Abstract
Xeroderma pigmentosum (XP) is a DNA repair disease that predisposes to early skin cancers as cutaneous melanoma. Melanoma microenvironment contains inflammatory mediators, which would be interesting biomarkers for the prognosis or for the identification of novel therapeutic targets. We used a PCR array to evaluate the transcriptional pattern of 84 inflammatory genes in melanoma tumors obtained from XP patients (XP-Mel) and in sporadic melanoma (SP-Mel) compared to healthy skin. Commonly expressed inflammatory genes were further explored via GTEx and GEPIA databases. The differentially expressed inflammatory genes in XP were compared to their expression in skin exposed to UVs, and evaluated on the basis of the overall survival outcomes of patients with melanoma. Monocyte subsets of patients with SP-Mel, XP and healthy donors were also assessed. PCR array data revealed that 34 inflammatory genes were under-expressed in XP-Mel compared to SP-Mel. Differentially expressed genes that were common in XP-Mel and SP-Mel were correlated with the transcriptomic datasets from GEPIA and GTEx and highlighted the implication of KLK1 and IL8 in the tumorigenesis. We showed also that in XP-Mel tumors, there was an overexpression of KLK6 and KLK10 genes, which seems to be associated with a bad survival rate. As for the innate immunity, we observed a decrease of intermediate monocytes in patients with SP-Mel and in XP. We highlight an alteration in the immune response in XP patients. We identified candidate biomarkers involved in the tumorigenesis, and in the survival of patients with melanoma. Intermediate monocyte’s in patients at risk could be a prognostic biomarker for melanoma outcome.
Subject terms: Melanoma, Monocytes and macrophages, Nucleotide excision repair, Biomarkers
Introduction
Melanoma is among the most lethal forms of skin cancers. Its incidence in Tunisia is relatively low (0.5–0.7 per 100,000 inhabitants per year)1. The risk of developing melanoma is greater 1000-fold in Xeroderma Pigmentosum (XP), a rare genetic disorder with an incidence of 1/1,000,000 in Europe2. This risk of cancer is greater than that observed in the general population. Indeed, more than 67% of XP patients develop a cutaneous cancer at an average age of 8 years old3,4. Metastatic malignant melanoma and squamous cell carcinoma are the two most common causes of death in XP patients3–5. In detail, XP is an autosomal recessive disease caused by a defect in one out of the 8 genes; 7 involved in the nucleotide excision repair pathway (XPA to XPG) and Variant (XP-V), that it is caused by mutations in the POLH gene, which codes for the translesion DNA polymerase6. All these genes greatly predisposes affected individuals to melanoma and other skin tumors in sun-exposed areas4,7. This rare pathology is, therefore, considered as an excellent model for studying cancer development, given the fact that, even with optimal protection against UV radiation, XP patients develop internal cancers8.
Decades of cancer research, supported by novel screening technologies, have revealed that tumor progression is caused by many interconnected pathways. Most cancer studies focus on studying melanoma tumors at advanced stages. However, little is known about genetic diseases predisposing to skin cancers as for the XP disease, which constitute an interesting model to study early stages of cancer development and progression. Indeed, the skin is impacted by the accumulation of genetic defects linked to poor DNA repair following repeated exposure to UVs over the years. Transcriptomic analysis for candidate genes with limited biological sampling is considered as an interesting approach in rare genetic diseases9, and PCR array is an example.
The mechanisms involved in melanoma cancer predisposition and development in XP patients have been mainly linked to the DNA repair defects. While, in other DNA repair syndromes that predispose to cancer such as Fanconi anemia and Bloom syndrome, severe immune alterations have been described10,11. In XP patients, very limited data are available regarding immune landscape12.
Different biological parameters, such as epidermal hyperplasia, inflammation, and oxidative stress, are important factors that contribute to the development and/or progression of skin cancers13,14. In particular, chronic inflammation is thought to be responsible for around 25% of all tumor types15. Several inflammatory mediators can cause DNA damages when it’s chronical16. Skin cancer has important inflammatory components contributing to each stage17. Inflammation is a self-limiting process, during tumorigenesis. It has been shown that alterations in its pathways can play both pro and anti-tumor effects18. Recently, it has been suggested that exploring the expression of different inflammatory mediators could help discover relevant biomarkers to stratify patients with cutaneous melanoma for immunotherapy and targeted therapy19. Although significant progress has been made in the field of melanoma immunotherapies, the identification of valuable biomarkers for monitoring melanoma prognosis or staging are limited to a few markers such as S100B protein in the serum20 or PDL1 who varies according to tumor infiltrating lymphocytes rate21.
Furthermore, the innate immunity plays an important role in preserving skin integrity against different external aggressions such as pathogen infections22 and UVs radiations. It regulates the reparation of DNA damage by generating inflammatory “sensor” signals such as cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), the RIG-I-like receptors (RLRs) and Mitochondrial antiviral-signaling protein (MAVS) pathways, which are responsible for cytosolic DNA and RNA sensing23,24. Monocytes are innate immune cells, they have the role of protecting and defending the body against foreign substances, pathogens and tumor cells. In the last few years, our understanding of this population has shifted from a simple observation of a homogeneous group to a heterogeneous cluster of cells that exhibit diverse responses to different antigens. Thus, they can be classified into three subsets according to the expression of CD14 and CD16 surface markers: classical monocytes (CD14 + + /CD16-), intermediate monocytes (CD14 + + /CD16 +), and non-classical monocytes (CD14 + /CD16 + +)25. Several studies, reported that phenotypically similar monocyte sub-populations might play opposite roles in cancer development26. Monocyte's phenotyping in patients with melanoma was limited to two studies: the first one explored monocyte subsets in patients with melanoma skin cancer in untreated stage IV, while the second one assessed the effect of ipilimumab treatment on ex-vivo monocyte phenotypes27,28. Identifying monocyte subsets phenotypes in DNA repair disorders, as for XP patients who are prone to develop skin cancers at early ages, will be important in understanding the first stages of cancerogenesis.
The aims of this study are to investigate the differential expression of 84 candidate inflammatory genes in melanoma tumors (sporadic and from XP patients), and to identify the monocyte subsets, which would be associated with the early development of melanoma cancer.
Results
Transcriptional profile of inflammatory genes in SP-Mel and XP-Mel
Immunity landscape in cutaneous melanoma microenvironment was investigated using a pathway-focused PCR array including 84 inflammatory-related genes. Three sporadic melanoma (SP-Mel) and 3 melanoma biopsies obtained from XP patients (XP-Mel) were analyzed compared to 3 healthy skin. The relative gene expression was measured in the RNA extracted from those tumors. The selected genes encode for different inflammatory pathways related to leukotriene, complement, interferon pathways, etc.…
As a result, and due to the rarity of samples, fold change analysis p-value did only give a significant over-expression for KLK6 (fold change = 5; p-value = 0.03) in XP-Mel group. In order to further explore the disparity in these samples, we set a threshold for the fold change and considered genes whose expression was above the two-fold as a differentially over-expressed gene and those whose expression was below 0.5 fold as a differentially under-expressed gene. As a first overview, 44 out of 84 studied genes showed altered expression in XP-Mel compared to healthy controls, while for the SP-Mel, 34 out of 84 genes were altered compared to healthy controls (Supplementary Table S1). In details comparative analysis between over and under-expressed genes in both groups suggested the existence of a common pathway via Fisher exact test (p value < 0.001).
We drew a Venn diagram for over and under-expressed genes from PCR array results of XP-Mel and SP-Mel tumors to delimit overlapping inflammatory gene expression. Among the examined genes, we noted in XP-Mel, the over-expression of 10 genes, while in SP-Mel group 25 genes were over expressed. Among theses over expressed genes, four were in common.
Regarding the under-expressed genes, 34 out of 84 genes in XP-Mel and 9 out of 84 genes in SP-Mel group were found, with 6 genes under-expressed in common. These data suggest the existence of a common altered inflammatory pathway in XP and SP-Mel (Fig. 1).
Figure 1.
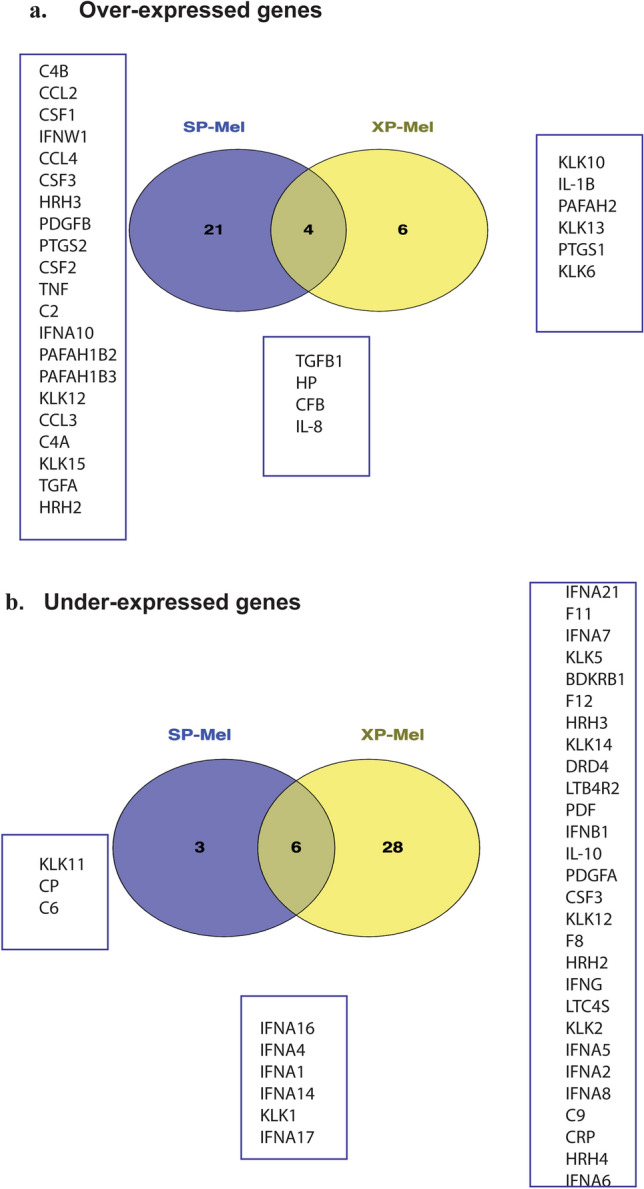
Venn diagram illustrating common and specific differentially expressed inflammatory genes in both groups XP-Mel and SP-Mel compared to healthy control. (a) over-expressed genes (Fold change > 2) (b) under-expressed genes (Fold change < 0.5).
Validation of random genes sets from the pre-screening results of PCR array using qPCR
Primers were designed for 3 randomly selected genes identified by PCR array, whose expression was taken into consideration (2 < fold change < 0.5) in both groups (KLK1), or specifically in XP-Mel (KLK13), or SP-Mel (HRH2) groups. Quantitative real-time PCR (q-PCR) was carried out in triplicate on the same samples as used for the PCR arrays and expression was normalized to healthy skin controls. Results obtained using real-time qPCR were similar to those obtained by PCR array (Fig. 2).
Figure 2.
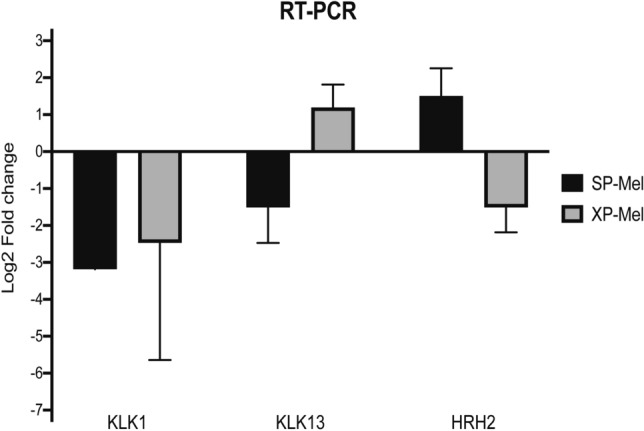
Validation of randomly selected inflammatory genes by RT-qPCR . Data are represented as Log2 fold change. Results are presented as mean ± standard error for RT-qPCR normalized to 2 housekeeping genes RPLP0 and PPIA (mean of triplicate analyses for XP-Mel (n = 3) and SP-Mel (n = 3).
Investigation of commonly altered (over or under expressed) inflammatory genes expression in PCR array and RNA-seq database
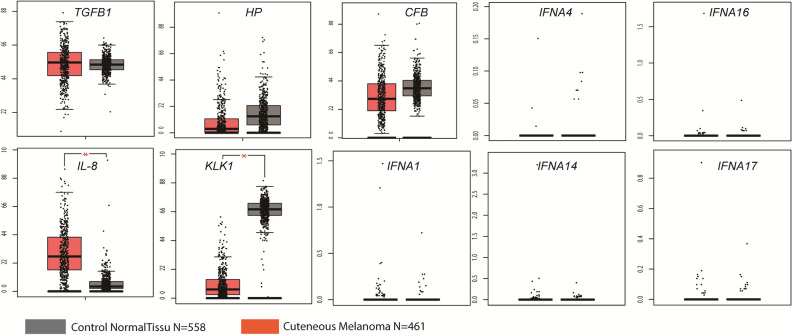
In order to explore the involvement of the commonly expressed inflammatory genes in XP-Mel and SP-Mel, we evaluated their expression in 461 samples of cutaneous melanoma that are included in the cancer genome atlas database (TGCA)29. In detail, we consulted the gene expression interactive analysis (GEPIA) online server that combines the data of TGCA, comparing the sporadic melanoma samples with those of 558 matched healthy skin from Genotype-Tissue Expression (GTEx) data sets30. Analysis of the different boxplots showed that the results obtained for TGFB1, IL-8, and KLK1 have the same trend as those obtained by PCR array. In particular, statistically significant over-expression of IL-8 was noted in cutaneous melanoma (TPM = 0.27 in healthy skin compared to melanoma tumors TPM = 4. 52 (adjusted p-value 6.39e−99 ); and a significant under-expression was obtained for KLK1 (TPM = 70.74 in healthy skin compared to Melanoma tumors TPM = 0.52 (adjusted p-value0.00e + 0). For the IFNA gene family, their expression was too low to be considered in both types of tissue (TPM < 10).
Two genes namely HP and CFB have different expression profiles compared to the results found using PCR array (Fig. 3).
Figure 3.
Exploration of 10 commonly expressed inflammatory genes in XP-Mel and SP-Mel via RNA-seq data of sporadic melanoma using GEPIA web server based on the TCGA and GTEx databases. Box plots represent the gene expression level in terms of log2 (TPM) in tumors (red, n = 461) and healthy skin (grey, n = 558) samples, respectively. Healthy skin is matched TCGA adjacent tissue and GTEx data. The method for differential analysis is one-way ANOVA (*p-value < 0.05).
Impact of sun sensitivity on the differentially expressed inflammatory genes in XP-Mel samples
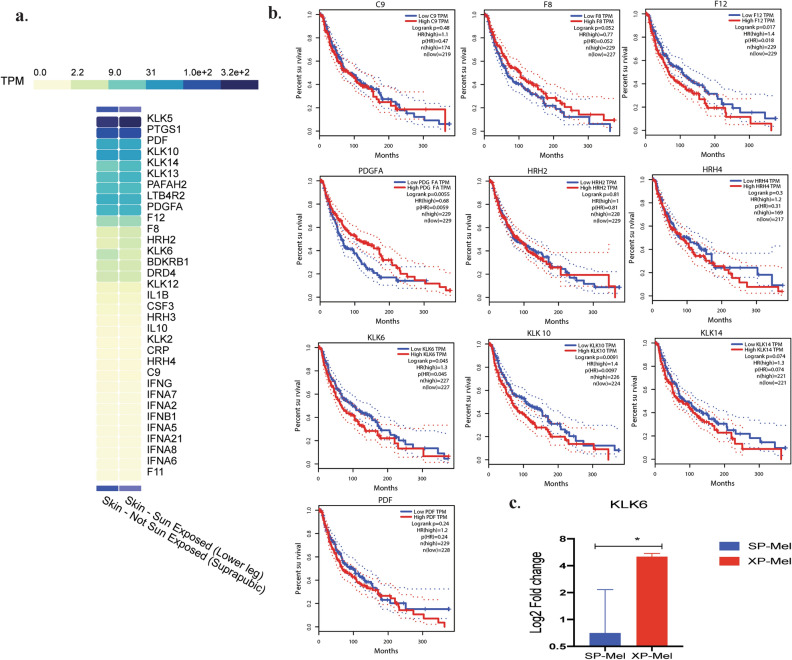
We explored specifically the 34 differentially expressed genes in the XP-Mel group to assess whether the difference in the expression of the inflammatory genes (XP-Mel versus healthy skin) is due specifically to the accumulation of UVs damage in XP tumors or due to defects in the NER pathway that affects the functioning of the immune system. For this purpose, the RNA-seq transcriptomic data in the GTEx database was explored for the expression of the 34 genes in skin exposed to solar radiation compared to non-exposed skin. Nine out of 34 genes (PTGS1, DRD4, CSF3, PAFAH2, BDKRB1, HRH3, LTB4R2, KLK13, KLK2) have the same expression trend as the results found using the PCR array in XP-Mel, which may be linked to the extreme sun sensitivity in these patients. While the 13 other genes were not detected or were not taken into account due to unavailable data in GTEx database (Fig. 4a).
Figure 4.
Exploration of the 34 specific inflammatory genes in XP-Mel tumors in GTEx and GEPIA databases: (a) Heatmap depicting the altered inflammatory genes via GTEx platform in healthy and sun-exposed skin samples; results are expressed in TPM (the number of Transcripts Per million Readings). (b) Overall survival outcomes of patients with sporadic melanoma with low and high gene expression of C9, F8, F12, PDGFA, HRH2, HRH4, KLK6, KLK10, KLK14, and PDF. Kaplan–Meier curves are plotted using the GEPIA online Tool, 95% confidence intervals are shown (c) KLK6 gene expression in SP-Mel and XP-Mel, normalized expression from the PCR array results. Hazard ratios (HR); (*p-value < 0.05).
As for the 12 genes whose expression in RNA-seq was different from the PCR array data (KLK10, KLK6, HRH4, C9, F8, F12, PDGFA, HRH2, KLK14, PDF, KLK5, KLK12), we speculate that their altered expression was related to the bad survival rate upon tumor development in XP patients. We, therefore, explored the GEPIA webserver to estimate how the level of expression for 10 of these genes was associated with the overall survival (OS) in sporadic forms (OS: time from cancer diagnosis to death for any cause). Kaplan–Meier analysis was used to compare between the subgroups with high and low gene expression (using the median, 50% quartile values of gene expression as cut-off points) in a cohort of patients with sporadic cutaneous melanoma, which was available in GEPIA database. The OS outcome was significantly associated with a high level of PDGFA and a low level of KLK6 and KLK10 expression in melanoma tumors (p-value = 0.0055, p-value = 0.045, and p-value = 0.0091, respectively). These genes were expressed in an opposite way in XP patients who developed melanoma at a young age (Fig. 4b).
It is interesting to note that only the KLK6 was significantly over-expressed in XP-Mel group according to the results of the PCR array (fold change 5 ± 0.5 p-value = 0.03) (Fig. 4c).
Phenotyping of monocyte subsets in sporadic melanoma and in XP patients
Flow cytometry analysis was performed to assess monocyte phenotypes, in the PBMC of 16 healthy donors (controls), 8 patients with sporadic melanoma (SP-Mel), and 6 XP (3 XP-C, 2XP-A and one XP-V) patients that did not develop cutaneous tumors at the time of the study.
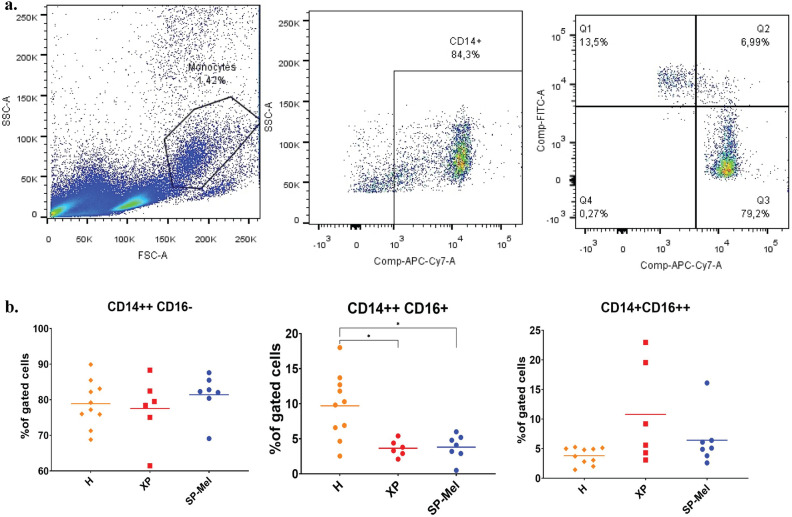
For classical monocytes, with a CD14 + + CD16- phenotype, there was no difference in both groups compared to healthy donors 80% ± 2. For intermediate monocytes, with CD14 + + CD16 + phenotype, there was a significant decrease in their percentage in patients with SP-Mel 3.8% ± 1.82 (p-value = 0.009) and in XP patients 3.6% ± 1.16 (p-value = 0.01) compared to healthy controls 9.7% ± 4.6. As for non-classical monocytes with a CD14 + CD16 + + phenotype, there was no significant difference in the three groups SP-Mel 6.42% ± 4.45, XP 10.78% ± 8.4 compared to healthy controls 3.8% ± 1.4 (Fig. 5).
Figure 5.
Characterization of blood monocyte’s subsets in healthy donors (n = 18), in patients with sporadic melanoma SP-Mel (n = 8), and in Xeroderma Pigmentosum patients (n = 6) (a) Gating strategy for the identification of the three monocyte’s subsets (b) Percentages of the different monocyte’s subsets CD14 + + CD16- classical CD14 + CD16 + intermediate and CS14 + CD16 + + non classical (*p-value < 0.05).
Discussion
Melanoma is a rare form of skin cancer especially in the Tunisian population1. It is the source of high mortality rates1. Sun exposure and UVs radiation are well-established risk factors. The exceptionally high incidence and early onset of melanoma in patients with XP indicate that DNA repair is involved in the etiology of melanoma. Advances in high-performance technology make it possible to test individuals at-risk for melanoma, leading to the discovery of new preventive methods in the general population, such as adequate use of sun protection and skin cancer screening at regular intervals, as well as the use of chemo-preventative agents31.
Skin cancer develops early in XP patients in whom the risk increases 10,000 times compared to the general population as in the case of the United States where XP patients develop melanoma at an average age of 22 years old32. Although North African XP patients develop mostly basal cell carcinomas, and more rarely melanomas, and spinocellular carcinoma33 we noted that the XP patients develop melanoma 42 years earlier than sporadic cases. In this work, the three Tunisian XP patients developed cutaneous melanoma at an average age of 17 years, which was in accordance with a previous study conducted in the Tunisian population1 . It is important also to note that this study was done through 5 years of monitoring in collaboration with the dermatology department and that we were able to reach only 3 XP patients who developed Melanoma. Extensive patient awareness of the harmful effects of UV exposure has reduced the number of skin cancers in XP patients and in particular melanoma4. This work includes two patients from XP-C complementation group and one XP-A.
High inflammation level observed via Hematoxylin and Eosin staining on paraffin section from the XP-A patient (data not shown) may be associated to the severe sunburn reaction observed in these patients34. XP-A and XP-C patients develop skin cancers with a similar average age ranging from 8 to 20 years old4 .
Although XP-A patients develop also neurological damage, they exhibit the same cutaneous histological manifestations. In our study, the expression of inflammatory genes was made only from cutaneous melanoma.
The mechanisms involved in the development of skin cancers in XP patients have been linked solely to the impact of DNA repair defects following exposure to UVs in most of the studies. However, in other DNA repair syndromes with similar underlying pathways such as Fanconi anemia and Bloom syndrome, investigations suggested an impact of the DNA repair impairment on the immune system. Indeed, patients with Bloom syndrome have low number of lymphocytes, with mild immunodeficiency affecting the class switch recombination process during B cell development11. While other reports on patients with Fanconi anemia, reported low NK-cell number and altered functions as well as a decrease in cytotoxic T-cell responses10,35.
Cancer susceptibility in XP patients is not solely due to persistent DNA lesions due to defective DNA repair after UV exposure, but could be the result of a more complex situation. In fact, XP patients protected from UVs exposure develop other internal cancers such as myeloid leukemia4,36,37 and the cause behind this is still unknown.
Recent studies highlighted the possibility of oxidative stress involvement in the development of cancers in DNA repair diseases, and in age-related pathologies38. This could influence immune responses. XP patients represent a form of skin aging even in mild forms39 , hence our hypothesis about the involvement of immunity and particularly inflammatory actors in the pathophysiology of skin cancer in XP patients.
As XP is a rare disease, few studies investigated the immune responses. Indeed, following clinical observations, several immunological defects have been reported in XP patients, including the reduced lytic activity of NK cells40, decreased production of interferons by lymphocytes41, decreased proportion of TCD4, and TCD8 lymphocytes42, as well as altered UV-induced cytokine production43.
More recently, studies of the tumor microenvironment of Basal cell carcinoma in XP patients also showed increased expression of apoptosis-related biomarkers in immune cells such as CD95, Bcl-2 and Bax compared to sporadic cases, which may explain the earlier establishment of cancer in these patients44. The most recent report also describes a dense infiltration of CD163 + macrophages with anti-inflammatory action, in the tumor microenvironment of basal cell carcinoma45.
The analysis of the inflammatory genes’ expression profiles in cutaneous melanoma of XP-Mel and SP-Mel shows a specific transcriptomic signature for each group. The singularities in gene expression patterns in XP-Mel and in SP-Mel have been previously found in BRAF and PTEN pathways, where it has been shown an increased frequency of PTEN mutations and activation of the mTOR pathway in the XP-Mel tumors compared to a lower frequency of BRAF mutations46. Several studies have focused on a large-scale analysis of inflammatory genes in cutaneous melanoma47, but none has addressed this aspect in XP patients.
PCR array is becoming one of the standard approaches measuring differential gene expression in many diseases such as cancer48 and autoimmune diseases49. This work represents the first report in the study of a rare genetic disease using pathway-focused PCR-based arrays for candidate genes involved in the immune response and inflammatory process. Genome-wide studies and microarray investigation could be a better strategy to study a more important set of genes, however, it represents low sensitivity for under-expressed genes50, like what we noted in the XP-Mel patients (34 out of 84 genes).
DNA damages caused by the UVs in skin cells activate several transcription factors51. Although it is well recognized that these changes can cause a shift in gene expression, transcriptional regulation following DNA damage is still poorly understood. Few reports suggest that UV-induced lesions in the transcribed strand of DNA form an obstacle to the RNA Polymerase activity translocation leading to its arrest which induces the activation of nucleotide excision repair system52. It is not then surprising that due to XP genetic defects, we found that the expression of some inflammatory genes appears to be altered in melanoma tumors compared to those of sporadic origin.
The selected candidate genes used in the PCR array experiments were mostly inflammatory mediators that are normally released upon UV-radiation of the skin, such as cytokines (e.g. IL-1a, IL-6, IL-8, TNF-alpha), growth factors (e.g. TGF-beta, VEGF, NGF) and vasoactive amines (e.g. histamine, bradykinin,)53. Following the PCR array experiments, we observed a higher number of inflammatory genes whose expression was altered in XP-Mel patients (51.16% of total). Indeed, the 34 inflammatory genes were under-expressed in XP-Mel tumors while only 9 were under-expressed in SP-Mel ones. This seems to be associated with an immunodeficiency status in these patients54.
Given the rarity of samples used in this study and the lack of data investigating the inflammatory response in XP patients, we consolidate the results via the consultation of larger sampling in melanoma patients through RNA-seq data exploring different databases. One of the common biomarkers in SP-Mel and XP-Mel implicate two genes namely IL-8 and KLK1. IL-8, also known as CXCL8, was one of the most expressed genes in the studied melanoma samples. This cytokine is not produced by healthy melanocytes and its overexpression in seems contributing to melanoma progression55,56.
As for tissue KLK1, it exerts a role in producing vasoactive kinins, which in itself carry out a wide range of biological activities, including vasodilation, lowering blood pressure, pain induction, and inflammation57. KLK1 expression was downregulated in both pancreatic and colon cancer in previous studies58. However, it was never linked to melanoma. In this work, we noted a low level of KLK1 expression in both XP-Mel and SP-Mel tumors as well as in RNA-Seq data. These findings require further investigations to determine its role in skin cancer development.
In addition, two genes HP (Haptoglobin) and CFB (Complement Factor B) had a different expression depending on the technique used, either PCR array or RNA-seq. Regarding HP, it is known for its systemic anti-inflammatory function. HP affects immune cells and its expression is increased in the mouse model of melanoma59, which is in accordance with our PCR array findings. As for CFB, little is known about its implication in skin cancers and in particular in melanoma60. The variable expression rate of these two genes could be due to the fact that the inflammatory status of the microenvironment of the melanoma tumors that we analyzed is different from that of the samples analyzed via RNA seq.
The analysis of inflammatory gene expression in RNA-seq database GTEx of healthy skin exposed to sunlight revealed that nine genes (PTGS1, DRD4, CSF3, PAFAH2, KLK13, BDKRB1, HRH3, LTB4R2, and KLK2) present the same expression profiles as for XP patients. As it is well established, XP patients exhibit extreme sensitivity to sunlight, triggering severe sunburn61. These genes are implicated in pathways of prostaglandins, bradykinin and histamine, which are well known as first interactors to induce inflammatory response during sunburn62. As for the genes related to the kallekrein families (KLK), it may represent potential targets for studies on severe sunburns.
In this work, we observed that the overall survival outcome was significantly associated with a high level of PDGFA and a low level of KLK6 and KLK10 expression in melanoma tumors and that these genes were expressed in an opposite way in XP patients who developed melanoma at a young age.
The KLKs represent a family of secreted serine proteases composed mainly of 15 genes. The peptidases of the kallikrein family (KLK) are proteases secreted by granular keratinocytes in the upper layer of the skin; they play an important role in the keratinocyte exfoliation process63. They also participate in the degradation of the extracellular matrix and tissue remodeling of many tissues64. Atypical KLK expression can disrupt skin barrier homeostasis, resulting in a variety of skin diseases such as Netherton syndrome (NS), atopic dermatitis (AD), rosacea, and psoriasis65. However, few studies explored the functionalities of KLKs in cancer initiation and progression66.
KLK6 was suggested to be involved in the epidermal proteolytic process that regulates the desquamation process67. It is one of the most highly upregulated genes in keratinocytes upon differentiation induction with vitamin D3 analogs68. Whereas UV-B rays are essential for endogenous production of vitamin D in the skin and that vitamin D deficiency was generally found in XP patients69, its overexpression noted in XP-Mel tumors is therefore unrelated to UVs sensitivity but rather due to tumorigenesis process. In support of this hypothesis, it was shown that KLK6 is highly expressed in primary melanoma, which points to its involvement in the neoplastic processes and malignant progression57. As for KLK10, it is expressed in the follicular dendritic cells that are essential for the maturation of B cells which suggest possible implications in the regulation of immune cells in lymphoid tissues70. As for the KLK10 gene, we found an overexpression in XP patients. Its increased expression in other types of cancers such as triple-negative breast cancer has been associated with poor prognosis71.Giving the Fact that KLK10 is also expressed in healthy skin66, we could also suspect an association with the development of melanoma which would require further exploration.
PDGF (platelet-derived growth factor) is a proangiogenic factor72. PDGF-AA (PDGFA), PDGF-BB (PDGFB), PDGF-CC (PDGFC), PDGF-DD (PDGFD), and the PDGF-AB heterodimer (PDGFAB) are the five PDGF ligands known to date73. Data of DNA sequencing from different types of tumors indicated that mutations in genes coding for PDGF ligands occur in approximately 20% of melanoma74. PDGF-AA is overexpressed in 50% of renal cancer, which is correlated with a good survival rate75. Through our study, we also suggest that the under-expression of PDGFA is associated with low survival risk in XP patients.
Given the significance of the interferon alpha family genes (IFNA) in the immunotherapy of cutaneous melanoma, we explored their level of expression via qPCR. In fact, IFNA are a group of glycoproteins secreted by immune cells during viral infections and microenvironmental stimuli76. It englobes 12 distinct proteins that differ in their specific activities. However, the function of each subset is still not well defined. The lack of IFNA genes expression in XP-Mel patients suggests their involvement in the DNA repair process. In accordance with this concept, recent studies suggest that DNA double-strand breaks activate the DNA damage response through IFNA expression77. Our results therefore suggest that the expression of IFNA subtypes is associated with the NER pathway since the majority of genes associated with the interferon signaling pathway tend to be dysregulated in XP-Mel patients. Moreover, previous work indicates that XP patients have a lack in interferon expression78, which is in line with our findings.
For IFNA family genes whose expression level were different or non-existent in the GTEx database, the hypothesis that this alteration is caused by defects in the DNA repair system appears therefore to be plausible. However, we were unable to establish a direct link. This highlights the need for further investigations to explore the link between DNA repair pathways and inflammatory responses.
Peripheral mononuclear cells are the main producer of IFNA79. It was suggested that the abundance of the different monocyte subsets in systemic lupus erythematosus is related to the level of expression of different IFN genes in particular, the classical and the intermediate monocytes, which are the main IFN responsive cells80. Therefore, the decreased expression of the IFNA genes in the tumor microenvironment could be related to the small number of intermediate monocytes in patients developing melanoma that we observed via flow cytometry.
A distinction between the different monocyte sub-groups in patients with melanoma was performed to evaluate the link with cancer development. In general, intermediate monocytes group (CD14 + + CD16 +) account for approximately 2–8% of circulating monocytes25. We found a major declines of this cell population in patients with melanoma. However, the role of these cells is not well established yet. Several studies mentioned their roles in the production of reactive species of oxygen during the immune response, antigen presentation, the involvement of lymphocyte-T proliferation and activation, inflammatory responses, and angiogenesis81. Studying the intermediate monocyte poses many challenges given the fact that no equivalent cells exists in animal models.
A low level of intermediate monocytes was reported in other types of cancers such as squamous cell carcinoma and oropharyngeal cell cancer and was correlated with poor clinical outcome82,83. Regarding classical monocytes, it was reported that a decreased number of these cells is associated to cutaneous melanoma stage IV by Chavan et al.27, while another study reported an increased number of non-classical monocytes in patients with cutaneous melanoma and associated it with a better response to immunotherapy28. The disparity between the results obtained in our work and that reported by Chavan et al27 can be explained by the fact that our study includes primary melanoma tumors. Moreover, as the intermediate monocytes subtype represents a transitional state between the classical and non-classical subtypes, makes its delineation variable depending on the gating strategies84–87.
It is important to note that the XP patients that we have enrolled in the phenotyping of monocytes had not developed cancers at the time of the study. They presented a low rate of intermediate monocytes as in patients with sporadic melanoma, which suggest that this sub-type could be a relevant biomarker for monitoring cancer development.
Although this work is done on a limited number of samples, it is the first study that has focused on the immune landscape in cutaneous melanoma in patients with DNA repair disorders as XP. It is likely that a small sample size could decrease the study's significance level and raises the margin of error, which can contribute to bias as it has been stated in other studies using the PCR array approaches88. However, the current research identified the same patterns of gene expression as shown in previous studies that investigated melanoma cancer such as IL8 and TGFB89 as well as in the RNA-seq databases that we investigated in this work. Further exploration with larger sample size is required to confirm the current findings and to examine the consequences of the altered transcriptional profile especially concerning KLK6 and KLK10 who may contribute to the worsening of Melanoma tumorigenesis.
It should also be noted that the gene expression pattern in XP patients could be related to the age differences between this group, the healthy group and the sporadic cases, since the XP patients were young. However, several studies that have conducted inflammatory gene expression have linked this difference to photo-aging of the skin, particularly for the set of genes associated to senescence-associated secretory phenotype (SASP) (IL1b, IL8 , CCL2, CCL3, MCF.)90. Moreover, the Xeroderma pigmentosum has recently been considered as a potential model for on premature human aging studies8.
Finally, we observed a decrease of intermediate monocytes number in patients with sporadic melanoma and in XP patients that could be considered as a prognostic marker for skin cancer. Further studies exploring the functions of this monocytes subtype in melanoma would be important to investigate.
Methods
Patients and healthy donors
The study was carried out in accordance with the Helsinki principles and approved by Institute Pasteur Ethics Committee in Tunisia under the ethical accord number (reference PCI/22/2012/v2).
After obtaining written informed consent from patients (over 18 years old) and from patient’s parents (for minors), melanoma biopsies were obtained from the Department of Dermatology (Hospital Charles Nicolle, Tunis).
Melanoma samples from 3 XP patients (N = 2 XP-C; 1 XP-A forms) and 3 sporadic melanoma tumors were collected for PCR array analysis. For the XP patients, biopsies of 5 mm were taken from the tumors, a half was snap-frozen in liquid nitrogen, and the other part was embedded in paraffin and stained with hematoxylin/eosin to confirm the melanoma diagnosis and for further histological investigations. For healthy skin samples, pieces of tissue (5–8 mm) that are next to seborrheic keratosis or epidermal cysts was obtained from 3 donors. Each recovered healthy skin biopsy was examined by a pathologist following hematoxylin/eosin staining, who verified that there is no inflammatory infiltrate, before proceeding to RNA extraction. More details about the samples we used are described in Table 1.
Table 1.
Detailed PCR array cohort description
| Code | Healthy donors (control) | SP-Mel | XP-Mel | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Age | 33 | 35 | 40 | 59 | 87 | 72 | 9 | 28 | 16 |
| Sexe | F | M | M | M | M | F | F | F | F |
| Pathology | Seborrheic keratosis | Epidermal cysts | Seborrheic keratosis | ALM Sporadic melanoma | SSM Sporadic melanoma | ALM Sporadic melanoma | SSM Melanoma in XP patient | LMM Melanoma in XP patient | LMM Melanoma in XP patient |
| Genetic mutation | NA | NA | NA | NA | NA | NA | XPC V548A fs X572 | XPC V548A fs X572 | XPA R228X |
| Localization | Scalp | Face | Face | Left heel | Toe | Foot sole | Cheek | Cheek | Nose |
| Inflammation level | Low | Low | Low | high | Low | Low | Low | Low | High |
NA = Not Applicable.
Blood samples were also collected from 18 healthy donors, 8 patients with sporadic melanoma, and 6 XP patients (3 XP-C, 2 XP-A forms and one XP-V) for flow cytometry analysis.
PCR array
RNA is isolated from melanoma biopsies and from healthy skin using trizol extraction method via Rnaeasy kit (Qiagen). RNA concentration and purity were determined by using a NanoDrop. Then, 1000 ng of RNA were reverse transcribed to single-stranded cDNA in a total volume of 20 µL, using reverse transcriptase (Superscript II, #18064014, Invitrogen), according to the manufacturer’s protocol.
Analysis of 84 inflammatory gene’s expression, as well as of eight housekeeping genes (HKG), was performed using PCR array for human inflammation signaling pathways (anygenes, #IF1H1), whose gene list is presented in (Supplementary Table S2). The qPCR was done according to the manufacturer’s protocol and Ct values were measured via LightCycler 480 software (Roche). Data analysis of the raw data was carried out using the ∆∆CT method and Ct were normalized to the mean values of three stable HKG (PPIA, ACTB, and RPLP0). Fold-change calculations were done using the Anygenes sign array data analysis excel sheet available online (https://www.anygenes.com/home/resources/data-analysis-tools), which automatically calculates the fold-change in gene expression between the melanoma patients and the healthy donors group.
qPCR
We tested the expression of KLK-13, HRH2, KLK1 genes using the SYBR Green-based qPCR technique. Primers were selected from the Primer bank database (https://pga.mgh.harvard.edu/primerbank/) and ordered from sigma life science (Table 2). Relative quantification Ct values were obtained from the threshold cycle number of a triplicate test and normalized to the healthy skin. PPIA and RLPO were used as housekeeping genes.
Table 2.
List of qPCR Primers.
| Primer | Sequence 5' to 3' |
|---|---|
| HRH2-fw | CGTGTCCTTGGCTATCACTGA |
| HRH2-rw | GGCTGGTGTAGATATTGCAGAAG |
| KLK1-fw | CCCGATTCAGTCCCGGATTG |
| KLK1-rw | AGCTGGTAATTGTCGCTGATG |
| KLK13-fw | TTGGCCTTGTCAGGAGGTG |
| KLK13-rw | AGGACCCATTTGGGGTGGA |
The LightCycler 480 SYBR Green I Master Mix (# 04 707 516 001, Roche) was used according to the supplier’s recommendations. Q-PCR was performed on LightCycler 480 System (Roche Diagnostics). The program consisted of an initial denaturation at 95 °C for 10 min, followed by 45 amplification cycles (95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s). This was followed by a melting program of 95 °C for 5 s and 65 °C for 1 min and 97 °C with continuous monitoring of the fluorescence. The final step consisted of cooling at 2.2 °C/s to 40 °C. Threshold cycle (Ct) was used to calculate relative gene expression by the 2-ΔΔCT method.
Flow cytometry analysis
Blood samples from 18 healthy donors, 8 patients with sporadic melanoma, and 6 XP patients (3 XP-C, 2 XP-A forms and one XP-V) were collected. Written informed consent was obtained from all donors. Peripheral Blood Mononuclear Cells (PBMC) were extracted from heparinized blood samples using density gradient centrifugation on a Ficoll cushion.
Monocyte phenotyping according to surface markers expression was performed using monoclonal antibodies: anti-CD14 labeled with APC-Cy7.5 (clone MP9, # 557831, BD Biosciences) and anti-CD16 labeled with FITC (clone 3G8, # 555406, BD Biosciences). Plot acquisition was done on flow cytometer FACSCanto™ II (BD Biosciences) for at least 10.000 events and analyzed using flowjo software.
Differential inflammatory gene expression and survival analysis from the GTEx and GEPIA databases in melanoma skin cancer
Expression of common inflammatory genes of interest, identified in the in sporadic melanoma SP-Mel and XP-Mel biopsies using PCR array were further tested in RNA-seq-melanoma dataset (n = 461) with matched control samples (n = 558). Data was acquired from the GEPIA webserver (http://gepia.cancer-pku.cn/).
As for the genes of interest whose expression was specific to XP-Mel patients, it was compared to the GTEx dataset of sun-exposed and non-exposed skin (https://www.gtexportal.org/home/). The XP-Mel genes of interest were also examined in terms of overall survival in Melanoma patients through the GEPIA database.
Statistical analysis
Differences in the fold change expression between XP-Mel, SP-Mel and healthy controls following qPCR, were tested by the Mann–Whitney U test. The level of significance was set at 0.05 (p-value < 0.05). The analyses were performed using GraphPad software version 7.00 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com.
Ethical approval
The study was carried out in accordance with the Helsinki principles and approved by Institute Pasteur Ethics Committee in Tunisia under the ethical accord number (reference PCI/22/2012/v2).
Supplementary Information
Acknowledgements
The authors would like to thank the patients and their families as well as the patients’ support group “Helping Xeroderma pigmentosum Children” (http://www.xp-tunisie.org.tn/) for their collaboration.
Author contributions
A.C. did PCR array, flow cytometry, analyzed the data and drafted the manuscript. M.J. did clinical investigation and skin biopsies of XP patients. T.R. did qPCR experiments and revised the manuscript. M.B.A. helped in flow cytometry analyses. C.N. helped in samples recruitment. S.B. confirmed the histological diagnosis of melanoma. M.Z helped for XP patients follow-up. R.K, S.A. helped in study conception and revised the manuscript. H.Y.Y. did the study conception, helped in experiments, supervised the study and revised the manuscript.
Funding
This work was supported by the Tunisian Ministry of Public Health, the Ministry of Higher Education and Scientific Research (LR16IPT05), the “Projet Collaborative Interne” (PCI_Melanoma, IPT) and by the Slovenian Research Agency (ARRS) program (grants P1-0104, P1-0390).
Data availability
All processed data have been provided in the manuscript. The corresponding author upon reasonable request could provide raw data, generated for this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-17928-z.
References
- 1.Naouali C, et al. Epidemiological trends and clinicopathological features of cutaneous melanoma in sporadic and xeroderma pigmentosum Tunisian patients. Int. J. Dermatol. 2016;56:1–40. doi: 10.1111/ijd.13448. [DOI] [PubMed] [Google Scholar]
- 2.Kleijer WJ, et al. Incidence of DNA repair deficiency disorders in western Europe: Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. DNA Repair. 2008;7:744–750. doi: 10.1016/j.dnarep.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Emmert S, Kraemer KH. Do not underestimate nucleotide excision repair: It predicts not only melanoma risk but also survival outcome. J. Investig. Dermatol. 2013;133:1713–1717. doi: 10.1038/jid.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jerbi M, et al. Clinical, genealogical and molecular investigation of the xeroderma pigmentosum type C complementation group in Tunisia. Br. J. Dermatol. 2016;174:439–443. doi: 10.1111/bjd.14046. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 6.Stary A, Kannouche P, Lehmann AR, Sarasin A. Role of DNA polymerase η in the UV mutation spectrum in human cells. J. Biol. Chem. 2003;278:18767–18775. doi: 10.1074/jbc.M211838200. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum: cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch. Dermatol. 1987;123:241. doi: 10.1001/archderm.1987.01660260111026. [DOI] [PubMed] [Google Scholar]
- 8.Rizza ER, et al. Xeroderma pigmentosum: A model for human premature aging. J. Investig. Dermatol. 2021;141:976–984. doi: 10.1016/j.jid.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gülbakan B, et al. Discovery of biomarkers in rare diseases: Innovative approaches by predictive and personalized medicine. EPMA J. 2016;7:24. doi: 10.1186/s13167-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers KC, et al. Impaired immune function in children and adults with Fanconi anemia. Pediatr. Blood Cancer. 2017;64:e26599. doi: 10.1002/pbc.26599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenaker MH, et al. Immunodeficiency in bloom’s syndrome. J. Clin. Immunol. 2018;38:35–44. doi: 10.1007/s10875-017-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein B, Khilnani P, Lapey A, Cleaver JE, Rhodes AR. Combined immunodeficiency associated with xeroderma pigmentosum. Pediatr. Dermatol. 1990;7:132–135. doi: 10.1111/j.1525-1470.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 13.Hölzel M, Tüting T. Inflammation-induced plasticity in melanoma therapy and metastasis. Trends Immunol. 2016;37:364–374. doi: 10.1016/j.it.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Bisevac JP, et al. Association between oxidative stress and melanoma progression. J. Med. Biochem. 2018;37:12. doi: 10.1515/jomb-2017-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perwez Hussain S, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int. J. Cancer. 2007;121(11):2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 16.Kawanishi S, Ohnishi S, Ma N, Hiraku Y, Murata M. Crosstalk between DNA damage and inflammation in the multiple steps of carcinogenesis. Int. J. Mol. Sci. 2017;18:1808. doi: 10.3390/ijms18081808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang L, Wang K. Chronic inflammation in skin malignancies. J Mol Signal. 2016;11:2–2. doi: 10.5334/1750-2187-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neagu M, et al. Inflammation: A key process in skin tumorigenesis. Oncol. Lett. 2019;17:4068–4084. doi: 10.3892/ol.2018.9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson MJ, et al. Inflammation and apolipoproteins are potential biomarkers for stratification of cutaneous melanoma patients for immunotherapy and targeted therapy. Cancer Res. 2021;81:2545–2555. doi: 10.1158/0008-5472.Can-20-2000. [DOI] [PubMed] [Google Scholar]
- 20.Guo HB, Stoffel-Wagner B, Bierwirth T, Mezger J, Klingmüller D. Clinical significance of serum S100 in metastatic malignant melanoma. Eur. J. Cancer. 1995;31:924–928. doi: 10.1016/0959-8049(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 21.Weber JS, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/s1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 22.Coates M, Blanchard S, MacLeod AS. Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PLoS Pathog. 2018;14:e1007353. doi: 10.1371/journal.ppat.1007353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatzinikolaou G, Karakasilioti I, Garinis GA. DNA damage and innate immunity: Links and trade-offs. Trends Immunol. 2014;35:429–435. doi: 10.1016/j.it.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Härtlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Ziegler-Heitbrock L, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 26.Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J Leukoc Biol. 2019;106:309–322. doi: 10.1002/JLB.4RI0818-311R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavan R, et al. Untreated stage IV melanoma patients exhibit abnormal monocyte phenotypes and decreased functional capacity. Cancer Immunol. Res. 2014;2:241–248. doi: 10.1158/2326-6066.CIR-13-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano E, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc. Natl. Acad. Sci. 2015;112:6140–6145. doi: 10.1073/pnas.1417320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jager MJ, Brouwer NJ, Esmaeli B. The cancer genome atlas project: An integrated molecular view of uveal melanoma. Ophthalmology. 2018;125:1139–1142. doi: 10.1016/j.ophtha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens MC, Seebode C, Lehmann J, Emmert S. Photocarcinogenesis and skin cancer prevention strategies: An update. Anticancer Res. 2018;38:1153. doi: 10.21873/anticanres.12334. [DOI] [PubMed] [Google Scholar]
- 32.Bradford PT, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: Long term follow-up characterises the role of DNA repair. J. Med. Genet. 2011;48:168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zghal, M., Fazaa, B., Abdelhak, S. & Mokni, M. in Annales de Dermatologie et de Vénéréologie Vol. 145, pp. 706–722 (Elsevier, 2018). [DOI] [PubMed]
- 34.Sethi M, et al. Patients with xeroderma pigmentosum complementation groups C, E and V do not have abnormal sunburn reactions. Br. J. Dermatol. 2013;169:1279–1287. doi: 10.1111/bjd.12523. [DOI] [PubMed] [Google Scholar]
- 35.Giri N, et al. Immune status of patients with inherited bone marrow failure syndromes. Am. J. Hematol. 2015;90:702–708. doi: 10.1002/ajh.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolaev, S., Yurchenko, A. A. & Sarasin, A. Increased Risk of Internal Tumors in DNA Repair-Deficient Xeroderma Pigmentosum Patients: Analysis of Four International Cohorts (2021). [DOI] [PMC free article] [PubMed]
- 37.Oetjen KA, et al. Predisposition to hematologic malignancies in patients with xeroderma pigmentosum. Haematologica. 2020;105:e144. doi: 10.3324/haematol.2019.223370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–197. doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chikhaoui A, et al. Identification of a ERCC5 c.2333T>C (L778P) variant in two tunisian siblings with mild xeroderma pigmentosum phenotype. Front. Genet. 2019;10:111. doi: 10.3389/fgene.2019.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norris PG, et al. immune function, mutant frequency, and cancer risk in the DNA repair defective genodermatoses xeroderma pigmentosum, cockayne's syndrome, and trichothiodystrophy. J. Investig. Dermatol. 1990;94:94–100. doi: 10.1111/1523-1747.ep12873952. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H, et al. Impaired ultraviolet-B-induced cytokine induction in xeroderma pigmentosum fibroblasts. J. Investig. Dermatol. 2001;117:1151–1155. doi: 10.1046/j.0022-202x.2001.01525.x. [DOI] [PubMed] [Google Scholar]
- 42.Wysenbeek AJ, Weiss H, Duczyminer-Kahana M, Grunwald MH, Pick AI. Immunologic alterations in xeroderma pigmentosum patients. Cancer. 1986;58:219–221. doi: 10.1002/1097-0142(19860715)58:2<219::AID-CNCR2820580203>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Mariani E, et al. Immune defects in families and patients with xeroderma pigmentosum and trichothiodystrophy. Clin. Exp. Immunol. 1992;88:376–382. doi: 10.1111/j.1365-2249.1992.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abid K, et al. Xeroderma pigmentosum skin: An immune privilege site for tumor development. J. Cutan. Pathol. 2010;37:452–459. doi: 10.1111/j.1600-0560.2009.01401.x. [DOI] [PubMed] [Google Scholar]
- 45.Furudate S, Fujimura T, Tojo G-I, Haga T, Aiba S. Basal cell carcinoma arising from xeroderma pigmentosum: A case report and an immunohistochemical study. Case Rep. Dermatol. 2013;5:64–68. doi: 10.1159/000350182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masaki T, et al. High frequency of PTEN mutations in nevi and melanomas from xeroderma pigmentosum patients. Pigment Cell Melanoma Res. 2014;27:454–464. doi: 10.1111/pcmr.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y, Kim AL, Du R, Liu L. Transcriptome analysis identifies the dysregulation of ultraviolet target genes in human skin cancers. PLoS ONE. 2016;11:e0163054. doi: 10.1371/journal.pone.0163054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Y, Zhang H-W, Cao W-H, Mao Y, Cheng R-C. Exploration of the potential biomarkers of papillary thyroid cancer (PTC) Based on RT2 profiler PCR arrays and bioinformatics analysis. Cancer Manag. Res. 2020;12:9235. doi: 10.2147/CMAR.S266473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennet SM, et al. Altered intestinal antibacterial gene expression response profile in irritable bowel syndrome is linked to bacterial composition and immune activation. Neurogastroenterol. Motil. 2018;30:e13468. doi: 10.1111/nmo.13468. [DOI] [PubMed] [Google Scholar]
- 50.Xu S. Transcriptome profiling in systems vascular medicine. Front. Pharmacol. 2017;8:563. doi: 10.3389/fphar.2017.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrlich P, et al. The mammalian UV response: Mechanism of DNA damage induced gene expression. Adv. Enzyme Regul. 1994;34:381–395. doi: 10.1016/0065-2571(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 52.Gregersen LH, Svejstrup JQ. The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 2018;43:327–341. doi: 10.1016/j.tibs.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clydesdale GJ, Dandie GW, Muller HK. Ultraviolet light induced injury: Immunological and inflammatory effects. Immunol. Cell Biol. 2001;79:547–568. doi: 10.1046/j.1440-1711.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- 54.Gennery A, Cant A, Jeggo P. Immunodeficiency associated with DNA repair defects. Clin. Exp. Immunol. 2000;121:1. doi: 10.1046/j.1365-2249.2000.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Payne AS, Cornelius LA. The role of chemokines in melanoma tumor growth and metastasis. J. Investig. Dermatol. 2002;118:915–922. doi: 10.1046/j.1523-1747.2002.01725.x. [DOI] [PubMed] [Google Scholar]
- 56.Singh S, Singh AP, Sharma B, Owen LB, Singh RK. CXCL8 and its cognate receptors in melanoma progression and metastasis. Future Oncol. 2010;6:111–116. doi: 10.2217/fon.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krenzer S, et al. Expression and function of the kallikrein-related peptidase 6 in the human melanoma microenvironment. J. Investig. Dermatol. 2011;131:2281–2288. doi: 10.1038/jid.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yousef GM, et al. In-silico analysis of kallikrein gene expression in pancreatic and colon cancers. Anticancer Res. 2004;24:43–52. [PubMed] [Google Scholar]
- 59.Kumar DM, et al. Proteomic identification of haptoglobin α2 as a glioblastoma serum biomarker: Implications in cancer cell migration and tumor growth. J. Proteome Res. 2010;9:5557–5567. doi: 10.1021/pr1001737. [DOI] [PubMed] [Google Scholar]
- 60.Roumenina LT, Daugan MV, Petitprez F, Sautès-Fridman C, Fridman WH. Context-dependent roles of complement in cancer. Nat. Rev. Cancer. 2019;19:698–715. doi: 10.1038/s41568-019-0210-0. [DOI] [PubMed] [Google Scholar]
- 61.Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J. Rare Dis. 2011;6:1–6. doi: 10.1186/1750-1172-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerra, K. C., Urban, K. & Crane, J. S. Sunburn. (2018).
- 63.McGovern J, et al. Attenuated kallikrein-related peptidase activity disrupts desquamation and leads to stratum corneum thickening in human skin equivalent models. Br. J. Dermatol. 2017;176:145–158. doi: 10.1111/bjd.14879. [DOI] [PubMed] [Google Scholar]
- 64.Paliouras M, Diamandis EP. The kallikrein world: an update on the human tissue kallikreins. Biol. Chem. 2006;387:643–652. doi: 10.1515/BC.2006.083. [DOI] [PubMed] [Google Scholar]
- 65.Chen J-Q, Liang B-H, Li H-P, Mo Z-Y, Zhu H-L. Roles of kallikrein-related peptidase in epidermal barrier function and related skin diseases. Int. J. Dermatol. Venereol. 2019;2:150–155. doi: 10.1097/JD9.0000000000000036. [DOI] [Google Scholar]
- 66.Nauroy P, Nyström A. Kallikreins: Essential epidermal messengers for regulation of the skin microenvironment during homeostasis, repair and disease. Matrix Biol. Plus. 2020;6:100019. doi: 10.1016/j.mbplus.2019.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borgoño CA, et al. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J. Biol. Chem. 2007;282:3640–3652. doi: 10.1074/jbc.M607567200. [DOI] [PubMed] [Google Scholar]
- 68.Lu J, et al. Transcriptional profiling of keratinocytes reveals a vitamin D-regulated epidermal differentiation network. J. Investig. Dermatol. 2005;124:778–785. doi: 10.1111/j.0022-202X.2005.23641.x. [DOI] [PubMed] [Google Scholar]
- 69.Mohamed A, Bhargava A, Chaurasia S. Vitamin D supplementation in patients with xeroderma pigmentosum. Indian J. Ophthalmol. 2019;67:308–309. doi: 10.4103/ijo.IJO_1319_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Filippou PS, et al. Kallikrein-related peptidases protein expression in lymphoid tissues suggests potential implications in immune response. Clin. Biochem. 2020;77:41–47. doi: 10.1016/j.clinbiochem.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 71.Liu, Y. et al. Kallikrein-related peptidases in triple negative breast cancer: prognostic value of quantitatively assessed KLK8, KLK10 and KLK11 mRNA expression (2021).
- 72.Clark R, Folkvord J, Hart C, Murray M, McPherson J. Platelet isoforms of platelet-derived growth factor stimulate fibroblasts to contract collagen matrices. J. Clin. Investig. 1989;84:1036–1040. doi: 10.1172/JCI114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen P-H, Chen X, He X. Platelet-derived growth factors and their receptors: Structural and functional perspectives. Biochim. et Biophys. Acta (BBA) Proteins Proteomics. 2013;1834:2176–2186. doi: 10.1016/j.bbapap.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farooqi AA, Siddik ZH. Platelet-derived growth factor (PDGF) signalling in cancer: Rapidly emerging signalling landscape. Cell Biochem. Funct. 2015;33:257–265. doi: 10.1002/cbf.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Honda M, et al. Mesothelioma cell proliferation through autocrine activation of PDGF-ββ receptor. Cell. Physiol. Biochem. 2012;29:667–674. doi: 10.1159/000176386. [DOI] [PubMed] [Google Scholar]
- 76.Tarhini AA, Gogas H, Kirkwood JM. IFN-α in the treatment of melanoma. J. Immunol. 2012;189:3789–3793. doi: 10.4049/jimmunol.1290060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morales AJ, et al. A type I IFN-dependent DNA damage response regulates the genetic program and inflammasome activation in macrophages. Elife. 2017;6:e24655. doi: 10.7554/eLife.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaspari AA, Fleisher TA, Kraemer KH. Impaired interferon production and natural killer cell activation in patients with the skin cancer-prone disorder, xeroderma pigmentosum. J. Clin. Investig. 1993;92:1135–1142. doi: 10.1172/JCI116682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nyman TA, Kalkkinen N, Tölö H, Helin J. Structural characterisation of N-linked and O-linked oligosaccharides derived from interferon-α2b and interferon-α14c produced by Sendai-virus-induced human peripheral blood leukocytes. Eur. J. Biochem. 1998;253:485–493. doi: 10.1046/j.1432-1327.1998.2530485.x. [DOI] [PubMed] [Google Scholar]
- 80.Han S, et al. Differential responsiveness of monocyte and macrophage subsets to interferon. Arthrit. Rheumatol. 2020;72:100–113. doi: 10.1002/art.41072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sampath P, Moideen K, Ranganathan UD, Bethunaickan R. Monocyte subsets: Phenotypes and function in tuberculosis infection. Front. Immunol. 2018;9:1726. doi: 10.3389/fimmu.2018.01726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakakura K, et al. Immunological features of circulating monocyte subsets in patients with squamous cell carcinoma of the head and neck. Clin. Immunol. 2021;225:108677. doi: 10.1016/j.clim.2021.108677. [DOI] [PubMed] [Google Scholar]
- 83.Takahashi H, et al. Prognostic significance and population dynamics of peripheral monocytes in patients with oropharyngeal squamous cell carcinoma. Head Neck. 2019;41:1880–1888. doi: 10.1002/hed.25625. [DOI] [PubMed] [Google Scholar]
- 84.Patel AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong KL, et al. The three human monocyte subsets: implications for health and disease. Immunol. Res. 2012;53:41–57. doi: 10.1007/s12026-012-8297-3. [DOI] [PubMed] [Google Scholar]
- 86.Thomas GD, et al. Human blood monocyte subsets: a new gating strategy defined using cell surface markers identified by mass cytometry. Arterioscler. Thromb. Vasc. Biol. 2017;37:1548–1558. doi: 10.1161/ATVBAHA.117.309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zawada AM, et al. Monocyte heterogeneity in human cardiovascular disease. Immunobiology. 2012;217:1273–1284. doi: 10.1016/j.imbio.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 88.Yuan JC, Yogarajah T, Lim SK, Yvonne Tee GB, Khoo BY. Pilot study and bioinformatics analysis of differentially expressed genes in adipose tissues of rats with excess dietary intake. Mol. Med. Rep. 2020;21:2063–2072. doi: 10.3892/mmr.2020.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elias EG, Hasskamp JH, Sharma BK. Cytokines and growth factors expressed by human cutaneous melanoma. Cancers. 2010;2:794–808. doi: 10.3390/cancers2020794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ho CY, Dreesen O. Faces of cellular senescence in skin aging. Mech. Ageing Dev. 2021;198:111525. doi: 10.1016/j.mad.2021.111525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All processed data have been provided in the manuscript. The corresponding author upon reasonable request could provide raw data, generated for this study.