of the central nervous system in wild felids which has previously been demonstrated in humans only (Song et al. 2021; Meinhardt et al. 2021). Previously, one SARS-CoV-2-infected captive Asiatic lion died at Chennai, India (unpublished); however, necropsy examination could not be conducted to ascertain the exact cause and comorbidities.
Currently, there is negligible role of wildlife in epidemiology of SARS-CoV-2; however emergence of a novel reservoir of SARS-CoV-2 infection in wildlife will pose challenge for control and elimination of the pathogen (Delahay et al. 2021). Infection in free-living wildlife would be having adverse implications for wildlife conservation activities. In the present report, SARS-CoV-2 was detected in the free-ranging leopard. The forest range in Bijnor is a protected area but shares a border with human habitation. On an average, 10–15 leopards die, either due to natural causes or diseases, and there is no alarming increase in the mortality in the area; hence, it seems to be a focal case of SARS-CoV-2 infection. Similarly, high level of viral circulation in the human population was associated with natural SARS-CoV-2 infection in kept ferrets but lack of virus-positive ferrets at resampling suggested that small ferret populations are unable to maintain prolonged virus circulation (Gortázar et al. 2021).
Most cases of natural infection of SARS-CoV-2 detected in animals have been linked to transmission from infected humans. However, transmission from infected humans to free-living wildlife could be associated with human contamination of the environment (e.g., feces in wastewater), supplemental food, and urban waste or fomites (Delahay et al. 2021). Leopards are relatively less shy when compared to other wild felids which may provide opportunity for spill over events in this species. Detection of SARS-CoV-2 in free-ranging leopard when incidences of human COVID-19 have dropped to significantly lower level underlines the necessity to intensify the screening and check for development of carrier status in wild felids.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the Bijnor Forest Division for submitting the carcass for necropsy examination.
Funding
We are thankful to ICAR, NASF, and DST-SERB for providing funds and infrastructure to take up the work.
Data availability
Annotated complete genome sequences are available in NCBI (GenBank accession: OL691925). Raw sequencing data are available at NCBI BioSample (Accession; PRJNA786974).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sonalika Mahajan, Mathesh Karikalan and Vishal Chander contributed equally.
References
- Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, VandeWoude S, Ragan IK, Maison RM, Bowen RA (2020) Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA 20;117(42):26382–26388. 10.1073/pnas.2013102117 [DOI] [PMC free article] [PubMed]
- Caramaschi S, Kapp ME, Miller SE, Eisenberg R, Johnson J, Epperly G, Maiorana A, Silvestri G, Giannico GA. Histopathological findings and clinicopathologic correlation in COVID-19: a systematic review. Mod Pathol. 2021;34(9):1614–1633. doi: 10.1038/s41379-021-00814-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool K, Gaudreault NN, Morozov I, Trujillo JD, Meekins DA, McDowell C, Carossino M, Bold D, Mitzel D, Kwon T, Balaraman V, Madden DW, Artiaga BL, Pogranichniy RM, Roman-Sosa G, Henningson J, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg Microbes Infect. 2022;11(1):95–112. doi: 10.1080/22221751.2021.2012528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahay RJ, de la Fuente J, Smith GC, Sharun K, Snary EL, Flores Girón L, Nziza J, Fooks AR, Brookes SM, Lean FZX, Breed AC, Gortazar C. Assessing the risks of SARS-CoV-2 in wildlife. One Health Outlook. 2021;3:7. doi: 10.1186/s42522-021-00039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortázar C, Barroso-Arévalo S, Ferreras-Colino E, Isla J, de la Fuente G, Rivera B, Domínguez L, de la Fuente J, Sánchez-Vizcaíno JM. Natural SARS-CoV-2 infection in kept ferrets. Spain Emerg Infect Dis. 2021;27(7):1994–1996. doi: 10.3201/eid2707.210096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, Ehrlich M, Grieser J, Winston J, Lombardi D, Gibson S, Saif L, Killian ML, Lantz K, Tell RM, Torchetti M, Robbe-Austerman S, Nelson MI, Faith SA, Bowman AS. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602(7897):481–486. doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Reid TJ. Animals and SARS-CoV-2: species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound Emerg Dis. 2021;68(4):1850–1867. doi: 10.1111/tbed.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikalan M, Chander V, Mahajan S, Deol P, Agrawal PK, Nandi S, Rai SK, Mathur A, Pawde A, Singh KP, Sharma GK (2021) Natural infection of Delta mutant of SARS-CoV-2 in Asiatic lions of India Transbound Emerg Dis 17. 10.1111/tbed.14290.10.1111/tbed.14290 [DOI] [PMC free article] [PubMed]
- Meinhardt J, Radke J, Dittmayer C, Franz J et al (2021) Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19 Nat Neurosci 24(2):168-175. 10.1038/s41593-020-00758-5 [DOI] [PubMed]
- Mishra A, Kumar N, Bhatia S, Aasdev A, Kanniappan S, Sekhar AT, Gopinadhan A, Silambarasan R, Sreekumar C, Dubey CK, Tripathi M, Raut AA, Singh VP. SARS-CoV-2 Delta variant among Asiatic lions. India Emerg Infect Dis. 2021;27(10):2723–2725. doi: 10.3201/eid2710.211500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas S, Yadav PD, Shete A, Nyayanit D, Sapkal G, Lole K, Gupta N (2021) SARS-CoV-2 Delta variant pathogenesis and host response in Syrian hamsters. Viruses 5;13(9):1773. 10.3390/v13091773. [DOI] [PMC free article] [PubMed]
- Molenaar RJ, Vreman S, Hakze-van der Honing RW, Zwart R, de Rond J, Weesendorp E, Smit LAM, Koopmans M, Bouwstra R, Stegeman A, van der Poel WHM. Clinical and pathological findings in SARS-CoV-2 disease outbreaks in farmed Mink (Neovison vison) Vet Pathol. 2020;57(5):653–657. doi: 10.1177/0300985820943535. [DOI] [PubMed] [Google Scholar]
- Munir K, Ashraf S, Munir I, Khalid H, Muneer MA, Mukhtar N, Amin S, Ashraf S, Imran MA, Chaudhry U, Zaheer MU, Arshad M, Munir R, Ahmad A, Zhao X. Zoonotic and reverse zoonotic events of SARS-CoV-2 and their impact on global health. Emerg Microbes Infect. 2020;9(1):2222–2235. doi: 10.1080/22221751.2020.1827984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G, Colona VL, Pandolfi PP. A focus on the spread of the delta variant of SARS-CoV-2 in India. Indian J Med Res. 2021;153(5&6):537–541. doi: 10.4103/ijmr.ijmr_1353_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piplani S, Singh PK, Winkler DA, Petrovsky N (2021) In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin. Sci Rep. 24;11(1):13063. 10.1038/s41598-021-92388-5. Erratum in: Sci Rep 11(1):18610 [DOI] [PMC free article] [PubMed]
- Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman OE, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Kazmi SAJ, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas JL, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A (2021) Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med 1;218(3):e20202135. 10.1084/jem.20202135 [DOI] [PMC free article] [PubMed]
- Sun Y, Ge L, Udhane SS, Langenheim JF, Rau MJ, Patton MD, Gallan AJ, Felix JC, Rui H (2021) Sensitive and specific immunohistochemistry protocol for nucleocapsid protein from all common SARS-CoV-2 virus strains in formalin-fixed, paraffin embedded tissues. Methods Protoc 10;4(3):47. 10.3390/mps4030047 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Annotated complete genome sequences are available in NCBI (GenBank accession: OL691925). Raw sequencing data are available at NCBI BioSample (Accession; PRJNA786974).