Introduction
Complete resection of colorectal liver metastases (CRLMs) is considered potentially curative, but recurrence is common1. Vauthey and colleagues2 were among the first to shift from reporting on the prognostic impact of clinicopathological features to focusing on KRAS as a marker of tumour biology in CRLMs. Little attention has been paid to potential differences between the specific mutations that give rise to the activated oncoproteins with respect to their different prognostic impact. In fact, data on the three levels that categorize KRAS mutations (exon, codon, and specific nucleotide mutations) are not only scarce, but also controversial3–6. Thus, the current practice of using KRAS mutations as a binary variable may be problematic. This study aimed to test this hypothesis by analysing the prognostic impact of various mutations based on their location in different exons or codons and, even more specifically, the effect of the different point mutations within each codon in patients who underwent surgery for CRLMs.
Methods
The International Genetic Consortium for Colorectal Liver Metastasis database, which was built to include only patients with CRLMs and known KRAS mutational status, was queried for adult patients who underwent surgery between 2000 and 2017. Patients were excluded if the codon and point mutation status were unknown. Further details on mutational testing, the inclusion of patients with extrahepatic disease, and variables included in the analyses can be found in the Supplementary material. The study was conducted in accordance with the ethical standards of the participating institutions, and was approved by the appropriate ethical committees of the leading institution (Johns Hopkins University).
Survival was defined as the time between liver resection and death or last follow-up. To generate risk groups, KRAS point mutations were ranked based on associated median overall survival rates. Mutations associated with median overall survival below that of the overall KRAS-mutated group were classified as high risk, whereas those with a higher survival rate comparable to that of the overall KRAS group were classified as low risk. Median overall survival for the three patients with G13R mutations was not reached, and these were classified as high risk based on poor survival of one patient and early recurrence in two.
Statistical analyses were performed using SPSS® version 25 (IBM, Armonk, NY, USA) and R version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria). Further details can be found in Supplementary material.
Results
Among the 1567 patients included, median follow-up was 58.4 (i.q.r. 95 per cent c.i., 54.8–62.1) months. Median and 5-year overall survival were 55.3 (51.0–59.6) months and 46.6 per cent respectively. KRAS mutations were found in 562 patients (35.9 per cent). Exon 3 mutation was omitted from the analysis, as few patients harboured it. Clinicopathological and treatment characteristics, as well as outcomes, are summarized in Table 1 and Supplementary material.
Table 1.
Clinicopathological and treatment characteristics of patients with low- or high-risk KRAS mutations who underwent liver resection for colorectal liver metastases
| Wild-type (n = 1005) | High risk (n = 254) | Low risk (n = 308) | P† | |
|---|---|---|---|---|
| Age (years)* | 61.2 (53.0–68.9) | 63.0 (55.0–71.0) | 62.0 (54.0–71.0) | 0.031‡ |
| Sex | 0.056 | |||
| M | 357 (36) | 109 (43) | 124 (40) | |
| F | 647 (64) | 145 (57) | 184 (60) | |
| T3–4 primary tumour | 0.179 | |||
| No | 125 (13) | 42 (18) | 39 (13) | |
| Yes | 838 (87) | 198 (82) | 259 (87) | |
| Primary lymph node status | 0.014 | |||
| Negative | 338 (34) | 106 (43) | 124 (41) | |
| Positive | 646 (66) | 142 (57) | 179 (59) | |
| Primary tumour location | <0.001 | |||
| Right colon | 224 (22) | 93 (37) | 125 (41) | |
| Left colon | 422 (42) | 85 (34) | 72 (24) | |
| Rectum | 352 (35) | 73 (29) | 107 (35) | |
| Synchronous CRLM | 0.724 | |||
| No | 426 (49) | 111 (49) | 142 (51) | |
| Yes | 449 (51) | 115 (51) | 134 (49) | |
| CEA (ng/mL)* | 7.1 (3.1–23.0) | 7.6 (3.5–29.2) | 9.1 (3.6–25.8) | 0.117‡ |
| Tumour size (cm)* | 2.5 (1.6–4.0) | 2.4 (1.5–3.7) | 2.5 (1.5–3.9) | 0.425‡ |
| Chemotherapy before hepatic resection | 0.221 | |||
| No | 383 (38) | 111 (44) | 125 (41) | |
| Yes | 621 (62) | 142 (56) | 181 (59) | |
| Extrahepatic disease | 0.502 | |||
| No | 896 (89) | 221 (87) | 269 (87) | |
| Yes | 109 (11) | 33 (13) | 39 (13) | |
| No of tumours* | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 0.619‡ |
| Bilobar disease | 0.094 | |||
| No | 621 (62) | 174 (69) | 201 (66) | |
| Yes | 376 (38) | 77 (31) | 105 (34) | |
| Intraoperative concurrent ablation | 0.084 | |||
| No | 787 (88) | 184 (82) | 239 (86) | |
| Yes | 110 (12) | 40 (18) | 40 (14) | |
| R0 resection | 0.013 | |||
| No | 729 (74) | 201 (81) | 239 (80) | |
| Yes | 258 (26) | 46 (19) | 61 (20) | |
| Adjuvant chemotherapy | 0.001 | |||
| No | 491 (51) | 105 (46) | 110 (39) | |
| Yes | 473 (49) | 123 (54) | 173 (61) | |
| Recurrence-free survival | 0.006 | |||
| 1 year | 354 (36) | 76 (31) | 78 (26) | |
| 3 years | 642 (64) | 172 (69) | 222 (74) | |
| Site of recurrence | <0.001 | |||
| Intrahepatic | 301 (51) | 55 (39) | 66 (32) | |
| Extrahepatic | 170 (29) | 45 (32) | 72 (35) | |
| Both | 122 (21) | 42 (30) | 66 (32) |
Values in parentheses are percentages unless indicated otherwise; *values are median (i.q.r.). CRLM, colorectal liver metastasis; CEA, carcinoembryonic antigen. †χ2 or Fisher’s exact test, except ‡Mann Whitney U test.
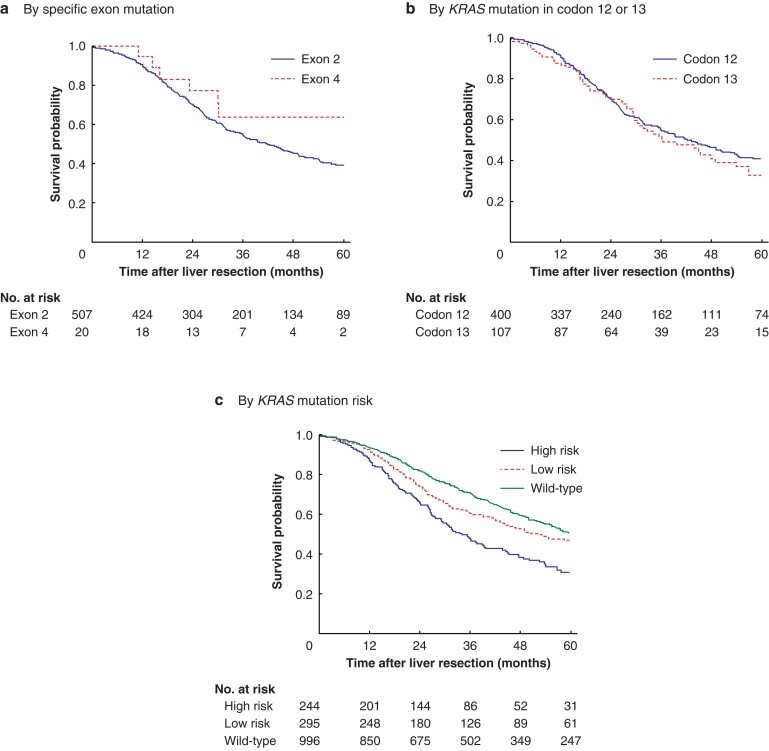
There was no difference in survival when mutations were analysed according to location in exons 2 versus 4 (P = 0.374) (Fig. 1a). Overall survival was also similar for patients with exon 2 codon 12 and 13 mutations (P = 0.282) (Fig. 1b), with median overall survival rates of 42.4 (34.9–49.9) and 36.5 (24.3–48.8) months respectively.
Fig. 1.
Overall survival curves for patients who underwent liver resection for colorectal liver metastases according to KRAS mutation type
Overall survival according to a specific KRAS exon mutations, b KRAS mutation in codon 12 or 13, and c KRAS mutation risk versus wild-type. a P = 0.374, b P = 0.282, c P < 0.001 (log rank test).
Overall survival by mutation-based subgroups based on the amino acid change is shown in Table S1. Median overall survival was 34.2 (28.7–39.8) months among the 244 patients with high-risk KRAS mutations, compared with 53.1 (41.8–64.3) months in the 295 patients with low-risk KRAS mutations, and 60.8 (55.1–66.6) months in the 996 patients with KRAS wild-type tumours (P < 0.001) (Fig. 1c). On multivariable analysis, KRAS risk groups were identified as independent prognostic factors (Table S2).
Of note, after recurrence, median survival was longer for patients with wild-type and low-risk KRAS mutations than for those with high-risk KRAS mutations (34.8, 26.5, and 22.0 months respectively (P < 0.001) (Fig. S1). High-risk patients were less often treated with curative intent (high risk: 78 (48.1 per cent); low risk: 130 (61.3 per cent); wild-type: 441 (70.1 per cent); P < 0.001). High-risk patients also underwent significantly fewer re-resections of metastases (high risk: 53 (35.1 per cent); low risk: 99 (47.1 per cent); wild-type: 327 (54.5 per cent); P < 0.001).
Discussion
Although there was no significant survival difference associated with exon 2 versus 4 mutations, the Kaplan–Meier curves did separate suggesting that patients with exon 4 mutations may have a distinct and less aggressive tumour biology. Saadat and co-workers5 similarly found that, although there was no significant survival difference associated with exon 2, 3, and 4 mutations, the Kaplan–Meier curves separated. In contrast to a previous report7, overall survival was also similar for patients with exon 2 codon 12 and 13 mutations. Of note, the previous study7 on codon-specific mutations in CRLM included only 67 and 24 patients with codon 12 and 13 mutations respectively, whereas the present study included 415 and 111 patients respectively.
The clinical heterogeneity among KRAS variants was ultimately confirmed at the point-mutation level. In this cohort, G12V, G13D, and G12D mutations were most common and represented 70 per cent of all KRAS mutations, which is consistent with the current literature. G12V was associated with the worst survival across all point mutation groups at 31.7 months, whereas patients with a G12D mutation had a median survival of 49.2 months. The association of G12V with poor survival is consistent with previous studies in patients with non-metastatic colorectal cancer7–9.
In contrast, the present findings for patients with G13D and G12S mutations are not consistent with existing studies. For example, another study7 reported poor survival of patients with G12S mutations, although their sample size was only 7. By comparison, the present study included 34 patients with G12S mutations, who had a median survival of 80.3 months. This was not only higher than that of any other point-mutation group, but even higher (albeit not significantly) than the median survival of patients with wild-type tumours. Although this is the first study to suggest that certain KRAS subgroups may serve as positive prognosticators, similar findings have been found for BRAF mutations10,11.
Patients with left-sided primary tumours were more likely to have high-risk mutations, whereas those with right-sided disease were more likely to have low-risk mutations. These differences may explain why others12 found that KRAS status is prognostic for patients with left-sided but not for right-sided primary tumours.
Clinically, the differences in overall survival rates between patients with high- versus low-risk KRAS mutations were partially driven by inferior postrecurrence survival of the former group. Indeed, patients with high-risk recurrences were less likely to receive treatment with curative intent. Interestingly, this cannot be attributed to different patterns of recurrence as the rates of intrahepatic, extrahepatic, and intrahepatic and extrahepatic recurrences were similar in the high- and low-risk KRAS groups. Instead, it can be speculated that patients with high-risk KRAS mutations experience recurrences with a higher tumour burden that are not amenable to potentially curative therapies. The limitations of the study are documented in the Supplementary material.
This study has demonstrated that the prognosis of patients with KRAS mutations is not as uniform as previously believed. Instead, patients can be stratified into those with low- and high-risk mutations, which carry a 28 and 72 per cent increase in risk of death respectively compared with that associated with wild-type tumours. Aside from the obvious refinement of prognostication, the findings from this study may inform future trials. Finally, the need for knowledge regarding the different effects of specific mutations will inevitably increase as new drugs targeting mutated KRAS are developed13.
Supplementary Material
Acknowledgements
P.B.O. and S.B. are joint first authors of this article. The authors thank N. Bampatsikou for technical support. As this is a multi-institutional study, the sharing of individual-patient data from each participating centre is subject to the policy and procedures of the institutions. This study was not preregistered with an analysis plan in an independent institutional registry.
Disclosure. Dr I.M.L. reported receiving 2 personal fees from Bayer, Roche, and Servier outside the submitted work. Dr I.B. reported receiving grants from Taiho Pharmaceutical Co Ltd, Ono Pharmaceutical Co Ltd, and MSD Pharmaceuticals; and receiving personal fees from Taiho Pharmaceutical Co Ltd, Ono Pharmaceutical Co Ltd, Takeda Pharmaceutical Co Ltd, Chugai Pharmaceutical Co Ltd, and Eli Lilly Japan outside the submitted work. Dr C.L.W. reported receiving personal fees from Healthcore outside the submitted work. The authors declare no other conflict of interest.
Contributor Information
Pim B Olthof, Department of Surgery, Erasmus University, Rotterdam, the Netherlands.
Stefan Buettner, Department of Surgery, Erasmus University, Rotterdam, the Netherlands.
Nikolaos Andreatos, Division of Medical Oncology, Mayo Clinic, Rochester, Minnesota, USA.
Jane Wang, Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Inger Marie Løes, Department of Clinical Science, University of Bergen, and Department of Oncology, Haukeland University Hospital, Bergen, Norway.
Doris Wagner, Department of General Surgery, Medical University of Graz, Graz, Austria.
Kazunari Sasaki, Department of General Surgery, Digestive Disease Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Andrea Macher-Beer, Department of Pathology, Medical University of Vienna, Vienna, Austria.
Carsten Kamphues, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany.
Ioannis Pozios, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany.
Hendrik Seeliger, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany.
Daisuke Morioka, Department of Gastroenterological Surgery, Yokohama City University Graduate School of Medicine, Yokohama, Japan.
Katsunori Imai, Department of Gastroenterological Surgery, Yokohama City University Graduate School of Medicine, Yokohama, Japan.
Klaus Kaczirek, Department of General Surgery, Medical University of Vienna, Vienna, Austria.
Timothy M Pawlik, Department of Surgery, Ohio State University Wexner Medical Center, Columbus, Ohio, USA.
George Poultsides, Department of Surgery, Stanford University School of Medicine, Stanford, California, USA.
Richard Burkhart, Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Itaru Endo, Department of Gastroenterological Surgery, Yokohama City University Graduate School of Medicine, Yokohama, Japan.
Hideo Baba, Department of Gastroenterological Surgery, Graduate School of Medical Sciences, Kumamoto University, Kumamoto, Japan.
Peter Kornprat, Department of General Surgery, Medical University of Graz, Graz, Austria.
Federico N Aucejo, Department of General Surgery, Digestive Disease Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Per Eystein Lønning, Department of Clinical Science, University of Bergen, and Department of Oncology, Haukeland University Hospital, Bergen, Norway.
Katharina Beyer, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany.
Matthew J Weiss, Department of Surgery, Zucker School of Medicine at Hofstra, Northwell Health Cancer Institute, Lake Success, New York, USA.
Christopher L Wolfgang, Department of Surgery, New York University School of Medicine, New York, New York, USA.
Martin E Kreis, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany.
Georgios A Margonis, Department of General and Visceral Surgery, Charité Campus Benjamin Franklin, Berlin, Germany; Department of Surgery, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
Funding
G.A.M is supported by the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748. The funding sources had no role in study design, implementation, analysis, or reporting.
Supplementary material
Supplementary material is available at BJS online.
References
- 1. Stangl R, Altendorf-Hofmann A, Charnley RM, Scheele J. Factors influencing the natural history of colorectal liver metastases. Lancet 1994;343:1405–1410 [DOI] [PubMed] [Google Scholar]
- 2. Vauthey JN, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, Andreou A et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013;258:619–626; discussion 626–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brudvik KW, Kopetz SE, Li L, Conrad C, Aloia TA, Vauthey JN. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg 2015;102:1175–1183 [DOI] [PubMed] [Google Scholar]
- 4. Frankel TL, Vakiani E, Nathan H, DeMatteo RP, Kingham TP, Allen PJ et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer 2017;123:568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saadat LV, Boerner T, Goldman DA, Gonen M, Frankel TL, Vakiani E et al. Association of RAS mutation location and oncologic outcomes after resection of colorectal liver metastases. Ann Surg Oncol 2021;28:817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res 2012;18:4753–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Margonis GA, Kim Y, Spolverato G, Ejaz A, Gupta R, Cosgrove D et al. Association between specific mutations in KRAS codon 12 and colorectal liver metastasis. JAMA Surg 2015;150:722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter ‘RASCAL’ study. J Natl Cancer Inst 1998;90:675–684 [DOI] [PubMed] [Google Scholar]
- 9. Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 2001;85:692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cremolini C, Di Bartolomeo M, Amatu A, Antoniotti C, Moretto R, Berenato R et al. BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis. Ann Oncol 2015;26:2092–2097 [DOI] [PubMed] [Google Scholar]
- 11. Margonis GA, Buettner S, Andreatos N, Kim Y, Wagner D, Sasaki K et al. Association of BRAF mutations with survival and recurrence in surgically treated patients with metastatic colorectal liver cancer. JAMA Surg 2018;153:e180996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasaki K, Margonis GA, Wilson A, Kim Y, Buettner S, Andreatos N et al. Prognostic implication of KRAS status after hepatectomy for colorectal liver metastases varies according to primary colorectal tumor location. Ann Surg Oncol 2016;23:3736–3743 [DOI] [PubMed] [Google Scholar]
- 13. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020;383:1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.