Abstract
Even when polygenic risk scores (PRSs) are trained in African ancestral populations, Kamiza and colleagues showed that genetic and environmental variation within sub-Saharan African populations impacts prediction performance, highlighting the challenges of clinical implementation of PRSs for risk assessment.
Even when polygenic risk scores (PRSs) are trained in African ancestral populations, Kamiza and colleagues showed that genetic and environmental variation within sub-Saharan African populations impacts prediction performance, highlighting the challenges of clinical implementation of PRSs for risk assessment.
Main text
Common DNA variation across hundreds of thousands of individuals has been used in genome-wide association studies (GWASs) to identify thousands of associations between genetic variants and complex traits. From GWASs, we have learned that the genetic architectures for most common, complex diseases are polygenic, meaning that many DNA variants each additively play a small role in determining phenotype variability.1 Polygenic risk scores (PRSs) are the sum of GWASs effect sizes multiplied by an individual’s allele dosages. It may be possible to predict disease susceptibility with PRSs.2 However, most GWASs to date have been performed in European ancestries, and PRSs show reduced prediction performance when transferred to individuals of other ancestries.3 Therefore, clinical implementation of PRSs could widen health disparities between European and non-European populations.
When using genotypes to classify and cluster individuals, human geneticists commonly refer to groups by discrete continental ancestry groups, even though genetic ancestry is multi-dimensional and continuous. All individuals have multiple ancestries depending on the timescale; thus, as recently recommended, we prefer to use the plural “ancestries.”4 Continental ancestries do not accurately represent human genetic diversity, and referring to populations by continental ancestry in the singular risks supporting the misconception of race as a biological attribute.4 As such, the genetic ancestries of individuals within GWASs are an important factor when building and interpretating PRSs. Furthermore, distinct complex traits have different heritability estimates, which quantify how much of the trait variability is due to genetic differences. Therefore, PRS accuracy, transferability, and clinical applicability will depend on the phenotype of interest.5 To demonstrate these challenges, Kamiza and colleagues6 compared PRS accuracy and transferability in two sub-Saharan African populations using scores estimated from African American, European, or multiancestry (African American, European, and Hispanic) cohorts for four lipid traits: low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, and total cholesterol. According to their findings, PRS performance was greater in three out of four lipid traits in the South African Zulu cohort when using scores derived from African American GWAS summary statistics, which highlights how ancestral differences can affect PRS performance. However, the authors noted that African American-derived PRSs did not perform as well in the Ugandan cohort as they had the best performance in only one out of the four lipid traits tested. More importantly, prediction performance in the Ugandan cohort was lower than in South African Zulu with PRS models tested from all ancestries.
We do not expect performance of PRSs trained in African American populations to perform the same in South African Zulu and Uganda populations, given the distance between these populations in genotype principal component space.6 African American populations often comprise individuals with varying proportions of West African and European ancestries, while West African Yoruba and Mende populations cluster closer to the South African Zulu than the Ugandan population discussed in Kamiza et al.6 Given that African populations harbor the highest levels of genetic diversity among all populations in the world,7 it is not surprising that PRSs cannot be easily transferred between them. Indeed, variants’ effect sizes obtained from one African population might not accurately work in PRS assessment for other populations of the same geographic region, given the high levels of intra-continental genetic diversity in Africa.8
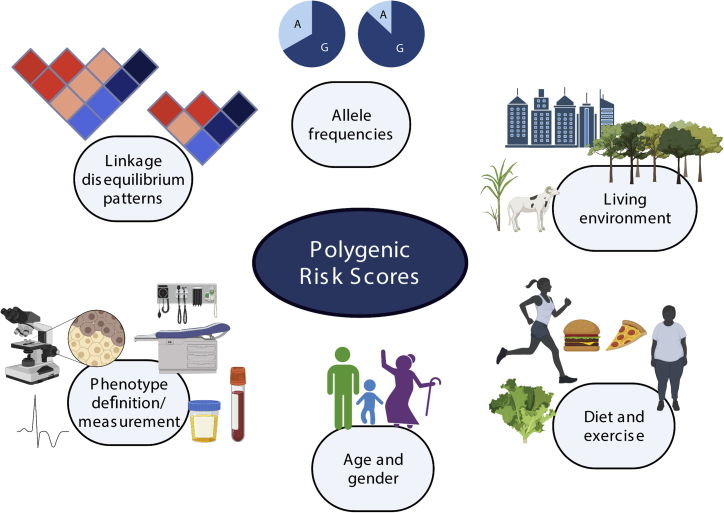
In addition to genetic factors, we must be mindful of other factors that can affect PRS transferability (Figure 1). Environmental factors and other characteristics, such as age and gender, have been shown to affect PRS prediction accuracy even within ancestral groups, where variation due to genetic factors such as linkage disequilibrium (LD) and allele frequency has been minimized.9 In their work, Kamiza et al.6 postulated that, in addition to allele frequency differences, genetic interactions with living conditions (urban versus rural environments), age, and BMI also limited the PRS transferability in sub-Saharan Africa populations.
Figure 1.
Factors that influence the transferability of polygenic risk scores (PRSs) between populations
Genetic factors, including differences in linkage disequilibrium patterns and minor allele frequencies and their interactions with environmental factors like urban versus rural living, differences in diet and exercise, and differences in age and gender may affect PRS transferability. Also, how the phenotype was measured may attenuate PRS transferability. Created with BioRender.com.
To realize the promise of precision medicine, how can we work toward a more inclusive and accurate applicability of PRSs? Given the current sampling bias in GWASs, it is imperative to increase diverse ancestral representation in future studies, which will not only contribute to potentially reducing health disparities, but also more immediately improve fine-mapping methods that identify which DNA variants are likely to cause complex traits.10 Because allele frequencies and LD patterns vary between populations that have been geographically and culturally separated, combining information across populations increases fine-mapping resolution through harnessing these differences in LD and increases power through larger sample size. Fine-mapping across ancestries will lead to better resolution when causal variants are shared across ancestries, which can only be true when variants are present in all ancestries examined.10 Since we know some genetic variation is unique to particular ancestries at appreciable frequency, a combination of cross-ancestral and ancestral-matched fine-mapping may be necessary to optimize PRS performance. Kamiza et al.6 elegantly demonstrate the challenges of implementing precision medicine equitably, and more studies like it, where the primary authors have primary appointments in Africa, should be funded to support this goal of diversifying GWASs.
Addressing transferability challenges and working toward overcoming them is the only way to ensure that PRSs can be properly implemented for clinical use without increasing health disparities. While evaluating disease risk through PRSs seems promising, especially for the most heritable traits, clinicians should also be aware of the wide range of genetic and environmental factors that can influence PRS performance. PRSs should not substitute for conventional clinical factors but rather could become another tool that clinicians use to evaluate disease risk.
Acknowledgments
This work is supported by the NIH National Human Genome Research Institute Academic Research Enhancement Award R15HG009569.
Declaration of interests
The authors declare no competing interests.
References
- 1.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis A.C.F., Molina S.J., Appelbaum P.S., Dauda B., Di Rienzo A., Fuentes A., Fullerton S.M., Garrison N.A., Ghosh N., Hammonds E.M., et al. Getting genetic ancestry right for science and society. Science. 2022;376:250–252. doi: 10.1126/science.abm7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis C.M., Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamiza A.B., Toure S.M., Vujkovic M., Machipisa T., Soremekun O.S., Kintu C., Corpas M., Pirie F., Young E., Gill D., et al. Transferability of genetic risk scores in African populations. Nat. Med. 2022;28:1163–1166. doi: 10.1038/s41591-022-01835-x. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez F., Hirbo J., Tishkoff S.A. Genetic variation and adaptation in Africa: Implications for human evolution and disease. Cold Spring Harbor Perspect. Biol. 2014;6:a008524. doi: 10.1101/cshperspect.a008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury A., Brandenburg J.-T., Chikowore T., Sengupta D., Boua P.R., Crowther N.J., Agongo G., Asiki G., Gómez-Olivé F.X., Kisiangani I., et al. Meta-analysis of sub-Saharan African studies provides insights into genetic architecture of lipid traits. Nat. Commun. 2022;13:2578. doi: 10.1038/s41467-022-30098-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mostafavi H., Harpak A., Agarwal I., Conley D., Pritchard J.K., Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. Elife. 2020;9 doi: 10.7554/eLife.48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W.J., Khera A.V., Okada Y., Furukawa Y., Morisaki T., Murakami Y., et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54:450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]