Abstract
Invasive Salmonella has been reported to induce apoptosis in a fraction of infected macrophages within 2 to 14 h from the time of infection by a mechanism involving the type III secretion machinery encoded by the Salmonella pathogenicity island 1 (SPI-1). Here, we show that bacteria in the transition from logarithmic to stationary phase cause 90% of the macrophages to undergo phagocytosis-independent, caspase-mediated apoptosis within 30 to 60 min of infection. The ability of Salmonella to induce this rapid apoptosis was growth phase regulated and cell type restricted, with epithelial cells being resistant. Apoptosis induction was also abrogated by disruption of the hilA gene (encoding a regulator of SPI-1 genes) and by the expression of a constitutively active PhoPQ. hilA itself and a subset of SPI-1 genes were transiently expressed during aerobic growth in liquid medium. Interestingly, however, hilA was found to be required only for the expression of the prgH gene, while sipB, invA, and invF were expressed in a hilA-independent manner. The expression of SPI-1 genes and the secretion of invasion-associated proteins correlated temporally with the induction of apoptosis and are likely to represent its molecular basis. Thus, growth phase transition regulates the expression and secretion of virulence determinants and represents the most efficient environmental cue for apoptosis induction reported to date.
Infection of mice with Salmonella typhimurium results in a systemic disease similar to typhoid. In humans, S. typhimurium is one of the most common causes of food poisoning. Otherwise-healthy individuals develop a gastroenteritis which might become chronic (the infected persons continue to shed bacteria for more than 1 year), but in immunocompromised patients S. typhimurium can cause a serious systemic disease. In the host’s intestine, Salmonella adheres to specialized epithelial cells (M cells [6, 17, 29]). The bacteria cause cytoskeletal and membrane rearrangements that result in their uptake by the host cells (10). By destroying the infected M cells (29), the bacteria gain access to the mesenteric lymph follicles, where they encounter and infect macrophages. Recently, the induction of macrophage cytotoxicity and/or apoptosis by invasive Salmonella has been documented in vitro (4, 21, 25) and in vivo (32). In two cases, components of the SPI-1-encoded type III secretion system have been shown to be necessary for efficient apoptosis induction (4, 25). The type III secretion system is a specialized, evolutionarily conserved protein secretion apparatus required for the export of virulence determinants which plays an important role in the invasion and destruction of the epithelial cells by Salmonella (12, 29). Several genes coding for putative components of type III secretion systems map to a contiguous region at centisome 63 of the Salmonella chromosome, the Salmonella pathogenicity island 1 (SPI-1). SPI-1 genes encode secreted proteins required for epithelial cell invasion, like SipA, -B, -C, and -D, SpaO, and SpaN, as well as proteins regulating the expression of invasion genes, like HilA (1, 2). Environmental signals, like oxygen concentration, osmolarity, and the growth state of the bacteria, influence the expression of HilA and the secretion of invasion-associated proteins (1). HilA expression can be regulated by two members of the phosphorylated response regulator superfamily, namely, SirA (the positive regulator [16]) and PhoP (the negative regulator [1]).
In this paper, we show that changes in growth phase have a profound effect on the ability of Salmonella to induce apoptosis in macrophages. The ability to induce apoptosis is restricted to a narrow, specific bacterial growth phase, namely, the transition from the exponential to the stationary phase. In this particular growth phase, Salmonella induces the most rapid and complete apoptosis ever reported (more than 90% of the macrophages are dead 30 min after infection). Transposon insertion mutagenesis and analysis of existing mutants confirm the pivotal importance of the invasion genes for this process. Furthermore, we find that the expression of several invasion genes is also growth phase regulated and correlates with apoptosis induction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Unless otherwise stated, the bacteria were generally grown overnight (16 to 20 h) at 37°C in 5 ml of Luria-Bertani (LB) broth with shaking at 150 rpm. For reinoculation, 50 μl of an overnight culture was added to 5 ml of fresh LB broth and incubated for the time periods indicated.
Transposon-induced mutagenesis.
Transposon mutagenesis was performed as described by Tsolis and Heffron (36). For screening of generated mutants for loss of cytotoxicity, single colonies were grown in 100 μl of LB broth in microtiter plates for 5 h after reinoculation from an overnight culture. One microliter of the reinoculated culture was used to infect macrophages as described below.
Characterization of the transposon insertion sites.
Isolated S. typhimurium mutants unable to kill macrophages were further analyzed by PCR. Genomic DNA was isolated from the mutants and digested with EcoRV, which cleaves once in Tn10d(Tc). EcoRV-digested DNA was ligated at a low concentration to generate circular DNA. The genomic DNA flanking the transposon on either side was amplified by using two different sets of primers homologous to the Tn10d(Tc) transposon. The amplified fragments were sequenced with the AmpliCycle sequencing kit from Perkin-Elmer.
Analysis of gene expression by RT-PCR.
RNA was isolated from S. typhimurium strains at different growth stages. One microgram of RNA was used in the reverse transcription (RT) with Superscript at 42°C for 60 min in the presence of 1 pmol of downstream primer. One-twentieth of the RT mixture was used in the PCR. Different amounts of chromosomal DNA were used to ensure that the amplification was in a linear range.
Analysis of the profile of secreted proteins.
The bacterial cultures (3 ml) were chilled on ice for 30 min before the bacteria were pelleted. The supernatants were filtered through 0.45-μm-pore-size filters prior to precipitation with trichloracetic acid. The precipitate was dissolved in sample buffer and run on a sodium dodecyl sulfate–10% polyacrylamide gel. Proteins were visualized with Coomassie brilliant blue R-250.
Cell culture and infection.
BAC-1.2F5 cells (26) and bone marrow-derived macrophages from C57BL/6J mice were cultured in minimum essential medium (MEM)-α supplemented with 10% fetal calf serum (FCS) and 20% L-cell conditioned medium as a source of colony-stimulating factor 1 (35). J774A.1 cells (30), MDCK cells (31), and Caco-2 cells (9) were grown in Dulbecco’s modified minimal essential medium with 10% FCS, and HeLa cells (13) were grown in RPMI containing 10% FCS. The cells (50,000/well) were seeded into 96-well plates 16 h prior to infection with bacteria diluted in cell culture medium without antibiotics (with a constant multiplicity of infection [MOI] of 25 in all experiments). The plates were centrifuged at 300 × g for 5 min to synchronize the infection and incubated for 30 min at 37°C (5% CO2).
In selected experiments, a peptide inhibitor of caspases (general inhibitor) (Z-VAD-CH2F; Enzyme System Products, Dublin, Calif.) was added to the cells 45 min prior to infection with Salmonella.
Assessment of host cell viability and detection of Salmonella-induced apoptosis.
Host cell viability was determined either by trypan blue dye exclusion or by crystal violet staining of viable cells. For crystal violet staining, the cells were washed three times with phosphate-buffered saline prior to the addition of 100 μl of crystal violet solution (0.2% crystal violet in 20% methanol). The cells were then incubated for 15 min at 37°C and washed extensively with water to remove excess dye. The absorbance was measured at 620 nm. The absorbance of uninfected cells was set to 100%. The absorbance of cells infected with bacteria deficient in apoptosis induction or treated with inhibitors of apoptosis prior to infection exceeds the 100% value due to the staining of the infecting bacteria.
RESULTS AND DISCUSSION
Growth phase-dependent induction of macrophage apoptosis by Salmonella.
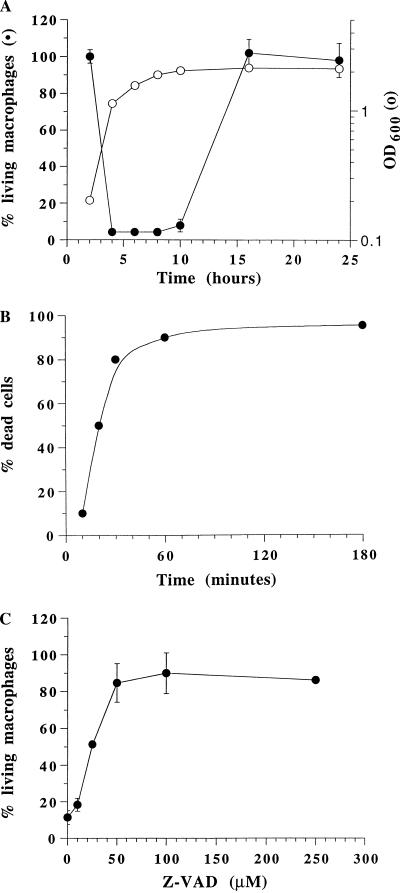
Infection of BAC-1.2F5 cells with S. typhimurium grown under different conditions resulted in the death of the macrophages (data not shown). To determine whether the ability of some of the cultures to induce macrophage cytotoxicity correlated with a specific growth phase, the bacteria were grown at 37°C with agitation. Aliquots were taken at different time points and used to infect BAC-1.2F5 cells in 96-well plates with a constant MOI of 25. The survival of the macrophages was assessed 30 min after infection by crystal violet staining. Interestingly, S. typhimurium acquires and retains the capability of inducing macrophage death only in a narrow growth phase, namely, in the transition between logarithmic and stationary growth phases (Fig. 1A). A culture in this specific growth phase will be referred to from now on as activated. Marked cellular damage was already morphologically evident 10 min after infection (data not shown). Previous reports of Salmonella-induced macrophage apoptosis (4, 21, 25) described cellular damage and DNA fragmentation in a fraction of the infected cells (20 to 60%) 2 to 14 h after incubation with invasive S. typhimurium. To determine the time course and the extent of the cytotoxic effect in our system, BAC-1.2F5 cells were infected with an activated S. typhimurium culture and trypan blue was added to the cells at different time points after infection. Cell death proceeded extremely rapidly, and 80 to 90% of the macrophages were stained with trypan blue 30 min after infection (Fig. 1B).
FIG. 1.
Effect of bacterial growth phase on Salmonella-induced macrophage apoptosis. BAC-1.2F5 cells were seeded into 96-well plates (50,000 cells/well) and either left uninfected or infected with S. typhimurium LT2 (34) in different growth phases. Macrophages were infected with an activated culture with a constant MOI of 25. The bacteria were spun onto the macrophages to synchronize infection. (A) Thirty minutes after infection, the number of viable cells was determined by crystal violet staining. The percentage of viable cells (uninfected controls = 100%) and the optical density at 600 nm (OD600) of the bacterial culture used to infect the cells are plotted. The assays were carried out in triplicate. The vertical bars represent the standard deviations of the mean. (B) Time course of macrophage death induced by infection with an activated culture. The percentage of dead cells was determined by trypan blue dye exclusion. (C) Salmonella-induced apoptosis is caspase dependent. Macrophages were infected with Salmonella after preincubation (45 min) with different concentrations of a caspase inhibitor (Z-VAD-CH2F). Thirty minutes after infection, the number of viable cells was determined by crystal violet staining.
The cytotoxic effect of S. typhimurium on BAC-1.2F5 cells is the fastest reported to date. To determine its nature, we preincubated the macrophages with different concentrations of a cell permeant general caspase inhibitor (Z-VAD-CH2F). Caspases are a family of cysteine proteases which play a central role in apoptosis but not in necrosis (20, 27). The inhibitor blocked Salmonella-induced cell death by 85% at the concentration of 50 μM (Fig. 1C). Protection of the macrophages from the cytotoxic effect of S. typhimurium by the caspase inhibitor unequivocally establishes that cell death in our system occurs via apoptosis. Yersinia enterocolitica (33)- and Shigella flexneri (5)-induced macrophage apoptosis can also be blocked by caspase inhibitors, indicating that bacterial components must be able to cross talk with the mechanisms of activation of this group of enzymes.
Macrophages undergoing Salmonella-induced apoptosis showed the characteristic morphological features of this process (nuclear shrinkage and chromatin condensation in the typical crescent shape [data not shown]); in addition, their DNA was fragmented and displayed the nucleosomal ladder that correlates with apoptosis (data not shown).
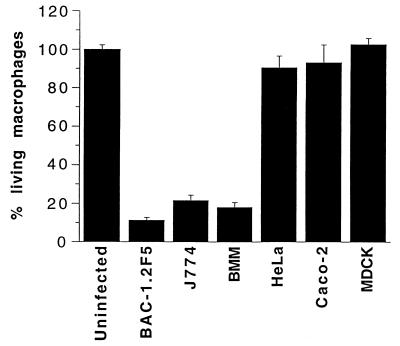
Salmonella-induced apoptosis was not restricted to BAC-1.2F5 cells. A second mouse macrophage-like cell line, J774, and primary bone marrow-derived macrophages were also efficiently killed, with the same fast kinetics. In contrast, three epithelial cell lines, HeLa (human), Caco-2 (human), and MDCK (canine), did not undergo apoptosis upon infection with an activated S. typhimurium culture (Fig. 2). Epithelial cells are resistant to Salmonella-, Shigella (22)- and Yersinia (24)-induced apoptosis (Fig. 2), although they are able to undergo caspase-mediated apoptosis when treated with anti-Fas antibodies or tumor necrosis factor alpha (7). It is possible that epithelial cells, in contrast to macrophages, might lack a component of the signal transduction pathway targeted by the putative Salmonella factor(s) triggering apoptosis (e.g., the surface receptors). Alternatively, they might contain an inhibitor(s) of this specific apoptotic pathway. Several Salmonella strains (Salmonella dublin, Salmonella enteritidis, and Salmonella bongori) were tested to determine their ability to induce apoptosis. All tested strains were cytotoxic for BAC-1.2F5 cells when grown to the appropriate growth phase (data not shown). Cytotoxicity was independent of the presence of the virulence plasmid in two different strains: ST12/75, compared with its plasmid-cured derivative, STM8c (14); and S. dublin SD2229, compared with its plasmid-cured derivative, SDM173c (reference 14 and data not shown). In addition, the two strains of S. bongori from the Salmonella Reference Collection C (3) efficiently induced macrophage apoptosis (data not shown), despite their lack of at least part of the recently described SPI-2 (28).
FIG. 2.
Salmonella induces apoptosis in macrophages but not in epithelial cells. Macrophages (BAC-1.2F5 and J774 cells and bone marrow-derived macrophages [BMM]) or epithelial cells (HeLa, MDCK, or Caco-2 cells) were seeded into 96-well plates (50,000/well) and infected with activated Salmonella cultures in the appropriate growth phase (MOI of 25). Thirty minutes after infection, the number of viable cells was determined by crystal violet staining. The results are expressed as percentage of viable cells (uninfected controls = 100%). The assays were carried out in quadruplicate. The vertical bars represent the standard deviations of the mean.
Mutations affecting Salmonella-induced macrophage apoptosis.
Recent reports have indicated the importance of the ompR-envZ regulon (21) and of the components of the type III secretion system (4, 25) in Salmonella-induced macrophage cytotoxicity. In our system, the ompR mutant (CJD359 [8]) was still cytotoxic, whereas a mutation in the invA gene (SB111 [11]) abrogated the effect (data not shown), indicating that a functional type III secretion apparatus was necessary for macrophage killing.
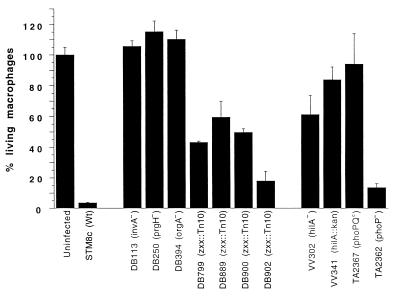
In support of this, 6 of 10 apoptosis-deficient mutants identified by Tn10d(Tc) insertion mutagenesis mapped to three SPI-1 genes (invA, prgH, and orgA). The remaining four mutants did not map to the sequenced portions of SPI-1 or SPI-2 (data not shown) and displayed some residual cytotoxic activity (Fig. 3). These mutants are currently being characterized in more detail.
FIG. 3.
Effects of different Salmonella mutant strains on macrophage viability. BAC-1.2F5 cells were seeded into 96-well plates (50,000/well) and either left uninfected or infected with different mutant strains of Salmonella in the appropriate growth phase. Infection was performed as described in the legend to Fig. 1A. Thirty minutes after infection, the number of viable cells was determined by crystal violet staining. The results are expressed as percentage of viable cells (uninfected controls = 100%). The assays were carried out in quadruplicate. The vertical bars represent the standard deviations of the mean. Wt, wild type.
The results described above predicted that loss of function of the invasion gene regulator HilA would abrogate apoptosis induction (Fig. 3). Two independent hilA mutants were tested and found to induce apoptosis less efficiently than the wild-type strain. The phoPQ(Con) mutation (strain TA2367 [18]), which has been shown to inhibit the expression of HilA and of HilA-regulated genes (17), was also severely impaired in apoptosis induction; by contrast, phoP mutant bacteria (strain TA2362 [18]) behaved like the wild-type strain (Fig. 3). The experiments shown in Fig. 3 reaffirm the importance of the gene products encoded by the SPI-1 and of their regulators in Salmonella-induced macrophage apoptosis. It should be noted, however, that the effects of both the hilA and the phoPQ(Con) mutations on apoptosis induction were less severe than those of the prgH, invA, and orgA mutations.
Expression of a subset of SPI-1 genes and secretion of invasion-associated proteins are growth phase regulated.
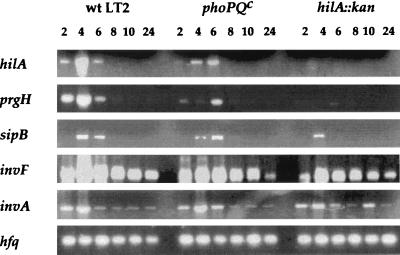
The experiments summarized above prompted us to determine whether the expression pattern of the genes implicated in the induction of apoptosis was influenced by the growth phase. Gene expression was monitored by RT-PCR in RNA samples isolated at different time points of the growth curve. A subset of genes related to the SPI-1-encoded type III secretion system (hilA, prgH, and sipB) were shown to be tightly growth regulated, with maximum expression coinciding with the apoptosis-inducing phenotype. The expression of prgH and sipB followed that of hilA, confirming its role as a master regulator of genes needed for invasion (17). In contrast, the invA and invF transcript could be detected throughout the growth curve. The expression of hfq was constant throughout the growth curve and is shown as a control (Fig. 4, wild type).
FIG. 4.
Analysis of invasion gene expression in wild-type (wt), phoPQ(Con) (phoPQc) and hilA::kan Salmonella strains. Wild-type, phoPQ(Con), and hilA-deficient Salmonella strains were grown at 37°C under continuous agitation. At the time points indicated, RNA was extracted and subjected to RT-PCR analysis with the appropriate primers (see Materials and Methods).
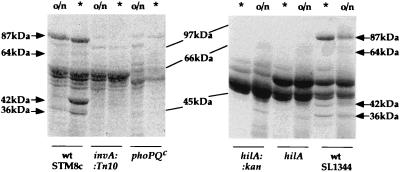
Analysis of the profiles of secreted, invasion-associated proteins revealed at least four protein bands that were absent in the invA and hilA mutants but could be routinely identified in the supernatants of the wild-type cultures. In good agreement with the growth phase-regulated expression of a subset of invasion genes, these proteins (87, 64, 42, and 36 kDa, respectively) were much more abundant in the supernatant of the activated culture than in those of the stationary-phase culture (Fig. 5). Thus, both secretion of invasion-associated proteins and expression of a subset of invasion genes are growth phase regulated.
FIG. 5.
Secreted-protein profiles of wild-type and mutant Salmonella strains. Trichloroacetic acid-precipitated proteins from supernatants (3 ml) of activated (∗) or overnight (o/n) cultures were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining for each of the strains indicated. The molecular masses of invasion-associated proteins are indicated next to the arrows. The hilA mutations were in SL1344 background (15). wt, wild type.
The sharp peak of hilA expression could result either from a positive regulation of the transcript during its induction or from its repression at the other time points or from a combination of both. The mRNAs of the hilA regulators sirA and phoPQ are present throughout the growth curve (data not shown). Therefore, if one of these genes is responsible for the behavior of the hilA transcript, they must be regulated posttranscriptionally or, assuming that the proteins behave like the respective transcripts, posttranslationally. PhoP, PhoQ, and SirA are members of two-component systems and are therefore expected to be subjected to posttranslational regulation (16, 23). Contrary to our expectations, the phoPQ(Con) mutation that suppresses hilA induction by low oxygen, low pH, and high osmolarity (17) did not block growth phase-regulated hilA expression. A reduction of hilA expression could, however, be observed, and peak induction occurred 6 instead of 4 h after reinoculation of this mutant (Fig. 4); consistent with a role of hilA in prgH expression (1), the amount of this transcript was reduced in the phoPQ(Con) mutant, and its kinetics of accumulation paralleled those observed for hilA. The causal link between prgH and hilA was further confirmed by the finding that prgH expression was abrogated in an hilA-deficient strain (Fig. 4). The PrgH protein is a component of the needle complex, essential for the transfer of secreted proteins into eukaryotic cells (19). The decrease [in the case of the phoPQ(Con) strain] or the absence (in the case of the hilA-deficient strain) of prgH expression therefore likely accounts for the severe reduction in apoptosis induction (Fig. 3) and in the secretion of invasion-associated proteins observed in these two mutants (Fig. 5). In agreement with this hypothesis, but surprising in view of the dominant role ascribed to HilA in previous publications, the expression of other invasion genes was only marginally (sipB, coding for a secreted protein) or not at all (invA and invF) affected by either the phoPQ(Con) or the hilA mutation (Fig. 4). Thus, our data confirm that the phoPQ(Con) mutation can negatively influence Salmonella virulence by repressing hilA expression (1). However, in contrast to previous reports (1, 2), under our culture conditions an absolute requirement for HilA exists in the case of the prgH gene but not in the cases of the sipB, invA, or invF genes.
Whether this discrepancy is due to differences in the assay used (expression of invasion gene-lacZY fusions was used in the previous studies, while we assay mRNA level directly) or to the conditions used to induce expression (low oxygen, low pH, and high osmolarity were the stimuli used previously) remains to be determined.
Salmonella-induced macrophage cytotoxicity, with some features of apoptosis, has been reported by several groups (4, 21, 25). In all cases, induction of the cytotoxicity depended on the use of specific bacterial culture conditions. Lindgren et al. (21) and Monack et al. (25) grew the bacteria under reduced aeration, while Chen et al. (4) used hyperosmolarity (0.3 M NaCl) to stimulate apoptosis induction by Salmonella.
We show here that the transition from logarithmic to stationary phase alone, which all bacteria are likely to undergo, represents the most potent stimulus for apoptosis induction reported to date (Fig. 1).
Shigella (5), Yersinia (33), and Salmonella (Fig. 1C) require the caspase cascade to effect macrophage apoptosis. The unusual rapidity of the apoptotic processes indicates that Salmonella, rather than suppressing the activity of molecules that normally prevent apoptosis, triggers the cell death pathway of the macrophages directly, possibly by interacting with the caspase cascade. The task will now be to identify the Salmonella virulence determinants which interact with the caspase cascade and to unravel their mechanism of activation.
ACKNOWLEDGMENTS
Ursula Vinatzer and Daniela Berdnik made equal contributions to this work.
The skillful technical assistance of Julia Katzenbeisser is gratefully acknowledged. We are indebted to Thomas Decker, Andreas Meinke, and Pavel Kovarik, Vienna Biocenter, for many helpful discussions and for critically reading the manuscript. We also thank Alistair Lax (Institute for Animal Health, Compton, Nr Newbury, United Kingdom), Catherine Lee (Department of Microbiology and Molecular Genetics, Harvard Medical School, Boston, Mass.), Kenneth Sanderson (Department of Biological Sciences, University of Calgary), and Mikael Rhen (Center for Tumor Biology, Karolinska Institute) for their generous gifts of strains.
This work was supported by grant P10766-MED of the Austrian Research Fund (to M.B.) and by grant 70008/2-Pr/4/95 of the Austrian Federal Ministry of Science, Transport and the Arts (to A.V.G.).
REFERENCES
- 1.Bajaj V, Lucas R L, Hwang C, Lee C A. Coordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Boyd E F, Wang F-S, Whittam T S, Selander R K. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 6.Clark M A, Jepson M A, Simmons N L, Hirst B H. Preferential interaction of Salmonella typhimurium with mouse Peyer’s patch M cells. Res Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Shenk T. Protection against apoptosis by the vaccinia virus SPI-2 (B13R) gene product. J Virol. 1996;70:6479–6485. doi: 10.1128/jvi.70.9.6479-6485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman C J, Chatfield S, Higgins C F, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989;57:2136–2140. doi: 10.1128/iai.57.7.2136-2140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogh J, Fogh J M, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59:221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 10.Francis C L, Starnbach M N, Falkow S. Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol Microbiol. 1992;6:3077–3087. doi: 10.1111/j.1365-2958.1992.tb01765.x. [DOI] [PubMed] [Google Scholar]
- 11.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galán J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 13.Gey G O, Coffman W D, Kubicek M T. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–265. [Google Scholar]
- 14.Guilloteau L A, Wallis T S, Gautier A V, MacIntyre S, Platt D J, Lax A J. The Salmonella virulence plasmid enhances Salmonella-induced lysis of macrophages and influences inflammatory responses. Infect Immun. 1996;64:3385–3393. doi: 10.1128/iai.64.8.3385-3393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 16.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 17.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:7–9. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kier L D, Weppelman R M, Ames B N. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol. 1979;138:155–161. doi: 10.1128/jb.138.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 20.Lemaire C, Andreau K, Souvannavong V, Adam A. Inhibition of caspase activity induces a switch from apoptosis to necrosis. FEBS Lett. 1998;425:266–270. doi: 10.1016/s0014-5793(98)00252-x. [DOI] [PubMed] [Google Scholar]
- 21.Lindgren S W, Stojiljkovic I, Heffron F. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:4197–4201. doi: 10.1073/pnas.93.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantis N, Prévost M-C, Sansonetti P. Analysis of epithelial cell stress response during infection by Shigella flexneri. Infect Immun. 1996;64:2474–2482. doi: 10.1128/iai.64.7.2474-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller S I. PhoP/PhoQ: macrophage-specific modulators of Salmonella virulence? Mol Microbiol. 1991;5:2073–2078. doi: 10.1111/j.1365-2958.1991.tb02135.x. [DOI] [PubMed] [Google Scholar]
- 24.Mills S D, Boland A, Sory M-P, van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan C, Pollard J W, Stanley E R. Isolation and characterization of a cloned growth factor dependent macrophage cell line, BAC1.2F5. J Cell Physiol. 1987;130:420–427. doi: 10.1002/jcp.1041300316. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 28.Ochman H, Groisman E A. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Noninvasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer’s patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 30.Ralph P, Nakoinz I. Phagocytosis and cytolysis by a macrophage tumour and its cloned cell line. Nature. 1975;257:393–394. doi: 10.1038/257393a0. [DOI] [PubMed] [Google Scholar]
- 31.Richardson J C, Scalera V, Simmons N L. Identification of two strains of MDCK cells which resemble separate nephron tubule segments. Biochim Biophys Acta. 1981;673:26–36. [PubMed] [Google Scholar]
- 32.Richter-Dahlfors A, Buchan A M J, Finlay B B. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruckdeschel K, Roggenkamp A, Lafont V, Mangeat P, Heesemann J, Rouot B. Interaction of Yersinia enterocolitica with macrophages leads to macrophage cell death through apoptosis. Infect Immun. 1997;65:4813–4821. doi: 10.1128/iai.65.11.4813-4821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanderson K E, Roth J R. Linkage map of Salmonella typhimurium, edition VI. Microbiol Rev. 1983;47:410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley E R. The macrophage colony-stimulating factor, CSF-1. Methods Enzymol. 1985;116:564–587. doi: 10.1016/s0076-6879(85)16044-1. [DOI] [PubMed] [Google Scholar]
- 36.Tsolis R M, Heffron F. Mutagenesis and variant selection in Salmonella. Methods Cell Biol. 1994;45:79–106. doi: 10.1016/s0091-679x(08)61847-6. [DOI] [PubMed] [Google Scholar]