Abstract
Transposon mutagenesis of Mycobacterium smegmatis mc2155 enabled the isolation of a mutant strain (called LGM1) altered in the regulation of piperidine and pyrrolidine utilization. The complete nucleotide sequence of the gene inactivated in mutant LGM1 was determined from the wild-type strain. This gene (pipR) encoded a member of the GntR family of bacterial regulatory proteins. An insertion element (IS1096), previously described for M. smegmatis, was detected downstream of the gene pipR. Three additional open reading frames were found downstream of IS1096. The first open reading frame (pipA) appeared to encode a protein identified as a cytochrome P450 enzyme. This gene is the first member of a new family, CYP151. By a gene replacement experiment, it was demonstrated that the cytochrome P450 pipA gene is required for piperidine and pyrrolidine utilization in M. smegmatis mc2155. Genes homologous to pipA were detected by hybridization in several, previously isolated, morpholine-degrading mycobacterial strains. A gene encoding a putative [3Fe-4S] ferredoxin (orf1) and a truncated gene encoding a putative glutamine synthetase (orf2′) were found downstream of pipA.
Rapidly growing species of the genus Mycobacterium play an important role in the metabolism of a variety of recalcitrant organic molecules, including vinyl chloride (20), polycyclic aromatic hydrocarbons (6, 14, 24, 26), halogenated phenols (52, 53), isonicotinate (30), aromatic compounds (10, 45, 48, 50), and secondary amines (8, 11, 28, 29, 40). Morpholine, pyrrolidine, and piperidine are closely related secondary amines used in industry and eventually released in the environment. The latter two compounds can also be synthesized by different organisms. Recently, we isolated several mycobacterial strains able to degrade morpholine and demonstrated that a cytochrome P450 enzyme was involved in the degradation of this amine in all these strains (41). All these bacteria, like nearly all the morpholine degraders described in the literature, belong to the genus Mycobacterium. The induction of a heme-containing monooxygenase was also noted when these bacteria were grown on pyrrolidine and, for some of them, on piperidine. It was shown that morpholine-nondegrading mycobacterial strains (Mycobacterium fortuitum and Mycobacterium smegmatis mc2155) produced a cytochrome P450 monooxygenase during growth on piperidine and pyrrolidine (41).
Cytochromes P450 are heme-containing enzymes which play a central role in the oxidative metabolism of organic compounds. These proteins seem to be important for mycobacteria, since 22 genes encoding putative cytochromes P450 have been detected in the genome of Mycobacterium tuberculosis (13).
In this study, we have isolated, sequenced, and analyzed the genes encoding the piperidine-inducible cytochrome P450 (PipA) and its regulatory protein (PipR) from the M. smegmatis strain mc2155. The isolation of a mutant, in which pipR was inactivated by transposition mutagenesis, clearly demonstrated the involvement of the protein PipR in the regulation of piperidine and pyrrolidine metabolism. It was shown, by gene replacement experiment, that pipA was required for piperidine and pyrrolidine utilization in strain mc2155.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in liquid medium or on solid Luria-Bertani (L) medium containing ampicillin (50 μg ml−1), gentamicin (20 μg ml−1), kanamycin (20 μg ml−1), or streptomycin (50 μg ml−1). M. smegmatis mc2155 was grown in Middlebrook liquid 7H9 medium or solid 7H10 medium (Difco, Fisher Scientific, Elancourt, France) supplemented with Bacto Middlebrook ADC Enrichment supplement and 0.05% Tween 80, at 37°C except for transposition mutagenesis. With this strain, gentamicin and kanamycin were used at 5 and 20 μg per ml, respectively. Other mycobacterial strains were grown in liquid medium or on solid L medium at 30°C.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | F′::Tn10 proA+B+lacIq Δ(lacZ)M15 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 | Stratagene |
| M. smegmatis mc2155 | Highly transformable mutant | 47 |
| M. smegmatis LGM1 | Tn611 insertion mutant of strain mc2155 | This work |
| M. aurum MO1 | Morpholine-degrading strain | 11 |
| Mycobacterium sp. | ||
| BM01 | Morpholine-, piperidine-, and pyrrolidine-degrading strain | 41 |
| BM04 | Morpholine- and pyrrolidine-degrading strain | 41 |
| BM05 | Morpholine- and pyrrolidine-degrading strain | 41 |
| BM06 | Morpholine- and pyrrolidine-degrading strain | 41 |
| FM10 | Morpholine- and pyrrolidine-degrading strain | 41 |
| FM30 | Morpholine- and pyrrolidine-degrading strain | 41 |
| LM20 | Morpholine- and pyrrolidine-degrading strain | 41 |
| LM32 | Morpholine- and pyrrolidine-degrading strain | 41 |
| LM40 | Morpholine- and pyrrolidine-degrading strain | 41 |
| RP1 | Morpholine-, piperidine-, and pyrrolidine-degrading strain | 40 |
| Plasmids | ||
| pBluescript II KS(+) | E. coli cloning vector; Ampr | Stratagene |
| pJQ200 | Cloning vector with sacB and aacC1 (Gmr) genes; nonreplicative vector in mycobacteria | 42 |
| pCG79 | Mycobacterium and E. coli shuttle plasmid, thermosensitive for replication or maintenance in mycobacteria, carries an Strr gene expressed in E. coli and the Tn611 transposon containing the Kanr gene from Tn903 which is expressed in mycobacteria and in E. coli | 19 |
| pLGM20 | pBluescript with 5.8-kb EcoRI fragment of M. smegmatis containing pipR, IS1096, pipA, orf1, and truncated orf2′ | This work |
| pLGM21 | pBluescript with 2,177-bp SacI-EcoRI fragment of pLGM20 | This work |
| pLGM22 | pLGM21 with a Kanr cassette in the blunt-ended MluI site | This work |
| pLGM23 | pJQ200 with XbaI-PstI fragment of pLGM22 in SmaI site | This work |
In gene replacement experiments, transformants were selected on solid L medium containing gentamicin and kanamycin (LGK). Ten percent sucrose was added to solid L medium containing kanamycin (LKS) to select allelic-exchange events.
Mineral salts (MS) medium which contained (per liter) 1 g of KH2PO4, 1 g of K2HPO4, 0.04 g of MgSO4 · 7H2O, 0.004 g of FeCl3 · 6H2O, and 1 g of (NH4)2SO4 (the last compound was omitted when the amines were used as the carbon, nitrogen, and energy sources) was used to grow mycobacterial strains on piperidine, pyrrolidine, or glucose. Noble agar (Difco) was added at 1.5% (wt/vol) to prepare solid MS medium. The pH was adjusted to 7.0 with HCl after the addition of the amine. Piperidine (C5H11N) and pyrrolidine (C4H9N) were purchased from Fluka (Sigma Aldrich Sarl, St. Quentin Fallavier, France) and used at 10 mM. Bacterial growth was determined by monitoring the optical density of the cultures at 600 nm.
Degradation of piperidine and pyrrolidine by M. smegmatis resting cells.
Cells growing in 7H9 medium or MS medium amended with piperidine were harvested, at the end of the exponential phase, by centrifugation at 6,000 × g for 10 min at 20°C. The supernatant was discarded, and the pellet was washed twice with MS medium and resuspended in 8 ml of this medium (circa 1010 cells per ml). Cells were incubated with 5 mM piperidine or pyrrolidine at 37°C with magnetic agitation and aeration (2 liters of air per h). Samples (110 μl) were taken at regular intervals and immediately centrifuged at 20,000 × g for 2 min, and the concentration of the amine was determined.
Analytical methods.
The piperidine and pyrrolidine concentrations were estimated spectrophotometrically by the method of Stevens and Skov (49) as modified by Knapp et al. (28). The protein concentration was determined by the method of Bradford (7).
Spectrophotometric analysis of cytochrome P450.
Glucose-grown M. smegmatis cells were harvested by centrifugation (6,000 × g for 10 min) at 4°C. Cells were broken by three passages through a French pressure cell (SLM-Aminco) at 18,000 lb in−2. The crude extract was treated as previously described (40).
DNA manipulations.
Restriction and modification enzymes were purchased from Eurogentec (Eurogentec S.A., Seraing, Belgium). Plasmid DNA isolation was performed with a Qiagen plasmid extraction kit (Qiagen S.A., Courtaboeuf, France). DNA fragments of interest were purified from agarose gel by using the Geneclean III kit (Bio 101, Ozyme, St. Quentin, France). Standard recombinant DNA techniques were carried out as described by Sambrook et al. (43).
Electroporation of bacteria.
Electrocompetent E. coli cells were prepared and electroporated by the method of Dower et al. (17). Cells of M. smegmatis mc2155 were made electrocompetent and used according to the method described by Pelicic et al. (38).
Tn611 transposon mutagenesis.
The transposon mutagenesis of M. smegmatis mc2155 was performed as described by Guilhot et al. (19) by using the thermosensitive plasmid pCG79. This vector, carrying Tn611, was introduced into M. smegmatis by electroporation, and transformants were selected on 7H10 medium containing kanamycin at 30°C. A randomly chosen clone was grown for 72 h at 30°C in 5 ml of 7H9 medium supplemented with kanamycin. Antibiotic-free 7H9 medium was then inoculated with this preculture and incubated for 24 h at 39°C. Various dilutions were spread on 7H10 medium supplemented with kanamycin and incubated at 39°C. Eight thousand clones were taken at random and replica plated on solid MS medium containing 10 mM piperidine plus kanamycin and on 7H10 medium containing kanamycin.
Isolation of genomic DNA, gene cloning, and sequencing.
M. smegmatis genomic DNA was isolated from a 10-ml culture (7H9 medium) as follows. Cells were pelleted by centrifugation (6,000 × g for 10 min), resuspended in 5 ml of 7H9 medium supplemented with 50 μg of d-cycloserine ml−1 and 100 μg of lysozyme ml−1, and incubated overnight at 37°C. After centrifugation, the cells were resuspended in 500 μl of solution I (25% glucose, 50 mM Tris-HCl [pH 8.0], 50 mM EDTA, 500 μg of lysozyme ml−1) and incubated for 1 h at 37°C. Then, 500 μl of solution II (100 mM Tris-HCl [pH 8.0], 50 mM EDTA, 400 μg of proteinase K ml−1) was added and the mixture was incubated for 3 h at 55°C. DNA was extracted twice with phenol-chloroform and once with chloroform and then was ethanol precipitated. The pellet was dissolved in 1× TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and treated with RNase A (50 μg ml−1) for 1 h at 37°C. Proteinase K (50 μg ml−1) was added to the DNA solution, and incubation was continued for 1 h at 37°C. DNA was extracted with phenol-chloroform and chloroform and then concentrated by ethanol precipitation. The genomic DNA of the other mycobacterial strains was isolated as previously described (41).
The site of the Tn611 insertion in the chromosome of mutant LGM1 was determined by using marker rescue as described by Billman-Jacobe et al. (5). Genomic DNA of strain LGM1 was digested with the restriction enzyme EcoRI (a unique site in pCG79), diluted, self-ligated, and transformed into E. coli. Transformants were selected on solid L medium supplemented with streptomycin and checked for their ability to grow in kanamycin-containing medium. An EcoRI-HindIII DNA fragment (759 bp) of M. smegmatis chromosomal origin was subcloned into pBluescript II KS(+) (Stratagene, Ozyme, St. Quentin, France) for sequence analysis. The HindIII restriction site is situated in IS6100a (position 3,300) of Tn610 (GenBank accession no. X53635). Tn611 is a derivative of Tn610 in which the sulfonamide resistance (sul3) gene has been replaced by the Km resistance (Kmr) gene of Tn903 (31). The 759-bp DNA fragment was used in hybridization experiments (probe I). Total DNA of wild-type M. smegmatis mc2155 was digested overnight with different restriction endonucleases and separated by electrophoresis by using a 0.8% agarose gel. Southern blots (neutral membrane; Appligene Oncor, Illkirch, France) were hybridized with the digoxigenin-labeled probes. The labeling method, prehybridization and hybridization steps, and detection procedure were performed as recommended by the manufacturer (Boehringer Mannheim S.A., Meylan, France). Prehybridization and hybridization were performed at 42°C by using 50% formamide in solutions. DNA fragments of interest were purified from the agarose gel and cloned into pBluescript II KS(+). Double-stranded DNA sequencing was carried out with an automated sequencer (Genome Express, Paris, France). The French server BISANCE (15) was used to perform computer-assisted sequence analyses. The homologies between our sequences and the database sequences were determined by using the FASTA (36) and BLASTP (1) programs.
Construction of pLGM23.
The plasmid pLGM21 was linearized with the restriction enzyme MluI (which acts on a single site in the pipA gene), treated with Klenow fragment to fill in the ends of the restriction site, and ligated to a blunt-ended PstI fragment (approximately 1.1 kb) containing the Tn903 Kmr cassette (aph) to obtain the pLGM22 vector. A 2.8-kb XbaI-PstI fragment containing the pipA::Km region of plasmid pLGM22 was blunt ended and ligated into the SmaI site in pJQ200 to generate the suicide vector pLGM23 (Table 1).
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this study have been assigned accession no. AF102509 and AF102510 by GenBank.
RESULTS AND DISCUSSION
Pyrrolidine and piperidine utilization by M. smegmatis mc2155.
M. smegmatis mc2155 is able to grow in liquid MS medium containing piperidine or pyrrolidine as the sole sources of carbon, nitrogen, and energy. Piperidine and pyrrolidine were degraded quite rapidly, and after 80 h of incubation, no secondary amines could be detected in the culture medium. The values for turbidity of the cultures increased from 0.02 to 1.00 and 0.84 in piperidine-containing medium and pyrrolidine-containing medium, respectively (data not shown).
Isolation and characterization of a transposon-induced piperidine utilization mutant of M. smegmatis mc2155.
In order to isolate M. smegmatis mutants altered in piperidine metabolism, Tn611 transposon mutagenesis was performed as described by Guilhot et al. (19). A total of 8,000 thermoresistant clones randomly selected from the library were replica plated on MS agar medium containing 10 mM piperidine and on 7H10 medium. Both types of medium were supplemented with kanamycin. After five days of incubation at 39°C on piperidine-containing MS agar medium, a colony growing faster than other ones was obtained. This clone (called strain LGM1) was isolated for further analysis.
The kinetics of piperidine and pyrrolidine degradation by M. smegmatis mc2155 and strain LGM1 resting cells were determined (data not shown). When mc2155 cells were grown in MS medium containing piperidine, the degradation of this amine began rapidly. All of this compound disappeared from the medium within 60 min. Over the same period, the degradation of piperidine by M. smegmatis mc2155 cells grown in 7H9 medium was very slow. The results are consistent with the fact that the piperidine degradation pathway of strain mc2155 is inducible. In contrast, cells of strain LGM1 pregrown in 7H9 medium could degrade piperidine and pyrrolidine with the same efficiency as piperidine-grown mc2155 cells. The enzymes associated with the ability to degrade piperidine and pyrrolidine in M. smegmatis mc2155 need not be induced in strain LGM1.
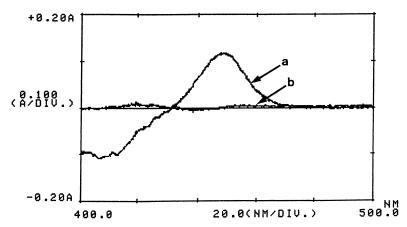
A spectrophotometric analysis of reduced extracts of mycobacterial cells (strains mc2155 and LGM1) grown in MS medium supplemented with glucose was recorded. In the spectrum of CO-treated minus nontreated reduced extracts of strain LGM1 cells, an absorption maximum at about 450 nm was observed, indicating the presence of a cytochrome P450 (Fig. 1). This peak was not detected in strain mc2155 cell extracts. Extracts prepared from glucose-grown LGM1 cells, piperidine-grown LGM1 cells, and piperidine-grown strain mc2155 cells all contained similar amounts of cytochrome P450 (125 pmol per mg of protein). Thus, a constitutive expression of a cytochrome P450 in mutant LGM1 was noted. This monooxygenase could correspond to the cytochrome P450 involved in the metabolism of piperidine in M. smegmatis mc2155. Therefore, a gene encoding a protein involved in the negative regulation of the synthesis of this cytochrome P450 was probably disrupted by the insertion of Tn611.
FIG. 1.
Reduced CO difference spectra of crude extracts of glucose-grown M. smegmatis cells. Spectrum a, Tn611 insertion mutant LGM1; spectrum b, wild-type, M. smegmatis mc2155 strain. The protein concentrations were 10 mg ml−1.
Site of insertion of transposon Tn611 in strain LGM1.
Tn611 has been shown to transpose by a replicative mechanism (31). During this transposition event, plasmid pCG79 containing Tn611 was integrated into the chromosome of the bacteria and one of the two copies of IS6100 was duplicated. Thus, by digesting the genomic DNA of the Tn611 insertional mutant bacteria by EcoRI (which acts on a unique site in pCG79) it was possible to rescue a plasmid containing the origin of replication for pUC18 and the Strr and/or the Kmr markers (depending on which IS6100 copy was duplicated). The rescued plasmid obtained after strain LGM1 genomic DNA digestion with the restriction endonuclease EcoRI was introduced into E. coli. This plasmid conferred only Strr to E. coli. Consequently, during the process of transposition of Tn611 in M. smegmatis IS6100a was duplicated since the rescued plasmid obtained after IS6100b duplication would have conferred Strr plus Kmr to E. coli. The DNA sequence of chromosomal origin was subcloned from the rescued plasmid and sequenced on one strand (data not shown). In this nucleotide sequence, 737 bp belong to M. smegmatis genomic DNA and 22 bp belong to IS6100a of transposon Tn611. The deduced polypeptide had similarity to proteins of the GntR family of bacterial regulatory proteins (21).
Cloning of the gene encoding a regulatory protein of the piperidine metabolism pathway.
The 737-bp fragment from M. smegmatis LGM1 chromosome (probe I) was used to probe Southern blots of completely digested M. smegmatis mc2155 DNA in order to isolate the entire gene. An EcoRI fragment of about 5.8 kb was detected, cloned in pBluescript II KS(+) (pLGM20), and selected by colony hybridization. Southern blot hybridization analysis of plasmid pLGM20 DNA, digested with several restriction endonucleases, was carried out with probe I (data not shown). The gene encoding the regulatory protein was located on one end of the 5.8-kb EcoRI fragment (Fig. 2). Starting from this end, a 1,215-bp region was sequenced on both strands. Sequence analysis of this DNA fragment revealed the presence of an open reading frame (ORF) (designated pipR) encoding a protein of 245 amino acids with a calculated molecular mass of 26,331 Da (Fig. 3). This ORF begins with a GUG initiation codon, is preceded by the sequence AAGGAG, a putative ribosome binding site, and is followed by a sequence potentially forming a stable transcription termination hairpin structure. The use of GUG as the translational initiation codon of PipR could allow the autoregulation of the expression of this protein as demonstrated for the cam repressor (CamR) of the cytochrome P-450cam hydrolase operon (2). The pipR-encoded protein shows significant similarity to members of the GntR bacterial regulatory family, as follows: a probable transcriptional regulator (SCI35_36) of Streptomyces coelicolor (35) (31% identity in 234 amino acids), a putative lactate operon regulator (LctR) of E. coli (16) (30% identity in 227 amino acids), a pyruvate dehydrogenase complex repressor (PdhR) of Rhodobacter capsulatus (54) (28% identity in 218 amino acids), and an Uxu operon regulator (UxuR) of Haemophilus influenzae (18) (27% identity in 238 amino acids). In members of this family (21), a conserved helix-turn-helix DNA-binding domain was found in the N-terminal part of the protein ([LIVAPKR]-[PILV]-X- [EQTIVMR]-X2-[LIVM]-X3-[LIVFT]-[DNGSTK]-[RGT LV]-X-[STAIVP]-[LIVA]-X2-[STAGV]-[LIVMFYH]-X2-[LMA]). This PROSITE (3) consensus sequence (PROSITE accession no. PS00043) was detected in the N-terminal region of PipR protein, between residues 38 and 62, but with two differences, at positions 51 (A instead of [DNGSTK]) and 52 (P instead of [RGTLV]). Many members of this family act as transcriptional repressors and possess, in addition to the DNA-binding domain, an effector-binding domain.
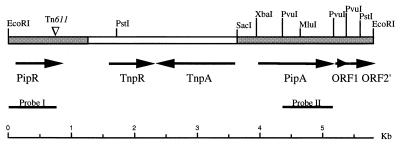
FIG. 2.
Map of the 5.8-kb EcoRI DNA region of M. smegmatis mc2155 containing the piperidine-inducible cytochrome P450 (pipA) and its regulatory gene (pipR). The DNA regions sequenced on both strands are indicated in shaded boxes. The other ORFs shown correspond to the putative resolvase (TnpR) and transposase (TnpA) of IS1096 insertion element and to the putative ferredoxin (ORF1) and glutamine synthetase (truncated ORF2′) of M. smegmatis mc2155. Only the restriction sites mentioned in the text are indicated. The position of Tn611 insertion (triangle) into the pipR gene, determined from sequence analysis, is shown.
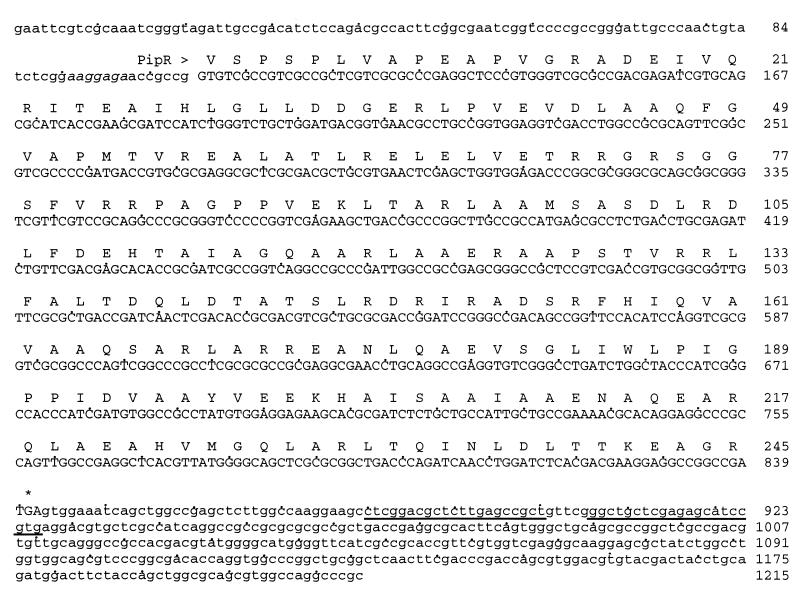
FIG. 3.
Nucleotide sequence of the 1,215-bp fragment containing the regulatory protein (PipR) gene. The presumptive ribosome binding site aaggaaga (italic), the stop codon (asterisk), and the nucleotides potentially involved in the formation of a stem-loop structure (underlined) are indicated. The GenBank accession number is AF102509. Bases shown in lowercase letters are noncoding sequences.
Identification of putative functional ORFs.
The DNA sequence downstream of pipR was partially sequenced (data not shown). It is identical to the nucleic acid sequence encoding the putative resolvase of the insertion element IS1096 (GenBank accession no. M76495) isolated from M. smegmatis mc26 by Cirillo et al. (12). This insertion element is 2,276 bp in length. Strain mc2155 is an efficient plasmid transformation mutant of M. smegmatis mc26.
The 2,177-bp SacI-EcoRI fragment located downstream of IS1096 was subcloned (pLGM21), and its nucleotide sequence was determined (Fig. 4). In this sequence, three ORFs (designated pipA, orf1, and orf2′) were found in the same orientation. The last ORF (orf2′) was truncated. The deduced amino acid sequence of pipA (400 amino acids; Mr, 44,747) matches the PROSITE consensus motif, F-[SGNH]-X-[GD]-X-[RHPT]-X-C-[LIMVFAP]-[GAD], for cytochromes P450 (PROSITE accession no. PS00086). PipA shows similarity to bacterial cytochromes P450, as follows: 31% identity to a putative cytochrome P450 (MTV023_25) of M. tuberculosis (13), 31% identity to a cytochrome P450-like protein governing hydroxylation and epoxidation in mycinamicin II biosynthesis of Micromonospora griseorubida (22), and 36% identity (but in a 291-amino-acid overlap only) to a cytochrome P450 of Streptomyces hygroscopicus (32). PipA clearly has less than 40% identity with known P450 proteins, which indicates that it represents the first member of a new family, CYP151 (33). Upstream of pipA, two imperfect repeats, which could constitute a DNA-binding motif, were detected. An imperfect 9-bp inverted repeat sequence spanned the putative −35 region (TTGACA) of pipA, and a 7-bp imperfect inverted repeat was localized between the putative −10 region (TAGAGT) and the putative ribosome binding site (GGAGG) of pipA. Bacterial cytochromes P450 are often substrate inducible, and the expression of some of them has been shown to be negatively regulated at a transcriptional level. The expression of the cytochrome P450BM-3 gene of Bacillus megaterium ATCC 14581 (46) and of the cytochrome P-450cam of Pseudomonas putida PpG1 ATCC 17543 (2) were demonstrated to be negatively regulated through the interaction of a repressor with an operator (inverted repeats) located upstream of these genes. In both these cases, the genes encoding cytochrome P450 and their regulatory proteins were adjacent and divergently oriented.
FIG. 4.
Nucleotide sequence of the 2,177-bp SacI-EcoRI region encoding a cytochrome P450 (PipA), a putative ferredoxin (ORF1), and a putative glutamine synthetase (truncated ORF2′). The two sets of possible inverted repeats (underlined) in the putative promoter region of pipA, the putative ribosome binding sites ggagg (italic), and stop codons (asterisks) are indicated. The first 182 nucleotides of this sequence are identical to the end of the IS1096 insertion element (positions 2086 to 2268) except for one nucleotide: the base A, in position 2147 in IS1096, is missing. The GenBank accession number is AF102510. Bases shown in lowercase letters are noncoding sequences.
The presence of sequences homologous to the M. smegmatis pipA gene was checked, with the internal PvuI fragment of pipA (788 bp) used as a probe (probe II), in the genomes of the following mycobacterial strains: Mycobacterium sp. strains BM01, BM04, BM05, BM06, FM10, FM30, LM20, LM32, LM40, and RP1 and Mycobacterium aurum MO1. These different strains were isolated from activated sludge, soils, or sediments for their ability to degrade morpholine (11, 40, 41). They are also able to degrade pyrrolidine and eventually piperidine. The degradation of these cyclic amines required the involvement of a cytochrome P450 in all these bacteria. A positive signal was obtained with all the DNAs tested in the experiment, indicating that closely related cytochrome P450 genes are present in these strains (data not shown). The bacteria tested in this study are not representative of all the rapidly growing mycobacterial strains but represent several clusters as defined by the phylogenetic analysis of Pitulle et al. (39). Thus, it is tempting to conclude that a homologous gene encoding a cytochrome P450 involved in the metabolism of cyclic amines is present in rapidly growing mycobacteria.
An ORF encoding a protein of 62 amino acids with an Mr of 6,760 (orf1) was identified in the region downstream of pipA. This ORF begins 46 bp after the stop codon (UGA) of pipA. The two genes are in the same reading frame. The amino acid sequence of the protein was 44, 41, and 40% identical to the reported primary structures of ferredoxin 2 and ferredoxin 1 (Fd-1 and Fd-2) from Streptomyces griseolus (34) and a putative ferredoxin (AE001094_1) from Archaeoglobus fulgidus (27), respectively. As shown in Fig. 5, the three cysteine residues (Cys-11, Cys-17, and Cys-56 of Fd-1 and Cys-10, Cys-16, and Cys-55 of Fd-2) supposed to be involved in ligating a [3Fe-4S] cluster in each of Fd-1 and Fd-2 (34) are conserved in the protein encoded by orf1 (Cys-10, Cys-16, and Cys-53). Type II cytochrome P450 systems, which are found in bacteria, use a flavin-containing reductase and a small iron- and sulfur-containing redox protein (ferredoxin) to transfer electrons to the terminal cytochrome P450. However, no ferredoxin reductase gene was detected downstream of pipA. In the genome of M. tuberculosis (13), two genes encoding putative ferredoxin proteins (MTCY369_08c and MTV049_08) linked to cytochrome P450 genes (MTCY369_09c and MTV049_07c) were found. The sequences of these ferredoxins are similar to that of the ferredoxin of M. smegmatis. The cytochrome P450 systems cloned from S. griseolus (34) and Streptomyces griseus (51) contain a cytochrome P450 gene and, downstream, the ferredoxin one. Cytochrome P450 systems present in Streptomyces and Mycobacterium strains are similarly organized.
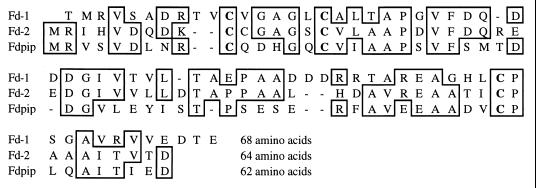
FIG. 5.
Amino acid sequence comparisons of S. griseolus (Fd-1 and Fd-2) and M. smegmatis (Fdpip) ferredoxins. The three cysteine residues, which are supposed to be involved in the attachment of a [3Fe-4S] cluster and are conserved in all three proteins, are indicated in bold. Hyphens indicate gaps introduced to optimize alignments. Residues that are identical in at least two of the three sequences are boxed.
A truncated ORF (orf2′) was detected, on the SacI-EcoRI DNA sequence, 6 bp downstream of the UGA stop codon of orf1. The polypeptide deduced from this incomplete gene shows significant homology with different glutamine synthetases (GlnA) from Archaea and Eubacteria. It is 28, 31, and 24% identical to the putative GlnA (U67574_10) of Methanococcus jannaschii (9), the putative GlnA (MTV003_4) of M. tuberculosis (13), and the GlnA (SSGLNA_2) of Sulfolobus solfataricus (44), respectively. Glutamine synthetase catalyzes the formation of glutamine by condensation of glutamate and ammonia and therefore plays a key role in the metabolism of nitrogen. A consensus pattern, [FYWL]-D-G-S-S-X6,8-[DENQSTAK]-[SA]-[DE]-X2-[LIVMFY], is usually present in the N-terminal section of these enzymes (PROSITE accession no. PS00180). The putative GlnA of M. jannaschii and the GlnA of S. solfataricus contain this consensus sequence, but it is missing in the putative GlnA of M. tuberculosis and M. smegmatis mc2155. The three ORFs (pipA, orf1, and orf2′) are closely linked and could form part of an operon.
Disruption of the pipA gene.
Experiments were performed to replace the functional allele pipA with an inactivated copy (pipA::Km) in M. smegmatis mc2155. A two-step method for gene replacement, as described by Pelicic et al. (38), was performed by using the suicide vector pLGM23 and M. smegmatis mc2155. This plasmid carries the aacC1 and sacB genes, from the pJQ200 vector (42), which confer to mycobacteria gentamicin resistance and sucrose sensitivity, respectively (37). An individual clone was randomly chosen from those obtained on LGK plates, was propagated overnight in 7H9 medium containing kanamycin, and was spread on LKS plates. The clones resulting from this experiment were expected to contain the interrupted allele of the pipA gene. A Southern blot analysis was performed on 10 randomly chosen clones by using probe II (data not shown). The internal pipA probe hybridized to a 3.9-kb fragment of the PstI-digested DNA of M. smegmatis mc2155, as expected. All the clones selected on medium containing kanamycin plus sucrose showed hybridization to a 5-kb fragment. An additional hybridization fragment was observed in one clone (clone 4) which could have been the result of a single recombination event in the pipA gene. The plasmid pLGM23 was still present in the chromosome of this bacterium, since this clone was resistant to gentamicin. The resistance to sucrose could be due to a mutation in sacB gene. All these clones showed a shift in the size of the fragment hybridizing to the internal pipA probe (from 3.9 to 5 kb) corresponding to the length of the aph cassette. The ability of these clones to grow on piperidine- or pyrrolidine-containing MS medium or to degrade these compounds was tested. None of these bacteria could grow on these amines or degrade them (data not shown). These results clearly indicated the involvement of CYP151 in the piperidine and pyrrolidine degradative pathway. However, growth was noted in piperidine- and pyrrolidine-containing medium with clone 4 when incubation had been prolonged for several days. The interrupted allele of the pipA gene could have been eliminated by a deletion-recombination event in some bacteria of this clone.
The cytochrome P450 PipA could catalyze the C-N bond cleavage of piperidine and pyrrolidine rings, as has been suggested for morpholine metabolism by Mycobacterium sp. strain RP1 (40), but further experiments have to be done to confirm this. Jacoby and Fredericks (23) have studied the metabolism of pyrrolidine by Pseudomonas fluorescens ATCC 13430. No pyrrolidine-inducible cytochrome P450 was described for this strain, but it is worth noting that their experiments were done in 1959. The authors proposed a pathway for pyrrolidine deg-radation (pyrrolidine→Δ1-pyrrolidine→γ-aminobutyralde-hyde→γ-aminobutyric acid→succinic acid semialdehyde→succinate) by this strain. They demonstrated that the enzymes induced by pyrrolidine metabolism were also detected during putrescine utilization. The linearized compounds formed during piperidine or pyrrolidine metabolism by M. smegmatis mc2155 could be further transformed by enzymes involved in the cadaverine or putrescine utilization pathways.
Cytochromes P450 have been shown to play a central role in many anabolic and catabolic reactions performed by bacteria, particularly by actinomycetes. Among rapidly growing mycobacteria, the cytochrome P450 systems induced in Mycobacterium chlorophenolicum by pentachlorophenol (53), in M. fortuitum CG-2 by halogenated phenols (52), and in M. fortuitum KCTC 1062 by steroids (25) have not yet been genetically characterized. At least 20 cytochrome P-450 genes were found in the genome of M. tuberculosis (13). The physiological function of these proteins is still unknown, but one of them (CYP51-like P450) was able to convert, in vitro, dihydrolanosterol to its 14α-demethylated product (4). Some of these cytochrome P450 enzymes could be involved in the synthesis of the complex cell wall components and therefore could constitute potential targets for antimycobacterial drugs.
ACKNOWLEDGMENTS
We thank S. N. Snapper and W. R. Jacobs for the gift of M. smegmatis mc2155, C. Guilhot and B. Gicquel for supplying plasmids pCG79 and pJQ200, and J. S. Cech for providing M. aurum MO1.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myersand E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aramaki H, Sagara Y, Kabata H, Shimamoto N, Horiuchi T. Purification and characterization of a cam repressor (CamR) for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J Bacteriol. 1995;177:3120–3127. doi: 10.1128/jb.177.11.3120-3127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamine A, Mangla A T, Nes D, Waterman M R. Is the Mycobacterium tuberculosis CYP51-like P450 a 14α-demethylase?, abstr. IC-11. In: Durst F, Kelly S L, editors. Program and abstracts of the Fourth International Symposium on P450 Biodiversity and Biotechnology, Strasbourg, France. 1998. [Google Scholar]
- 5.Billman-Jacobe H, Sloan J, Coppel R L. Analysis of isoniazid-resistant transposon mutants of Mycobacterium smegmatis. FEMS Microbiol Lett. 1996;144:47–52. doi: 10.1111/j.1574-6968.1996.tb08507.x. [DOI] [PubMed] [Google Scholar]
- 6.Boldrin B, Tiehm A, Fritzsche C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1993;59:1927–1930. doi: 10.1128/aem.59.6.1927-1930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brown V R, Knapp J S. The effect of withdrawal of morpholine from the influent and its reinstatement on the performance and microbial ecology of a model activated sludge plant treating a morpholine-containing influent. J Appl Microbiol. 1990;69:43–53. [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J D, Geoghagen N S, Weidman J F, Fuhrmann J L, Nguyen D T, Utterback T, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B B, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Burback B L, Perry J J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl Environ Microbiol. 1993;59:1025–1029. doi: 10.1128/aem.59.4.1025-1029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cech J S, Hartman P, Slosarek M, Chudoba J. Isolation and identification of a morpholine-degrading bacterium. Appl Environ Microbiol. 1988;54:619–621. doi: 10.1128/aem.54.2.619-621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cirillo J D, Barletta R G, Bloom B R, Jacobs W R., Jr A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J Bacteriol. 1991;173:7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Dean-Ross D, Cerniglia C E. Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol. 1996;46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- 15.Dessen P, Fondrat C, Valencien C, Mugnier C. BISANCE: a French service for access to biomolecular sequence databases. Comput Appl Biosci. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 16.Dong J M, Taylor J S, Latour D J, Iuchi S, Lin E C. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high-voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 19.Guilhot C, Otal I, Van Rompaey I, Martín C, Gicquel B. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmans S, Kaptein A, Tramper J, de Bont J A M. Characterization of a Mycobacterium sp. and a Xanthobacter sp. for the removal of vinyl chloride and 1,2-dichloroethane from waste gases. Appl Microbiol Biotechnol. 1992;37:796–801. [Google Scholar]
- 21.Haydon D J, Guest J R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;76:291–296. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 22.Inouye M, Takada Y, Muto N, Beppu T, Horinouchi S. Characterization and expression of a P-450-like mycinamicin biosynthesis gene using a novel Micromonospora-Escherichia coli shuttle cosmid vector. Mol Gen Genet. 1994;245:456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby W B, Fredericks J. Pyrrolidine and putrescine metabolism: γ-aminobutyraldehyde dehydrogenase. J Biol Chem. 1959;234:2145–2150. [PubMed] [Google Scholar]
- 24.Jimenez I Y, Bartha R. Solvent-augmented mineralization of pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1996;62:2311–2316. doi: 10.1128/aem.62.7.2311-2316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kand H K, Lee S S. Heterogeneous natures of the microbial steroid 9α-hydrolase in nocardioforms. Arch Pharm Res. 1997;20:519–524. doi: 10.1007/BF02975204. [DOI] [PubMed] [Google Scholar]
- 26.Kleespies M, Kroppenstedt R M, Rainey F A, Webb L E, Stackebrandt E. Mycobacterium hodleri sp. nov., a new member of the fast-growing mycobacteria capable of degrading polycyclic aromatic hydrocarbons. Int J Syst Bacteriol. 1996;46:683–687. doi: 10.1099/00207713-46-3-683. [DOI] [PubMed] [Google Scholar]
- 27.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 28.Knapp J S, Callely A G, Mainprize J. The microbial degradation of morpholine. J Appl Bacteriol. 1982;52:5–13. [Google Scholar]
- 29.Knapp J S, Brown V R. Morpholine biodegradation. Int Biodeterior. 1988;25:299–306. [Google Scholar]
- 30.Kretzer A, Andreesen J R. A new pathway for isonicotinate degradation by Mycobacterium sp. INA1. J Gen Microbiol. 1991;137:1073–1080. doi: 10.1099/00221287-139-11-2763. [DOI] [PubMed] [Google Scholar]
- 31.Martín C, Timm J, Rauzier J, Gómez-Lus R, Davies J, Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 32.Molnar I, Aparicio J F, Haydock S F, Khaw L E, Schwecke T, Konig A, Staunton J, Leadlay P F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, D. R. 1998. Personal communication.
- 34.O’Keefe D P, Gibson K J, Emptage M H, Lenstra R, Romesser J A, Litle P J, Omer C A. Ferredoxins from two sulfonylurea herbicide monooxygenase systems in Streptomyces griseolus. Biochemistry. 1991;30:447–455. doi: 10.1021/bi00216a021. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill J, Barrell B G, Rajandream M A. Streptomyces coelicolor sequencing project. 1998. Unpublished data. [Google Scholar]
- 36.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 37.Pelicic V, Reyrat J M, Gicquel B. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol. 1996;178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelicic V, Reyrat J M, Gicquel B. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 39.Pitulle C, Dorsch M, Kazda J, Wolters J, Stackebrandt E. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol. 1992;42:337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- 40.Poupin P, Truffaut N, Combourieu B, Besse P, Sancelme M, Veschambre H, Delort A M. Degradation of morpholine by an environmental Mycobacterium strain involves a cytochrome P-450. Appl Environ Microbiol. 1998;64:159–165. doi: 10.1128/aem.64.1.159-165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poupin, P., J. J. Godon, E. Zumstein, and N. Truffaut. Degradation of morpholine, piperidine, and pyrrolidine by mycobacteria: evidence for the involvement of a cytochrome P450. Can. J. Microbiol., in press. [PubMed]
- 42.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanangelantoni A M, Barbarini D, Di Pasquale G, Cammarano P, Tiboni O. Cloning and nucleotide sequence of an archaebacterial glutamine synthetase gene: phylogenetic implications. Mol Gen Genet. 1990;221:187–194. doi: 10.1007/BF00261719. [DOI] [PubMed] [Google Scholar]
- 45.Semba H, Mukouyama M, Sakano K. A para-site-specific hydroxylation of various aromatic compounds by Mycobacterium sp. strain 12523. Appl Microbiol Biotechnol. 1996;46:432–437. doi: 10.1007/s002530051047. [DOI] [PubMed] [Google Scholar]
- 46.Shaw G C, Fulco J. Barbiturate-mediated regulation of expression of the cytochrome P450BM-3 gene of Bacillus megaterium by Bm3R1 protein. J Biol Chem. 1992;267:5515–5526. [PubMed] [Google Scholar]
- 47.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 48.Spiess T, Desiere F, Fischer P, Spain J C, Knackmuss H J, Lenke H. A new 4-nitrotoluene degradation pathway in a Mycobacterium strain. Appl Environ Microbiol. 1998;64:446–452. doi: 10.1128/aem.64.2.446-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens W H, Skov K. A rapid spectrophotometric method for determining parts per million of morpholine in boiler water. Analyst. 1965;90:182–183. [Google Scholar]
- 50.Tay S T L, Hemond H F, Polz M F, Cavanaugh C M, Dejesus I, Krumholz L R. Two new Mycobacterium strains and their role in toluene degradation in a contaminated stream. Appl Environ Microbiol. 1998;64:1715–1720. doi: 10.1128/aem.64.5.1715-1720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trower M K, Lenstra R, Omer C, Buchholz S E, Sariaslani F S. Cloning, nucleotide sequence determination and expression of the genes encoding cytochrome P-450soy (soyC) and ferredoxinsoy (soyB) from Streptomyces griseus. Mol Microbiol. 1992;6:2125–2134. doi: 10.1111/j.1365-2958.1992.tb01386.x. [DOI] [PubMed] [Google Scholar]
- 52.Uotila J S, Kitunen V H, Saastamoinen T, Coote T, Häggblom M M, Salkinoja-Salonen M S. Characterization of aromatic dehalogenases of Mycobacterium fortuitum CG-2. J Bacteriol. 1992;174:5669–5675. doi: 10.1128/jb.174.17.5669-5675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uotila J S, Salkinoja-Salonen M S, Apajalahti J H A. Degradation of pentachlorophenol by membrane bound enzymes from Rhodococcus chlorophenolicus. Biodegradation. 1992;2:68–75. doi: 10.1007/BF00122422. [DOI] [PubMed] [Google Scholar]
- 54.Zeilstra-Ryalls J H, Gabbert K, Mouncey N J, Kaplan S, Kranz R G. Analysis of the fnrL gene and its function in Rhodobacter capsulatus. J Bacteriol. 1997;179:7264–7273. doi: 10.1128/jb.179.23.7264-7273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]