Abstract
Renal cell carcinoma (RCC) with venous tumor thrombus (VTT) arising from the primary tumor occurs in approximately 10% of cases and is thought to represent more advanced disease. The intravascular nature of VTT suggests that it may serve as a source for hematogenous metastases. RCC with VTT and distant metastasis provides unique opportunities to examine the origins and emergence timing of these distinct tumor lesions, and to identify molecular correlates with disease state. We performed multi-region exome and RNA-sequencing analysis of 16 patients with RCC with VTT, with eight patients also having sequenced metastasis, to identify genomic alterations, biological pathways, and evolutionary processes contributing to VTT and metastasis, and to ask whether metastasis arises directly from or independent of VTT. No specific genomic alterations were associated with VTT. Hallmark copy-number alterations (deletions of 14q, 8p, and 4q) were associated with metastasis and disease recurrence, and secondary driver alterations tended to accumulate in metastatic lineages. Mismatch repair mutational signatures co-occurred across most tumors, suggesting a role for intracellular DNA damage in RCC. Robust phylogenetic timing analysis indicated that metastasis typically emerged before VTT, rather than deriving from it, with the earliest metastases predicted to emerge years before diagnosis. As a result, VTT in metastatic cases frequently derived from a metastatic lineage. Relative to the primary tumor, VTT upregulated immediate-early genes and transcriptional targets of the TNFα/NF-κB pathway, whereas metastases upregulated MTOR and transcriptional targets downstream of mTORC1 activation.
Implications:
These results suggest that VTT and metastasis formation occur independently, VTT presence alone does not necessarily imply more advanced disease with inevitably poor prognosis.
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney cancer and is one of the most prevalent neoplasms worldwide (1). RCC exhibits local vascular invasion in up to 10% of cases, and the resulting venous tumor thrombus (VTT), which invades the renal vein and can eventually progress up the inferior vena cava (IVC) to the heart, impacts treatment options and may present challenges for surgical intervention (2, 3). Recent work has made strides toward genomic characterization of clear cell RCC (ccRCC) with VTT and has sought to identify genomic features distinguishing VTT tumors from non-VTT tumors (4, 5). For example, ccRCC tumors with VTT exhibit modestly greater tumor mutational burden, possible higher genomic instability, and higher tumor proliferative indices than tumors without VTT (4, 6).
Several studies have used multi-region sequencing (MR-seq) to explore patterns of tumor evolution in RCC for cases both with and without VTT (5–9). RCC tumors exhibit branched evolution leading to high intratumoral heterogeneity and substantial subclonal diversity. MR-seq has also shed some light on the evolutionary patterns that give rise to VTT. Warsow and colleagues (5) demonstrated that although multiple VTT regions typically derive from a single common ancestor, they likely originate from subclones common to primary tumor regions. Turajlic and colleagues (6) found similar patterns, with VTT seemingly derived directly from the most recent common ancestor of all tumor clones in the greatest proportion of assayed cases. However, the relationship between VTT and metastases remains unclear. Many patients with VTT present with metastasis (20%–50%), but VTT formation and metastatic spread may not necessarily be linked (10, 11). It is possible that either metastases could be derived directly from the VTT lineage via hematogenous spread, or that metastases are derived from separate lineages that may originate earlier or later than the VTT. As VTT physically emerges from the primary tumor, its genomic composition provides a point of comparison for the relative timing and phylogenetic origin of distant metastasis from the primary tumor.
The timing of metastatic spread is an important issue for clinical care (12) and has been explored in other cancers (13, 14). Many patients with RCC present with metastases at the time of diagnosis (20%–30%); however, in some patients metastases only become evident over time (15). Knowledge of the relevant timelines is critical to inform surveillance regimens and treatment decisions. Therefore, quantitative approaches for estimating the timing of metastasis in RCC from genomic evidence are increasingly critical for improving prognosis, treatment algorithms, and ultimately clinical outcomes (16). Similarly, understanding which biological pathways contribute to the formation of new lesions and their spread from the primary tumor is important to inform therapeutic development and treatment selection. To these ends, we performed MR-seq analysis of RCC with VTT and metastases—including both mutation and gene expression characterization—to estimate the timing of their clonal emergence and to identify biological pathways associated with each type of lesion. We further hypothesized that the resulting phylogenomic patterns might shed light on the clinical pictures of these patients and inform RCC treatment more broadly.
Materials and Methods
Clinical sample identification and pathology analysis
Cases clinically diagnosed as RCC with VTT that contained banked tissue for research were identified. All patient identifiers were reassigned to protect anonymity. All participants or caregivers provided written informed consent, and the study was conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule). Institutional Review Board approval was obtained from the University of California San Francisco (Protocol Numbers: 10-02288 and 11-06240). A surgical pathologist reviewed all slides associated with each case, established that primary tumor, metastatic tumor, and thrombus regions were available based on the gross description and confirmed diagnoses based on morphologic and immunohistochemical findings. Viable tumor content (percentage of viable tumor/total epithelial surface area), stromal abundance (low, moderate, or high) and presence of cystic change were estimated for each case by an independent pathologist. Retrospective chart review was performed to identify and capture relevant clinical and demographic information. Clinical case history information and sample details are provided in Supplementary Tables S1–S3 and Supplementary Fig. S1.
Generation and processing of sequencing data
DNA and RNA were jointly extracted from FFPE tissue using the AllPrep DNA/RNA FFPE Kit (Qiagen), following deparaffinization using xylene. In this protocol, solubilized RNA was heated to partially reverse formalin crosslinks, then purified using RNeasy MinElute spin column with on-column DNase I digestion. Insufficient DNA for Exome-seq was recovered from a vertebral metastasis from PT4, and RNA was unrecoverable from the VTT sample from PT7 due to a broken slide—these samples were excluded from further analysis.
Exome sequencing libraries were created with Agilent SureSelectXT. SureSelect All Exon V6 bait probes were used to perform targeted capture. Exome sequencing yielded an average of 90 million 100bp paired-end reads, and an average depth (before removing duplicate reads) of 150X per sample. RNA-seq libraries were generated using the RNA Access platform. RNA-seq yielded an average of 60 million 50 bp paired-end reads per sample. Library preparation and sequencing was performed at the Broad Institute for PT1, PT2, PT3, and PT4, and at Q2 Solutions for the remainder. Sequencing data are available from EGA under accession number EGAS00001005897.
Exome and RNA-seq FASTQ files were assessed for quality using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and resulting statistics can be found in Supplementary Table S4. The FASTQ files were then trimmed to remove adapter contamination and low-quality trailing sequences, and then mapped to the GRCh38 reference genome with GSNAP v2013–11–01 (17). Alignment statistics are reported for WES and RNA-seq in Supplementary Table S5 and Supplementary Table S6, respectively.
Indels were realigned with GATK IndelRealigner v3.5 (18). Mutations were then called using both Lofreq2 v2.1.2 (19) and Strelka v1.0.14 (20), with the union of both outputs retained. As the libraries were not treated with UDG, further filtering was performed to reduce the contribution of spurious mutations identified due to formalin fixation. Mutations were required to have a minimum VAF of 0.05 and coverage of 20 in at least one sample in which they were called, maximum VAF of 0.01 in the normal sample, and a maximum ExAC (21) global frequency of 0.01. Somatic copy-number alterations (SCNA) and tumor purity estimates were determined using TitanCNA v1.7.1 (22). RNA-seq FASTQ files were trimmed to remove adapter contamination and low-quality trailing sequences, and then mapped to GRCh38 using GSNAP v2013-11-01.
Driver alterations and evolutionary analysis
Driver alterations were identified as putative loss-of-function mutations in previously reported ccRCC and pRCC driver genes, including VHL, PBRM1, SETD2, PIK3CA, MTOR, PTEN, KDM5C, CSMD3, BAP1, TP53, TSC1, TSC2, ARID1A, TCEB1, MET, NF2, KDM6A, SMARCB1, FAT1, STAG2, NFE2L2, and CDK2NA, as well as previously established arm-level driver SCNAs: chr 3p loss, 5q gain, 4q loss, 8p loss, and 14q loss (7, 23–25). Loss-of-function mutations were defined as indels, nonsense SNVs, splice site dinucleotide variants, and putatively deleterious missense SNVs according to SIFT (26), PolyPhen (27), or CONDEL (28). For samples with apparently missing driver alterations, raw alt/ref base counts were examined to confirm absence, which led to several apparently missing mutations being recovered.
Tumor phylogenetic trees were constructed from binarized (present/absent) mutations across samples using maximum parsimony (29). Trees were reconstructed 100 additional times on nonparametric bootstrap samples of the input mutation sets (30). We used the phangorn (31) and APE R packages (32) for tree construction and bootstrapping, respectively.
Clonality analysis was performed with PyClone (33) using the quality-filtered somatic mutation calls (restricted to autosomes), SCNA, and tumor purity estimates in multi-sample mode with all samples per tumor, and separately in single-sample mode with each tumor sample. Cancer cell fractions from the multi-sample and single-sample results were compared and overlaid with the phylogenies to identify clones associated with the trunk, clonal expansions between trunk and specific lesions, and parallel evolution.
Lesion emergence ordering and timing analysis
Bootstrapped phylogenetic trees generated as described above were leveraged to assess the relative order of tumor lesion emergence. The sums of the internal branch lengths from the root to the parent node associated with a tumor lesion were compared in each bootstrap replicate. To estimate absolute timing of lineages associated with specific tumor lesions, we adapted the linear mixed modeling approach of Mitchell and colleagues (16), using the lme4 package (ref. 34; see Supplementary Methods for further details). In brief, patient age was regressed against sample mutation burden. The resulting model was used along with estimated mutation burden at the parent nodes of VTT and metastatic lesions to generate predictions and associated confidence intervals for the age at which those branching events occurred.
Mutational signature analysis
The sample frequencies of SNV trinucleotide motifs were extracted using the SomaticSignatures R package (35). Previously identified signatures (36, 37) were then fit to the resulting motif spectra using non-negative least squares with the MutationalPatterns R package, using the fit_to_signatures_bootstrapped function with method set to “strict” (38). Signatures with a non-zero median contribution over 100 bootstrap replicates were retained as robustly fit.
Differential gene expression analysis
RNA-seq reads within gene coding regions were quantified using HTseqGenie (39), and differential expression was assessed using DESeq2 (40). Genes were considered differentially expressed if the comparison had Padjusted ≤ 0.05, and the sign of the log-fold change was consistent among individual regions in more than half of the patients. The relevant pairwise comparisons were performed while controlling for patient-specific expression as recommended by the DESeq2 authors. Gene set overlap analysis of differentially expressed genes was performed using Enrichr (41), which performs a hypergeometric test, and gene set enrichment analysis (GSEA) was performed using the R package fGSEA (42). For the latter, all genes included in the differential expression testing were ranked by the Wald statistic derived from the comparison.
Histological analysis
PanCK and CD8 duplex chromogenic IHC were performed as previously described (29). Semiquantitative histologic analysis of the 78 tumor regions revealed variable stromal abundance within tumors that was relatively consistent across tumor regions in any one patient. Very little intrastromal inflammatory response or desmoplasia was detected (Supplementary Fig. S2).
Results
Integrative evolutionary analysis reveals complex evolutionary modes, with secondary driver accumulation in metastases but no genomic alterations associated with VTT
We investigated a cohort of 16 patients with RCC, 14 with ccRCC, and two with pRCC (Supplementary Fig. S1, Supplementary Table S1). A total of 78 distinct tumor regions were whole-exome sequenced, including multiple primary regions and at least one VTT region from each patient (Fig. 1A), and one metastatic region from eight patients (Supplementary Table S2). Somatic point mutation analysis identified a range of unique mutations/Mb per patient (3.6–25.2), and per sample (1.8 ± 0.24 to 8 ± 2.4, Supplementary Table S7). Tumors in this cohort exhibited a wide range of truncality (the percentage of mutations in any given sample that were detected in all samples from the same tumor (6.5%–81%, Supplementary Table S7). The ccRCC cases exhibited a median of 48% truncal mutations, somewhat higher than in previous reports (8).
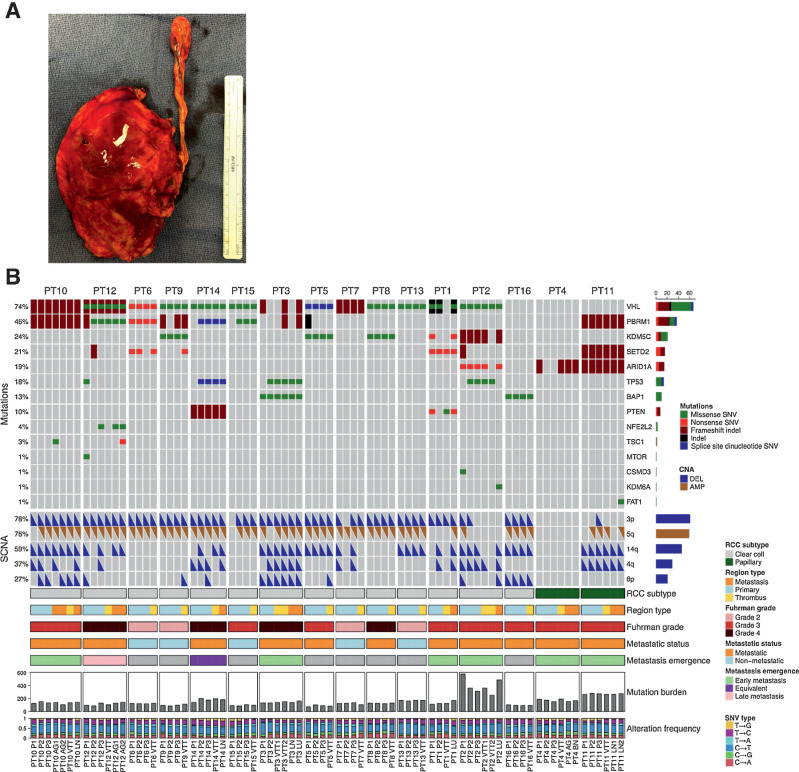
Figure 1.
A, Patient kidney and VTT post-nephrectomy with IVC thrombectomy. B, Genomic alteration landscape across tumor regions in 16 patients with RCC with VTT. An oncoprint showing the most prevalent somatic mutations and SCNAs found in this cohort. The prevalence of each alteration is indicated at left. The following are the tumor lesion labels: P, primary tumor; VTT, venous tumor thrombus; AG, adrenal gland metastasis; LN, lymph node metastasis; LU, lung metastasis; BN, brain metastasis.
We performed integrative evolutionary analysis of the 16 RCC tumors, overlaying genomic alteration profiles with tumor mutational phylogenies and mutation clonality across tumor regions (Supplementary Figs. S3–S18, Supplementary Table S8). All patients exhibited truncal (i.e., present in all regions) inactivating mutations in at least one previously identified RCC-driver gene (Fig. 1B, see Materials and Methods). All patients with ccRCC exhibited either truncal VHL mutation (13/14) or truncal loss of chromosome 3p (13/14), with co-occurrence of these truncal driver alterations in 12/14 patients. Consistent with previous observations (23), PBRM1 was the second-most commonly mutated driver gene, harboring truncal mutations in six patients with ccRCC and branch mutations in a further three. For the two papillary cases, PT11 exhibited mutations in multiple ccRCC driver genes (PBRM1, SETD2, and ARID1A), whereas patient PT4 exhibited only subclonal mutation of ARID1A. Expression of canonical RCC driver genes was generally consistent with the expected behavior for tumor suppressors (downregulated in tumor relative to matched normal), except for FAT1, MET, SMARCB1, and TP53 (Supplementary Fig. S19). Tumors in this cohort generally fell into two groups based on their phylogenetic patterns of major driver alteration (those with at least 5% prevalence in this cohort): Eight patients displayed relatively simple patterns with major driver alterations being largely truncal (PT5, PT7, PT8, PT10, PT11, PT13, PT15, and PT16), whereas in the other eight patients there was frequent secondary accumulation of driver alterations along internal branches. Among the latter group, five tumors showed parallel mutation of major RCC drivers in separate tumor regions (Supplementary Table S9), suggesting selective pressure for specific phenotypes later in tumor evolution (8). In ccRCC metastatic tumors, we observed an enrichment in regions with SCNA losses in chr14q, 4q, or 8p relative to regions of non-metastatic tumors (54% versus 15%, P < 10–6), consistent with the more advanced disease state. In addition, driver clonality analysis revealed both accumulation of secondary driver mutations and clonal expansions involving hallmark drivers along the lineage leading to metastasis (e.g., see PT1, PT2, PT3, PT12, and PT14 in Supplementary Figs. S3–S18). We did not identify any recurrently mutated genes associated with the VTT samples aside from genes known to be frequent false positives due to having long coding sequences or multiple paralogs (Supplementary Table S10).
We looked for evidence of clonal selection (tendency toward fixation in one or more tumor regions of a mutation cluster that was subclonal in the trunk) or polyclonal evolution (subclonality present in the trunk that persists in one or more regions) and any associations with VTT or metastasis. We observed possible clonal selection in six tumors, polyclonality in eight tumors, and both in five tumors (the two modes are mutually exclusive on a per-region basis). Overall, there were three broad patterns of tumor evolution: (i) clonal trunk with secondary accumulation of additional hallmark driver alterations and subclonal diversification (PT6, PT9-12, PT14-16), (ii) stable polyclonality with secondary accumulation of hallmark driver mutations in specific clones (PT1, PT8, PT13), and (iii) multi-modal with secondary accumulation of hallmark driver mutations and/or subclonal diversification (PT2-5, PT7). Despite this variety of evolutionary patterns, there was no clear association of VTT or metastasis (or tumor grade or clinical stage) with any specific pattern.
To further investigate whether these tumors followed prescribed patterns of evolution, we used tumor phylogenies annotated with canonical driver alterations (Supplementary Figs. S3–S18) to assign the patients with ccRCC to previously described evolutionary subtypes (7). We note that we were unable to consider the methylation status of driver genes; only loss-of-function mutations in driver genes and driver SCNAs were included. We were able to assign five of the 14 ccRCC cases (36%) to three evolutionary subtypes: PT6 and PT9 were “PBRM1 → SETD2,” PT7 and PT13 were “VHL mono driver,” and PT10 was “PBRM1 → PI3K” (Table 1). Three additional cases exhibited similarities to one or more subtypes (PT12, PT14, and PT16), but could not be confidently assigned to a specific subtype. The remaining six cases did not readily fit within the seven TRACERx subtypes.
Table 1.
Summary of evolutionary analyses of the 16 RCC VTT cases.
| Patient | Evolutionary subtype | Trunk/early | Branch/late | Parallel mutation | Dominant mutational signature(s) | Dominant signature etiology | Metastasis emergence | Earliest branching region | Possible clonal selection | Polyclonality | Pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PT1 | Undetermined | VHL, SETD2, 3p loss | KDM5C, PTEN, VHL, 5q gain, 4q loss, 14q loss | KDM5C, PTEN, 14q, 4q | SBS39 | Unknown | Early | P1/LU | P1, P2, VTT, LU | Stable polyclonal trunk, secondary fixation of PTEN + KDM5C | |
| PT2 | Undetermined | VHL, KDM5C, 4q loss | ARID1A, KDM6A, SETD2, CSMD3, 3p loss, 5q gain, 8p loss, 14q loss | ARID1A | SBS15 | MMR deficiency | Early | LU | P2 | P1, P3 | Multi-modal, secondary fixation of ARID1A |
| PT3 | Undetermined | VHL, PBRM1, 3p loss, 5q gain, 14q loss | TP53, BAP1, 4q loss, 8p loss | BAP1 | SBS26 | MMR deficiency | Early | P1 | P1, VTT2, LU | P2, VTT1, LN | Multi-modal, secondary convergence to BAP1 mut |
| PT4 | Na (papillary RCC) | ARID1A | SBS39 | Unknown | Early | P3 | P1, P3, BN | VTT, AG | Multi-modal, secondary subclonal diversification | ||
| PT5 | Undetermined | VHL, KDM5C, 3p loss, 5q gain, 14q loss | PBRM1, 8p loss | SBS1,SBS39 | 5mC deamination (age/mitotic clock), Unknown | P1 | P1? | P2, P3, VTT | Multi-modal, secondary fixation of PBRM1 + 8p | ||
| PT6 | Subtype 4 (PBRM1 → SETD2) | VHL, PBRM1, 3p loss, 5q gain | SETD2 | SETD2 | SBS39 | Unknown | VTT | Clonal trunk, secondary subclonal diversification | |||
| PT7 | Subtype 7 (VHL monodriver) | VHL, 3p loss, 5q gain | 14q loss, 4q loss | SBS1, SBS29, SBS39,SBS6 | 5mC deamination (age/mitotic clock), Tobacco chewing, Unknown, MMR deficiency | VTT | VTT? | P1, P2, P3 | Multi-modal, secondary fixation of 14q, 4q | ||
| PT8 | Undetermined | VHL, KDM5C, 3p loss, 5q gain | SBS5 | Unknown (possibly age) | P3 | P1, P2, P3, VTT | Stable polyclonal trunk, secondary subclonal diversification | ||||
| PT9 | Subtype 4 (PBRM1 → SETD2) | VHL, PBRM1, 3p loss, 5q gain | KDM5C, SETD2, 8p loss | KDM5C | SBS15, SBS39 | MMR deficiency, Unknown | P2/VTT | Clonal trunk, secondary subclonal diversification | |||
| PT10 | Subtype 5 (PBRM1 → PI3K) | VHL, PBRM1, 3p loss, 5q gain, 14q loss | TSC1, 4q loss, 8p loss | SBS39 | Unknown | Early | AG1/AG2 | Clonal trunk, secondary subclonal diversification | |||
| PT11 | NA (papillary RCC) | PBRM1, SETD2, ARID1A, 4q loss, 14q loss | FAT1, 3p loss, 5q gain | SBS39 | Unknown | Early | LN1/LN2 | LN2? | Clonal trunk, secondary subclonal diversification | ||
| PT12 | Undetermined | VHL, SETD2, 3p loss, 5q gain | PBRM1, NFE2L2, SETD2, MTOR, TSC1, TP53, 4q loss, 8p loss, 14q loss | PBRM1, SETD2, 14q | SBS24,SBS39 | Aflatoxin, Unknown | Secondary | P1 | Clonal trunk, secondary subclonal diversification | ||
| PT13 | Subtype 7 (VHL monodriver) | VHL, 3p loss, 5q gain, 14q loss | SBS19,SBS25,SBS26,SBS6 | Unknown, Chemotherapy, MMR deficiency, MMR deficiency | P2 | P1, P2, P3, VTT | Stable polyclonal trunk, secondary subclonal diversification | ||||
| PT14 | Undetermined | VHL, PTEN, 3p loss, 5q gain, 4q loss | TP53, PBRM1, 8p loss, 14q loss | SBS1,SBS31, SBS4,SBS5, SBS8 | 5mC deamination (age/mitotic clock), Patient chemotherapy, Smoking, Unknown (possibly age),Unknown | Equivalent | P1 | Clonal trunk, secondary subclonal diversification | |||
| PT15 | Undetermined | VHL, PBRM1, 3p loss, 5q gain | SBS39 | Unknown | equiv. | Clonal trunk, secondary subclonal diversification | |||||
| PT16 | Undetermined | BAP1, 3p loss, 5q gain, 8p loss, 14q loss | SBS6 | MMR deficiency | P3 | Clonal trunk, secondary subclonal diversification |
Note: Phylogenetic placement of driver alterations (trunk/early vs. branch/late) and assignment of each tumor using previously described evolutionary subtypes (6). In addition to the assigned subtypes listed, PT12 had potential overlap with both “PBRM1 → SETD2” and “PBRM1 → PI3K” subtypes, PT14 showed the reverse pattern of the “PBRM1 → PTEN” subtype, and PT16 had some overlap with the “BAP1 alone” subtype but lacked VHL mutation. Tumor regions with possible clonal selection (based on region-specific fixation of a clone that appeared subclonal in the trunk) or persistent polyclonality are listed (see Supplementary Figs. S3–S18). Dominant mutational signatures (see Fig. 2) and metastasis emergence class (see Fig. 3) are also listed for each tumor.
Although our RCC cohort was too small and heterogeneously treated for formal clinical outcome association analysis, five patients presented with non-metastatic disease and their disease never recurred and the other 11 patients had some recurrence (Supplementary Table S1). These recurrence groupings correlated with disease clinical stage, Fuhrman grade, and maximum tumor size. There was no association of disease recurrence with TMB or clonal TMB, and the only hallmark RCC driver alterations that were associated with recurrence (CNA deletions of chr14q, 8p, 4q) were also associated with metastatic disease at diagnosis. There was a tendency toward secondary driver accumulation or parallel mutations associating with tumor recurrence (8/10 such tumors recurred), although there were also clear exceptions to this such as PT6 and PT9. On the basis of phylogenetic analysis, there was a potential association of early VTT lineage branching in tumors that did not recur (3/5 cases) versus later VTT branching in tumors that did recur (11/11 cases; Supplementary Fig. S20A and S20B). However, due to sample size this trend did not reach statistical significance (P = 0.08, Fisher's exact test). There was also a trend toward non-recurring patients showing immune-inflamed status and higher levels of CD8 T-cells by IHC (Supplementary Fig. S20C and S20D).
DNA repair deficiency mutational signatures are prevalent in the RCC VTT cohort
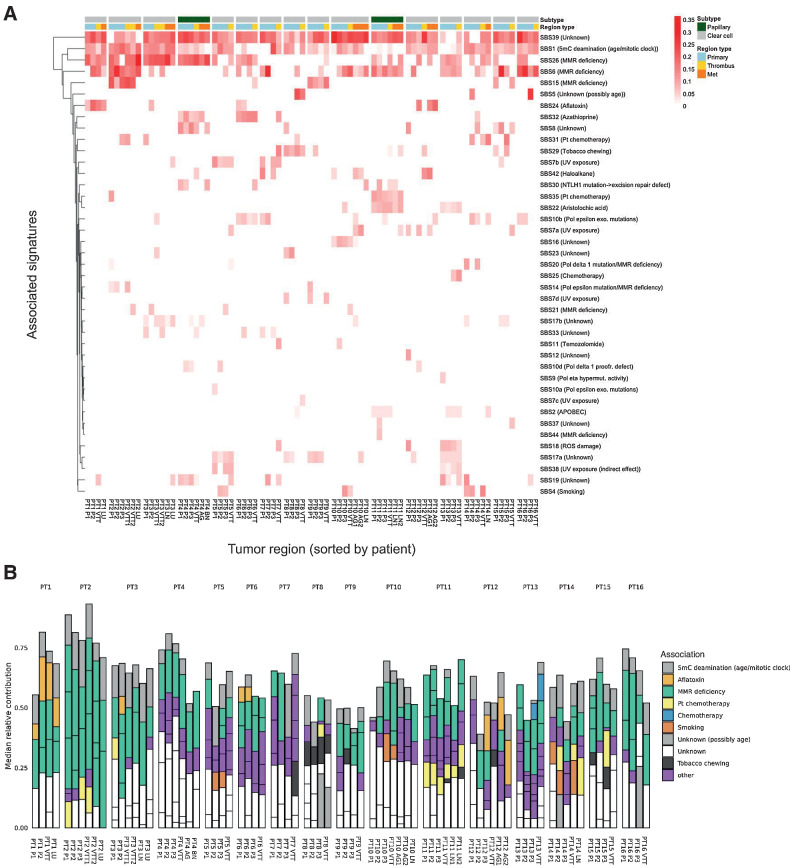
We next asked whether specific mutational signatures were identified in patients with VTT. We examined somatic SNV substitution patterns, along with their trinucleotide context, and compared the resulting spectra with 49 previously identified mutational signatures (37), using stringent fitting criteria and a bootstrap approach to assess the robustness of the fit (Supplementary Fig. S21). Determination of the relative contribution of known signatures to the observed spectra revealed a range of heterogeneity within and among tumors, with a relatively high mean sample-to-sample cosine distance (0.72), indicating generally similar signature abundances within patients (Fig. 2A, Supplementary Table S11). Extraction of the signatures contributing to each sample's mutational spectra revealed an enrichment of signatures associated with mismatch repair deficiency (SBS6, SBS15, and SBS26), along with signatures associated with age (SBS1), and signatures of unknown etiology (SBS39) across most tumors (Fig. 2B).
Figure 2.
SNV mutational signature analysis. A, Heatmap of previously established SNV mutational signatures contributing to the mutational spectra of each tumor region across all patients. The median values of 100 bootstrap replicates are displayed. B, Contribution of previously established SNV mutational signatures to the mutational spectra of each region across all patients. Included signatures are restricted to those with a nonzero median contribution across 100 bootstrap replicates.
The most prevalent and robustly fit signatures in our cohort were signatures SBS1, SBS6, SBS26, and SBS39. SBS1, which is associated with patient age and is known to contribute to the mutational landscape of RCC (36, 37), was robustly observed in all 16 patients and in 64/78 samples (Supplementary Fig. S22). Signatures SBS6, SBS15, and SBS26 are related to DNA mismatch repair (MMR) deficiency, and were observed in a total of 44, 12, and 44 samples, across 15, 4, and 13 tumors, respectively. Collectively, MMR deficiency-associated signatures were observed in all tumors, and in 68 of 78 sequenced tumor regions. A particularly striking enrichment of these signatures was observed in PT2. This tumor harbored a truncal nonsense mutation in MLH1, which is known to cause MMR defects but is rare in RCC (43). Moreover, this tumor exhibited the most mutations per sample (7.9 mutations/Mb compared with an average of 2.8 mutations/Mb for the other patients), and the most total unique mutations (25.2 mutations/Mb compared with an average of 5.7 for the other patients). MMR mutations were not observed in other tumors exhibiting MMR deficiency-associated signatures, apart from an MSH6 intronic variant in one sample from PT11. The presence of MMR deficiency-associated signatures in RCC has previously been reported (36), but is somewhat surprising given the relatively low mutational burden of RCC. Notably, SBS39 was present in most tumors and regions (16 and 72, respectively), but has no known etiology. This C→G transition-rich signature has been frequently observed in ovarian, pancreatic, and medullary cancer, but not in RCC.
Mutational signature fitting is sensitive to the number and identity of input known mutational signatures, and the number of known cancer signatures has recently expanded (37). We therefore applied the same bootstrap approach (Supplementary Fig. S23) to a more limited set of signatures identified in a foundational pan-cancer signatures study (Supplementary Fig. S24; ref. 36). In this smaller set, which notably lacked SBS39, age- and MMR deficiency–associated signatures were also common. We also identified SBS3, which is associated with BRCA1/2 mutation and was previously observed in another RCC VTT cohort (5). Almost all tumors (14/16) exhibited some degree of signature SBS3 (50/78 regions, Supplementary Fig. S25), despite only two tumors having potentially deleterious BRCA1/2 alterations. PT8 had a truncal frameshift mutation in BRCA1, and PT4 had a truncal splice mutation in ZAR1L, which is immediately upstream of BRCA2. The presence of signature SBS3 in the absence of BRCA mutations is nonetheless a previously described phenomenon (44, 45). The only case lacking the SBS3 “BRCA-ness” signature was patient PT2, who instead displayed a strong signature SBS6 (associated with DNA mismatch repair deficiency), consistent with the results obtained with the full set of 49 signatures. Interestingly, with the smaller signature set, SBS3 and MMR deficiency–associated signatures (SBS14, SBS15, SBS20, and SBS21) co-occurred in at least one region in 13/16 patients (38 regions total).
Phylogenetic timing analysis suggests that VTT and metastasis are independent, with relatively early emergence of metastatic lineages in some patients
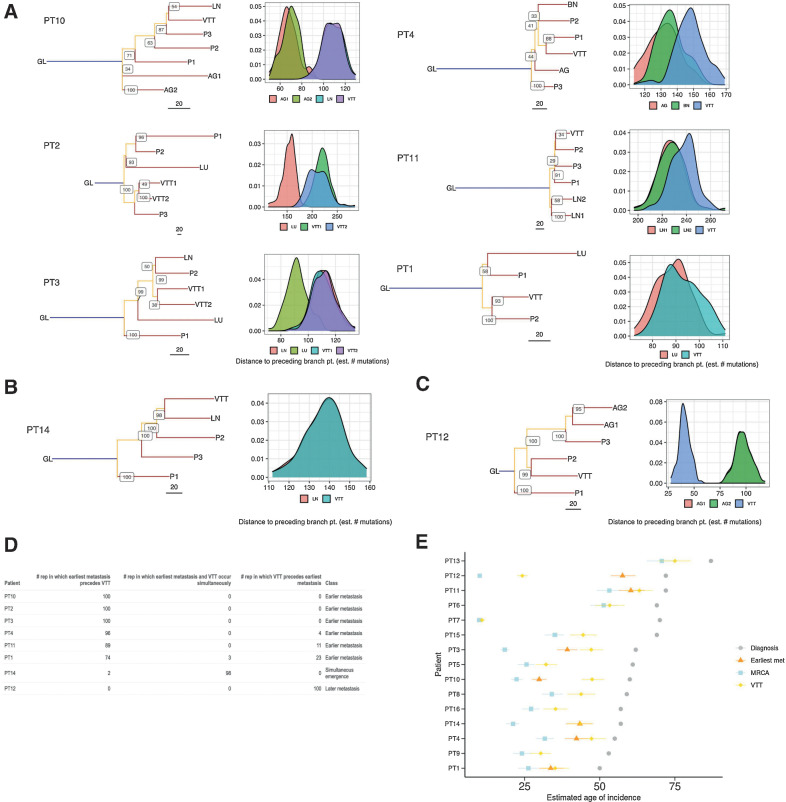
We next asked whether VTT acts as a reservoir for hematogenous metastases. If this were true, the VTT lineage should typically emerge before metastases and the metastases should share clonal ancestry with the VTT. To test this hypothesis, we performed phylogenetic lineage tracing for the eight patients with sequenced primary, VTT, and metastasis lesions (Fig. 3). In seven of eight patients, the VTT had a distinct mutational ancestry from the earliest metastasis (PT10, PT2, PT3, PT4, PT11, PT1, PT12). In PT14, the VTT and LN metastasis were the latest lineages to emerge and shared an immediate ancestor. PT10 was a more complex case, wherein the VTT shared an immediate ancestor with the LN metastasis, but the two adrenal gland metastases arose earlier from a distinct ancestor. To establish the certainty of the phylogenetic reconstruction and to establish the confidence in placement of specific nodes, we performed non-parametric bootstrapping of the mutational tree (30). Overall, across the eight patients, the prior assertions of distinct clonal origins for VTT and metastasis were well supported, with nodal bootstrap values consistently above 70 (Fig. 3A–C, top). However, PT4 had lower bootstrap values for the two key nodes (44 and 41), suggesting less certainty in the phylogenetic placement of the clonal ancestors for the AG, BN, P1, P2, and VTT regions. These results suggest that in most patients VTT did not directly seed metastases.
Figure 3.
Phylogenetic timing analyses of eight metastatic RCC tumors with VTT. A–C, Mutational phylogenies and reconstruction robustness for eight RCC tumors with VTT and distant metastasis. Internal node labels on the phylogenetic trees display the number of bootstrap replicates supporting each node (i.e., the certainty out of 100 that the placement of a particular node is supported). Density plots illustrate the distribution of path lengths to the parent nodes of all non-primary regions derived from 100 bootstrap replicates of the tree building process. The following are the region labels: GL, germline; P, primary tumor; VTT, venous tumor thrombus; AG, adrenal gland metastasis; LN, lymph node metastasis; LU, lung metastasis; BN, brain metastasis. A, Patients with early emergence of metastases relative to VTT. B, Patient with near-simultaneous emergence of VTT and the earliest assayed metastases, with both emerging from the same branch. C, Patient with late emergence of metastases relative to VTT. D, Table showing the number of bootstrap replicates in which phylogenetic tree reconstruction resulted in earlier, simultaneous, or later emergence of the earliest metastasis relative to the VTT. E, Timing estimates generated with linear mixed effects models of the emergence of MRCA, the immediate ancestor of the earliest metastasis, and the immediate ancestor of the VTT, relative to the age at diagnosis. Error bars indicate 95% confidence intervals generated by parametric bootstrapping of the i.i.d errors.
The relative timing of a given phylogenetic node can be approximated by the sum of the internal branch lengths leading up to that node. However, a single “representative” tree does not fully capture the inherent uncertainty in these branch lengths. To make our nodal confidence analysis more quantitative, we developed a novel approach for quantifying total internal branch lengths that also captured the uncertainty in the phylogenetic reconstruction of the clonal ancestors of the VTT or of the metastasis. For each tumor, we quantified the total internal branch lengths across 100 bootstrap trees and plotted the resulting empirical distribution of distances from the root node to branch points immediately preceding either VTT or metastasis. These root-to-node distance distributions then provide a measure of certainty regarding which region emerged first (Fig. 3A–C, bottom). The frequency with which one branch-point precedes another in the bootstrap sampled trees provides an estimated P value (Fig. 3D). For example, in PT10 the AG1 and AG2 metastases showed clearly lower root-to-node distance distributions than the VTT and LN metastasis, suggesting that AG1 and AG2 emerged significantly earlier in mutational time. Similarly, in PT4 the AG and BN metastases showed consistently lower root-to-node distances than the VTT, with 96/100 bootstrap trees supporting the earlier emergence of the metastases (Fig. 3A and D). Taken together, these results suggest that not only does the VTT typically not give rise to metastases, but many metastatic lineages emerge earlier than VTT.
This analysis revealed three categories of RCC tumors: (i) early emergence of at least one metastasis before VTT emergence, (ii) late emergence of the earliest metastases after VTT emergence, and (iii) similar timing of VTT and the earliest metastasis emergence due to their divergence from the same branch. Patients PT2, PT3, PT4, and PT10 fell clearly into the early metastasis class (P < 0.05, Fig. 3A). Patient PT14 exhibited similar timing of relative emergence due to the VTT and LN metastasis sharing the same immediate ancestor (P < 0.05, Fig. 3B). Patient PT12 fell clearly into the late metastasis class (P < 0.01, Fig. 3C). We note that PT14 had recurrences in brain and bone that were not sequenced, and PT12 had a pleural lesion recurrence in the lungs that was not sequenced. Therefore, it is possible that metastases may have been seeded earlier in these cases than we were able to detect. Patients PT1 and PT11 were most consistent with the early metastasis class, but there was lower certainty (P ≅ 0.26 and P ≅ 0.11, respectively) in distinguishing the root-to-node distance distributions for VTT and metastasis owing to the small numbers of mutations distinguishing their immediate ancestors (Fig. 3D). Remarkably, in three patients (PT2, PT10, and PT11), the earliest metastatic lineages emerged before diversification of the earliest primary tumor lineages.
Estimating the absolute timing of metastatic lineage emergence could have important implications for clinical care. Having established the relatively early emergence of metastatic lineages relative to VTT, we sought to determine the age of the patients at which metastases emerged, as well as the number of years between metastasis emergence and clinical diagnosis. We adapted the modeling approach of Mitchell and colleagues (16) by generalizing it to encompass any node on the tumor phylogeny. This allowed us to infer the absolute timing of metastatic lineage emergence relative to the most recent common ancestor (“MRCA” or phylogenetic trunk), to the VTT lineage, and to age at diagnosis. Using the average of the root-to-node branch lengths from each patient's bootstrap sampled trees, we predicted the patient ages at which the earliest metastasis and VTT branching events occurred (Fig. 3E). PT2 was excluded due to the MLH1 mutation and associated high contribution of MMR deficiency-associated signatures to the mutation landscape. This timing analysis suggested that metastases emerged 11.5 to 30.3 years before diagnosis (Supplementary Table S12) in the seven assessed patients. We further evaluated the relationship between mutational signature contribution and sample mutation burden while controlling for patient age and found that no signatures were significantly associated with mutation burden after adjustment for multiple testing (Supplementary Fig. S26, Supplementary Table S13, Supplementary Methods: “Estimating lesion emergence timing”). Together with the relative similarity of estimates of sample mutation rate, this suggests that any deviations from linear mutation accumulation within tumor lineages were likely modest.
This analysis largely recapitulated the observations from the phylogenetic trees, with the early-metastasis patients also exhibiting earlier absolute estimates of metastasis emergence as compared with VTT emergence. Three of these patients appeared to show nearly contemporaneous emergence of the VTT and metastasis immediate ancestors (PT1, PT4, and PT11), and the other two patients had significant, multi-year lags between the emergence of the VTT and metastasis immediate ancestors (PT3 and PT10). In PT12, the sole “late metastasis” patient, the VTT lineage emerged a striking 33 years before the emergence of the adrenal gland metastases.
Immediate-early genes are upregulated in VTT, and MTOR pathway targets are upregulated in metastasis
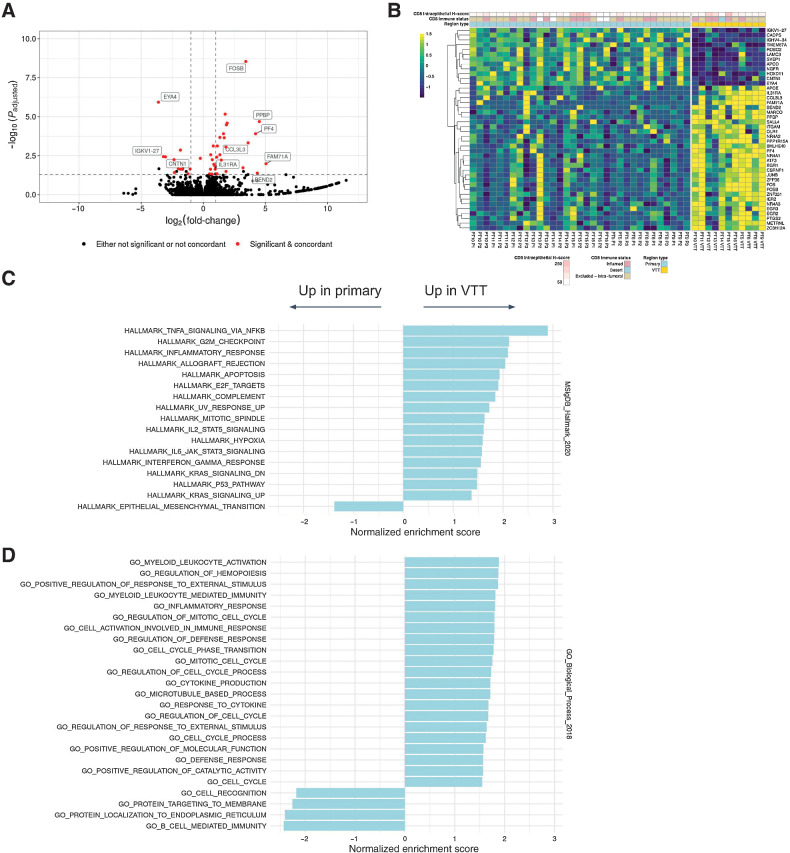
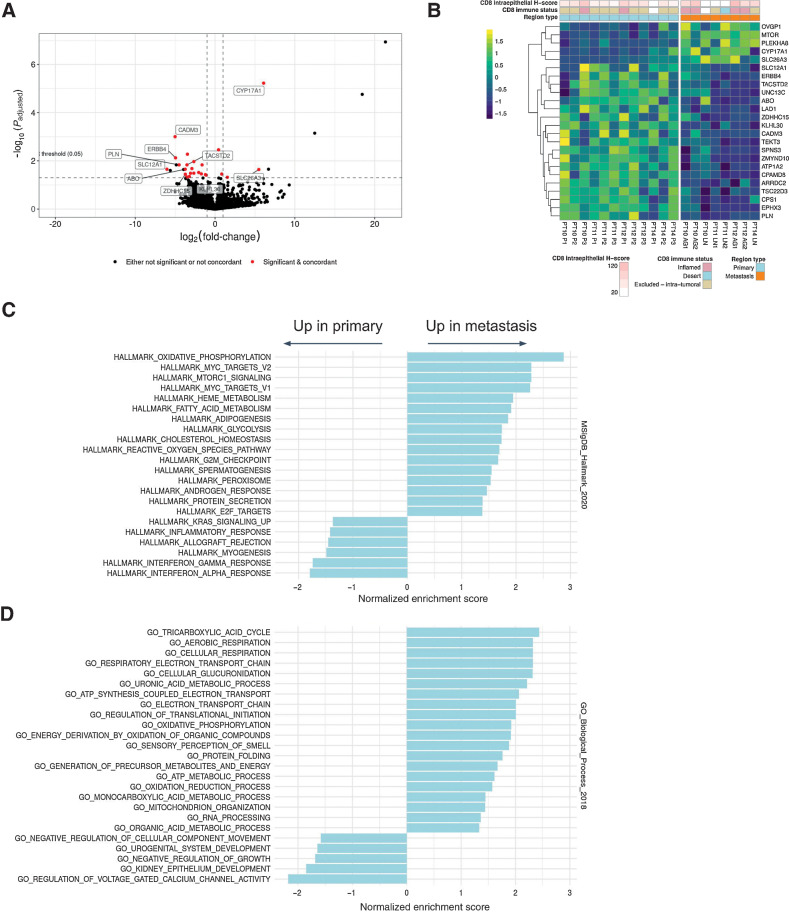
To explore whether distinct molecular programs govern the outgrowth of VTT versus metastasis from primary tumor, we performed paired differential gene expression analysis on 12 patients (RNA-seq was not available for PT1-PT4). We first compared primary tumor regions with VTT regions while accounting for patient-specific variability in gene expression. We identified 30 genes as significantly upregulated in VTT relative to primary tumor, and 12 as significantly downregulated (Fig. 4A and B). Several immediate-early genes, including FOS, FOSB, JUN, NR4A1, NR4A2, IER2, and EGR1-3, were upregulated in VTT. Gene set overlap analysis using Enrichr (41) revealed highly significant overlap of VTT-upregulated genes with the TNFα signaling via NF-κB Hallmark gene set (17/42 genes, Padjusted < 10–6, Supplementary Fig. S27A). This gene set consists of validated downstream targets of NF-κB after activation by TNFα (43), and 150/200 genes were upregulated in VTT (log2FC > 0). Among the upregulated genes were the AP-1 family transcription factors (FOS, FOSB, and JUNB), known downstream effectors of TNFα signaling (46). This suggests the possibility that the expression of the immediate-early genes was driven by TNFα via NF-κB. However, the expression of core receptors and kinases in TNF signaling pathways was not significantly perturbed, and our observations constitute only indirect evidence of pathway activation.
Figure 4.
Differential gene expression analysis of primary tumor versus VTT. A, Volcano plot showing log-fold changes and Padjusted values for differentially expressed genes (primary vs. VTT). Genes highlighted in red were both significantly different and exhibited concordance in the directionality of their changes across regions and patients. A few non-concordant outliers have been removed for clarity of plotting. B, Heatmap showing expression values of differentially expressed genes between primary and VTT regions. Expression values are normalized per gene and per patient. C, Significantly enriched or depleted Hallmark pathways in VTT relative to primary regions as determined by GSEA. D, Significantly enriched GO Biological Process terms enriched or depleted in VTT relative to primary regions as determined by GSEA. Significance is defined as the Padjusted value of ≤0.05 (Benjamini–Hochberg).
GSEA with the full set of detected genes (see Materials and Methods) revealed multiple significantly upregulated Hallmark gene sets, including TNFα signaling and inflammatory response (Fig. 4C). Interestingly, the sole significantly downregulated Hallmark gene set was epithelial-to-mesenchymal transition (EMT). GSEA further revealed multiple significantly enriched gene ontology biological process (GOBP) terms (Fig. 4D; ref. 47). The upregulated GOBP terms also included inflammatory response, with the notable additions of immune-related processes, cell-cycle regulation, and response to external stimuli. An overlap of our differentially expressed gene list with genes identified by a previous study (4) resulted in three genes in common: MARCO, PF4, and PPBP. These genes as well as others upregulated in VTT (e.g., APOE) are associated with activated macrophages and platelets.
To address the possibility that these differences in gene expression were driven by specific cell types, we performed histologic analyses and gene expression deconvolution analysis. The histology analysis confirmed that significant neutrophilic infiltrate was not detected in VTT or primary tumors, and IHC analysis suggested no increase in CD8 immune infiltration of VTT as compared with the primary tumor (Fig. 4B). We then used xCell (48) to assess the enrichment of gene expression signatures associated with 67 immune and stromal cell types. We normalized the scores within each cell type and patient, and compared them across tumor region types (Supplementary Fig. S28). No cell type was significantly different across regions after correction for multiple testing. This suggests that if present, TNFα signaling or inflammation likely does not arise from lymphocytic or other specific immune cells in the tumor.
We next compared primary tumor regions to metastases, and identified 24 differentially expressed genes (5 genes up in metastasis, 19 genes down; Fig. 5A and B). Patterns of differential expression were consistent across metastases in distinct anatomical locations (i.e., lymph nodes vs. adrenal glands). Although no specific biological pathways were enriched in these 24 genes alone, GSEA with the full set of detected genes revealed upregulation of mTORC1 downstream transcriptional targets in metastases, along with MYC targets and oxidative phosphorylation (Fig. 5C and D). These results are consistent with previous observations that mutations that increase mTOR activity are associated with metastatic RCC (49). Although we did not observe consistent mutations in mTOR pathway genes (except TSC1 mutations in PT10 AG1 and PT12 AG2), MTOR itself was notably upregulated in metastases, which could explain the observed enrichment of Hallmark mTORC1 transcriptional targets.
Figure 5.
Differential gene expression analysis of primary tumor versus metastasis. A, Volcano plot showing log-fold changes and Padjusted values for differentially expressed genes (primary vs. metastasis). B, Heatmap showing expression values of differentially expressed genes between primary and metastatic regions. Expression values are normalized per gene and per patient. C, Significantly enriched or depleted Hallmark pathways in metastases relative to primary regions as determined by GSEA. D, Significantly enriched GO Biological Process terms enriched or depleted in metastases relative to primary regions as determined by GSEA. Significance is defined as the Padjusted value of ≤0.05 (Benjamini–Hochberg).
Finally, we compared gene expression in metastasis versus VTT and identified nine differentially expressed genes (four genes upregulated in metastasis, five genes down; Supplementary Fig. S29A). This analysis was limited to the four patients for which we had metastasis samples and RNA-seq data, and so may be under-powered.
Discussion
In this study, we analyzed 16 RCC cases with VTT by MR-seq to inform how and when new lesions emerge, to test whether VTT seed metastases directly, and to characterize the genomic and evolutionary landscape of tumors with VTT. The unique nature of these cases—each with distinct extension from the primary tumor and in some cases with separate metastases—also enabled us to investigate how and when new lesions arise from the primary tumor. Although only eight patients had sequenced VTT and metastasis samples, our study surveyed a diverse variety of metastatic sites within these cases.
As expected, we observed early branching evolution with frequent, extensive internal branches in most ccRCC cases (Supplementary Figs. S3–S18). Evolutionary analysis revealed three broad patterns (clonal trunk, stable polyclonality, multi-modal), yet no pattern associated with tumor clinical characteristics. In addition, 36% of tumors exhibited driver alteration patterns consistent with previously described evolutionary subtypes (7), whereas the others had distinct alteration patterns (Table 1). Notably, 37% of the TRACERx RCC cases were considered unclassifiable. We did not identify recurrently mutated genes associated with VTT, suggesting that tumor-intrinsic epigenetic factors or tumor-extrinsic factors may lead to VTT formation. Metastatic tumors were enriched for hallmark SCNAs and secondary driver alterations, which also tended to accumulate in lineages leading to metastasis (Fig. 1A, Supplementary Figs. S3–S18). Much of the heterogeneity among driver mutations tended to be due to secondary accumulation and parallel mutation occurring in distinct tumor regions (Supplementary Figs. S3–S18, Supplementary Table S9). This suggests that early RCC may give rise to multiple lineages (branching evolution), and there is frequently strong selection for specific phenotypes later in their development, perhaps to enable their persistence. Early RCC clones may begin in “fits and starts,” with relatively low fitness (e.g., VHL is an essential gene in most non-kidney tissues), and only when additional hallmark pathways are altered do tumors become fit enough to metastasize.
Despite substantial genetic heterogeneity in our cases, mutational signatures were relatively concordant across regions of most tumors (mean cosine distance >0.7 in 11 of 16 patients, Supplementary Table S11), with notable exceptions such as PT7 and PT12, in whom substantial contributors to some regions were unobserved in others. We did not observe any markedly distinct signature contributions in VTT or metastatic regions that distinguished them from primary tumor regions. The prevalence of MMR deficiency-associated signatures in our cohort, albeit at low levels in most cases, is seemingly at odds with the relatively low mutation burden of RCC (36). Given that many RCC tumors respond to immunotherapy, future studies should explore whether the level of MMR deficiency signatures constitutes a potential biomarker. In an analysis fitting only a subset of established signatures, SBS3 was identified in a substantial proportion of patients, as was previously observed (5). However, this result was not seen with the full set of 49 signatures, where instead SBS39 was prevalent. Given the overlap of SBS3 and SBS39 mutational spectra, we could not rule out some contribution from SBS3-related processes (i.e., BRCA-ness or homologous repair deficiency). A potential biological rationale for the presence of SBS3 exists: Genes required for homologous recombination and mismatch repair are downregulated by VHL inactivation, presumably via the resulting stabilization of HIF-1α (50). The different results obtained from these two signature sets are likely a consequence of the difficulty in inferring the ground truth mutational signature composition using a large number of potential component signatures, a number of which are highly similar to one another (37). Nonetheless, our observation of prevalent age- and MMR deficiency-associated signatures is consistent with previous reports (5, 36).
By phylogenetic analysis, most metastatic tumors in our cohort (6/8) displayed a clear earlier emergence of metastases relative to VTT. This effectively ruled out the possibility of direct hematogenous seeding of metastases from the VTT and suggested that VTT and metastasis formation occur independent of one another. (We cannot be certain that VTT or metastatic competence was attained at the time of these phylogenetic branching events—they may have occurred later via subsequent mutations or epigenetic processes.) We also noted that, in metastatic cases, the VTT often arose from a lineage that had already given rise to metastasis (6/8 tumors; PT1 and PT12 were the exceptions). At the same time, VTT lineage branching tended to occur rather early in non-metastatic tumors. These results can be explained by a simple model: VTT formation occurs in RCC at some frequency and is independent of metastasis, and in tumors that are “born to be bad,” metastatic lineages tend to arise early and thus often overlap with the VTT lineage. This model is consistent with clinical experience wherein aggressive surgical intervention despite VTT can result in long-term survival, many patients with VTT present without metastatic disease (7/16 in our cohort), and probability of metastatic presentation is related to primary tumor size whereas probability of VTT presence is not (51).
Our timing analysis suggested that in all patients the earliest metastases (i.e., the set of clones that ultimately constituted the earliest metastases) emerged well before diagnosis—as long as three decades in one patient. This early emergence is consistent with recent findings that metastases in colorectal, breast, and lung cancer are disseminated early relative to time of diagnosis (13, 14). Our approach for estimating the absolute timing of metastatic lineage emergence assumes that the tumor mutation rate is not substantially different from the background somatic mutation rate, and that mutation rates do not vary substantially across tumor regions. In our patients, the presence of multiple non–age-associated mutation signatures suggests that the former assumption may be violated. Indeed, three mutational signatures identified have a proposed etiology of MMR deficiency (SBS6, SBS15, and SBS26) and therefore imply higher SNV mutation rates (52). However, upon fitting the model, the patient-specific slope estimates were relatively similar to one another, with the exception of PT2 (Supplementary Fig. S30). After excluding PT2, minimal relationship across samples and patients was observed between mutational signature contribution and sample mutation burden, when controlling for age (Supplementary Fig. S26, Supplementary Table S13). Given that many RCC tumors initially present with metastasis, early detection studies are needed to examine which cases may develop RCC metastases well before the disease is clinically evident.
VTT regions showed consistent upregulation of downstream transcriptional targets of TNFα signaling via NF-κB and related inflammatory processes relative to primary tumor regions. However, the association with TNFα or inflammation is indirect. It is possible that upregulation of immediate-early genes represents a more general tumor response to the environment of the renal vein or IVC. There was no clear evidence from IHC analysis or analysis of immune and stromal cell type gene expression signatures that these differences were due to immune infiltration or stromal content of VTT. Our data also cannot fully rule out an immune or stromal source for TNFα—indeed, RCC tumors are known to have an angiogenic phenotype and substantial myeloid involvement (and some myeloid genes were upregulated in VTT in our cohort). However, without a clear source for TNF or pro-inflammatory cytokines, we can only speculate on the causes and consequences of these upregulated genes and their relationship to VTT. Further studies are needed to distinguish a more general stress response from pro-survival signaling downstream of secreted factors in the kidneys or associated vasculature.
The sole significantly depleted Hallmark pathway in the VTT identified by GSEA was EMT. This could indicate that the VTT is in general less competent to generate metastases than the primary tumor, rendering it a less frequent source of hematogenous metastases than might otherwise be expected. Although the genes we identified as differentially expressed between primary and VTT regions had minimal overlap (only three genes) with those from a previous study (4), the two analyses did identify similar pathways as upregulated (e.g., inflammatory response) and downregulated (e.g., EMT). A smaller number of genes were observed to be differentially expressed between primary and metastatic sites with MTOR notably upregulated in metastases. Perhaps as a consequence of this, downstream transcriptional targets of mTORC1 were also upregulated, as determined by GSEA. mTOR signaling is thought to drive invasiveness in RCC and other cancers, and the use of mTORC1 inhibitors has seen some clinical success (49). Inactivating or putatively deleterious mutations in PTEN and TSC1, known negative regulators of mTORC1 activity (53, 54), were identified in a number of patients (Fig. 1), but these were either not metastasis-restricted (e.g., PT14) or not detected in all metastases (e.g., PT10). Intriguingly, we observed mutation of NFE2L2 in both sequenced metastatic regions and a single primary region in PT12. NFE2L2 has been reported to be a direct transcriptional activator of the MTOR gene (55), and its mutation could conceivably contribute to MTOR upregulation in this patient. However, future studies are needed to definitively determine the basis of MTOR upregulation and activity in metastases.
Taken together, our data reveal new dimensions of RCC VTT and metastasis biology. They highlight the need for earlier detection of RCC and suggest that metastatic clones can emerge and remain largely dormant for years before becoming clinically detectable. They also suggest potential expression and mutational biomarkers of RCC patient prognosis and treatment response.
Supplementary Material
Acknowledgments
We thank our colleagues at Genentech and UCSF for helpful discussions. We particularly thank Suchit Jhunjhunwala and Richard Bourgon for their guidance and support of this project, and Eric Collisson for facilitating our collaboration. We also thank Christina Curtis (Stanford) for helpful discussions.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
A. Wallace, D. Oreper, and C. Havnar report employment with Genentech and shareholders of Roche. S.P. Porten reports grants and other support from Photocure, and personal fees from Coloplast outside the submitted work. A.A. Lo reports employment and stockholder at Genentech and Roche. X. Pechuan-Jorge reports employment with Genentech. O.A. Zill employment with Genentech, and shareholder of Roche, founder and owner of init.bio, Inc. No disclosures were reported by the other authors.
Authors' Contributions
A. Wallace: Data curation, software, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. S.P. Porten: Conceptualization, resources, data curation, investigation, methodology, writing–original draft, project administration, writing–review and editing. A.A. Lo: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing. D. Oreper: Software, formal analysis, methodology, writing–review and editing. N. Lounsbury: Software, formal analysis, methodology. C. Havnar: Resources, investigation. X. Pechuan-Jorge: Software, visualization, methodology. O.A. Zill: Conceptualization, software, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M.V. Meng: Conceptualization, resources, supervision, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, et al. Epidemiology of renal cell carcinoma. World J Oncol 2020;11:79–87–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Quencer KB, Friedman T, Sheth R, Oklu R. Tumor thrombus: incidence, imaging, prognosis and treatment. Cardiovasc Diagn Ther 2017;7:S165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Psutka SP, Leibovich BC. Management of inferior vena cava tumor thrombus in locally advanced renal cell carcinoma. Ther Adv Urol 2015;7:216–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang XM, Lu Y, Song YM, Dong J, Li RY, Wang GL, et al. Integrative genomic study of Chinese clear cell renal cell carcinoma reveals features associated with thrombus. Nat Commun 2020;11:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warsow G, Hübschmann D, Kleinheinz K, Nientiedt C, Heller M, Van Coile L, et al. Genomic features of renal cell carcinoma with venous tumor thrombus. Sci Rep 2018;8:7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 2018;173:581–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 2018;173:595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitson JM, Reese AC, Meng MV. Population based analysis of survival in patients with renal cell carcinoma and venous tumor thrombus. Urol Oncol 2013;31:259–63. [DOI] [PubMed] [Google Scholar]
- 11. Jason AE, Spiess Philippe E, Vitaly M, Master Viraj A, Michael M, Kamran ZS, et al. Cytoreductive nephrectomy for renal cell carcinoma with venous tumor thrombus. J Urol 2017;198:281–8. [DOI] [PubMed] [Google Scholar]
- 12. Gong J, Maia MC, Dizman N, Govindarajan A, Pal SK. Metastasis in renal cell carcinoma: biology and implications for therapy. Asian J Urol 2016;3:286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 2019;51:1113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu Z, Li Z, Ma Z, Curtis C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat Genet 2020;52:701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brufau BP, Cerqueda CS, Villalba LB, Izquierdo RS, González BM, Molina CN. Metastatic renal cell carcinoma: radiologic findings and assessment of response to targeted antiangiogenic therapy by using multidetector CT. Radiographics 2013;33:1691–716. [DOI] [PubMed] [Google Scholar]
- 16. Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O'Brien T, et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 2018;173:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Auwera GAV er, O'Connor BD. Genomics in the Cloud: using Docker, GATK, and WDL in Terra [Internet]. 1st Edition. O'Reilly Media, Inc.; 2020[cited 2021 Dec 19]. Available from: https://www.oreilly.com/library/view/genomics-in-the/9781491975183/. [Google Scholar]
- 19. Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 2012;40:11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 21. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ha G, Roth A, Khattra J, Ho J, Yap D, Prentice LM, et al. TITAN: inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res 2014;24:1881–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ricketts CJ, De Cubas AA, Fan H, Smith CC, Lang M, Reznik E, et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep 2018;23:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016;374:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Gordon Robertson A, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;76:7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan X, Bai J, Zhang J, Yang L, Duan J, Li Y, et al. CONDEL: detecting copy number variation and genotyping deletion zygosity from single tumor samples using sequence data. IEEE/ACM Trans Comput Biol Bioinform 2020;17:1141–53. [DOI] [PubMed] [Google Scholar]
- 29. Lo AA, Wallace A, Oreper D, Lounsbury N, Havnar C, Pechuan-Jorge X, et al. Indication-specific tumor evolution and its impact on neoantigen targeting and biomarkers for individualized cancer immunotherapies. J Immunother Cancer 2021;9:e003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evol Int J Org Evol 1985;39:783–91. [DOI] [PubMed] [Google Scholar]
- 31. Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics 2011;27:592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paradis E, Claude J, Strimmer K. APE: analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004;20:289–90. [DOI] [PubMed] [Google Scholar]
- 33. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48. [Google Scholar]
- 35. Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 2015;31:3673–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pau G, Reeder J. HTSeqGenie: an NGS analysis pipeline. [Internet]. Bioconductor version: Release (3.13); 2021[cited 2021 Jun 7]. Available from: https://bioconductor.org/packages/HTSeqGenie/.
- 40. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021;060012. [Google Scholar]
- 43. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017;49:1476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bodily WR, Shirts BH, Walsh T, Gulsuner S, King M-C, Parker A, et al. Effects of germline and somatic events in candidate BRCA-like genes on breast-tumor signatures. PLoS One 2020;15:e0239197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chu WM. Tumor necrosis factor. Cancer Lett 2013;328:222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lieberthal W, Levine JS. The role of the mammalian target of rapamycin (mTOR) in renal disease. J Am Soc Nephrol 2009;20:2493–502. [DOI] [PubMed] [Google Scholar]
- 50. Scanlon SE, Hegan DC, Sulkowski PL, Glazer PM. Suppression of homology-dependent DNA double-strand break repair induces PARP inhibitor sensitivity in VHL-deficient human renal cell carcinoma. Oncotarget 2017;9:4647–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thompson RH, Hill JR, Babayev Y, Cronin A, Kaag M, Kundu S, et al. Metastatic renal cell carcinoma risk according to tumor size. J Urol 2009;182:41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pećina-Šlaus N, Kafka A, Salamon I, Bukovac A. Mismatch repair pathway, genome stability and cancer. Front Mol Biosci 2020;7:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A 2007;104:16158–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K–Akt signaling through downregulation of PDGFR. J Clin Invest 2003;112:1223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bendavit G, Aboulkassim T, Hilmi K, Shah S, Batist G. Nrf2 transcription factor can directly regulate mTOR. J Biol Chem 2016;291:25476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.