Abstract
The Mycobacterium tuberculosis hmp gene encodes a protein which is homologous to flavohemoglobin in Escherichia coli. Northern blotting analysis demonstrated that hmp transcription increased when a microaerophilic culture became oxygen limited as it entered stationary phase at 20 days. There was a fivefold increase of the hmp transcripts during early stationary phase compared with the value which was observed in the exponential growth phase. This induction of hmp transcription was not due to changes in the mRNA stability since the half-life of hmp mRNA was very short in a 20-day microaerophilic culture. No induction of hmp mRNA was observed during entry into stationary phase when the culture was continuously aerated. hmp transcription was induced after a short exposure of a late-exponential-phase culture to anaerobic conditions. These data indicate that oxygen limitation is the trigger for hmp gene transcription. In addition, when a microaerophilic culture entered into the stationary phase at 20 days, transcription of hmp increased to a small extent after exposure to S-nitrosoglutathione (a nitric oxide [NO] releaser) and sodium nitroprusside (an NO+ donor) and decreased after exposure to paraquat (a superoxide generator) and H2O2. In log phase (4 days) and late stationary phase (40 days), the transcription of hmp was unaffected by nitrosative and oxidative stress. Three primer extension products were observed. The −10 region is 100% identical to that of promoter T3 in mycobacteria and shows a strong similarity to the −10 sequence of hmp and rpoS promoters in E. coli. These observations of hmp mRNA induction in response to O2 limitation and nitrosative stress suggest that the hmp gene of M. tuberculosis may have a role in protection of the organism from NO killing under microaerophilic conditions.
Mycobacterium tuberculosis is the single most important infectious pathogen in the world and causes over 3 million deaths annually (23). M. tuberculosis persists in a dormant form in the body, by resisting the effects of the human immune response and chemotherapy. It has been reported that one-third of the human population may be latently infected with dormant tubercle bacilli which can reactivate, causing tuberculosis later in life (40). If it were possible to eradicate the latent organisms, the control of tuberculosis would be improved. An understanding of the molecular basis by which M. tuberculosis becomes dormant is a prerequisite for the identification of molecules in dormant bacteria which can be targeted by new drugs.
It is thought that low oxygen tension in the lesions may be responsible for M. tuberculosis entering a stationary-phase-like dormant state (5, 36, 39). If animals which have experimental tuberculosis are housed in low-oxygen-tension conditions, the level of the disease in the tissues is reduced (36); also the localization of adult pulmonary disease in subapical lesions may be explained by an O2 tension higher than that in the lower parts of the lungs (39). An in vitro dormancy model (43, 45) in which tubercle bacilli settle through an oxygen-depletion gradient in unagitated broth cultures and undergo an orderly metabolic shiftdown with a change from oxygen-dependent metabolic pathways to the glyoxylate cycle (44) has been developed. M. tuberculosis is able to adapt to a nonreplicating persistent stage when the rate of dissolved O2 is reduced in a controlled manner (46). This adaptation to microaerophilic and anaerobic conditions is associated with a marked increase in nitrate reduction (47). It is not known which M. tuberculosis molecules respond to a change in oxygen tension. For other microorganisms, a hemoglobin-like protein which is controlled by changes in O2 concentration has been described. It has been reported that an increase in cell density and a decrease in oxygen tension result in a 50-fold induction of Vitreoscilla single-domain hemoglobin (VtHB) (4). Overexpression of the Vitreoscilla globin (vgb) gene in Escherichia coli enhances growth under microaerophilic conditions, suggesting that the protein may have roles in O2 storage or facilitation of O2 transport (22). Transcriptional fusions of the vgb promoter in E. coli exhibit a five- to sevenfold induction in the reporter gene expression under microaerophilic conditions, suggesting that the vgb promoter is regulated by oxygen tension (11). Limitation of oxygen supply also causes a 20-fold increase in intracellular flavohemoprotein concentration in Alcaligenes eutrophus (35). Overexpression of VtHB protein in E. coli terminal oxidase mutants restored the enzyme activity of the cells, suggesting that VtHB protein may have oxidase activity (12). The expression of the hmp gene of Bacillus subtilis, encoding flavohemoglobin, is strongly induced in response to O2 limitation and by nitrite (24). These observations indicate that hmp may be involved in anaerobic metabolism. The hmp expression in E. coli was not induced by low O2 tension but was stimulated by nitrate, nitrite, and nitric oxide (NO), suggesting that hmp may participate in metabolism of nitrogen compounds (34). Unlike the single-domain VtHB protein, flavohemoglobin proteins usually have two domains. The N terminus contains the heme domain, which binds oxygen to form an oxygenated complex (21, 32), and the C-terminal domain is homologous to the ferredoxin NADP+ reductase protein with potential binding sites for flavin adenine dinucleotide and NAD(P)H (1). It has been suggested that flavohemoglobin in E. coli might act as an oxygen sensor based on the observation that flavin reduction increases upon deoxygenation of the heme domain during oxygen limitation (33).
In addition to its putative roles in oxygen diffusion and metabolism, flavohemoglobin is also proposed to function in protection of microorganisms from oxidative and nitrosative stress. It has been reported that a strain of Saccharomyces cerevisiae in which the hmp gene is deleted becomes sensitive to oxidative stress (49). In E. coli, hmp expression was induced in the presence of paraquat (a superoxide generator) (28), NO, S-nitrosoglutathione (GSNO; an NO releaser), or sodium nitroprusside (SNP; an NO+ donor) (29, 30). This induction is independent of the SoxRS regulon which is activated by superoxide-generating agents and NO (30). The hmp gene mutant is very sensitive to these nitrogen oxide species (30). It has been demonstrated that the E. coli flavohemoglobin may function as an NO dioxygenase which participates in S-nitrosothiol and NO metabolism and protects the bacterium against NO killing (16, 18). In Salmonella typhimurium, growth of hmp gene deletion mutants was not affected by paraquat and H2O2 but was sensitive to NO generators, acidified nitrite, and S-nitrosothiols (10). These observations suggest that flavohemoglobin of these microorganisms may play an important role in protection against oxidative and nitrosative stress.
The complete genome sequence of M. tuberculosis (7) has revealed an open reading frame which shows 30.8% identity in a 211-amino-acid overlap to E. coli hmp-encoded protein. The hmp gene of M. tuberculosis has been allocated in the category of 171 miscellaneous oxidoreductases and oxygenases in the genome sequence. Taking advantage of the complete M. tuberculosis genome sequence, we studied the transcription of the M. tuberculosis hmp gene under different conditions of oxygen supply and growth phases in order to investigate the possible link between hmp expression and differential gene expression associated with microaerophilic adaptation to stationary phase by using an in vitro microaerophilic model of dormancy (43, 45). We have recently reported that M. tuberculosis protein synthesis is shut down in this model (20). We also examined hmp transcription in response to various oxidative and nitrosative agents to investigate the possible roles of hmp in protection of M. tuberculosis from oxidative and nitrosative stress. Such data contribute to a better understanding of the molecular mechanisms of stationary-phase adaptation and survival in M. tuberculosis.
MATERIALS AND METHODS
Bacteria and culture.
M. tuberculosis H37Rv was grown at 37°C in Middlebrook 7H9 medium containing 0.05% Tween 80 supplemented with 10% albumin dextrose complex (Difco Laboratories). Samples of a 10-day mid-log-phase culture were stored at −70°C. They were thawed and subcultivated once for 10 days before being inoculated 1:10 in fresh medium to form the experimental cultures. Microaerophilic growth was achieved by incubating 10-ml cultures in 28-ml screw-cap bottles without shaking for up to 60 days. Anaerobic cultures were obtained by incubating 3 ml of the cultures with loosened caps in a jar (GasPak 150 system; Becton Dickinson) in which H2 and CO2 were generated with GasPaks (Oxoid) and checked with anaerobic indicators (Oxoid). Aerated growth was obtained by incubating 10-ml cultures on a shaker (Vibrax; Kikalabortechnik) at 150 rpm; at 24-h intervals, the bottles were opened once in order to ensure sufficient oxygen supply. Estimation of viability as CFU was performed by the method described previously (20).
Estimation of oxygen consumption.
Dissolved O2 was measured in a sealed and undisturbed culture in a glass incubation chamber of the oxygen electrode system (Digital model 10) with the electrodes (a platinum working electrode and an Ag-AgCI reference electrode) mounted at the bottom of the chamber (Rank Brothers Ltd., Cambridge, United Kingdom). This method was used only to measure the oxygen consumption of the bacilli at the bottom of the container. Dissolved O2 was also measured in a continuously disturbed culture which was stirred with a mini-magnetic bar at 100 rpm. At 1- to 2-day intervals, the incubation chamber was opened in order to ensure sufficient oxygen supply.
DNA manipulations, sequencing, and analysis.
DNA isolation, ethanol precipitation of DNA, electrophoresis of DNA in agarose, and transformations were performed by standard procedures (37). The TOPO TA cloning kit (Invitrogen) was used for cloning of PCR products. DNA for sequencing was isolated with a plasmid miniprep kit (Qiagen). Sequencing reactions were carried out on double-stranded plasmid DNA with the T7 Sequenase version 2.0 sequencing kit (U.S. Biochemical) in accordance with the manufacturer’s instructions, based on the dideoxy chain termination method (38) with α-35S-dATP (specific activity, >1,000 Ci/mmol; Amersham) as the radioactive label. Computer-aided analysis of the DNA sequence was performed by using the GCG sequence analysis software package (University of Wisconsin Biotechnology Center, Madison).
PCR amplification of DNA.
PCR was used to generate a 620-bp sequence which began at 421 bp upstream and ended at 196 bp downstream of the TTG start codon of the hmp gene for sequencing the DNA ladder used in primer extension experiments. The primers used were 5′-TGCACGCCGACGATTGAGC-3′ and 5′-TACGCTCGCTGGGCACGC-3′. The probes for detecting the hmp mRNA (EMBL-GenBank accession no. Z92774, coding sequence nucleotide [nt] 17664 to 18741), cspA mRNA (EMBL-GenBank accession no. Z95436, coding sequence nt 28425 to 28628), ftsZ mRNA (EMBL-GenBank accession no. Z95388, coding sequence nt 15919 to 17058), and 16S rRNA (EMBL-GenBank accession no. mtu16srn) were also prepared by PCR. PCR amplification was performed with 1 to 5 ng of M. tuberculosis chromosomal DNA in a final volume of 50 μl containing 1 μM (each) primers; 200 μM (each) dATP, dCTP, dGTP, and dTTP; 1 U of Taq polymerase (Promega); and a buffer supplied with the enzyme. Amplification was carried out for 30 cycles as follows: denaturation for 1 min at 94°C, primer annealing for 2 min at 55°C, and primer extension at 72°C for 3 min. Primers which were designed to be in the coding regions of the transcripts were as follows: hmp (5′-TCACGGTCAAACGAACCGCC-3′ and 5′-GGGTTGTGGGGACGAAGTTG-3′; for position, see Fig. 5), cspA (5′-GAGAAGGGGTTCGGCTTTAT-3′, nt 28595 to 28576, and 5′-CTGGTTTTCTTCAAGGGTGC-3′, nt 28491 to 28510), ftsZ (5′-GTCGTGGGTATCGGTGGTGG-3′, nt 17022 to 17003, and 5′-ATCTCGTCCTTGGCGTCCTC-3′, nt 16802 to 16821), and 16S (5′-GCCTGGGAAACTGGGTCTAA-3′, nt 109 to 128, and 5′-TCTCCACCTACCGTCAATCC-3′, nt 427 to 446).
FIG. 5.
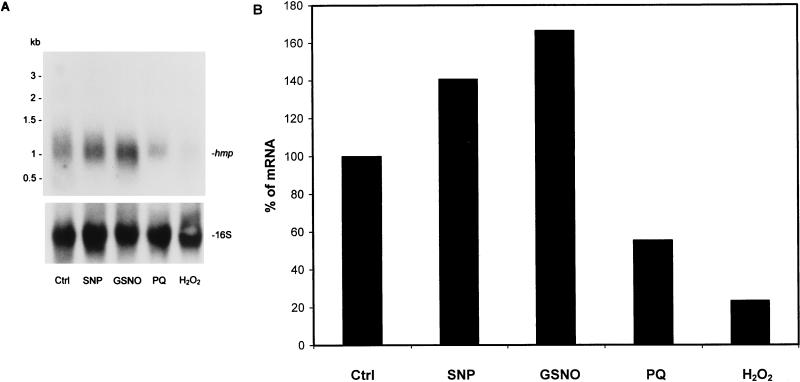
Northern blot analysis of the hmp mRNA level in response to nitrosative and oxidative stresses. RNA was extracted from a 20-day microaerophilic culture after exposure to different stress conditions described in Materials and Methods and subjected to Northern blot analysis. (A) Northern blot hybridization of hmp mRNA. The same blots were stripped and hybridized with the 16S rRNA-specific probe. (B) Densitometric analysis of hmp mRNA level in response to stresses. The values shown are the averages of two independent experiments. Ctrl, control; PQ, paraquat.
RNA extraction.
Total RNA extraction from cultures was carried out by the method of Mangan et al. (25). After isopropanol precipitation, the RNA pellets were treated with RNase-free DNase I (Life Technologies), phenol and chloroform extracted, and reprecipitated. RNA concentration was determined spectrophotometrically at 260 nm.
Northern blotting analysis.
Northern blotting analysis was performed by fractionation of RNA samples on a 1.2% agarose gel containing 6.5% formaldehyde, followed by transfer to a Hybond-N filter (Amersham) in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (37). The same amount of total RNA (20 μg) was loaded into each well of the gel. Sizes were determined with an RNA ladder (Sigma) as the molecular size standard. The probe which was purified with QIAquick PCR purification kits (Qiagen) was labelled with [α-32P]dCTP (specific activity, >3,000 Ci/mmol; Amersham) by the random priming method according to the instructions of the manufacturer (Amersham). Generally, 1 × 105 to 5 × 105 cpm/ml was used in hybridization for probes with a specific activity of more than 108 cpm/μg. After prehybridization for 3 h at 42°C in a buffer containing 5× Denhardt’s solution, 5× SSC, 0.2% sodium dodecyl sulfate (SDS), 50% formamide, and 100 μg of salmon sperm DNA (Sigma) per ml, the filters were hybridized overnight at 42°C with 32P-labelled probes in the same buffer and washed at high stringency (6× SSC–1% SDS, 1× SSC–1% SDS, and 0.1× SSC–1% SDS at 68°C for 45 min, respectively). The same filters were stripped and reprobed for 16S rRNA to verify equal loading of RNA. The filters were exposed to X-ray films. The films, exposed for various periods, were scanned with a high-resolution laser Personal Densitometer SI (Molecular Dynamics) linked to ImageQuant software (Molecular Dynamics).
Chemical half-life determination.
Total RNA was isolated from an early-stationary-phase (20 days) microaerophilic culture at selected intervals after transcription initiation was inhibited by the addition of 100 μg of rifampin (Sigma) per ml, and the half-life was determined by Northern blotting analysis. The incorporation of [3H]uridine (activity, 31 to 56 Ci/mmol; Amersham) into trichloroacetic-acid-precipitable RNA was rapidly reduced to 2% of that of the drug-free control (data not shown) after addition of 100 μg of rifampin per ml, showing that transcription initiation was blocked with this concentration of rifampin.
Analysis of RNA accumulation under stress conditions.
A series of 10-ml cultures were used for the determination of stress responses. For oxidative stress, H2O2 and paraquat were added to the cultures at the final concentrations of 10 mM and 200 μM, respectively, for 30 min. For nitrosative stress, GSNO and SNP were added to the cultures at the final concentrations of 1 and 10 mM, respectively, for 1 h. RNA was extracted after exposure of the cells to these stress conditions and analyzed by Northern blotting.
Primer extension.
The synthetic oligonucleotide PE1 (5′-GCCGAGTGGCTCGTCTCCAA-3′, 12 to 31 nt downstream of hmp gene TTG start codon) and PE2 (5′-TCGTCGACGAAACCGACG-3′, 62 to 79 nt downstream of hmp gene TTG start codon) were 5′ end labelled with [γ-32P]ATP (specific activity, >3,000 Ci/mmol; Amersham) by using Ready-To-Go T4 polynucleotide kinase (Pharmacia Biotech). Prior to primer extension, the total RNA was analyzed by Northern blotting and hybridized with the 16S rRNA-specific probe in order to verify that equal amounts of total RNA were used for each time point. Total RNAs (40 μg) from different growth phases were annealed with 1 μl of 5′-end-labelled primer (approximately 105 cpm) in 5× reverse transcriptase buffer (Life Technologies) at 72°C for 20 min and then slowly cooled to room temperature in 30 min. Primer extension was performed in the same solution for 1 h at 42°C containing 500 μM (each) dATP, dCTP, dGTP, and dTTP; 40 U of RNasin (Promega); 5 mM dithiothreitol; and 200 U of Superscript II reverse transcriptase (Life Technologies). The primer extension products were precipitated with ethanol and sodium acetate at −70°C, washed with 70% ethanol, and dried. The pellets were resuspended in an appropriate amount of formamide dye solution (U.S. Biochemical) and then separated on a 6% polyacrylamide sequencing gel containing 8 M urea adjacent to a DNA sequence generated with the same primer.
Nucleotide sequence accession number.
The nucleotide sequence accession number for the M. tuberculosis hmp gene is EMBL-GenBank Z92774 (coding sequence nt 17664 to 18741); the gene name is Rv3571 (7).
RESULTS
hmp gene transcription in different growth phases.
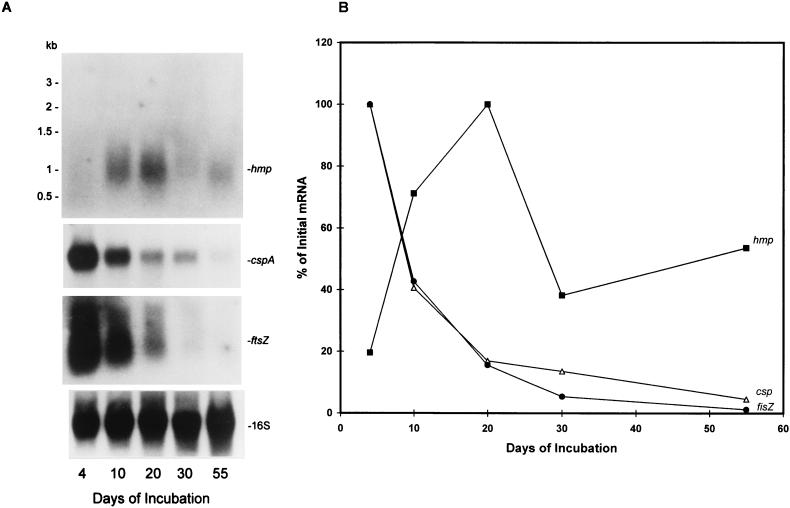
In order to determine if hmp gene transcription was dependent on the growth phase of a culture, RNA was isolated from bacilli taken at different growth phases under unagitated microaerophilic conditions across the growth curve (Fig. 1, growth curve a) and was analyzed by Northern blotting. In our model (Fig. 1, growth curve a), entry into stationary phase, defined as slowing of log-phase growth, began at about 10 to 20 days and ended at 30 to 40 days when the bacilli entered the stationary phase. Oxygen concentration, which was measured at the bottom of the culture, decreased from 20% (air-saturated 7H9 medium) to 0.1% during the first 24 h of incubation and remained at <0.1% for the rest of the incubation period. This indicates that in these cultures the oxygen gradient is established within 24 h, after which a low concentration of oxygen is maintained in the bottom of the culture and a higher but gradually decreasing oxygen concentration is maintained in the top of the culture (43, 44). In log-phase-growth cultures, the majority of the bacilli are dispersed throughout the culture, but in unagitated cultures, they gradually settle to the bottom of the container over several days where they enter the stationary growth phase. As shown in Fig. 2A (top panel), the hmp RNA was weakly expressed during exponential growth (4 days) when there was an abundance of oxygen in the top layer of the medium. As the culture entered the late exponential growth period at 10 days, when oxygen tension began to decrease as a result of bacterial consumption, hmp transcription increased and reached its maximum level at early stationary phase (20 days). After this, the transcript level decreased while the bacilli adapted to the late stationary phase. The length of the hmp transcript is approximately 1.1 kb, and this matches the prediction of the hmp mRNA length based on DNA sequence and primer extension analysis (see below). The blots were stripped and reprobed to detect 16S rRNA, cspA mRNA, and ftsZ mRNA. The purpose of examining cspA and ftsZ mRNA in these samples was to compare the transcription of other genes with that of hmp. cspA encodes cold shock protein A, which has 73.4% amino acid identity to the cold shock protein of Arthrobacter globiformis (3). ftsZ encodes a cell division protein having 77.3% amino acid identity to the ftsZ gene product of Streptomyces coelicolor (26). As shown in Fig. 2A, the levels of cspA and ftsZ mRNA were maximal at log-phase growth (4 days) and then gradually decreased as the bacilli reached stationary phase, in agreement with our previous finding that total protein synthesis showed substantial reduction in the progress to stationary phase (20). The level of 16S rRNA (Fig. 2A, bottom panel) was relatively constant throughout different growth phases, indicating that equal amounts of total RNA were loaded into each well of the gel. Densitometric analysis of the autoradiographs shown in Fig. 2A and others from two independent experiments revealed that there was a 5- ± 0.02-fold increase of the hmp transcript in early stationary phase relative to the values observed in the early exponential phase. In contrast, there were 22- ± 0.07-fold and 23- ± 0.07-fold decreases of the cspA mRNA and ftsZ mRNA, respectively, in late stationary phase compared with the value measured in log phase (Fig. 2B).
FIG. 1.
Growth curves of M. tuberculosis H37Rv. The bacilli were grown in 7H9 medium containing 0.05% Tween 80 supplemented with 10% albumin dextrose complex without shaking and with shaking as described in Materials and Methods. Viability was estimated as CFU per milliliter at 0, 4, 10, 20, 30, 40, 50, 55, and 60 days of unagitated incubation (black squares; curve a) and at 0, 4, 10, and 30 days of agitated incubation (white squares; curve b). The values shown are the averages of three independent experiments.
FIG. 2.
Northern blotting analysis of steady-state levels of the hmp mRNA in M. tuberculosis. (A) RNA was extracted at 4, 10, 20, 30, and 55 days of microaerophilic incubation across growth curve a (Fig. 1) and analyzed by formaldehyde-agarose gel electrophoresis and Northern blotting as described in Materials and Methods. The filter was hybridized with an hmp gene-specific probe. The blot was stripped and reprobed with cspA-, ftsZ mRNA-, and 16S rRNA-specific probes. (B) Densitometric analysis of the autoradiographs and two other independent experiments showing the steady-state levels of the hmp, cspA, and ftsZ transcripts. The quantification was based on several exposures of different periods. The signal obtained from each band for each mRNA was divided by the corresponding signal of 16S rRNA. The corrected data of the bands for each mRNA were plotted against the days of incubation and expressed relative to maximal value.
Determination of the hmp mRNA half-life in a microaerophilic culture.
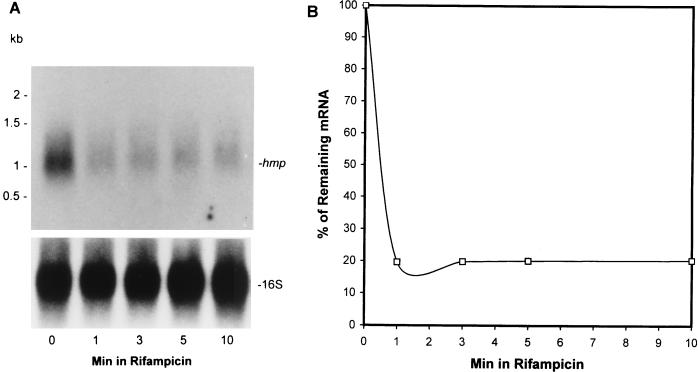
Since the steady-state level of any mRNA species is determined by the rate of transcriptional initiation and the rate of decay, we determined the hmp mRNA half-life in a 20-day stationary-phase culture after transcription initiation was inhibited with 100 μg of rifampin per ml. The autoradiograph shown in Fig. 3A (top panel) revealed that the hmp mRNA was very unstable, with a half-life of less than 1 min determined by densitometric analysis (Fig. 3B). This indicates that the high level of hmp mRNA which is present at 20 days is due to an increase in transcriptional initiation and not due to enhanced stability of the mRNA. The blot was stripped and reprobed to detect 16S rRNA (Fig. 3A, bottom panel) in order to ensure that an equal amount of total RNA was loaded into each lane. Determination of the hmp half-life in log-phase and late-stationary-phase cultures was not feasible since the steady-state levels of the hmp mRNA in those cultures were very low (Fig. 2A) and the decay of the mRNA after treatment with rifampin was below the level of detection.
FIG. 3.
Northern blot analysis of decay of hmp mRNA in 20-day-old stationary-phase bacilli. (A) Total RNA was extracted from the bacilli at 0, 1, 3, 5, and 10 min after addition of 100 μg of rifampin per ml. (B) Densitometric analysis of decay of hmp mRNA. The quantification is based on several exposures of the autoradiographs (A) and two others from independent experiments for different periods. The signals of the bands were plotted against the times of the RNA isolation and expressed as the percentages of the initial values. The half-life calculated from the blots was 0.8 ± 0.05 min.
hmp gene transcription under aerobic and anaerobic conditions.
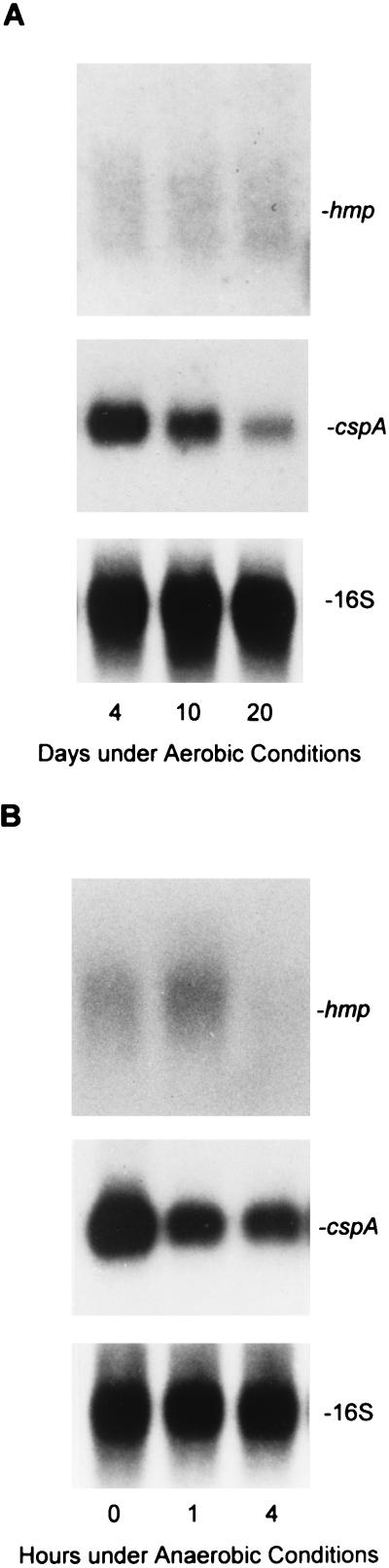
We then posed the question whether hmp induction, during entry into the stationary phase, was due either to the decrease in oxygen tension caused by bacterial consumption or to growth rate changes independent of O2 level. We thus measured the transcription of the hmp gene under aerobic conditions. Total RNA was prepared from samples taken after 4, 10, and 20 days of aerobic incubation across the growth curve (Fig. 1, growth curve b) and subjected to Northern blotting analysis. O2 concentration measured at the bottom of the culture with continuous stirring decreased from 20 to 18% over 30 days of the incubation, showing no significant change. As shown in Fig. 4A (top panel), hmp was expressed at low levels throughout different growth phases, showing no induction of the mRNA when the bacilli entered the stationary phase with respect to continuous oxygen availability. The blot was stripped and rehybridized to detect cspA. The steady-state level of cspA mRNA exhibited the similar pattern as that in microaerophilic growth, in which an approximately 6.7- ± 0.04-fold decrease was observed in stationary phase compared to log-phase growth (Fig. 4A, middle panel). The blot was stripped and reprobed with the 16S rRNA probe and showed that an equal amount of total RNA was loaded in each lane (Fig. 4A, bottom panel).
FIG. 4.
Effects of aerobic and anaerobic conditions on transcription of the hmp gene. (A) RNA was extracted at 4, 10, and 20 days of aerobic incubation (Fig. 1, growth curve b) and analyzed by Northern blotting. The filter was hybridized to detect hmp mRNA (upper panel) and then reprobed to detect cspA mRNA (middle panel) and 16S rRNA (lower panel) after being stripped. (B) RNA was extracted from a mid-log-phase culture (7 days) (Fig. 1, growth curve b) after exposure to anaerobic conditions for 0, 1, and 4 h and subjected to Northern blotting analysis. Hybridization of hmp mRNA (upper panel) and rehybridization of cspA mRNA (middle panel) and 16S rRNA (lower panel) were performed as described for panel A. These results have been independently confirmed in two additional experiments.
To further investigate the effects of growth phase and oxygen tension on the hmp expression, we took a mid-log-phase microaerophilic culture (7 days) and exposed it to anaerobic conditions to examine if hmp is up-regulated by low oxygen in mid-log-phase bacilli rather than the early-stationary-phase bacilli. As shown in Fig. 4B, after exposure of the culture to anaerobiosis for 1 h, the hmp mRNA increased approximately 1.45- ± 0.05-fold and decreased rapidly after 4 h of anaerobic incubation (top panel). Exposure to anaerobiosis did not kill the bacilli since CFU counts remained constant after 24 h of anaerobic incubation. In contrast, anaerobic incubation for 1 and 4 h resulted in 2.2- ± 0.05-fold and 2.5- ± 0.05-fold reduction of cspA mRNA, respectively (Fig. 4B, middle panel). The equal loading of total RNA was confirmed by the same intensity of the 16S rRNA bands shown in Fig. 4B, bottom panel. These observations suggest that oxygen limitation rather than growth rate change is the real trigger for hmp induction.
hmp gene transcription under oxidative and nitrosative stress conditions.
In order to determine if the hmp expression was affected by oxidative and nitrosative stress, we exposed a 20-day microaerophilic culture to H2O2, paraquat, GSNO, and SNP and analyzed the transcription of hmp by Northern blotting. As shown in Fig. 5A (top panel) and 5B, compared with the steady-state level of the mRNA (control), transcription of hmp was increased 1.4- ± 0.01-fold after exposure to SNP and 1.6- ± 0.04-fold after exposure to GSNO. The level of hmp mRNA decreased after exposure to hydrogen peroxide and paraquat. The blot was stripped and reprobed to detect 16S rRNA (Fig. 5A, bottom panel) in order to ensure that an equal amount of total RNA was loaded into each lane. The same experiments were also performed with log-phase (4-day) and late-stationary-phase (40-day) cultures. No changes in the level of hmp mRNA were observed in response to any of the stress conditions (data not shown). Viability of the bacilli was assessed by CFU counts after exposure of the organisms to the stress conditions described above. The CFU counts were unaltered by any of these stress conditions.
Primer extension analysis.
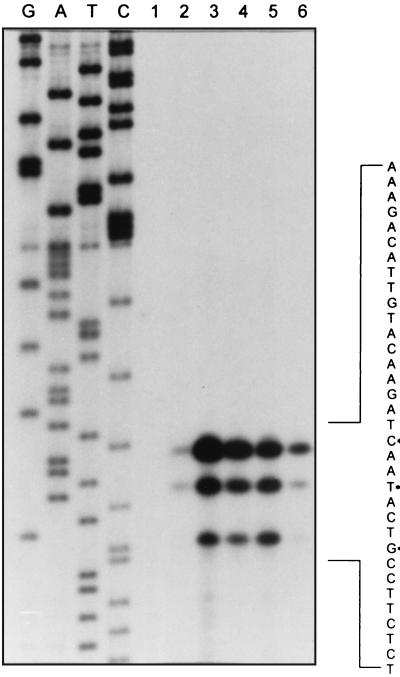
Total RNA was harvested at various times from the microaerophilic growth cultures (Fig. 1, growth curve a) and was subjected to primer extension analysis by using the oligonucleotide primer PE1 in order to identify the transcription start sites for the hmp mRNA. As shown in Fig. 6, three primer extension products of 58, 55, and 51 nt which correspond to G, A, and C positions 25, 22, and 18 nt (Fig. 7) upstream of the TTG start codon were found. This identifies the potential 5′ end of hmp mRNA of about 1.1 kb in size. This result was independently confirmed by using the second primer, PE2 (Materials and Methods). As shown in Fig. 7, when the sequence centered around −10 and −35 regions upstream from the transcription start sites was examined for promoter-like sequences, we found that the −10 region (TAACAT) contains a highly conserved sequence identical to the mycobacterial promoter T3 which is one of the promoters randomly selected by a shuttle vector (2) and shows a clear similarity to the −10 region (TAAGAT) of the E. coli hmp promoter (27) and the −10 sequence (TATCAT) of the E. coli katF (ςS gene) promoter (31). The sequence in the −35 region, assuming an optimal spacing of 17 bp between −10 and −35, has a 3- of 6-nt match to the E. coli −35 consensus sequence (TTGACA) but has no similarity to that of mycobacterial promoters, which was consistent with previous findings (2). A highly conserved Shine-Dalgarno sequence is centered 8 bp upstream of the TTG initiation codon. The upstream noncoding region was amplified by PCR, cloned, and sequenced and was identical to the DNA sequence (GenBank accession no. Z92774) from the M. tuberculosis chromosome.
FIG. 6.
Determination of the hmp transcription initiation site. Primer extension analysis was performed as described in Materials and Methods with total RNA prepared from cultures at different growth phases. Lanes G, A, T, and C contained sequence reactions generated with the same primer. Lanes 2, 3, 4, 5, and 6 indicate the primer extension products with the RNA isolated from the bacilli after 4, 15, 20, 25, and 30 days of unagitated incubation (Fig. 1, growth curve a), respectively. A reaction without RNA is shown in lane 1. Primer extension points are marked by asterisks in the DNA sequence shown on the right, which is the nontranscribed strand and is the complement of the sequence that is readable from the sequencing ladder.
FIG. 7.
DNA sequence of the hmp gene regulatory region. Shown are the partial region encoding hmp and a 146-bp upstream sequence. The three primer extension points which correlated with nucleotides G, A, and C are indicated by asterisks. The putative promoter regions are highlighted by boxes and labelled −10 and −35. The putative ribosome-binding site is marked with a double underline and is labelled SD. The start codon is in boldface. The primers used for generating the Northern blotting probe are underlined.
Primer extension analysis was also used to examine the temporal patterns of hmp gene transcription (Fig. 6). The signals were much stronger during entry into the stationary phase. Densitometric analysis revealed that the temporal patterns of the three bands were very similar and closely matched the patterns in Northern blotting.
DISCUSSION
We have shown that oxygen tension modulates M. tuberculosis hmp gene transcription. Northern blotting analysis revealed that transcription of the hmp gene increased when a microaerophilic culture became oxygen limited as it reached stationary phase. This was not due to the changes in the mRNA stability since the hmp mRNA was very unstable in a 20-day stationary-phase culture. This increase in hmp transcription was not observed throughout different growth phases when the cultures were continuously aerated, indicating that oxygen abundance inhibits hmp expression. Furthermore, hmp transcription was significantly induced after a short exposure of an exponential-growth-phase culture to anaerobic conditions, showing that an increase in hmp expression is associated with a decrease in oxygen tension. Our previous studies have demonstrated that M. tuberculosis, under anaerobic conditions, rapidly reduces or completely switches off protein synthesis in order to achieve a shutdown of cellular metabolic activity (20). hmp mRNA disappears quickly under further anaerobic incubation. A possible explanation for this might be the metabolic shutdown which is seen during anaerobiosis. The transient increase in the level of hmp mRNA during entry into stationary growth phase when O2 became limiting was in contrast to the mRNA levels of other genes such as cspA and ftsZ mRNAs, whose transcription decreased during the same period. These observations suggest that oxygen limitation triggers hmp induction. In addition, we have observed that transcription of the hmp gene in early-stationary-phase cultures was slightly increased in response to GSNO and SNP, suggesting that the hmp gene product may be involved in the protection of the organisms from nitrosative stress.
The primer extension analysis reveals three termination points which may be representative of 5′ ends of the mRNA. However, these three signals do not necessarily mean that there are three transcription start sites, since the two shorter products may result from either incomplete extension of the primer or the degradation or processing of the mRNA species. It is possible that the longest extension product reveals the potential hmp transcription start site. Further experiments with S1 nuclease mapping will help to resolve these possibilities. The primer extension results together with Northern blotting data suggest that the unit of transcription might be monocistronic, which is consistent with the findings for E. coli (27). The hmp gene also contains a potential ribosome-binding site which is just upstream of the TTG start codon and is correctly located to initiate the translation of an open reading frame. Hence, the most likely translational start site is the TTG codon. In M. tuberculosis, TTG start codons have been predicted for approximately 4% of the coding regions (7, 31a). The putative promoter sequences of the M. tuberculosis hmp gene show a similarity to that of the E. coli hmp gene. In E. coli, induction of hmp in stationary phase is dependent on ςS (27); also, ςS is the dominant regulator of hmp expression in the presence of paraquat during stationary phase (28). It is not known which ς factor controls the expression of the hmp gene. Recently, it has been demonstrated that the transcription of the M. tuberculosis sigB gene is induced during transition from log phase to stationary phase and under stress conditions, which suggests that the sigB gene may encode an alternative sigma factor (19).
Hemoglobin-like genes are present in many microorganisms including E. coli (1, 21, 41), Vitreoscilla sp. (4, 11, 12, 22, 42), A. eutrophus (8, 35), Erwinia chrysanthemi (13), B. subtilis (24), Rhizobium meliloti (17), S. typhimurium (10), and S. cerevisiae (9, 49). It is still not clear what role the proteins play in the microorganisms, although several possible functions have been proposed, including oxygen transport and storage, oxidase and reductase activities, and oxygen sensing (reviewed in reference 32). The hmp gene may also be involved in anaerobic metabolism. In B. subtilis, hmp gene induction under anaerobic conditions is dependent on ResDE (two-component signal transduction proteins), FNR (anaerobic regulator), and NarGHJI (respiratory nitrate reductases) (24). In E. coli, hmp expression is negatively regulated by FNR under anaerobic conditions, is induced by nitrite and NO, and may participate in anaerobic metabolism of nitrogen compounds (34). No binding sites identical to E. coli FNR protein were found in the region of the hmp promoter in M. tuberculosis, indicating a possible difference in regulation mechanisms or functions of hmp between the two species. It is not known what role the M. tuberculosis hmp gene plays in response to O2 limitation, and its involvement in anaerobic metabolism requires further investigation. Recently, a body of evidence has arisen from the discovery that the bacterial hmp gene might function in response to nitrosative or oxidative stress (10, 16, 18, 28–30). A defined hmp gene mutant of E. coli is hypersensitive to nitrosating agents and NO-related species (30). Hmp protein may act as an NO dioxygenase which converts NO to NO3− (16, 18). The hmp gene deletion mutant of S. typhimurium showed an increasing sensitivity to acidified nitrite and S-nitrosathiols, but the growth of the mutant was not inhibited by oxidative stress (10). These observations strongly suggest that the hmp gene plays an important role in protection of bacteria from NO attack. We found that the hmp gene in M. tuberculosis was induced to a small extent by nitrosating agents under microaerophilic conditions. M. tuberculosis is capable of replicating and persisting within mononuclear phagocytic cells. Activated macrophages generate reactive nitrogen intermediates (RNI) including NO, NO2−, and NO3−, which play an important role in controlling the bacteria in the host (6, 14). The ability of M. tuberculosis to survive RNI attack involves an array of proteins which are synthesized in response to this stress. The 16-kDa α-crystalline-like protein, which is a stationary-phase-associated protein (48), is induced under RNI stress (15). The induction of hmp mRNA in response to these NO generators indicates that the Hmp protein might have a role in the protection of M. tuberculosis from nitrosative stress when O2 tension decreases, which might help the organism to persist in inflammatory and necrotic lesions of the human host. The hmp mRNA level was reduced by oxidative stress, which was consistent with the finding for S. typhimurium (10), suggesting that the regulation of the response to NO is different from that of the response to oxidative stress in M. tuberculosis and S. typhimurium. Gardner et al. suggested that the induction of (flavo)hemoglobins under microaerophilic conditions might be beneficial for bacteria to survive NO killing since the cellular level of NO dioxygenase increased when O2 became limiting (16). It is not known if this is the case in M. tuberculosis. Future work will aim to construct an hmp gene deletion mutant of M. tuberculosis which will help to elucidate the role that hmp may play in response to oxygen limitation and nitrosative stress.
ACKNOWLEDGMENTS
We thank the British Medical Research Council for support and thank The Wellcome Trust for support of M.-A. Rajandream.
We thank Julian Parkhill, Sanger Centre, Hinxton, Cambridgeshire, United Kingdom, for his advice about the start codon usage in M. tuberculosis.
REFERENCES
- 1.Andrews S C, Shipley D, Keen J N, Findlay J B C, Harrison P M, Guest J R. The haemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal shares homology with ferredoxin NADP+ reductases. FEBS Lett. 1992;302:247–252. doi: 10.1016/0014-5793(92)80452-m. [DOI] [PubMed] [Google Scholar]
- 2.Bashyam M D, Kaushal D, Dasgupta S K, Tyagi A K. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger F, Normand P, Potier P. capA, a cspA-like gene that encodes a cold acclimation protein in the psychrotrophic bacterium Arthrobacter globiformis SI55. J Bacteriol. 1997;179:5670–5676. doi: 10.1128/jb.179.18.5670-5676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerman S J, Webster D A. Control of heme content in Vitreoscilla by oxygen. J Gen Appl Microbiol. 1982;28:35–43. [Google Scholar]
- 5.Canetti G. The tubercle bacillus in the pulmonary lesion of man. The histobacteriogenesis of tuberculous lesions: experimental studies. New York, N.Y: Spring Publishing Company, Inc.; 1955. pp. 87–90. [Google Scholar]
- 6.Chan J, Xing Y, Magliozzo R S, Bloom B R. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Cramm R, Siddiqui R A, Friedrich B. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J Biol Chem. 1994;269:7349–7354. [PubMed] [Google Scholar]
- 9.Crawford M J, Sherman D R, Goldberg D E. Regulation of Saccharomyces cerevisiae flavohemoglobin gene expression. J Biol Chem. 1995;270:6997–7003. doi: 10.1074/jbc.270.12.6991. [DOI] [PubMed] [Google Scholar]
- 10.Crawford M J, Goldberg D E. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J Biol Chem. 1998;273:34028–34032. doi: 10.1074/jbc.273.51.34028. [DOI] [PubMed] [Google Scholar]
- 11.Dikshit K L, Dikshit R P, Webster D A. Study of Vitreoscilla globin (vgb) gene expression and promoter activity in E. coli through transcriptional fusion. Nucleic Acids Res. 1990;18:4149–4155. doi: 10.1093/nar/18.14.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikshit R P, Dikshit K L, Liu Y X, Webster D A. The bacterial hemoglobin from Vitreoscilla can support the aerobic growth of Escherichia coli lacking terminal oxidases. Arch Biochem Biophys. 1992;293:241–245. doi: 10.1016/0003-9861(92)90391-9. [DOI] [PubMed] [Google Scholar]
- 13.Favey S, Labesse G, Vouille V, Boccara M. Flavohemoprotein HMPX: a new pathogenicity determinant in Erwinia chrysanthemi strain 3937. Microbiology. 1995;141:863–871. doi: 10.1099/13500872-141-4-863. [DOI] [PubMed] [Google Scholar]
- 14.Flesch I E A, Kaufmann S H E. Mechanisms involved in mycobacterial growth inhibition by gamma interferon-activated bone marrow macrophages: role of reactive nitrogen intermediates. Infect Immun. 1991;59:3213–3218. doi: 10.1128/iai.59.9.3213-3218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbe T R, Hibler N S, Deretic V. Response to reactive nitrogen intermediates in Mycobacterium tuberculosis: induction of the 16-kilodalton α-crystallin homolog by exposure to nitric oxide donors. Infect Immun. 1999;67:460–465. doi: 10.1128/iai.67.1.460-465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner P R, Gardner A M, Martin L A, Salzman A L. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilles-Gonzalez M A, Ditta G S, Helinski D R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 18.Hausladen A, Gow A J, Stamler J S. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y M, Butcher P D, Sole K, Mitchison D A, Coates A R M. Protein synthesis is shut down in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol Lett. 1998;158:139–145. doi: 10.1111/j.1574-6968.1998.tb12813.x. [DOI] [PubMed] [Google Scholar]
- 21.Ioannidis N, Cooper C E, Poole R K. Spectroscopic studies on an oxygen-binding haemoglobin-like flavohaemoprotein from Escherichia coli. Biochem J. 1992;288:649–655. doi: 10.1042/bj2880649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khosla C, Bailey J E. The Vitreoscilla hemoglobin gene: molecular cloning, nucleotide sequence and genetic expression in Escherichia coli. Mol Gen Genet. 1988;214:158–161. doi: 10.1007/BF00340195. [DOI] [PubMed] [Google Scholar]
- 23.Kochi A. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle. 1991;72:1–6. doi: 10.1016/0041-3879(91)90017-m. [DOI] [PubMed] [Google Scholar]
- 24.LaCelle M, Kumano M, Kurita K, Yamane K, Zuber P, Nakano M M. Oxygen-controlled regulation of the flavohemoglobin gene in Bacillus subtilis. J Bacteriol. 1996;178:3803–3808. doi: 10.1128/jb.178.13.3803-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangan J A, Sole K M, Mitchison D A, Butcher P D. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick J R, Su E P, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 27.Membrillo-Hernández J, Cook G M, Poole R K. Roles of RpoS (ςS), IHF and ppGpp in the expression of the hmp gene encoding the flavohemoglobin (Hmp) of Escherichia coli K-12. Mol Gen Genet. 1997;254:599–603. doi: 10.1007/s004380050457. [DOI] [PubMed] [Google Scholar]
- 28.Membrillo-Hernández J, Kim S O, Cook G M, Poole R K. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by ςS. J Bacteriol. 1997;179:3164–3170. doi: 10.1128/jb.179.10.3164-3170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Membrillo-Hernández J, Coopamah M D, Channa A, Hughes M N, Poole R K. A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohaemoglobin) gene by the ’NO releaser’, S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the gly hmp intergenic region. Mol Microbiol. 1998;29:1101–1112. doi: 10.1046/j.1365-2958.1998.01000.x. [DOI] [PubMed] [Google Scholar]
- 30.Membrillo-Hernández J, Coopamah M D, Anjum M F, Stevanin T M, Kelly A, Hughes M N, Poole R K. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J Biol Chem. 1999;274:748–754. doi: 10.1074/jbc.274.2.748. [DOI] [PubMed] [Google Scholar]
- 31.Mulvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel ς transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Parkhill, J. (Sanger Centre). Personal communication.
- 32.Poole R K. Oxygen reactions with bacterial oxidases and globins: binding, reduction and regulation. Antonie Leeuwenhoek. 1994;65:289–310. doi: 10.1007/BF00872215. [DOI] [PubMed] [Google Scholar]
- 33.Poole R K, Ioannidis N, Orii Y. Reactions of the Escherichia coli flavohaemoglobin (Hmp) with oxygen and reduced nicotinamide adenine dinucleotide: evidence for oxygen switching of flavin oxidoreduction and a mechanism for oxygen sensing. Proc R Soc Lond Biol Sci. 1994;255:251–258. doi: 10.1098/rspb.1994.0036. [DOI] [PubMed] [Google Scholar]
- 34.Poole R K, Anjum M F, Membrillo-Hernández J, Kim S O, Hughes M N, Stewart V. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol. 1996;178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Probst I, Wolf G, Schlegel H G. An oxygen-binding flavohemoprotein from Alcaligenes eutrophus. Biochim Biophys Acta. 1979;576:471–478. doi: 10.1016/0005-2795(79)90422-7. [DOI] [PubMed] [Google Scholar]
- 36.Raffel S. Immunopathology of tuberculosis. Am. Rev. Tuberc. (August Suppl.) 1956. pp. 60–74. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith D T, Abernathy R S, Smith G B, Jr, Bondurant S. The apical localisation of reinfection pulmonary tuberculosis. I. The stream flow theory. Am Rev Tuberc. 1954;70:547. doi: 10.1164/art.1954.70.4.547. [DOI] [PubMed] [Google Scholar]
- 40.Sudre P, ten Dam G, Kochi A. Tuberculosis: a global overview of the situation today. Bull W H O. 1992;70:149–159. [PMC free article] [PubMed] [Google Scholar]
- 41.Vasudevan S G, Armarego W L, Shaw D C, Lilley P E, Dixon N E, Poole R K. Isolation and nucleotide sequence of hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet. 1991;226:49–58. doi: 10.1007/BF00273586. [DOI] [PubMed] [Google Scholar]
- 42.Wakabayashi S, Matsubara H, Webster D A. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. Nature. 1986;322:481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]
- 43.Wayne L G. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis. 1976;114:807–811. doi: 10.1164/arrd.1976.114.4.807. [DOI] [PubMed] [Google Scholar]
- 44.Wayne L G, Lin K Y. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wayne L G. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 46.Wayne L G, Hayes L G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wayne L G, Hayes L G. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:127–132. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- 48.Yuan Y, Crane D D, Barry C E., III Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X J, Raitt D, Burke P V, Clewell A S, Kwast K E, Poyton R O. Function and expression of flavohemoglobin in Saccharomyces cerevisiae. Evidence for a role in the oxidative stress response. J Biol Chem. 1996;271:25131–25138. doi: 10.1074/jbc.271.41.25131. [DOI] [PubMed] [Google Scholar]