Abstract
The lignin peroxidases of Phanerochaete chrysosporium are encoded by a minimum of 10 closely related genes. Physical and genetic mapping of a cluster of eight lip genes revealed six genes occurring in pairs and transcriptionally convergent, suggesting that portions of the lip family arose by gene duplication events. The completed sequence of lipG and lipJ, together with previously published sequences, allowed phylogenetic and intron/exon classifications, indicating two main branches within the lip family. Competitive reverse transcription-PCR was used to assess lip transcript levels in both carbon- and nitrogen-limited media. Transcript patterns showed differential regulation of lip genes in response to medium composition. No apparent correlation was observed between genomic organization and transcript levels. Both constitutive and upregulated transcripts, structurally unrelated to peroxidases, were identified within the lip cluster.
Lignin is second only to cellulose as the most abundant form of carbon, and its mineralization is a pivotal step in the carbon cycle. The white-rot basidiomycete Phanerochaete chrysosporium has become the model system for studying the physiology and genetics of lignin degradation (for reviews, see references 1, 12, and 31). Under nutrient limitation in defined media, P. chrysosporium secretes multiple isozymes of lignin peroxidase (LiP). In vitro depolymerization of lignin by LiP has been demonstrated previously (22, 23), although the role and interaction of individual isozymes remain uncertain.
The LiPs are encoded by a family of 10 structurally related genes, designated A to J (16). In 1992, four lip subfamilies were proposed based on the intron/exon structure of the five known P. chrysosporium lip sequences (21, 44). Segregation of restriction fragment length polymorphisms and allele-specific markers (16, 41, 42) demonstrated linkage of lipA, lipB, lipC, lipE, lipG, lipH, lipI, and lipJ. Southern blots of pulsed field gels supported the observed genetic linkage and localized lipD and lipF to chromosomes separate from each other and from the eight linked genes (13, 15, 16, 46). Mapping of cosmid (15) and λ clones (26) established precise distances and transcriptional orientation for lipA, lipB, and lipC, but the genomic organization of lipE, lipG, lipH, lipI, and lipJ has not been described.
When P. chrysosporium is grown on defined media containing limiting amounts of carbon or nitrogen (35, 36, 38, 47), lip genes are upregulated (3, 6, 7, 25, 27, 37, 49, 51). Quantitative transcript analyses has been limited to a subset of lip genes, and results have often been contradictory, perhaps due to differences in methodology. Nevertheless, as Northern blots first demonstrated for lipA and lipD (25), it is now firmly established that certain lip genes are differentially regulated in response to medium composition. To distinguish closely related transcripts, quantitative reverse transcription-PCR techniques were developed (8, 9, 46), and it was shown that lipI, lipC, and lipJ transcript levels are also differentially regulated under carbon versus nitrogen limitation (46). Nuclease protection assays showed lipE transcripts to be upregulated in C-limited media compared to N-limited media (43). Broda and coworkers found lipD transcripts dominating under all growth conditions examined (8, 9, 27).
To elucidate relationships between lip gene structure, organization, and transcriptional regulation, we have sequenced two lip genes, mapped the lip gene cluster, and systematically assessed relative transcript levels for all 10 lip genes under standard conditions of nitrogen and carbon starvation. In addition, we have identified constitutive and upregulated transcripts within the lip cluster.
MATERIALS AND METHODS
Fungal strains and culture conditions.
P. chrysosporium BKM-F-1767 was obtained from the Center for Forest Mycology Research, Forest Products Laboratory, Madison, Wis., and used throughout the study. Standard B3 salts media with limiting carbon or nitrogen were grown statically at 39°C as previously described (10, 32) and harvested on days 4 and 5, respectively. Mycelia were harvested by filtration through Miracloth (Calbiochem, La Jolla, Calif.), immediately immersed in liquid nitrogen, and stored at −90°C. LiP activities as measured by veratryl alcohol oxidation (48) were 7.3 and 12.8 nmol min−1 ml−1 in C-limited and N-limited cultures, respectively. Mycelia were also harvested from log-phase B3 cultures containing nonlimiting levels of carbon or nitrogen and lacked extracellular peroxidase activity.
DNA sequencing and analysis.
Nucleotide sequences were determined by using the ABI prism dye terminator cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.) with an ABI373 DNA sequencer. Nucleotide and amino acid sequence similarity searches used the BLAST method (2) on the National Center for Biotechnology Information databases. Nucleotide and amino acid sequences were analyzed and phylogenetic trees constructed by using DNASTAR software (DNASTAR, Madison, Wis.).
Genomic organization of lip genes.
Cosmid clones containing different lip genes were identified from a pWE15-based cosmid library (15) by using lip-specific probes. Preliminary restriction maps of the cosmids were constructed, and lip intergenic regions were PCR amplified by using the GeneAmp XL PCR kit (Perkin-Elmer, Foster City, Calif.) according to manufacturer’s recommendations. Specifically, each PCR mixture (100 μl) contained 20 to 50 ng of cosmid template, 1× XL buffer, 0.8 to 1.0 mM magnesium acetate, 40 pmol of each primer, 10 mM (each) deoxynucleoside triphosphate, 5% dimethyl sulfoxide, and 2 to 4 U of rTth polymerase. Cycling conditions after hot start were 94°C, 1 min, 1 cycle, followed by 16 cycles of 94°C for 30 s and 68°C for 10 min. These conditions were repeated for an additional 12 cycles, with an autoextension of 15 s/cycle. This was followed by a final extension at 72°C for 10 min. The primer pairs for intergenic regions were as follows: 5′-ATGGCGTCGGAAACCTGGGAACTT-3′ and 5′-TGAGGAGCATGTCCCGAGGTAGAG-3′ for lipA and lipE, 5′-CGCCATCGCTATCTCTCCCGC-3′ and 5′-AACACGAGCGATGATCTGGTCG-3′ for lipB and lipC, and 5′-GAGCGTCGGAGCGCGAGAACC-3′ and 5′-CTTTACCAGCCGATTACAGAGATG-3′ for lipG and lipH. PCR products were electroeluted, subcloned into pCRTOPO-XL, and transformed into Escherichia coli TOP10 (Invitrogen, Carlsbad, California) following the manufacturer’s recommendations.
RNA isolation.
Total RNA from P. chrysosporium was extracted from frozen mycelium and pelleted in CsCl (45, 50). Poly(A) RNA was extracted from total RNA by using a magnetic capture technique involving oligo(dT)25 Dynabeads (Dynal, Great Neck, N.Y.), following the manufacturer’s recommendations.
Competitive RT-PCR of lip genes.
To quantify lip transcripts, a competitive reverse transcription-PCR (RT-PCR) protocol was adapted from Gilliland et al. (19) as previously described (4, 46). Specifically, each RT reaction contained 2 μl of poly(A) RNA and was primed with 15 pmol of oligo(dT) 15-mers. Competitive PCRs (100 μl) contained 1.25 U of Taq DNA polymerase, 21 pmol of each primer, and competitive template added as 10-fold serial dilutions. Full-length lip genomic clones served as competitive templates, and lip-specific primers were as described previously (4). Reactions were cycled for 94°C (6 min), 54°C (2 min), and 72°C (40 min) for 1 cycle, followed by 94°C (1 min), 54°C (2 min), and 72°C (5 min) for 35 cycles and a final 72°C extension (15 min). Experiments quantifying lipA, lipD, lipI, and lipJ transcripts with various amounts of poly(A) template in RT-PCRs showed no evidence of reverse transcriptase inhibition (11).
PCR products were electrophoresed, and ethidium bromide-stained gel images were acquired by using Photoshop 3.0 (Adobe, San Jose, Calif.). National Institutes of Health (NIH) Image 1.61 software was used for image analysis and assigning equivalence points. The image was labeled by using Illustrator 7.0 (Adobe).
Transcript analyses of lip intergenic regions.
The intergenic regions between lipE and lipA, lipB and lipC, and lipG and lipH were XL-PCR amplified to aid in identifying areas of transcriptional activity. Fragments were size fractionated on 0.8% agarose gels, transferred to Nytran membranes (Schleicher and Schuell, Keene, N.H.), and probed with 32P-labeled cDNA prepared from C-limited, N-limited, and log-phase B3 media. Total cDNA was prepared from oligo(dT)-primed poly(A) RNA by using the Smart cDNA library kit (Clontech, Palo Alto, Calif.) and labeled by nick translation. Blots were hybridized and washed at high stringency and exposed to XAR film (Kodak) for 1 to 3 days. Film was scanned in Adobe Photoshop 3.0, and the image was labeled with Adobe Illustrator 7.0.
Nucleotide sequence accession number.
Nucleotide sequences for lipJ and lipG were assigned GenBank accession no. AF140062 and AF140063, respectively. Noncoding regions between lipI and lipG and between lipH and lipJ were assigned no. AF140064 and AF140065, respectively.
RESULTS
Structure and phylogeny of lip genes.
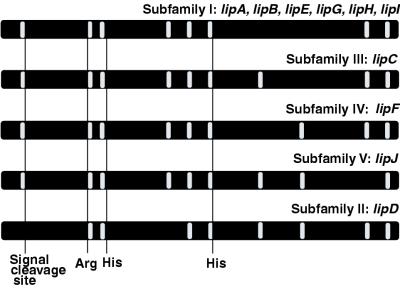
The complete nucleotide sequences of lipG and lipJ were determined, and all members of the lip family were then classified by intron/exon structure as proposed by Ritch and Gold (44). Five distinct subfamilies emerged (Fig. 1). The lipG intron positions were identical to lipA, lipB, lipE, lipH, and lipI. The number of introns varied within the lip family; lipD and the members of the lipG subfamily contained eight introns, and the others, lipC, lipF, and lipJ, contained nine. The positions of five introns were invariant among all lip genes. Two introns were missing in lipD but conserved among all other lip genes—one intron adjacent to the signal sequence and the other intron immediately preceding intron no. 4. All introns, except the second to the last, are conserved in lipJ. Intron-exon junctions of P. chrysosporium’s lip family conform to those of other filamentous fungi, specifically, PuPy (usually GT) at the 5′ end and AG at the 3′ end (21).
FIG. 1.
Classification of lip genes by intron/exon structure. Introns and exons are represented by white and black bars, respectively. The positions of conserved amino acid residues coordinating the heme (49) the relative positions of signal cleavage sites and are indicated.
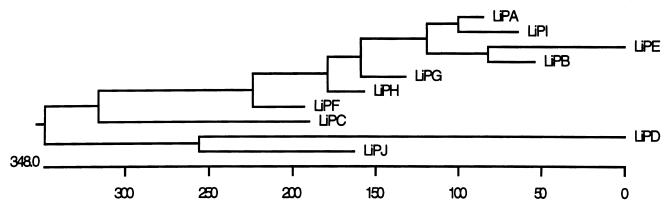
Cladistic analysis based on deduced amino acid sequences established two main branches within the LiP family (Fig. 2). The branch consisting of LiPD and LiPJ was the most divergent from all other LiPs, correlating with their unique intron structures. The second branch indicated that LiPA, LiPB, LiPE, LiPG, LiPH, and LiPI were the most recent members of the LiP family to have emerged. This result is further supported by the conserved intron/exon structures of the respective genes in this branch. The more distantly related LiPC and LiPF appear to have diverged much earlier from this branch.
FIG. 2.
Cladistic analysis of deduced amino acid sequences. The underlying scale shows the number of substitutions between sequences. Analysis was done with the Jotun Hein (24) algorithm on DNASTAR software.
Genomic organization of lip genes.
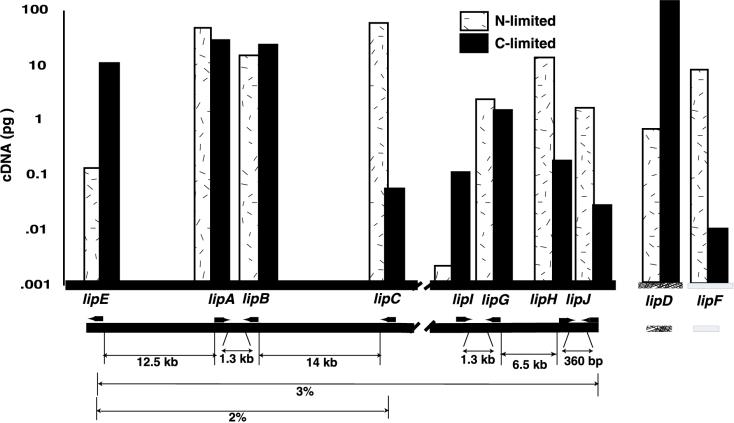
A detailed physical map of the genomic organization of the eight lignin peroxidase-encoding genes was constructed (Fig. 3). Four genes, lipA, lipB, lipC, and lipE, resided within a 35-kb region. The remaining genes, lipG, lipH, lipI, and lipJ, lie within a 15-kb region. Six genes were paired and transcriptionally convergent (lipA and lipB, lipG and lipI, and lipH and lipJ). The regions that separated paired genes were 1.3 kb or less but lacked significant nucleotide similarity to one another or to any database sequences. Of the eight genes, lipE and lipC appear to be the only unpaired members and flank the lipA and lipB pair by >10 kb.
FIG. 3.
Transcript levels and genomic organization of the lip genes. Genomic organization is shown under the graph. Thickened arrows indicate transcriptional orientation. Physical distances are in kilobases, and genetic distances are in percent recombination. Two unlinked genes, lipD and lipF, are detached to the right. Transcript levels were determined by competitive RT-PCR and are given in picograms of cDNA.
Competitive RT-PCR indicates lip genes are differentially regulated.
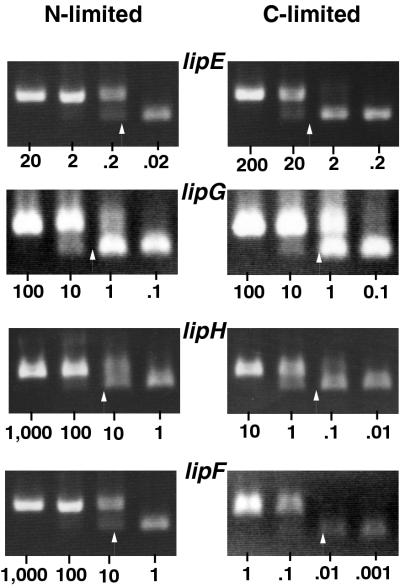
Transcript levels of all lip genes in chemically defined media were quantified by competitive RT-PCR (Fig. 4) and analyzed with respect to their genomic organization (Fig. 3). The paired genes lipA and lipB maintained similar transcript levels when limited for either carbon or nitrogen, suggesting coordinate regulation. However, transcript patterns of other lip genes, paired or otherwise, changed depending upon the limiting nutrient, indicating that most lip genes were differentially regulated. lipC had the highest transcript level of all lip genes under nitrogen limitation but one of the lowest transcript levels under carbon limitation. The dominant transcript under carbon limitation, lipD, had only moderate transcript levels under nitrogen-limiting conditions.
FIG. 4.
Competitive PCRs comparing four lip transcripts in samples harvested from nitrogen- or carbon-limited conditions. Transcript levels were estimated by determining the equivalence points (arrows) with competitive templates (46). The amount of plasmid competitive template added to each PCR is indicated below the gels and expressed in picograms. Ethidium bromide-stained gels were photographed with a Foto/Analyst Visionary system (Fotodyne, Hartland, Wis.) and scanned with a Microtek ScanMaker III and Adobe Photoshop 3.0.
Previous studies indicated lipE transcripts dominating in P. chrysosporium cultures under carbon limitation (43, 44). Our data show that lipA, lipB, and lipD transcript levels exceed those of lipE in carbon-limited media. Broda and coworkers, working with the closely related strain ME446, reported lipD to be the only member of the lip family to be highly transcribed in carbon- or nitrogen-limited cultures (8, 9, 27). In strain BKM, we found all lip genes to be transcribed under both culture conditions. However, lipD was the most abundant transcript in carbon-limited cultures.
Transcript analyses in lip intergenic regions.
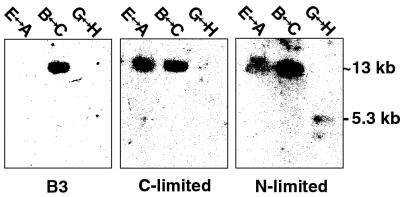
PCR-amplified intergenic regions were blotted and probed with labeled cDNA isolated from cultures grown under either ligninolytic or nonligninolytic conditions (Fig. 5). In the region between lipB and lipC, a substantial signal was observed in all media tested. In contrast, transcriptional activity between lipE and lipA was observed under carbon and nitrogen limitation but not under nonligninolytic B3 medium. Transcripts arising between lipG and lipH were barely detectable and only under nitrogen limitation (Fig. 5, 5.3-kb signal).
FIG. 5.
Southern blots of XL-PCR-amplified intergenic regions probed with 32P-labeled cDNA from log-phase B3, C-limited B3, and N-limited B3 media.
DISCUSSION
The lignocellulosic component of plant cells is comprised of lignin, cellulose, and hemicellulose. To attack these complex polymers, P. chrysosporium produces an array of enzyme families, including lignin peroxidases, manganese peroxidases, and cellulases. Why P. chrysosporium maintains multiple isozymes to catalyze presumably similar reactions remains unclear. Some substrate and kinetic differences between LiP isozymes have been observed previously (14, 20) and may indicate specific roles for individual LiPs during lignin depolymerization. Alternatively, it is possible that the majority of LiPs are redundant, having arisen through various chromosomal rearrangements such as duplications, translocations, or unequal crossover events during meiosis. The report of an insertion element that transcriptionally inactivates lipI2 indicates that not all alleles are necessary for efficient lignin depolymerization (17). In addition, there is growing evidence that redundant genes are maintained if they are not deleterious to the organism (18, 39).
The evolutionary origins of the lip family remain unclear despite various attempts to categorize them by intron/exon structure or deduced amino acid similarity. The full sequences of lipG and lipJ, presented here, allow for a comprehensive analysis of all known members of the lip family for the first time. Ritch and Gold first proposed dividing the lip genes into four subfamilies based on intron/exon structure (44). A fifth subfamily, consisting solely of lipJ, is now evident (Fig. 1). Interestingly, only subfamily I (consisting of lipA, lipB, lipE, lipH, lipI, and now lipG) has more than one member, suggesting that these genes are the most recent members of the lip family to have emerged. Cladistic analysis of deduced LiP amino acid sequences (Fig. 2) further supports the high degree of similarity and recent emergence of this subfamily. These six genes are tightly linked to lipC and lipJ (Fig. 3) but are more similar to the unlinked lipF. Thus, clustering of the lip genes does not appear to require sequence conservation.
The genomic organization of the lip cluster displays a striking pattern; six genes occurred in pairs and were transcriptionally convergent (Fig. 3). This unusual organization may indicate that these pairs arose by duplication events of an ancestral pair. The intergenic distance between paired lip genes is conserved at 1.3 kb except for the pair lipH and lipJ, which is separated by only 364 bp. These intergenic regions lack significant nucleotide similarity to one another and to database sequences.
It has been proposed that selective advantages may drive some gene families to cluster (30, 34). Such advantages include a greater likelihood for the entire family to be horizontally transferred or the simplicity of regulating a coordinately expressed cluster versus individual regulation of separated genes. Portions of the fungal secondary metabolic pathways of penicillin and cephalosporin may have been horizontally transferred from prokaryotes (40, 52), and coordinate expression of fungal gene clusters has been well documented (for a review, see reference 30). Information related to lip genomic organization in other white-rot fungi is limited, but two lip genes and a manganese peroxidase-encoding gene from Trametes versicolor are tandemly arranged within a 10-kb region (29), tentatively supporting a selective advantage in lip clustering. In P. chrysosporium two unlinked genes, lipD and lipF, were abundantly transcribed (Fig. 3), indicating that linkage is not essential for function.
Models for the origin and function of lip gene clustering must also take into account the presence of seemingly unrelated genes in the region (Fig. 5). For example, the signal consistently observed between lipB and lipC has been tentatively attributed to a gene with very high sequence similarity to Saccharomyces cerevisiae elongation factor G (BLASTX P = 2.7−136). Upregulated transcripts were also observed between lipA and lipE and between lipG and lipH. Although the precise genes have not yet been identified, Southern blots and sequence analysis exclude peroxidase sequences in these regions (data not shown).
To investigate the relationship between genomic organization and regulation, competitive RT-PCR was used to quantify transcript levels in defined media under carbon or nitrogen limitation (Fig. 4). Surprisingly, no correlation between lip genomic organization and transcript levels was apparent (Fig. 3). Only the lipA and lipB pair displays similar transcript patterns under both culture conditions, suggesting coordinate regulation. A clear pattern is not evident for the remaining lip genes, either in pairs or for general clusters. Furthermore, transcript levels do not correlate with intron/exon structure or with amino acid sequence comparisons. For example, transcript levels within intron subfamily I vary over a 1,000-fold range (carbon-limited levels of lipA versus lipJ), and LiPs that are over 90% similar at the amino acid level may vary over 10,000-fold (e.g., nitrogen-limited levels of lipA versus lipI transcripts). This observation may support the theory that individual lip genes are regulated for specific biological roles.
Recently, lip transcript levels from P. chrysosporium colonized wood chips and organopollutant-contaminated soil have been measured (4, 28, 33). The complexity of these substrates restricts observations concerning lip regulation, but the overall patterns reported are significantly different from those seen in defined media. This may reflect the occurrence of multiple layers of regulation in these substrates. As in defined media, transcript levels from solid substrates do not correlate with intron subfamilies, genomic organization, or amino acid sequence similarity (data not shown).
Although RT-PCR is an accurate method for quantifying transcript levels, it is not intended to gauge levels of active protein. It is conceivable that some lip transcripts are relatively unstable or are subject to other forms of posttranscriptional regulation that would not be detected by RT-PCR. However, competitive RT-PCR remains the most accurate method for assessing LiP transcripts, and in general, transcript levels have been shown to correlate well with enzyme activity (4, 5). The precise relationship between lip genes and specific isozymes is unclear except for lipA and lipD, which encode isozymes H8 and H2, respectively.
Differential regulation of the lignin peroxidases supports specific biological roles for individual isozymes. However, the possibility that some lip genes are redundant but have accumulated mutations altering their expression and physical properties cannot be excluded. The repeated pattern of genomic organization indicates that the lip family probably arose via a series of duplication events. Detailed physical examination of the regions surrounding the lip cluster may indicate if these areas are the result of duplications. Further analysis is also needed to identify regulatory sequences which must play a critical role orchestrating the expression of lip genes.
ACKNOWLEDGMENTS
This work was supported by Department of Energy grant DE-FG02-87ER13712.
We thank Jill Gaskell and Diane Dietrich for helpful discussions, technical assistance, and comments on the manuscript.
REFERENCES
- 1.Alic M, Gold M. Genetics and molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. In: Bennett J, Lasure L, editors. More gene manipulations in fungi. New York, N.Y: Academic Press; 1991. pp. 319–341. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Andrawis A, Pease E, Kuan I, Holzbaur E, Tien M. Characterization of two lignin peroxidase clones from Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1989;162:673–680. doi: 10.1016/0006-291x(89)92363-2. [DOI] [PubMed] [Google Scholar]
- 4.Bogan B, Schoenike B, Lamar R, Cullen D. Expression of lip genes during growth in soil and oxidation of anthracene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:3697–3703. doi: 10.1128/aem.62.10.3697-3703.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogan B, Schoenike B, Lamar R, Cullen D. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boominathan K, D’Souza T M, Naidu P S, Dosoretz C, Reddy C A. Temporal expression of the major lignin peroxidase genes of Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:3946–3950. doi: 10.1128/aem.59.11.3946-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boominathan K, Reddy C A. cAMP-mediated differential regulation of lignin peroxidase and manganese-dependent peroxidase production in the white-rot basidiomycete Phanerochaete chrysosporium. Proc Natl Acad Sci USA. 1992;89:5586–5590. doi: 10.1073/pnas.89.12.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broda P, Birch P, Brooks P, Sims P. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl Environ Microbiol. 1995;61:2358–2364. doi: 10.1128/aem.61.6.2358-2364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks P, Sims P, Broda P. Isozyme specific polymerase chain reaction analysis of differential gene expression: a general method applied to lignin peroxidase genes of Phanerochaete chrysosporium. Bio/Technology. 1993;11:830–834. doi: 10.1038/nbt0793-830. [DOI] [PubMed] [Google Scholar]
- 10.Brown A, Sims P F G, Raeder U, Broda P. Multiple ligninase-related genes from Phanerochaete chrysosporium. Gene. 1988;73:77–85. doi: 10.1016/0378-1119(88)90314-9. [DOI] [PubMed] [Google Scholar]
- 11.Chandler D, Wagnon C A, Bolton H. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl Environ Microbiol. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 13.D’Souza T M, Dass S B, Rasooly A, Reddy C A. Electrophoretic karyotyping of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Mol Microbiol. 1993;8:803–807. doi: 10.1111/j.1365-2958.1993.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 14.Farrell R L, Murtagh K E, Tien M, Mozuch M D, Kirk T K. Physical and enzymatic properties of lignin peroxidase isozymes from Phanerochaete chrysosporium. Enzyme Microb Technol. 1989;11:322–328. [Google Scholar]
- 15.Gaskell J, Dieperink E, Cullen D. Genomic organization of lignin peroxidase genes of Phanerochaete chrysosporium. Nucleic Acids Res. 1991;19:599–603. doi: 10.1093/nar/19.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaskell J, Stewart P, Kersten P, Covert S, Reiser J, Cullen D. Establishment of genetic linkage by allele-specific polymerase chain reaction: application to the lignin peroxidase gene family of Phanerochaete chrysosporium. Bio/Technology. 1994;12:1372–1375. doi: 10.1038/nbt1294-1372. [DOI] [PubMed] [Google Scholar]
- 17.Gaskell J, Vanden Wymelenberg A, Cullen D. Structure, inheritance, and transcriptional effects of Pce1, an insertional element within Phanerochaete chrysosporium lignin peroxidase gene lipI. Proc Natl Acad Sci USA. 1995;92:7465–7469. doi: 10.1073/pnas.92.16.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson T J, Spring J. Genetic redundancy in vertebrates: polyploidy and persistence of genes encoding multidomain proteins. Trends Genet. 1998;14:46–49. doi: 10.1016/s0168-9525(97)01367-x. [DOI] [PubMed] [Google Scholar]
- 19.Gilliland G, Perrin S, Bunn H. Competitive PCR for quantitation of mRNA. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols. New York, N.Y: Academic Press; 1990. pp. 60–69. [Google Scholar]
- 20.Glumoff T, Harvey P J, Molinari S, Goble M, Frank G, Palmer J M, Smit J D G, Leisola M S A. Lignin peroxidase from Phanerochaete chrysosporium: molecular and kinetic characterization of isozymes. Eur J Biochem. 1990;187:515–520. doi: 10.1111/j.1432-1033.1990.tb15333.x. [DOI] [PubMed] [Google Scholar]
- 21.Gold M, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammel K, Jensen K, Mozuch M, Landucci L, Tien M, Pease E. Ligninolysis by a purified lignin peroxidase. J Biol Chem. 1993;268:12274–12281. [PubMed] [Google Scholar]
- 23.Hammel K E, Moen M A. Depolymerization of a synthetic lignin in vitro by lignin peroxidase. Enzyme Microb Technol. 1991;13:15–18. [Google Scholar]
- 24.Hein J. Unified approach to alignment and phylogenies. Methods Enzymol. 1990;183:626–645. doi: 10.1016/0076-6879(90)83041-7. [DOI] [PubMed] [Google Scholar]
- 25.Holzbaur E, Tien M. Structure and regulation of a lignin peroxidase gene from Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1988;155:626–633. doi: 10.1016/s0006-291x(88)80541-2. [DOI] [PubMed] [Google Scholar]
- 26.Huoponen K, Ollikka P, Kalin M, Walther I, Mantsala P, Reiser J. Characterisation of lignin peroxidase-encoding genes from lignin-degrading basidiomycetes. Gene. 1990;89:145–150. doi: 10.1016/0378-1119(90)90218-g. [DOI] [PubMed] [Google Scholar]
- 27.James C M, Felipe M S S, Sims P F G, Broda P. Expression of a single lignin peroxidase-encoding gene in Phanerochaete chrysosporium strain ME446. Gene. 1992;114:217–222. doi: 10.1016/0378-1119(92)90577-c. [DOI] [PubMed] [Google Scholar]
- 28.Janse B, Gaskell J, Ahktar M, Cullen D. Phanerochaete chrysosporium genes encoding lignin peroxidases, manganese peroxidases and glyoxal oxidase in wood. Appl Environ Microbiol. 1998;64:3536–3538. doi: 10.1128/aem.64.9.3536-3538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson T, Nyman P. A cluster of genes encoding major isozymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene. 1996;170:31–38. doi: 10.1016/0378-1119(95)00846-2. [DOI] [PubMed] [Google Scholar]
- 30.Keller N P, Hohn T M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 31.Kirk T K, Cullen D. Enzymology and molecular genetics of wood degradation by white-rot fungi. In: Young R A, Akhtar M, editors. Environmentally friendly technologies for the pulp and paper industry. New York, N.Y: John Wiley and Sons; 1998. pp. 273–308. [Google Scholar]
- 32.Kirk T K, Schultz E, Conners W J, Lorentz L F, Zeikus J G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol. 1978;117:277–285. [Google Scholar]
- 33.Lamar R T, Schoenike B, Vanden Wymelenberg A, Stewart P, Dietrich D M, Cullen D. Quantitation of fungal mRNAs in complex substrates by reverse transcription PCR and its application to Phanerochaete chrysosporium-colonized soil. Appl Environ Microbiol. 1995;61:2122–2126. doi: 10.1128/aem.61.6.2122-2126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leisola M S A, Kozulic B, Meusdoerffer F, Fiechter A. Homology among multiple extracellular peroxidases from Phanerochaete chrysosporium. J Biol Chem. 1987;262:419–424. [PubMed] [Google Scholar]
- 36.Leisola M S A, Thanei-Wyss U, Fiechter A. Strategies for production of high ligninase activities by Phanerochaete chrysosporium. J Biotechnol. 1985;3:97–107. [Google Scholar]
- 37.Li D, Alic M, Gold M. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liwicki R, Paterson A, MacDonald M J, Broda P. Phenotypic classes of phenoloxidase-negative mutants of the lignin-degrading fungus Phanerochaete chrysosporium. J Bacteriol. 1985;162:641–644. doi: 10.1128/jb.162.2.641-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak M A, Boerlijst M C, Cooke J, Smith J M. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 40.Penalva M A, Moya A, Dopazo J, Ramon D. Sequences of isopenicillin N synthetase genes suggest horizontal gene transfer from prokaryotes to eukaryotes. Proc R Soc Lond. 1990;241:164–169. doi: 10.1098/rspb.1990.0081. [DOI] [PubMed] [Google Scholar]
- 41.Raeder U, Thompson W, Broda P. Genetic factors influencing lignin peroxidase activity in Phanerochaete chrysosporium ME446. Mol Microbiol. 1989;3:919–924. doi: 10.1111/j.1365-2958.1989.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 42.Raeder U, Thompson W, Broda P. RFLP-based genetic map of Phanerochaete chrysosporium ME446: lignin peroxidase genes occur in clusters. Mol Microbiol. 1989;3:911–918. doi: 10.1111/j.1365-2958.1989.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 43.Reiser J, Walther I, Fraefel C, Fiechter A. Methods to investigate the expression of lignin peroxidase genes by the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:2897–2903. doi: 10.1128/aem.59.9.2897-2903.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritch T G, Gold M H. Characterization of a highly expressed lignin peroxidase-encoding gene from the basidiomycete Phanerochaete chrysosporium. Gene. 1992;118:73–80. doi: 10.1016/0378-1119(92)90250-s. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 46.Stewart P, Kersten P, Vanden Wymelenberg A, Gaskell J, Cullen D. The lignin peroxidase gene family of Phanerochaete chrysosporium: complex regulation by carbon and nitrogen limitation, and the identification of a second dimorphic chromosome. J Bacteriol. 1992;174:5036–5042. doi: 10.1128/jb.174.15.5036-5042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tien M, Kirk T K. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–249. [Google Scholar]
- 48.Tien M, Kirk T K. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc Natl Acad Sci USA. 1984;81:2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tien M, Tu C-P D. Cloning and sequencing of a cDNA for a ligninase from Phanerochaete chrysosporium. Nature. 1987;326:520–523. doi: 10.1038/326520a0. [DOI] [PubMed] [Google Scholar]
- 50.Timberlake W E. Cloning and analysis of fungal genes. In: Bennett J W, Lasure L L, editors. More gene manipulations in fungi. New York, N.Y: Academic Press; 1991. pp. 51–85. [Google Scholar]
- 51.Walther I, Kaelin M, Reiser J, Suter F, Fritsche B, Saloheimo M, Leisola M, Teeri T, Knowles J K C, Fiechter A. Molecular analysis of a Phanerochaete chrysosporium lignin peroxidase gene. Gene. 1988;70:127–137. doi: 10.1016/0378-1119(88)90111-4. [DOI] [PubMed] [Google Scholar]
- 52.Wiegel G J, Burgett S G, Chen V J, Skatrud P L, Frolik C A, Queener S W, Ingolia T D. Cloning and expression in Escherichia coli of isopenicillin N synthetase genes from Streptomyces lipmanii and Aspergillus nidulans. J Bacteriol. 1988;170:3817–3826. doi: 10.1128/jb.170.9.3817-3826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]