Abstract
New therapeutics and combination regimens have led to marked clinical improvements for the treatment of a subset of colorectal cancer. Immune checkpoint inhibitors have shown clinical efficacy in patients with mismatch-repair–deficient or microsatellite instability–high (MSI-H) metastatic colorectal cancer (mCRC). However, patients with microsatellite-stable (MSS) or low levels of microsatellite instable (MSI-L) colorectal cancer have not benefited from these immune modulators, and the survival outcome remains poor for the majority of patients diagnosed with mCRC. In this article, we describe the discovery of a novel T-cell–dependent bispecific antibody (TDB) targeting tumor-associated antigen LY6G6D, LY6G6D-TDB, for the treatment of colorectal cancer. RNAseq analysis showed that LY6G6D was differentially expressed in colorectal cancer with high prevalence in MSS and MSI-L subsets, whereas LY6G6D expression in normal tissues was limited. IHC confirmed the elevated expression of LY6G6D in primary and metastatic colorectal tumors, whereas minimal or no expression was observed in most normal tissue samples. The optimized LY6G6D-TDB, which targets a membrane-proximal epitope of LY6G6D and binds to CD3 with high affinity, exhibits potent antitumor activity both in vitro and in vivo. In vitro functional assays show that LY6G6D-TDB–mediated T-cell activation and cytotoxicity are conditional and target dependent. In mouse xenograft tumor models, LY6G6D-TDB demonstrates antitumor efficacy as a single agent against established colorectal tumors, and enhanced efficacy can be achieved when LY6G6D-TDB is combined with PD-1 blockade. Our studies provide evidence for the therapeutic potential of LY6G6D-TDB as an effective treatment option for patients with colorectal cancer.
Introduction
Colorectal cancer is one of the most common cancers worldwide. In 2020, colorectal cancer accounted for 10% of cancer incidence and 9% of cancer-related deaths globally (1). In the United States, colorectal cancer is the second most common cause of cancer-related death, with a 5-year survival rate of 14% for those diagnosed with distant-stage colorectal cancer (2). Once standard chemotherapy regimens have been exhausted, patient survival is less than 6 months. Thus, colorectal cancer remains a high unmet medical need and requires the development of more effective and safer therapeutic interventions to improve this poor survival outcome.
Over the past decade, immune checkpoint inhibitors (ICIs) have revolutionized the field of oncology with proven clinical efficacy in multiple cancers, including a subset of colorectal cancer. ICIs have shown remarkable efficacy in patients with mismatch-repair–deficient (dMMR) or microsatellite instability–high (MSI-H) colorectal cancer, which led to the approvals of three ICI regimens for the treatment of MSI-H metastatic colorectal cancer (mCRC): Pembrolizumab, and nivolumab as monotherapy or in combination with the CTL-4 inhibitor ipilimumab (3–6). However, stage 4 dMMR–MSI-H tumors constitute only approximately 2% to 4% of all mCRC. Patients with microsatellite-stable (MSS) or microsatellite instability–low (MSI-L) colorectal cancer, who constitute the vast majority of patients with mCRC, have not benefited from ICI treatment. Differences in the tumor microenvironment between MSI-H and MSS/MSI-L colorectal cancer play a major role in the differential response to ICIs. The higher mutational burden and higher frequency of tumor-infiltrating lymphocytes, particularly CD8+ T cells, associated with MSI-H colorectal cancer make this subset more amenable to immune modulation. Meanwhile, the low tumor mutational burden and the lack of immune cell infiltration in the “immune-cold” MSS/MSI-L colorectal cancer make it challenging to show clinical efficacy of immunotherapy in these patients (7, 8).
A promising alternative approach to overcome the lack of pre-existing T-cell infiltration observed in MSS/MSI-L colorectal cancer is the use of T-cell–engaging bispecific antibodies to redirect effector T cells against tumor cells. These bispecific antibodies facilitate the formation of an immunological synapse between cancer cells and effector T cells, a prerequisite for target cell lysis by cytotoxic T cells. This is achieved by binding to tumor-associated antigen (TAA) on tumor cells and CD3 of the T-cell receptor (TCR) complex on T cells simultaneously, independently of TCR specificity, co-stimulation, or peptide antigen presentation (9). Clinical proof of concept for anti-TAA/CD3 bispecific antibodies has been shown in hematologic malignancies with the approval of blinatumomab, a bispecific T-cell engager (BiTE) targeting CD19 and CD3 for the treatment of relapsed/refractory B-cell acute lymphoid leukemia. However, the therapeutic potentials of T-cell engagers in solid tumors face a greater challenge. One of the obstacles is to identify TAAs that exhibit minimal expression in normal tissues, or TAAs that have a large differential in copy numbers between normal and tumor tissues to minimize normal tissue toxicity.
Here, we report the discovery of a T-cell–dependent bispecific antibody targeting lymphocyte antigen 6 family member G6D, LY6G6D-TDB, a novel TAA for colorectal cancer. LY6G6D belongs to a cluster of leukocyte antigens located in the MHC class III region on chromosome 6 and is a phosphatidylinositol (GPI)–anchored cell surface protein (10–12). We identified LY6G6D as a therapeutic target for colorectal cancer due to its differential expression in colorectal cancer and limited expression in normal tissues and other tumor types. In this report, we characterized LY6G6D expression in normal and tumor tissues using RNA-seq datasets and IHC, and evaluated in vitro and in vivo antitumor activity of LY6G6D-TDB, a full-length IgG1-bispecific antibody, in preclinical colorectal cancer models.
Materials and Methods
RNA-seq data analysis
Gene expression profile data analyzed in this report were obtained from the Cancer Genome Atlas (TCGA) at NCI Genomics Data Commons Data Portal (V15.0; GENCODE V22; duplicates removed), and the Genotype-Tissue Expression (GTEx) at dbGaP Accession phs000424.v6.p1. Processing and expression analysis of TCGA RNA-seq data were described previously (13).
IHC
Formalin-fixed paraffin-embedded (FFPE) tissue microarray blocks from normal human and cynomolgus monkeys were created from blocks of tissue procured from Discovery Life Sciences, Avaden Biosciences, and Capital Biosciences, with a minimum of two (range, 2–49) cases per tissue type. FFPE tissue blocks from human primary MSS colorectal cancer and liver metastases from patients with colorectal cancer were procured from the same vendors. Tissue type, tumor content, and tissue quality were confirmed by hematoxylin and eosin staining. IHC was performed on 4-μm-thick FFPE tissue sections mounted on glass slides. Primary IHC antibodies against LY6G6D were used in Ventana antibody diluent (90103, Ventana Medical Systems) and the staining was carried out on the Ventana Benchmark Ultra automated platform (Ventana Medical Systems). Sections were treated with ULTRA Cell Conditioning Solution-Ultra CC1 (Ventana Medical Systems) for 64 minutes. Specifically bound primary antibody was detected by incubating sections in OptiView DAB IHC Detection Kit (Ventana Medical Systems). The sections were counterstained with hematoxylin, dehydrated, and coverslipped. A minimum of 100 viable tumor cells was required for LY6G6D staining evaluation. Semiquantitative analysis of both the intensity of LY6G6D staining captured as either 0 (negative), 1 (low), 2 (moderate), 3 (high) or indeterminant and the percentage of positive tumor cells within each intensity category was performed by a pathologist in a blinded fashion. LY6G6D staining intensity scores incorporated both cytoplasmic and membrane staining given that staining in cytoplasmic and membrane fractions could not be meaningfully distinguished.
Antibody production
White New Zealand rabbits were immunized with human LY6G6D and single IgG+ huLY6G6D+ B cells were isolated and cultured as described previously (14). The B-cell culture supernatants were assayed by ELISA for binding to human LY6G6D. Variable regions (VH and VL) of each mAb from rabbit B cells were cloned into expression vectors as previously described (14). Individual recombinant rabbit antibodies were expressed in Expi293 cells and subsequently purified with protein A. Purified anti-LY6G6D antibodies were then subjected to functional activity assays and kinetic screening.
Mice were immunized with human LY6G6D in a similar manner and hybridomas were generated. The resulting hybridoma supernatants were assayed by ELISA, and positive samples were purified using protein A for subsequent functional and kinetic characterization.
Anti-LY6G6D and anti-CD3 antibodies were assembled as full-length IgG1 bispecific antibodies using the “knobs-into-holes” technology as previously described (15), with structural formulas in Supplementary Data. Antibodies were expressed either in Escherichia coli (E. coli) or in Chinese hamster ovary (CHO) cells. Antibodies expressed in E. coli were aglycosylated and thus effector-less. Antibodies expressed in CHO cells contain mutations (CH3 N297G) in the fragment crystallizable (Fc) regions to reduce binding to Fcγ receptors and, consequently, minimize Fc-effector function.
After production and purification, LY6G6D-TDB was characterized by mass spectrometry to show no residual half antibodies and <2% of homodimers, and size exclusion chromatography to show undetectable (<0.5%) amounts of aggregates. All preparations had a low endotoxin content (<0.5 EU/mg).
Epitope binning with Wasatch
An array-based SPR imaging system (Carterra) was used to epitope bin a panel of 96 mAbs as previously described (16). For epitope binning, human LY6G6D was first injected for 4 minutes at 50 nmol/L and was followed by a second 4 minutes injection of individual mAb at 10 μg/mL. The surface was regenerated with 10 mmol/L glycine pH1.5 between cycles. The experiment was performed at 25°C in HBS-T buffer. The epitope-binning data were processed using Wasatch binning software tool (Carterra).
Kinetic analyses for binding affinity of various LY6G6D and CD3 antibodies
The binding affinity of the antibodies was determined using the BIAcore T200 machine. For kinetics measurements, LY6G6D protein was coupled to BIAcore research grade CM5 chips according to the supplier's instructions to achieve approximately 100 response units in each flow cell. LY6G6D antibodies were expressed as chimera antigen-binding fragments (Fab) with rabbit or murine variable domain and human constant domain. Tenfold serial dilutions of Fabs were injected in HBS-P buffer at 37°C with a flow rate of 30 μL/min. Association rates (Ka) and dissociation rates (Kd) were calculated using a 1:1 Langmuir binding model (BIAcore T200 Evaluation Software version 2.0). The equilibrium dissociation constant (Kd) was calculated as the ratio Kd/Ka.
The binding affinity of CD3 antibodies was determined at 37°C following the same procedures as described above.
Cell lines
Colorectal cancer cell lines (LS1034, HT55, GP2D, and Colo320DM) were sourced from the Genentech cell line repository that was originally obtained from either the ATCC or ECACC. All cell lines were maintained in RPMI-1640 supplemented with 10% FBS (Sigma) and 2 mmol/L l-glutamine. Each cell line was authenticated as described before (17). All stocks were tested for Mycoplasma before and after cells are cryopreserved using Lonza Mycoalert and Stratagene Mycosensor to avoid false-positive/negative results. Cell lines were typically cultured for 2 months before thawing a new passage. HEK 293 cell line was purchased from Agilent Technologies. Transfection of huLY6G6D DNA into HEK 293 cells was performed using PolyFect Transfection Reagent (Qiagen) per the manufacturer's protocol.
Determination of LY6G6D binding sites
LY6G6D-binding sites on cell surface were determined using Quantum Simply Cellular anti-Human IgG antibody binding capacity kit (Bangs Laboratories).
In vitro target cell killing and T-cell activation
A total of 10 × 103 colorectal cancer cells per well were seeded in a 96-well plate and incubated at 37°C overnight for cell attachment. Human PBMCs were isolated from whole blood of healthy donors by Ficoll separation. CD3+ T cells were depleted using CD3 MicroBeads (Miltenyi Biotec). Fresh total PBMCs or CD3-depleted PBMCs were added to target cells at an effector and target cell ratio of 10:1 and incubated for 48-hours in the presence of the indicated TDB concentrations. At the end of incubation, the culture medium was transferred to a new 96-well plate to collect PBMCs for T-cell activation. Target cells were gently washed twice with PBS, and cell viability was measured using the CellTiter Glo reagent (Promega). PBMCs were stained with anti-CD8, CD4, CD69, and CD25 antibodies, and activated T cells were detected by CD69 and CD25 surface expression by flow cytometry. For intracellular granzyme B and Ki67 measurement, PBMCs were first stained with anti-CD8 and CD4 antibodies, then fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences) followed by staining with anti-granzyme B and Ki67 antibodies before flow cytometry. For cytokine release in co-culture of PBMC and target cell or culture of PBMC, culture medium was collected and centrifuged after 24-hour exposure to TDB. Cytokines in the supernatant were measured using Bio-Rad Pro Human multiplex immunoassay kit. All antibodies were purchased from BD Biosciences.
Expression of recombinant human LY6G6D
A construct encoding for human LY6G6D (Met1-Ser104 with native signal sequence Met1-Gly19) was synthesized (Genescript) and codon optimized for mammalian expression; the DNA was cloned into a proprietary target integrated stable vector containing an ampicillin resistant marker, a CMV promoter and a SV40 polyadenylation signal downstream of the gene.
Construct was expressed as a fusion protein with a human IgG1 fragment crystallizable (IgG1 Fc) affinity tag located at its C terminus along with a TEV protease cleavage site located between the protein and the Fc tag. Protein was expressed by a stable CHO pool using proprietary media in 14-day fed-batch process.
Sequence of the untagged construct used for structural studies is shown below:
NRMRCYNCGGSPSSSCKEAVTTCGEGRPQPGLEQIKLPGNPPVTLIHQHPACVAAHHCNQVETESVGDVTYPAHRDCYLGDLCNSGENLYFQ
Purification of LY6G6D
Human Fc-tagged LY6G6D (Met1-Ser104) extracellular domain (ECD) was purified over a protein A column (Thermo Fisher Scientific). The column was washed with 25 mmol/L Tris pH 8, 150 mmol/L NaCl (buffer A) and eluted with 0.3 mol/L acetic acid pH 3. Elution fractions were immediately buffered back to pH 7.5 with 1 mol/L Tris pH 7.5 (4 mL of Tris per 10 mL of elution). Elution fractions were further purified over a Superdex 200 16/60 gel filtration column (GE Healthcare) pre-equilibrated with buffer A. Fractions containing protein were pooled, incubated with 6xHis-tagged TEV protease (prepared in-house) and mutated overnight at 4°C. Untagged material was loaded onto a Ni-NTA column (Qiagen); flow through was collected and further purified over the protein A column. Flow through fraction was polished over a Superdex 75 16/60 column (GE Healthcare) pre-equilibrated with buffer A. Molecular weight of the protein was assessed by LC-MS and results suggest proteolytic cleavage of the first four N-terminal residues Asn20-Arg23.
Expression and purification of recombinant Fab
Constructs suitable for periplasmic expression of Fab in E. coli and containing sequence coding for Fab fragment of 1G4 were cloned; DNA was transformed into 34B8 E. coli cells and expressed at 30°C under control of the phoA promoter in CRAP phosphate-limiting autoinduction medium (18) supplemented with carbenicillin (50 μg/mL). After 24 hours, cells were harvested and resuspended in PBS supplemented with one complete EDTA-free Protease Inhibitor Cocktail tablet (Roche) per 50 mL of lysis buffer, lysozyme (0.1 mg/mL), and benzonase (0.01 mg/mL). The prepared suspension was microfluidized at 15,000 psi and clarified at 50,000 × g for 30 minutes at 4°C. The supernatant was then resolved on protein G Sepharose beads equilibrated with PBS, using 2-mL packed resin volume per original gram of cell paste. The column was washed extensively with PBS, and Fabs were eluted under mildly acidic conditions (0.56% glacial acetic acid pH 3.6). Eluted Fabs were immediately dialyzed overnight at 4°C against buffer containing 500 mmol/L NaCl, 10% glycerol, and 100 mmol/L Tris (pH 8.0). Fabs were further resolved on a Superdex 75 16/60 column (GE Healthcare) using PBS (pH 7.2) as the running buffer.
Structure determination for crystallization studies
1G4 Fab was incubated with a twofold molar excess of untagged Ly6G6D on ice for 30 minutes. The complex was passed over a Superdex 75 16/60 column (GE Healthcare) equilibrated in buffer A and peak fractions were collected and concentrated. Crystals appeared after 2 months using hanging drop vapor diffusion method at 20°C with 90 μL of reservoir solution (0.1 mol/L HEPES pH 7, 15% PEG 4000) and 0.1-μL drops containing equal volumes of protein (10 mg/mL) and reservoir. Crystals were cryoprotected with 25% glycerol added to the mother liquor, and a 2.2 Å resolution X-ray diffraction dataset was collected at the 12-2 beamline of the Stanford Synchrotron Radiation Lightsource. Crystallographic data were processed using XDS (19). The structure was solved by molecular replacement (four complexes per asymmetric unit) using PHASER (20), with the Fab fragment from the structure of trastuzumab (PDB code: 1FVE; ref. 21) used as search model. A large Fo − Fc positive difference density peak located in the vicinity of the CDRs was observed; a model consisting of residues Asp95-Cys96-Tyr97-Leu98-Gly99-Asp100-Leu101-Cys102-Asn103 from Ly6G6D sequence was built into the density using COOT (22) and refined in PHENIX (23). Electron density for residues Cys24-Arg94 was missing suggesting that this region might be disordered or that proteolysis had occurred during the crystallization experiment allowing only the region Asp95-Asn103 to crystallize. Pymol (24) was used to render the molecular graphics. Most of additional peaks were accounted for and refined; however, a few larger densities could not be fitted with buffer components and were assigned as multiple water molecules. The structure has 96% of its residues in the favored region of the Ramachandran plot; density around the four “outliers” residues was verified confirming that these residues adopt high energy conformations. Data collection and refinement statistics are summarized in Supplementary Table S1 (PDB code 7S4G).
SEC-MALS analysis
Samples were run on a Phenomenex Yarra SEC-X150 (1.8 μm, 150 × 4.6 mm) gel filtration column equilibrated in PBS in line with a Dawn HELEOS II (Wyatt Technologies) light-scattering detector connected to a Wyatt OptiLab T-rEX refractive index detector. Wyatt Technologies software (ASTRA) was used to determine the corresponding peaks' molecular weight based on the refractive index.
Glycosylation-engineered epitope mapping with Wasatch
The Wasatch array-based SPR imaging system (Carterra) was used to epitope map-purified antibodies glycosylation-engineered LY6G6D antigen (LY6G6D ECD with single glycosylation site mutations fused to human IgG1 Fc; Supplementary Table S2). Antigen was diluted at 10 μg/mL in 10 mmol/L sodium acetate buffer pH 4.5 and directly immobilized by amino coupling onto a CMD 200M chip (XanTec Bioanalytics) using a Continuous Flow Microspotter to create an array of glycosylation variants. For analysis, the IBIS MX96 SPRi was used to evaluate antibodies binding to the immobilized antigens. For epitope-mapping anti-LY6G6D antibodies were injected for 3 minutes at a single concentration of 100 nmol/L, followed by a dissociation period of 10 minutes. The surface was regenerated with 10 mmol/L glycine pH2.1 between cycles. The experiment was performed at 25°C in a running buffer of HBS-EP buffer (0.01 mol/L HEPES pH 7.4, 0.15 mol/L NaCl, 3 mmol/L EDTA, 0.005% surfactant P20). The binding data were processed using Carterra Kinetics Data Analysis software.
Mouse xenograft tumor models
All animal studies were carried out in compliance with NIH guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee (IACUC) at Genentech, Inc.
Seven- to 8-week-old NSG mice (The Jackson Laboratory) were inoculated with 5 million LS1034 or HT55 tumor cells in Hank's Balanced Salt Solution subcutaneously in a volume of 0.1 mL per mouse in the right unilateral-thoracic flank, followed by intraperitoneal injection of 10 million human PBMCs in sterile PBS either 1 day (HT55 model) or 8 days (LS1034 model) after cell inoculation. When tumors reached a mean volume of 100 to 200 mm3 (LS1034) or 150–250 mm3 (HT55), mice with similarly sized tumors were randomized into treatment groups (assigned as day 0). Tumor size and body weight were measured once or twice per week during the study.
Data availability
The data generated in this report are available within the article and its Supplementary Data Files. Coordinates and structure factor amplitudes of crystal structure were deposited in the PDB as entry 7S4G.
Data sharing
The data generated in this study are available upon request from the corresponding author.
Results
LY6G6D is differentially expressed in colorectal cancer
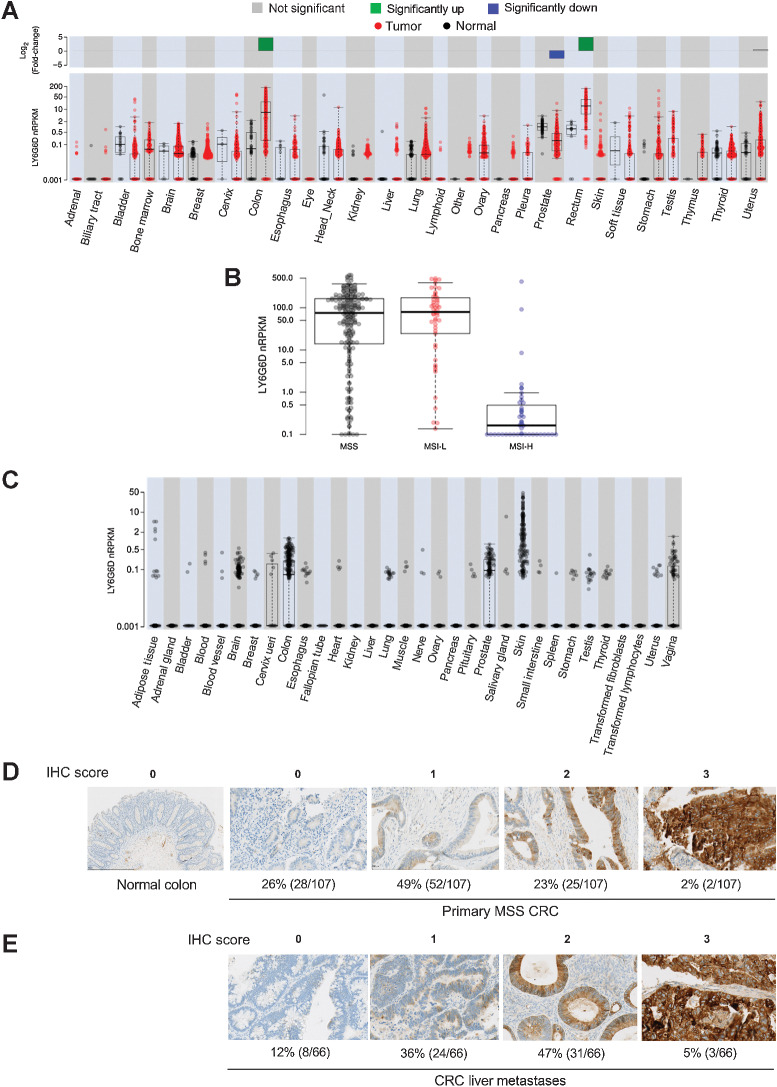
We performed differential gene expression analysis for LY6G6D using TCGA RNA-seq data that consisted of >11,000 human tissue samples across 20 normal tissue types and 28 tumor types. There was significant LY6G6D upregulation in colorectal tumors compared with normal colorectal tissues with log fold change = 6.8, P < 0.001 (n = 625 and 51 in sample sizes, respectively; Fig. 1A). A high prevalence of LY6G6D expression was observed in MSS and MSI-L colorectal cancer, whereas its expression in MSI-H colorectal cancer was low (Fig. 1B). Normal tissues and non-colorectal cancer tumors displayed minimal or low LY6G6D expression. Although normal prostate had the highest copy number of LY6G6D among normal tissues, the level was significantly lower than that in colorectal tumors, median nRPKM = 0.898 and 8.431, respectively. To further validate limited gene expression of LY6G6D in normal tissues, we analyzed normal tissue samples in the GTEx data that derived from 9,120 samples representing 32 normal tissue types from 549 donors (Fig. 1C). In GTEx cohort, all of the normal tissue types except colon and prostate had median nRPKM = 0.001 (Supplementary Table S3). Although a small percentage of samples in brain and prostate had relatively high copy numbers, all of the brain and prostate samples had nPRKM values <1. The skin samples displayed a wide range of nRPKM values, and around 9% of the samples (considered outliers) had nRPKM values >2. The median nPRKM values for brain (n = 1,331), colon (n = 367), prostate (n = 117), and skin (n = 639) were 0.001, 0.061, 0.085, and 0.001, respectively.
Figure 1.
LY6G6D is differentially overexpressed in colorectal cancer. A,LY6G6D differential expression in samples in TCGA RNA-seq dataset. Gene expression is displayed in normalized Reads Per Kilobase of exon model per Million mapped reads (nRPKM). B,LY6G6D expression in MSS/MSI-L/MSI-H colorectal cancer samples in TCGA dataset. MSS: n = 108; MSI-L: n = 36; MSI-H: n = 35. C,LY6G6D expression in human normal tissue samples in the GTEx RNA-seq dataset. A–C, Boxes represent the interquartile range (IQR, 25th to 75th percentile), the horizontal line is the median. Whiskers extend to the most extreme data point that is within 1.5*IQR of the 25th and 75th percentiles. Data points beyond the whiskers represent outliers. D, Representative images of LY6G6D IHC staining in normal colon and primary MSS colorectal cancer samples (n = 107). E, Representative images of LY6G6D IHC staining in colorectal cancer liver metastases (n = 66). Numbers of positive samples under each IHC score are derived at tumor cell expression cutoff of 50%, which requires that at least 50% of the tumor cells stained positive for LY6G6D at the denoted levels. IHC score: 0 = negative; 1+ = low signal; 2+ = moderate signal; 3+ = strong signal.
We conducted semiquantitative evaluation of LY6G6D protein expression in normal human tissues as well as primary and metastatic colorectal cancer tumors using an IHC-reactive rabbit mAb against human LY6G6D, with a minimum of two (range, 2–49) cases per normal tissue type. Screening of normal human tissues showed that all cases were negative (score = 0) for LY6G6D staining in 23 of 37 tissue types. Weak staining (1+) was present in scattered cells in five of 10 cases of lymph node, and moderate staining (2+) was present in three of three cases of tonsil. Equivocal staining was present in 13 tissue types (Supplementary Table S4A). Analysis of 36 normal cynomolgus monkey tissue types yielded similar findings, with positive staining (IHC 1+) detected only in some spleen and lymph node tissues (Supplementary Table S4B). In addition, we assessed LY6G6D expression in human immune cell subpopulations, including CD4+ and CD8+ T cells, natural killer cells, and monocytes from 12 healthy donors by flow cytometry. LY6G6D was primarily negative in these immune cells (Supplementary Fig. S1A–S1D; Supplementary Table S5). Screening of a total of 107 primary MSS colorectal cancer samples showed that the overall prevalence of LY6G6D expression was 74% (IHC 1+/2+/3+), with moderate to strong LY6G6D expression (IHC 2+/3+) in 25% of the cases (Fig. 1D), when setting a tumor cell expression cutoff value of 50%. The overall prevalence of LY6G6D expression in 66 colorectal cancer liver metastases was 88%, with moderate to strong LY6G6D expression in 52% of the cases (Fig. 1E). LY6G6D prevalence in primary MSS colorectal cancer and liver metastases when setting tumor cell expression cutoff value of 1%, 25%, 50%, and 75% is listed in Supplementary Table S6.
Discovery of LY6G6D-TDB
LY6G6D-TDB is a full-length IgG1-based bispecific antibody that binds to human CD3ε on T cells with one arm and to human LY6G6D antigen with the other arm. We characterized a panel of 96 anti-LY6G6D mAbs, including rabbit and murine clones, for binding affinity and epitope binning. The mAbs were sorted into 4 different epitope bins (bin1–bin4). To test the effect of affinity to CD3 on LY6G6D-TDB activity, we evaluated two anti-CD3 antibody clones as candidates to pair with LY6G6D arm: clone with high affinity to CD3 (CD3H, KD = 14.4 ± 0.42 nmol/L), and clone with low affinity to CD3 (CD3L, KD = 446 ± 46 nmol/L). The KD values were higher than the values we reported previously (25), due to BIAcore measurements taken at 37°C instead of room temperature.
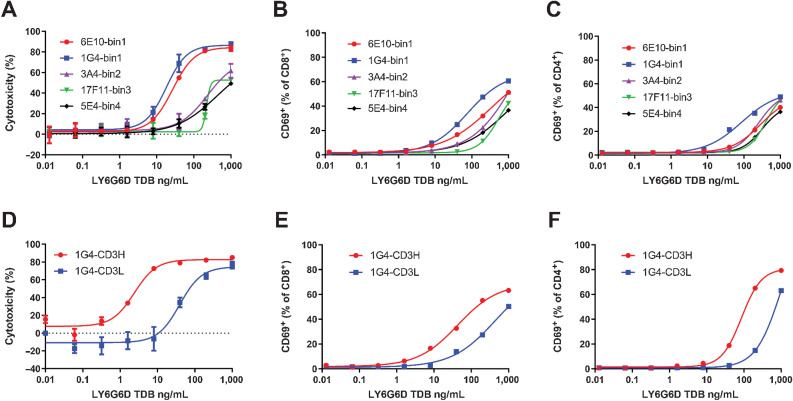
We compared TDBs of five anti-LY6G6D mAbs from different epitope bins that had similar binding affinity for human LY6G6D, KD ranging from 0.32 to 2.71 nmol/L, for the potency of target cell lysis. Murine clone 1G4 and rabbit clone 6E10 from bin1, rabbit clones 3A4 from bin2, 17F11 from bin3, and 5E4 from bin4 were assembled into TDBs with the low-affinity CD3 arm. TDBs of 1G4 and 6E10 differentiated themselves from TDBs harboring clones from other bins with the highest potency in lysis of LY6G6D-expressing colorectal cancer cell line HT55 (Fig. 2A). A slightly more robust T-cell activation was observed with 1G4-TDB (Fig. 2B and C). Despite similar binding affinity to LY6G6D and the same CD3 arm, the difference in potency of target cell lysis by the TDBs indicates that epitope can have a significant impact on LY6G6D-TDB potency.
Figure 2.
Selection of LY6G6D and CD3 arms for the optimized LY6G6D-TDB. A, Dose–response analysis of HT55 cell killing mediated by TDBs of anti-LY6G6D clones from different epitope bins paired with the low-affinity CD3 arm. Cell killing was measured by CellTiter-Glo reagent at 48-hour time point. B and C, CD8+ and CD4+ T-cell activation induced by TDBs of anti-LY6G6D clones from different epitope bins. T-cell activation was measured at 48-hour time point by flow cytometry. D, Killing of HT55 cells by 1G4-TDB paired with high- or low-affinity CD3 arm. 48-hour time point. EC50 values were 3.31 ± 1.91 ng/mL and 86.00 ± 60.61 ng/mL from 4 PBMC donors, respectively. E and F, CD8+ and CD4+ T-cell activation induced by 1G4-TDB paired with high- or low-affinity CD3 arm. A–C, The representative data of 3 PBMC donors. D–F, The representative data of 4 PBMC donors. Cell killing data are shown as means ± SD of triplicate wells.
We focused our optimization effort on the selection of a more effective CD3 arm by comparing the potency of 1G4/CD3L and 1G4/CD3H TDBs. In vitro cytotoxicity assay showed that high affinity to CD3 increased the potency by 26-fold in target cell killing and fourfold in T-cell activation (Fig. 2D–F), supporting the selection of the high-affinity CD3 arm for better LY6G6D-TDB activity.
Crystal structure analysis reveals membrane-proximal epitope bound by potent LY6G6D-TDB
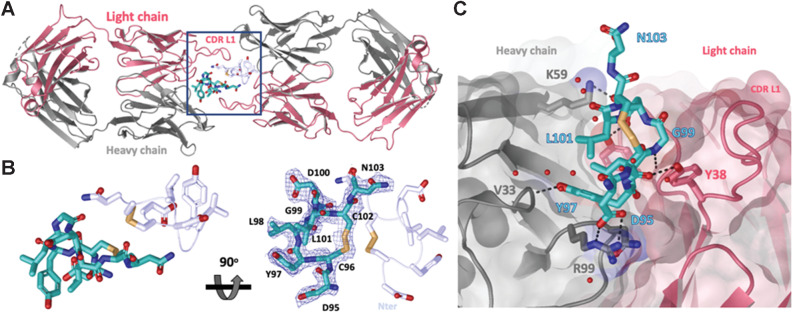
To further characterize the epitope recognized by clone 1G4, we solved a 2.2 Å X-ray structure of the complex between the recombinant ECD of LY6G6D and the Fab of 1G4 (Supplementary Table S1). A molecular replacement solution was obtained for 1G4 and a model for an 8-mer peptide consisting of residues Asp95-Asn103 was built into the positive density (Fig. 3A and B). The unstructured peptide is cyclized through a disulfide bond between Cys96 and Cys102. The LY6G6D-1G4 Fab interface reveals a network of hydrogen bonds and salt bridge interactions that stabilize the complex (Fig. 3C). Key interactions mediated by the 1G4 heavy chain include salt bridges between the carboxy group of Asp95 and the guanidinium moiety of HC.Arg99 and between the carboxy group of Asp100 and amine group of HC.Lys59 side chain along with a hydrogen bond pair between the hydroxyl group of Tyr97 and the backbone amide of HC.Val33. The light chain CDRs engage in the following hydrogen bond pairs: The carboxy group of Asp100 side chain with the hydroxyl group of LC.Tyr102, backbone carbonyl of Cys96 with the hydroxyl side chain of LC.Tyr38 and the backbone amide of Gly99 interacts with the backbone carbonyl of LC.Ser98. These contacts bury a total solvent-exposed surface area of approximately 500 Å2, with a similar contribution from both chains (257 Å2 from light chain versus 246 Å2 from the heavy chain) that is consistent with the average surface areas for peptide antigens (26).
Figure 3.
Crystal structure of LY6G6D–1G4 Fab complex at 2.2 Å resolution suggests Fab binds an epitope located close to the membrane. A, Ribbon diagram of one crystallographic dimer shown from a top view. The complex crystallized in space group P 1 21 1 with two dimers per asymmetric unit; the four complexes within the asymmetric unit superimpose with a root–mean–square deviation of less than 0.5 Å. The light and heavy chains from the Fab are colored in red and gray, respectively. The LY6G6D peptides (Asp95-Asn103) are shown in cyan and light blue and side chains are shown as sticks. Disulfides are shown in yellow (PDB accession pending). B, Close-up view of LY6G6D peptides within dimer (from highlighted box in A). mFo−DFc omit map for one LY6G6D peptide is shown in blue mesh contoured at 1σ. Both main and side chain atoms are shown; side chains density for most of the residues is clearly defined. For clarity, only the side chains of the symmetry-related peptide are shown in stick. Two views rotated by 90 degrees are depicted. C, Details of molecular interactions between the LY6G6D peptide (shown in sticks and colored in cyan) and its proximal Fab (surface representation, key CDR side chains involved in binding are shown in sticks). The peptide binds within a cleft between the light and heavy CDRs of the Fab. Water is represented as red spheres and contacts (hydrogen bonds and salt bridges) as black dotted lines.
Interestingly, the organization within the asymmetric unit arranges the LY6G6D complexes in pairs centered around a LY6G6D peptide dimer. In this dimer, we found that the long CDR L1 of the symmetry Fab extends across the dimer interface and potentially contributes an additional approximately 177 Å2 of buried surface area to the main complex (Supplementary Fig. S2). To assess whether this dimeric organization might be the result of a crystal packing artifact, we used size exclusion chromatography with multi-angle light scattering (SEC-MALS). The elution profile of recombinant LY6G6D reveals a single peak corresponding to a 9.6 kDa protein, suggesting that LY6G6D is also monomeric (Supplementary Fig. S3A). The complex eluted off the column as a single peak with an estimated molar mass of 56.2 kDa, suggesting a 1:1 stoichiometry (estimated molecular weight of the complex 57 kDa) and that the dimeric organization observed in the asymmetric unit is most likely a crystallographic artifact (Supplementary Fig. S3B).
Protein sequence analysis of human LY6G6D has shown that LY6G6D is a GPI-anchored cell surface protein of 133-amino acid in length, and Ser104 is predicted to be the GPI-anchoring site (10, 11). Our crystal structure revealed that the epitope recognized by 1G4 (residues Asp95-Asn103) immediately follows the GPI-anchoring sequence, suggesting that it is located proximal to the cell membrane. We also used a high-resolution, high-throughput glycosylation-engineered epitope mapping (GEM) method (27) to map the location of epitopes bound by mAbs from different bins. GEM results showed that the epitopes bound by 1G4 and 6E10 were the most membrane-proximal, meanwhile, the mAbs in the other bins bound to more membrane-distal epitopes (Supplementary Table S2).
LY6G6D-TDB is active against colorectal cancer cell lines with a broad range of target expression
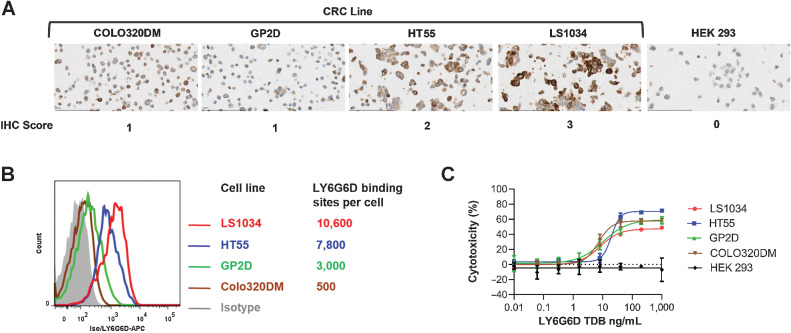
We evaluated the correlation between LY6G6D expression level and LY6G6D-TDB potency by selecting four colorectal cancer cell lines with surface LY6G6D ranging from 500 to 10,000 binding sites per cell. IHC was also used to determine LY6G6D expression in these cell lines, and the results of the two methods correlated well (Fig. 4A and B). The levels of LY6G6D in these cell lines correspond to those in colorectal cancer tumors having low to high levels of LY6G6D. Regardless of LY6G6D expression level, the four colorectal cancer cell lines showed comparable sensitivity to 1G4/CD3H–mediated cytotoxicity with EC50 values ranging from 7.11 to 19.49 ng/mL. No cell killing was observed in the LY6G6D-negative HEK 293 cells (Fig. 4C). These data suggest that target cell killing by LY6G6D-TDB is not strictly driven by the level of LY6G6D, and LY6G6D-TDB can mediate killing of colorectal cancer cell lines with a wide range of LY6G6D expression, including cell lines with low levels of the target.
Figure 4.
LY6G6D-TDB is active against colorectal cancer cell lines with a broad range of LY6G6D level. A, IHC staining on colorectal cancer cell pellets. B, LY6G6D-binding sites on cell surface determined by flow cytometry. C, Cell killing by 1G4/CD3H, 48-hour time point. Cell killing data are shown as means ±SD of triplicate wells. Representative data of assays with three PBMC donors.
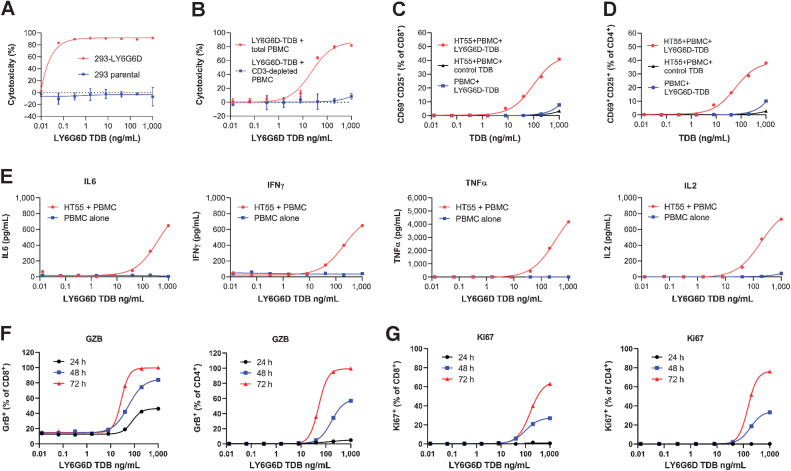
LY6G6D-TDB activity is target dependent
We conducted a series of in vitro experiments to assess the specificity of LY6G6D-TDB activity. First, there was a robust killing of an engineered LY6G6D-expressing 293 cell line when the cells were cultured with human PBMCs and 1G4/CD3H; meanwhile, no killing of the parental 293 cells was observed under the same condition (Fig. 5A). Although the engineered LY6G6D-expressing 293 cell line is not disease relevant as the level of LY6G6D is much higher than that in the endogenous colorectal cancer cell lines (>400,000 binding sites per cell, Supplementary Fig. S4A and S4B), the study shows that cell killing by LY6G6D-TDB requires the presence of LY6G6D target. Another experiment shows that the cytotoxicity of 1G4/CD3H is mediated by CD3+ T cells because depletion of CD3+ T cells from total PBMCs resulted in complete loss of target cell killing (Fig. 5B). Second, 1G4/CD3H induced T-cell activation when PBMCs were co-cultured with target cell HT55, but failed to activate T cells when PBMCs were cultured alone. Also, T cells were not activated by a control TDB comprised of CD3H paired with a non-binding arm (Fig. 5C and D). These data show that T-cell activation induced by LY6G6D-TDB is highly controlled and depends on the presence of LY6G6D-expressing target cells. Further evidence of target cell–dependent T-cell activation by LY6G6D-TDB came from the detection of several cytokines in the co-culture media of PBMC and HT55, whereas these cytokines were not detected in the culture media of PBMCs when exposed to 1G4/CD3H in the absence of HT55 cells (Fig. 5E).
Figure 5.
LY6G6D-TDB activity is target dependent. A, Killing of engineered LY6G6D-expressing 293 cells and parental cells. Forty-eight-hour time point. B, Killing of HT55 cells when co-cultured with either total PBMCs, or CD3+ T-cell–depleted PBMCs in the presence of 1G4/CD3H. 48-hour time point. C and D, CD8+ and CD4+ T-cell activation in co-culture of HT55 cells and PBMCs in the presence of either 1G4/CD3H, or a control TDB, or culture of PBMCs alone in the presence of 1G4/CD3H. 48-hour time point. E, Cytokine in culture media of co-culture of HT55 cells and PBMCs or culture media of PBMCs alone in the presence of 1G4/CD3H. 24-hour time point. F, Intracellular level of granzyme B in CD8+ T and CD4+ T cells in co-culture of PBMCs and HT55 in the presence of 1G4/CD3H at 24-, 48-, and 72-hour time points, measured by flow cytometry. G, Intracellular level of Ki67 in CD8+ T and CD4+ T cells in co-culture of PBMCs and HT55 in the presence of 1G4/CD3H at 24-, 48-, and 72-hour time points, measured by flow cytometry. Representative data of two to three donors.
We further assessed the kinetics of CD8+ and CD4+ T-cell activation in response to LY6G6D-TDB stimulation by measuring the levels of intracellular granzyme B and Ki67 in CD8+ and CD4+ T cells in co-culture of PBMC and HT55 cells during a 72-hour period by flow cytometry. The data showed that 1G4/CD3H induced upregulation of granzyme B and Ki67 in both CD8+ and CD4+ T cells in a dose-dependent and time-dependent manner (Fig. 5F and G). Although CD8+ and CD4+ T cells exhibited similar kinetics of Ki67 upregulation, there was delayed kinetics of granzyme B expression in CD4+ T cells compared with CD8+ T cells. At the 24-hour time point, approximately 46% of CD8+ T cells were positive for granzyme B, whereas only 5% of CD4+ T cells were positive at the highest TDB concentration. At 48-hour, approximately 84% of CD8+ T cells were positive compared with 57% in CD4+ T cells. Nevertheless, all CD8+ and CD4+ T cells were positive for granzyme B within 72 hours. This kinetic study shows that CD8+ T cells are the primary contributor to the T-cell effector function after LY6G6D-TDB treatment, and CD4+ T cells are able to contribute to target cell killing via granzyme B, albeit with a delay.
Antitumor efficacy of LY6G6D-TDB in vivo
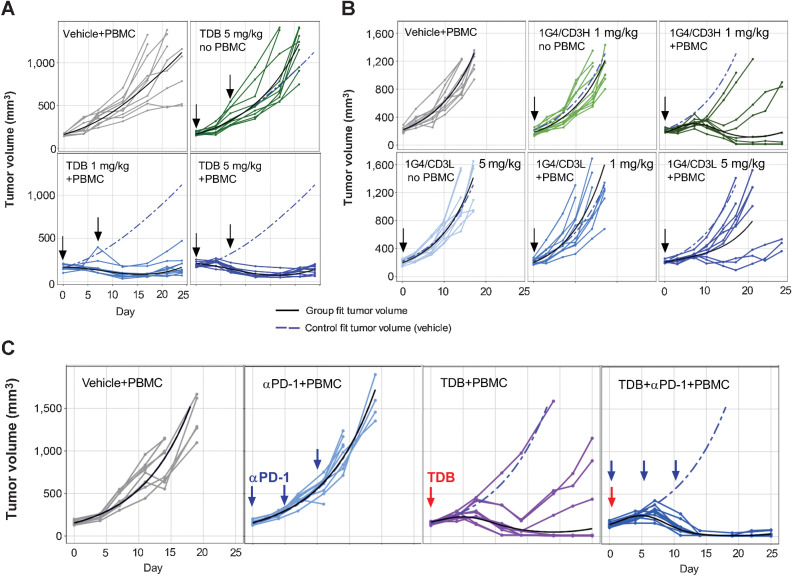
In vivo efficacy of LY6G6D-TDB in inhibiting colorectal cancer tumor growth was evaluated in established xenograft tumor models in NSG mice engrafted with human PBMCs. One model was LS1034 xenograft tumor model, an MSS colorectal cancer cell line (28) with strong LY6G6D-expression (IHC 3+) in xenograft tumors (Supplementary Fig. S5A). 1G4/CD3H demonstrated antitumor efficacy with a higher response rate in the 5 mg/kg dose group than that in the 1 mg/kg dose group (n = 10 per group), resulting in partial response (PR) in nine and five mice, or 90% and 50% of the treated mice, respectively. The control mice that received PBMCs and no TDB or TDB and no PBMCs exhibited uncontrolled tumor growth, indicating that the in vivo antitumor activity is mediated by human effector cells (Fig. 6A).
Figure 6.
LY6G6D-TDB is efficacious in inhibiting tumor growth in established xenograft tumor models in NSG mice. A,In vivo efficacy of 1G4/CD3H in an established LS1034 xenograft tumor model. After tumor establishment, mice received intravenous injections on days 0 and 7 of the following: (i) vehicle; (ii) 1G4/CD3H at 5 mg/kg; (iii) 1G4/CD3H at 1 mg/kg; (iv) 1G4/CD3H at 5 mg/kg. n = 10 per group B, Comparison of 1G4-TDBs with high- or low-affinity CD3 arm in established HT55 xenograft tumor model. After tumor establishment, mice received intravenous injection of either 1G4/CD3H at 1 mg/kg, or 1G4/CD3L at 1 mg/kg or 5 mg/kg on day 0. Control animals received either vehicle, or 1G4/CD3L at 5 mg/kg (without PBMCs), as labeled in the panel. n = 9 per group. C, Combination of 1G4/CD3H and anti–PD-1 antibody in established HT55 xenograft tumor model. After tumor establishment, mice received intravenous injection of the following: (i) vehicle; (ii) anti–PD-1 antibody at 10 mg/kg on days 0, 7, and 14; (iii) 1G4/CD3H at 1 mg/kg on day 0; (iv) co-administration of 1G4/CD3H at 1 mg/kg and anti–PD-1 antibody at 10 mg/kg on day 0, anti–PD-1 antibody on days 7 and 14. Some of the mice were euthanized before the end of the study due to tumor volumes ≥2,000 mm3 or losses in body weight ≥20% from their weight at the start of treatment per IACUC guidelines.
We evaluated the impact of affinity to CD3 on LY6G6D-TDB in vivo efficacy in HT55 xenograft tumor model, an MSS colorectal cancer cell line (28) with moderate LY6G6D-expression (IHC 2+) in xenograft tumors (Supplementary Fig. S5B). Tumor-bearing mice that received a single dose of 1G4/CD3H at 1 mg/kg showed better control of tumor growth than mice that received a single dose of 1G4/CD3L at 5 mg/kg, n = 9 per group, with PR in five mice (56% of the treated mice) versus PR in 1 mouse (11% of the treated mice) and tumor reduction (not reached PR) in 2 mice, respectively. 1G4/CD3L at 1 mg/kg failed to control tumor growth (Fig. 6B). The in vivo result was consistent with the in vitro finding, both facilitating the selection of the high-affinity CD3 arm for optimal antitumor activity of LY6G6D-TDB.
We next assessed the potential therapeutic benefit of combining LY6G6D-TDB with PD-1/PD-L1 blockade in the HT55 xenograft tumor model. An anti–PD-1 antibody was administered as a single agent and showed no effect on tumor growth at 10 mg/kg. In this study, two of the nine mice that received a single dose of 1G4/CD3H at 1 mg/kg had PR (22% of the treated mice), and four mice showed complete response (CR; 44% of the treated mice). Combination of 1G4/CD3H and anti–PD-1 antibody improved the antitumor activity of the TDB, resulting in tumor regression in all nine treated mice: seven PR (78% of the treated mice) and two CR (22% of the treated mice; Fig. 6C).
Pharmacokinetics (PK) of LY6G6D-TDB 1G4/CD3H were investigated in mice in two independent in vivo studies. In a single-dose PK study, the serum concentration-time profile exhibited a typical bi-exponential disposition. In a second study, serum exposure data in NSG mice in the context of the HT55 tumor xenograft efficacy model showed no apparent difference in exposure between the groups with or without PBMCs (Supplementary Fig. S6).
Discussion
Contrary to the successful development of bispecific T-cell engagers in treating hematological malignancies, efforts to bring this potent therapeutic modality to solid tumors have encountered many challenges. One key challenge is to identify “clean” TAAs that have minimal expression in healthy normal tissues, or TAAs that exhibit large differential expression between normal and tumor tissues. Many TAAs found in solid tumors can also be expressed by normal tissues, therefore, exposing patients to the risk of “on-target, off-tumor” toxicity.
In this report, we describe the discovery of a novel T-cell bispecific antibody targeting LY6G6D that can redirect T-cell effector function to colorectal tumors. Compared with other TAAs that have been selected for bispecific T-cell engagers for the treatment of colorectal cancer, such as P-cadherin and GUCY2C (29, 30), that can target a larger patient population due to higher and more abundant expression on colorectal cancer, LY6G6D exhibits the characteristics of a “clean” TAA with limited expression in normal tissues and elevated expression in colorectal cancer, therefore can potentially reduce the risk of “on-target, off-tumor” toxicity. Significant overexpression of LY6G6D in colorectal cancer, predominantly in MSS colorectal cancer, is corroborated by the works of Giordano and colleagues (31). Surface proteins with such large differential expression between normal and tumor tissues are rare, and thus far there are no reports of other therapeutics targeting LY6G6D.
One of the factors that can affect the functionality of a bispecific T-cell engager in T-cell redirection is the distance of the targeted epitope to the target cell membrane. A membrane-proximal epitope bound by anti-LY6G6D clone 1G4, a clone that demonstrated superior potency in the context of a TDB, is likely the determining factor in the higher potency of 1G4-TDB than TDBs of clones that bind to more membrane-distal epitopes. Our findings are consistent with the previous reports that show targeting a membrane-proximal epitope facilitates efficient synapse formation and is a crucial determinant for potent T-cell–mediated target cell lysis (32, 33). Unlike the previous reports that were based on targeting large proteins that may interfere with the formation of a tight synapse, therefore targeting an epitope close to the membrane could overcome this steric constraint, LY6G6D is quite small in size with 133 amino acids. Thus, targeting membrane-proximal epitope seems to be a generalized way to improve the potency of T-cell–engaging bispecific antibodies, regardless of the size of the target.
Optimal affinity to CD3 may vary for each unique T-cell bispecific antibody due to different properties of TAAs. In the development of a TDB targeting the acute myelogenous leukemia tumor antigen CLL-1 (CLEC12A), the anti–CLL-1/CD3 TDB with a low-affinity CD3 arm was found to be more efficacious and tolerable in vivo, contrary to the prediction from the in vitro studies (25). In the case of anti-HER2/CD3 TDB, T-cell–binding affinity had only limited impact on in vitro and in vivo antitumor activity (34). LY6G6D-TDB with a high-affinity CD3 arm showed increased antitumor activity both in vitro and in vivo. Because of the limited target expression in normal tissues, selection of a high-affinity CD3 arm may not pose serious safety concerns in the clinical setting for LY6G6D-TDB.
Our data on the correlation between LY6G6D expression level and LY6G6D-TDB potency add to the divergent results on correlation of the TAA level and the in vitro potency of IgG-like TDBs (25, 35, 36). Ellerman reviewed published data on the correlation between copy number of the TAA and the in vitro potency of different antibody formats (37). The data showed that the minimum copy number required for inducing an effective cell killing response varied among individual T-cell engagers, and the lack of correlation between antigen expression and potency could be indicative of a very potent molecule that only requires minimal target expression for supporting an effective cell killing.
Multiple strategies, including the use of T-cell–engaging bispecific antibodies, are currently under investigation to render “immune-cold” MSS/MSI-L colorectal cancers “immune-competent” and amenable for immunotherapy (8, 38). Encouraging clinical data have been reported in the ongoing phase Ia and Ib studies with the carcinoembryonic antigen (CEA) T-cell bispecific antibody (CEA-TCB) in advanced patients with colorectal cancer (39). The antitumor efficacy demonstrated by LY6G6D-TDB against the established colorectal cancer xenograft tumors suggests that LY6G6D-TDB can redirect immune effector cells into the vicinity of colorectal tumor cells and induce an antitumor response. The data presented in this report provide evidence that a novel T-cell–engaging bispecific antibody targeting LY6G6D may potentially provide clinical benefit to patients with colorectal cancer.
Supplementary Material
Acknowledgments
This work was supported by Genentech, Inc. We are grateful to the BioMolecular Resources and Research Material Groups for their support. We acknowledge use of synchrotron X-ray sources at the Stanford Synchrotron Radiation Lightsource supported by the Department of Energy’s Office of Science under contract DE-AC02-76SF00515. We would also like to thank Tiffany Wong and James Kiefer for their help and feedback. We thank Andres Paler Martinez for cytokine measurement. We thank Altaf Kassam for the help on the RNA-seq data analysis.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
C.P.C. Chiu reports patent 20210179715 pending and reports employment at Genentech. M. Dillon reports a patent for ARP200103463 pending, GC2020/41120 pending, JP2020-206866 pending, TW109143860 pending, US17/119753 pending, and WOPCT/US2020/064635 pending; reports employment at Genentech, Inc., is a member of the Roche Group, and may hold stock options. A.A. Lo reports employment and is a stockholder of Genentech/Roche. M. Das Thakur reports grants, personal fees, and other support from Roche during the conduct of the study. K.M. Zimmerman Savill reports personal fees from Genentech, Inc. during the conduct of the study and personal fees from Cardinal Health outside the submitted work. R. Piskol reports other support from Genentech, Inc. outside the submitted work. D. Ellerman reports a patent for US10323094B2 pending and reports authored articles before on this subject. C. Spiess reports a patent for AR P200103463 pending, GC 2020/41120 pending, JP 2020-206866 pending, TW 109143860 pending, US 17/119753 pending, and WO PCT/US2020/064635 pending; reports employment at Genentech, Inc.; is a member of the Roche Group; and may hold stock and options. No disclosures were reported by the other authors.
Authors' Contributions
P. Wang: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L.L. Sun: Conceptualization, formal analysis, supervision, validation, methodology, writing–review and editing. R. Clark: Formal analysis, validation, investigation, methodology. M. Hristopoulos: Formal analysis, validation, investigation, visualization, methodology, writing–review and editing. C.P.C. Chiu: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. M. Dillon: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. W.Y. Lin: Formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A.A. Lo: Conceptualization, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Chalasani: Investigation. M. Das Thakur: Conceptualization, formal analysis, supervision, writing–review and editing. K.M. Zimmerman Savill: Project administration. L. Rougé: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. P. Lupardus: Conceptualization, formal analysis, supervision, validation, methodology, writing–review and editing. R. Piskol: Formal analysis, investigation, writing–original draft, writing–review and editing. B. Husain: Resources. D. Ellerman: Resources, writing–original draft, writing–review and editing. V. Shivva: Conceptualization, formal analysis, supervision, visualization, methodology, writing–original draft. S.R. Leong: Resources, writing–original draft, writing–review and editing. M. Ovacik: Formal analysis, supervision, methodology. K. Totpal: Conceptualization, supervision, validation, methodology, writing–review and editing. Y. Wu: Conceptualization, supervision, methodology. C. Spiess: Conceptualization, supervision, methodology, writing–review and editing. G. Lee: Resources, project administration. D.D. Leipold: Project administration. A.G. Polson: Conceptualization, supervision, writing–original draft, writing–review and editing.
References
- 1. GLOBOCAN database, 2020: Available from:http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx#.
- 2. Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–64. [DOI] [PubMed] [Google Scholar]
- 3. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Overman MJ, Mcdermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Overman MJ, Lonardi S, Wong KYM, Lenz H-J, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair–deficient/microsatellite instability–high metastatic colorectal cancer. J Clin Oncol 2018;36:773–9. [DOI] [PubMed] [Google Scholar]
- 7. Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges, and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hermel D, Sigal D. The emerging role of checkpoint inhibition in microsatellite stable colorectal cancer. J Pers Med 2019;9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baeuerle PA, Reinhardt C. Bispecific T-cell–engaging antibodies for cancer therapy. Cancer Res 2009;69:4941–4. [DOI] [PubMed] [Google Scholar]
- 10. Mallya M, Campbell R, Aguado B. Transcriptional analysis of a novel cluster of LY-6 family members in the human and mouse major histocompatibility complex: five genes with many splice forms. Genomics 2002;80:113–23. [DOI] [PubMed] [Google Scholar]
- 11. Mallya M, Campbell RD, Aguado B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci 2006;15:2244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loughner CL, Bruford EA, Mcandrews MS, Delp EE, Swamynathan S, Swamynathan SK. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum Genomics 2016;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhakta S, Crocker LM, Chen Y, Hazen M, Schutten MM, Li D, et al. An anti-GDNF family receptor alpha 1 (GFRA1) antibody–drug conjugate for the treatment of hormone receptor–positive breast cancer. Mol Cancer Ther 2017;17:638–49. [DOI] [PubMed] [Google Scholar]
- 14. Lin WY, Liang W-C, Nguy T, Maia M, Tyagi T, Chiu C, et al. Rapid identification of anti-idiotypic mAbs with high affinity and diverse epitopes by rabbit single B-cell sorting-culture and cloning technology. PLoS ONE 2020;15:e0244158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atwell S, Ridgway JB, Wells JA, Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol 1997;270:26–35. [DOI] [PubMed] [Google Scholar]
- 16. Mai E, Chan J, Goon L, Ego BK, Bevers J, Wong T, et al. Development of an ultra-sensitive human IL-33 biomarker assay for age-related macular degeneration and asthma drug development. J Transl Med 2021;19:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu S-F, Lee DW, Zheng B, del Rosario G, Leipold D, Booler H, et al. An anti–CD22-seco-CBI-dimer antibody–drug conjugate (ADC) for the treatment of Non-Hodgkin lymphoma that provides a longer duration of response than auristatin-based adcs in preclinical models. Mol Cancer Ther 2020;20:340–6. [DOI] [PubMed] [Google Scholar]
- 18. Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, et al. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods 2002;263:133–47. [DOI] [PubMed] [Google Scholar]
- 19. Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007;40:658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eigenbrot C, Randal M, Presta L, Carter P, Kossiakoff AA. X-ray structures of the antigen-binding domains from three variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. J Mol Biol 1993;229:969–95. [DOI] [PubMed] [Google Scholar]
- 22. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- 23. Adams PD, Afonine PV, Bunakoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schrodinger LLC. The PyMOL Molecular Graphics System, Version 2.4.0. https://pymol.org/2/support.html?#citing.
- 25. Leong SR, Sukumaran S, Hristopoulos M, Totpal K, Stainton S, Lu E, et al. An anti-CD3/anti–CLL-1 bispecific antibody for the treatment of acute myeloid leukemia. Blood 2017;129:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramaraj T, Angel T, Dratz EA, Jesaitis AJ, Mumey B. Antigen–antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim Biophys Acta 2012;1824:520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lombana TN, Matsumoto ML, Iii JB, Berkley AM, Toy E, Cook R, et al. High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies. mAbs 2018;11:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Linnekamp JF, Hooff SRV, Prasetyanti PR, Kandimalla R, Buikhuisen JY, Fessler E, et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ 2018;25:616–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Root A, Cao W, Li B, LaPan P, Meade C, Sanford J, et al. Development of PF-06671008, a highly POTENT anti–P-cadherin/anti-CD3 bispecific DART molecule with EXTENDED half-life for the treatment of cancer. Antibodies 2016;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathur D, Root AR, Bugaj-Gaweda B, Bisulco S, Tan X, Fang W, et al. A novel GUCY2C-CD3 T-cell–engaging bispecific construct (PF-07062119) for the treatment of gastrointestinal cancers. Clin Cancer Res 2020;26:2188–202. [DOI] [PubMed] [Google Scholar]
- 31. Giordano G, Parcesepe P, D'Andrea MR, Coppola L, Raimo TD, Remo A, et al. JAK/Stat5-mediated subtype-specific lymphocyte antigen 6 complex, locus G6D (LY6G6D) expression drives mismatch repair proficient colorectal cancer. J Exp Clin Cancer Res 2019;38:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bluemel C, Hausmann S, Fluhr P, Sriskandarajah M, Stallcup WB, Baeuerle PA, et al. Epitope distance to the target cell membrane and antigen size determine the potency of T-cell–mediated lysis by BiTE antibodies specific for a large melanoma surface antigen. Cancer Immunol Immunother 2010;59:1197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J, Stagg NJ, Johnston J, Harris MJ, Menzies SA, Dicara D, et al. Membrane-proximal epitope facilitates efficient T-cell synapse formation by anti-FcRH5/CD3 and is a requirement for myeloma cell killing. Cancer Cell 2017;31:383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Staflin K, Zuch de Zafra CL, Schutt LK, Clark V, Zhong F, Hristopoulos M, et al. Target arm affinities determine preclinical efficacy and safety of anti-her2/cd3 bispecific antibody. JCI Insight 2020;5:e133757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Junttila TT, Li J, Johnston J, Hristopoulos M, Clark R, Ellerman D, et al. Antitumor efficacy of a bispecific antibody that targets HER2 and activates T cells. Cancer Res 2014;74:5561–71. [DOI] [PubMed] [Google Scholar]
- 36. Sun LL, Ellerman D, Mathieu M, Hristopoulos M, Chen X, Li Y, et al. Anti-CD20/CD3 T-cell–dependent bispecific antibody for the treatment of B-cell malignancies. Sci Transl Med 2015;7:287ra70. [DOI] [PubMed] [Google Scholar]
- 37. Ellerman D. Bispecific T-cell engagers: towards understanding variables influencing the in vitro potency and tumor selectivity and their modulation to enhance their efficacy and safety. Methods 2019;154:102–17. [DOI] [PubMed] [Google Scholar]
- 38. Ciardiello D, Vitiello PP, Cardone C, Martini G, Troiani T, Martinelli E, et al. Immunotherapy of colorectal cancer: challenges for therapeutic efficacy. Cancer Treat Rev 2019;76:22–32. [DOI] [PubMed] [Google Scholar]
- 39. Tabernero J, Melero I, Ros W, Argiles G, Marabelle A, Rodriguez-Ruiz ME, et al. Phase IA and IB studies of the novel carcinoembryonic antigen (CEA) T-cell bispecific (CEA CD3 TCB) antibody as a single agent and in combination with atezolizumab: preliminary efficacy and safety in patients with metastatic colorectal cancer (mCRC). J Clin Oncol 2017;35:3002.28644773 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this report are available within the article and its Supplementary Data Files. Coordinates and structure factor amplitudes of crystal structure were deposited in the PDB as entry 7S4G.