Abstract
Phaeocystis is a cosmopolitan, bloom-forming phytoplankton genus that contributes significantly to global carbon and sulfur cycles. During blooms, Phaeocystis species produce large carbon-rich colonies, creating a unique interface for bacterial interactions. While bacteria are known to interact with phytoplankton—e.g., they promote growth by producing phytohormones and vitamins—such interactions have not been shown for Phaeocystis. Therefore, we investigated the composition and function of P. globosa microbiomes. Specifically, we tested whether microbiome compositions are consistent across individual colonies from four P. globosa strains, whether similar microbiomes are re-recruited after antibiotic treatment, and how microbiomes affect P. globosa growth under limiting conditions. Results illuminated a core colonial P. globosa microbiome—including bacteria from the orders Alteromonadales, Burkholderiales, and Rhizobiales—that was re-recruited after microbiome disruption. Consistent microbiome composition and recruitment is indicative that P. globosa microbiomes are stable-state systems undergoing deterministic community assembly and suggests there are specific, beneficial interactions between Phaeocystis and bacteria. Growth experiments with axenic and nonaxenic cultures demonstrated that microbiomes allowed continued growth when B-vitamins were withheld, but that microbiomes accelerated culture collapse when nitrogen was withheld. In sum, this study reveals symbiotic and opportunistic interactions between Phaeocystis colonies and microbiome bacteria that could influence large-scale phytoplankton bloom dynamics and biogeochemical cycles.
Subject terms: Water microbiology, Microbial biooceanography, Microbial ecology, Molecular ecology
Introduction
Half of the photosynthesis on Earth is performed by phytoplankton, making them critical components of the global carbon cycle [1]. Like most organisms, phytoplankton do not act in isolation but instead interact with bacteria living in close proximity—either on phytoplankton cell surfaces or in the chemically influenced area surrounding phytoplankton (i.e., the phycosphere), a region analogous to the rhizosphere in plants [2]. While the phycosphere is not as extensively studied as the rhizosphere [3], it clearly hosts specialized, specific interactions with larger implications for marine ecosystem functioning and biogeochemical cycles [4]. For example, phycosphere bacteria can stimulate the growth of specific phytoplankton by producing growth hormones [5] or supplying a limiting nutrient, such as a B-vitamin [6–8] or fixed-nitrogen [9]. Conversely, phycosphere bacteria can manipulate phytoplankton communities by secreting algicidal molecules (e.g., roseobacticides) that selectively kill certain phytoplankton [10] or by competing for essential nutrients [11, 12]. Even small shifts in phytoplankton communities can alter how much organic carbon is recycled in the surface ocean or exported to deep water [13] since phytoplankton taxa vary in their C: N: P ratio [14], morphology, and sinking rates [15]. Therefore, phytoplankton-bacteria interactions are central to fully understanding phytoplankton dynamics and carbon cycling in the oceans. However, the stability, specificity, and currencies of exchange between phycosphere bacteria and diverse phytoplankton taxa are just beginning to be characterized.
The majority of our current knowledge regarding phytoplankton-bacteria interactions centers on interactions between diatoms and bacteria (reviewed in [16]) due to the ecological significance of diatoms on a global scale [17], as well as their experimental tractability (e.g. [5, 18]). Such studies have identified common bacterial inhabitants of phycospheres—including bacteria belonging to the orders Rhodobacterales, Flavobacteriales, Alteromonadales, and Oceanospirillales [18, 19]. Moreover, experiments with diatoms have demonstrated that phycosphere community assembly processes are mediated by both the phytoplankton and colonizing bacteria [20] and lead to highly reproducible bacterial community compositions [19]. However, it is unclear whether these traits extend to other phytoplankton taxa, particularly those that may be in competition with diatoms.
The Haptophyte genus, Phaeocystis, is an ideal system for extending studies of phytoplankton-bacterial interactions. Phaeocystis is a bloom-forming genus found from pole to pole. Notably, different bloom-forming species of Phaeocystis are specialized within latitudinal or thermal niches, often competing with diatoms [21]. P. globosa competes with diatoms to be the dominant spring bloom constituent in the North Sea, where it is considered a nuisance alga [22]. P. globosa is further distributed across temperate, subtropical, and tropical regions globally and has more recently started causing harmful algal blooms in Southeast Asia [23, 24]. The two polar species, P. pouchetii and P. antarctica, contribute to spring blooms in the Arctic / N. Atlantic oceans and the Southern Ocean, respectively [21]. In the N. Atlantic, P. pouchetii has begun to regularly replace diatoms in the spring bloom [25] and in the Southern Ocean, there is continued interannual variability as to whether P. antarctica or diatoms dominate spring blooms [26]. While not yet systematically studied, existing evidence supports the hypothesis that bacterial interactions could influence Phaeocystis blooms: consistent bacterial communities have been found in association with discrete samples of two P. globosa strains studied independently [27, 28] and P. antarctica blooms have been shown to strongly influence bacterioplankton community composition [29].
Bloom-forming Phaeocystis species share the ability to form large colonial structures (up to a centimeter in diameter) that are made up of thousands of cells embedded in a mucopolysaccharide matrix [30]. The carbon-rich colony substrate creates a unique physical and chemical interface for interaction between bacteria and Phaeocystis [31]. Bacteria are attracted to polysaccharides produced by phytoplankton [32] and the specific polysaccharides excreted by a phytoplankton cell contribute to which bacteria it encounters or is colonized by [18, 33, 34]. The concentrated availability of distinct polysaccharides in the Phaeocystis colony matrix, along with other chemical properties—such as the presence of amino sugars [35] and abundant dimethylsulfoniopropionate (DMSP) [36]—suggests a unique bacterial consortium would be attracted to and thrive in the colonial Phaeocystis microenvironment [18]. However, given the extensive functional redundancy among bacteria [37], it remains to be determined how specialized bacterial communities associated with Phaeocystis colonies are relative to other plankton and whether Phaeocystis microbiomes are stable-state systems.
In this study, we tested the specificity and stability of bacterial communities associated with Phaeocystis colonies by evaluating the microbiomes of individual colonies from replicate culture batches of four P. globosa strains that were isolated from globally wide-ranging locations across several decades. We further tested the reproducibility of P. globosa colony microbiome compositions by removing original microbiomes with antibiotics and evaluating compositions of recruited microbiomes after inoculation with bacterial communities in seawater. Finally, we tested two possible functional roles for P. globosa microbiomes by challenging axenic and nonaxenic cultures with nitrogen and B-vitamin stress. Nitrogen deprivation was chosen because nitrogen availability is a key factor believed to influence temperate and subtropical Phaeocystis blooms [25, 38] and because fixed-nitrogen is one currency of exchange between phycosphere bacteria and some diatoms [39]. B-vitamin limitation was chosen because (i) the availability of B-vitamins affects phytoplankton bloom dynamics [40, 41], (ii) P. globosa requires vitamins B1 and B12 to grow [42], and (iii) phycosphere bacteria can be sources of B-vitamins [7, 8, 43]. Overall, we found P. globosa colonies hosted remarkably consistent bacterial communities, both before antibiotic treatment (original microbiomes) and in the recruitment trial. These results demonstrate that, like many diatoms, Phaeocystis colonies regulate convergent bacterial community assembly towards a stable and reproducible core microbiome, albeit a different core microbiome than previously observed for other phytoplankton groups. Furthermore, the full consortia of co-cultured bacteria (in media and associated with colonies) differentially impact P. globosa growth depending on environmental conditions.
Methods
Phaeocystis globosa culture sources
We included four strains of Phaeocystis globosa in this study to investigate the consistency of Phaeocystis-associated bacterial community compositions across strains. Three strains—CCMP1528, CCMP629, and CCMP2710—were obtained from the National Center for Marine Algae and Microbiota (NCMA, Boothbay, ME, U.S.A.). We isolated the fourth strain in January 2019 from the Kuroshio current in the East China Sea (28.22 °N, 127.66 °E) by collecting surface water (5 m depth) and filtering it through 63 µm mesh-size nylon mesh before enriching with 10% L1–Si media [44]. Nutrient enrichment triggered a bloom of colonial P. globosa and individual P. globosa colonies were subsequently isolated, rinsed with sterile artificial seawater, and used to establish mono-algal cultures of a new P. globosa strain, referred to as “Kuroshio” herein. Together, these four strains represent global geographic diversity and have been maintained in culture for wide-ranging lengths of time, from several decades to just a few years (Fig. 1).
Fig. 1. Geographic origins of the four strains of Phaeocystis globosa included in the study.
The year that each strain was originally isolated is displayed below each strain’s name. “CCMP” strains were obtained from the National Center for Marine Algae and Microbiota in Boothbay, Maine, U.S.A. The Kuroshio strain was isolated for this study.
Evaluation of P. globosa colony and media microbiomes
Cultures of each P. globosa strain were synchronized by inoculating 35 ml of L1–Si media in triplicate 50 ml plastic culture flasks with 1 ml of maintenance culture, incubating for five days, and transferring 1 ml to a newly prepared culture flask. Cultures were incubated in a TOMY digital biology CLE-series plant growth chamber with cool-white fluorescent lighting (16 h light/8 h dark diurnal light cycle, ~50 µmol m−2 s−1 light) maintained at 22 °C. After five serial transfers, individual colonies and free-living cells were harvested for DNA extraction. Three colonies from each culture replicate were rinsed three times with sterile artificial sweater (ASW) before careful transfer (with <10 µl carry-over of ASW) to an Axygen Maxymum Recovery PCR tube with 30 µl 10% mass:volume Chelex 100 resin beads in PCR-grade water. The colony and Chelex mixtures were vortexed for 10 s, incubated at 96 °C for 20 min, vortexed again for 10 s, and placed on ice [45]. Free-living bacterial cells in Phaeocystis media were collected by gravity-filtering each culture replicate through 100-, 70-, and 40-µm mesh-size cell strainers to remove colonies and then filtering under gentle vacuum pressure through 3.0-µm (to remove free-living Phaeocystis cells) and 0.2-µm pore-size polytetrafluoroethylene (PTFE) filters (Millipore) to collect bacterial biomass. DNA was extracted from the 0.2-µm pore-size filters by following the manufacturer’s protocol for the DNeasy PowerSoil kit (Qiagen), including the optional heating step for enhanced cell lysis. The v3-v4 region of the 16S rRNA gene was amplified from extracted DNA from both extraction methods following the Illumina guide for “16S Metagenomic Sequencing Library Preparation” (https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf). The 16 S rRNA gene amplicon sequencing libraries were sequenced with 300 × 300-bp v3 paired-end chemistry on the MiSeq platform (Illumina).
Evaluation of recruited bacterial communities following antibiotic disruption of Phaeocystis microbiomes
To determine whether Phaeocystis strains reproducibly recruit specific bacteria to their microbiomes, the original microbiomes of each strain were disrupted and then each strain was exposed to bacterial assemblages in seawater before microbiome composition was assessed a second time. Original microbiomes were disrupted through antibiotic treatment: 1 ml of actively growing cultures were transferred to 50 ml culture flasks with 35 ml of L1–Si media and 1 ml of an antibiotic cocktail with 12,000 units/ml Penicillin G, 300 units/ml Polymyxin B, 50 µg/ml Chloramphenicol, 60 µg/ml Paromomycin and 6.25 mg/ml Cephalexin. After 48 h, 1 ml of each antibiotic-treated culture was transferred to L1–Si media without antibiotics and allowed to recover from antibiotic treatment for five five-day culture cycles (serial transfers). Microbiome disruption was checked by incubating aliquots of each antibiotic-treated culture with Hoechst 33342 DNA-binding dye and imaging each aliquot with confocal laser scanning fluorescence microscopy (Zeiss LSM 780). Additionally, aliquots of antibiotic-treated cultures were transferred to Difco Marine Broth 2216 (BD Biosciences) at the end of each ensuing culture cycle and monitored for bacterial growth by light microscopy (Olympus CKX53). No bacteria were observed in antibiotic-treated cultures with fluorescent microscopy, nor was bacterial growth detected with light microscopy in marine broth incubations. Henceforth, antibiotic-treated cultures were considered axenic.
Inoculating seawater for microbiome recruitment trials was collected outside the coral lagoon of Okinawa Island (Japan) by bucket cast and filtered through 63 µm-mesh-size nylon mesh before transport back to the lab. Inoculating seawater was further filtered through 3 µm- and 0.45 µm-pore-size PTFE filters. While this step undoubtedly decreased the diversity of the inoculating seawater—another study that similarly filtered inoculating seawater saw a 40% reduction of bacterial cell counts after filtration [46]—it was necessary to remove eukaryotic cells that could be grazers or photosynthetic competitors as well as cyanobacteria, such as Synechococcus, that would influence the outcome of the experiment. Two 50 ml replicates of the filtered seawater were filtered through 0.2 µm pore-size filters to assess the initial composition of bacterial communities inoculating Phaeocystis cultures. L1–Si media for recruitment trials was prepared with 50% inoculating seawater and 50% sterile artificial seawater. Culture replicates for microbiome recruitment trials were initiated with 1 ml of actively-growing axenic Phaeocystis cultures in triplicate flasks. The bacterial community was allowed to equilibrate over five consecutive five-day culture cycles (serial transfers were made every fifth day) before Phaeocystis colonies (five from each replicate) and free-living media-associated bacteria were harvested for DNA extraction and amplicon sequencing following the methods described above.
Bioinformatics and statistical analyses
Sequencing for this project was completed using two full MiSeq flow cells. Resulting sequencing reads were quality-filtered and amplicon sequence variants (ASVs) were identified using the DADA2 denoising algorithm [47], implemented within the QIIME 2 framework (v2020.6 [48]), which also removes chimera and singleton sequences. Sequencing reads from each of the MiSeq runs were denoised independently with results merged after denoising with ASVs clustered at 99% identity and, henceforth, referred to as OTUs (operational taxonomic units). Taxonomy was assigned to representative sequences with a naive Bayes classifier trained on the SILVA 99% 16S sequence database v138 [49] using the QIIME 2 feature-classifier plug-in [50]. The resulting OTU count table and taxonomic assignments were exported to the R statistical environment (v4.0.3 [51]) and further analyzed with the phyloseq (v1.34 [52]), CoDaSeq (v0.99.6 [53]), and vegan (v2.5–7 [54]) packages.
Effects of microbiomes on P. globosa growth under limiting culture conditions
To assess impacts of microbiomes on Phaeocystis growth, we monitored axenic and nonaxenic Phaeocystis cultures under nutrient-replete and two limiting conditions: nitrogen limitation and B-vitamin limitation. L1–Si media was prepared without nitrate to induce nitrogen limitation and L1–Si media was prepared without the L1 vitamin solution, which contains thiamine (vit. B1), biotin (vit. B7), and cobalamin (vit. B12), to induce B-vitamin limitation [44]. Triplicate 50 ml sterile plastic culture flasks were prepared with 35 ml of each media type (L1–Si replete, L1–Si with no nitrate, and L1–Si with no B-vitamins) in a laminar flow hood sterilized with UV light and cleaned with ethanol. One ml of actively growing axenic or nonaxenic P. globosa was transferred to each flask so there were three replicates of axenic and nonaxenic cultures for each of the four strains (total of 72 flasks per culture cycle). Growth was monitored daily by removing 1 ml from each flask, homogenizing by pipette mixing, and measuring in vivo chlorophyll a fluorescence with a Trilogy Fluorometer (Turner Designs). Serial transfers to fresh media were completed for five culture cycles (5–7 days each, depending on growth) for each treatment, except for nonaxenic cultures in L1–Si media without nitrate, as growth ceased after the third culture cycle. All transfers and aliquots were made inside the clean laminar flow hood. Following each culture cycle, an additional 100 µl aliquot was taken from each flask, transferred to 1 ml sterile marine broth in a 12-well cell-culture plate, and monitored for bacterial growth for 72 h. No bacterial growth was observed from aliquots taken from axenic cultures, whereas bacterial growth was always observed after 24 h in aliquots from nonaxenic cultures, thus confirming axenic and nonaxenic cultures, respectively.
Results
Sequence processing
Overall, amplicons from 100 samples (n = 34 original single-colony microbiomes, n = 12 original media microbiomes, n = 40 recruited single-colony microbiomes, n = 12 recruited media microbiomes, n = 2 inoculating seawater communities) were sequenced for this project, generating a total of 17,328,537 sequencing reads (median total sequences per sample = 147,351; Fig. S1). Raw sequencing data is available from the NCBI SRA under accession PRJN779092. Following quality-filtering, denoising, and chimera and singleton removal, 13,435,004 sequencing reads remained (median remaining sequences per sample = 121,035; Fig. S1). One concern when studying phytoplankton microbiomes is the proportion of sequencing resources allocated to non-target chloroplast 16S rRNA genes instead of target bacterial 16S rRNA genes. Lower input, single-colony samples yielded a larger proportion of chloroplast sequences than media or seawater samples: single colony samples had a mean of 22% bacterial sequences compared to 85% and 99.5% bacterial sequences for media and seawater samples, respectively (Fig. S2). However, following removal of chloroplast sequences, rarefaction curves for all samples reached OTU richness saturation (Fig. S3), indicating an adequate number of bacterial sequences remained for analysis due to sufficient sequencing depth. Chloroplast sequences were excluded from ensuing analyses.
Diversity of original and recruited P. globosa microbiomes
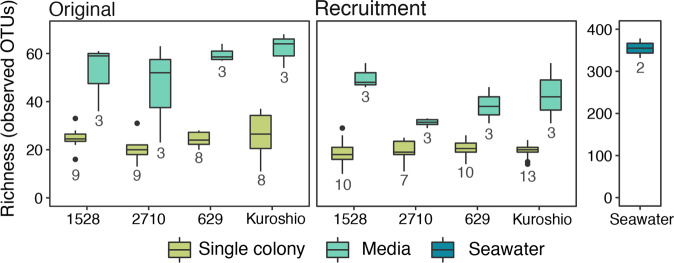
The inoculating bacterial community for the recruitment trial was significantly more diverse than original and recruited Phaeocystis microbiomes (Fig. 2; pairwise Wilcox test, p < 0.05). However, as expected, filtering the inoculating seawater to remove eukaryotic cells and cyanobacteria reduced the observed richness in the inoculating seawater by about half compared to unfiltered seawater from the same environment that was analyzed for another study [55]. In addition, original and recruited colony microbiomes were significantly less diverse than original and recruited media microbiomes (Fig. 2; pairwise Wilcox test, p < 0.001). In both these cases, colony microbiomes had about half the observed richness as media microbiomes.
Fig. 2. Observed richness of OTUs in original and recruited bacterial communities associated with individual Phaeocystis colonies, Phaeocystis culture media, and inoculating seawater used in recruitment trials.
Results from the four Phaeocystis globosa strains are plotted separately along the x-axis. The number of biological replicates (n) for each sample type is included below the corresponding box. Note that the observed richness in the inoculating seawater is plotted on a separate y-axis due to the much higher diversity in these samples. Boxes denote the 25th to 75th percentile ranges for each sample type; the center bar is the median; whiskers show 1.5 times the interquartile range above the 75th percentile and below the 25th percentile; outlier points are values outside 1.5 times the interquartile range above the 75th percentile and below the 25th percentile. Media-associated bacterial communities are significantly more diverse (higher richness) than colony-associated bacterial communities (pairwise Wilcox test, p < 0.001).
The beta diversity between different sample types was computed by normalizing OTU sequence read counts to a centered log-mean ratio (CLR) using the CoDaSeq package and calculating Euclidean distances between community compositions in each sample with the phyloseq package. This combination of CLR transformation and Euclidean distance is known as the Aitchison distance and minimizes bias inherent to compositional data, such as amplicon data [53]. A principal coordinates analysis (PCoA) performed on Aitchison distances revealed four clearly separated clusters: inoculating seawater communities, original media microbiomes, recruited media microbiomes, and colony-associated microbiomes (Fig. 3A). When sample points are colored by trial (original or recruitment), the colony-associated community cluster appears split by trial, but original and recruited colony microbiomes were not significantly different (Fig. 3A). Pairwise permutational multivariate analysis of variance (PERMANOVA) tests using the adonis2 function from the vegan package (999 permutations; [54]) showed original and recruited colony-associated communities were both significantly different from media and seawater-associated communities (p < 0.05) but not from each other, whereas recruited and original media-associated communities were both significantly different from the colony and seawater-associated communities as well as from each other (p < 0.05).
Fig. 3. Principal coordinates analysis (PCoA) of Aitchison distances between bacterial communities associated with individual Phaeocystis colonies, Phaeocystis media, and inoculating seawater.
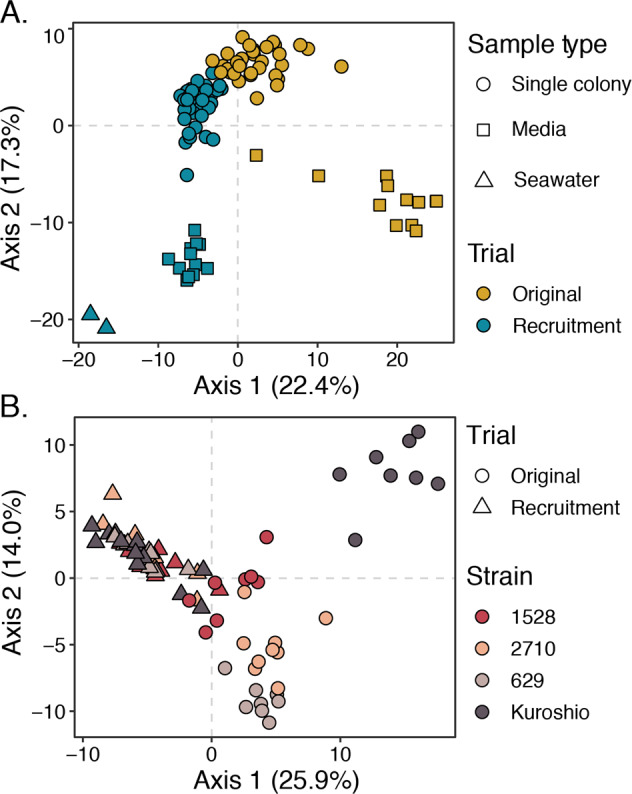
A PCoA of Aitchison distance between all samples with point shape representing sample type (single colony, media, or seawater) and point color representing trial (original or recruitment). Four main clusters are visible: colony-associated communities, original media-associated communities, recruited media-associated communities, and communities in seawater. Coloration reveals that the colony-associated community cluster is split by trial. B PCoA of Aitchison distances between single-colony samples only, with point shape representing trial (original or recruitment) and color representing P. globosa strains. Samples separate by strain among original colony-associated communities, especially the Kuroshio strain original colonies, but this differentiation collapses among recruited colony samples. Phaeocystis strain-level differences in colony microbiomes were significant among the original colony samples (p < 0.001, PERMANOVA, 999 permutations), but not among the recruitment trial colony samples (p = 0.117, PERMANOVA, 999 permutations).
To evaluate the consistency of colony microbiome compositions by host strain, data were subset to include only single-colony samples, and the Aitchison distances and PCoA were recomputed (Fig. 3B). This PCoA showed original colony microbiomes were differentiated by strain. Specifically, original colony microbiomes from the Kuroshio strain, which was isolated most recently and was not kept at a culture facility, cluster separately from the other original colony microbiomes. However, this separation by strain collapses when comparing microbiomes recruited to colonies after antibiotic treatment (Fig. 3B). Moreover, PERMANOVA tests by strain performed separately for original and recruited colony microbiomes showed strain-level differences were only significant (p < 0.001) among original microbiomes.
Composition of original and recruited P. globosa microbiomes
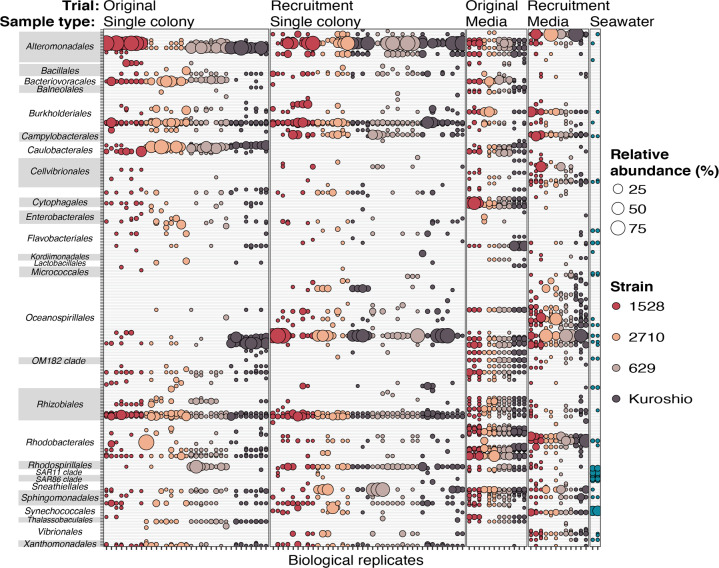
To further examine OTUs composing Phaeocystis colony and media microbiomes, data were subset to exclude OTUs only present in seawater samples. Subsetting reduced the total unique OTU count from 781 to 341. When the relative abundance of each remaining OTU was plotted for each sample, trends in original and recruited colony- and media-associated microbiomes were readily apparent (Fig. 4). Original and recruited colony microbiomes were remarkably consistent across strains, with most colony microbiomes including the same OTUs from the orders Alteromonadales, Burkholderiales, and Rhizobiales. Original colony microbiomes also consistently included Caulobacterales OTUs belonging to the genus Maricaulus, which are stalked, biofilm-forming bacteria that colonize gelatinous material in marine environments [56]. As particle-associated bacteria, Maricaulus spp. were probably excluded from the inoculating seawater due to the required filtration steps. Conversely, recruited colony microbiomes consistently included an Oceanospirillales OTU that was not in original colony or media-associated microbiomes but was in recruited media microbiomes and in inoculating seawater. In addition, the plot highlighted two OTUs in the order Oceanospirillales contributing to the Kuroshio strain’s distinct original colony microbiomes (Fig. 3B). Original media-associated microbiomes were most consistent, with almost all OTUs detected being found in at least 9 out of 12 samples. Notably, recruited OTUs were not always also detected in seawater. This is likely due to recruited OTUs representing bacteria with very low abundance in the inoculating seawater, especially since this seawater was filtered to remove particles and eukaryotic cells that likely harbor the recruited bacteria. Alternatively, original microbiomes might not have been completely removed during antibiotic treatment, but this would require that the remaining bacteria were sparse enough to evade detection with fluorescence microscopy and that they were unable to grow in marine broth tests. Overall, results demonstrate that the Phaeocystis colonial microbiome is a consistent subset of co-cultured bacteria in media and is primarily composed of bacteria belonging to orders Alteromonadales, Burkholderiales, and Rhizobiales.
Fig. 4. Relative abundance of OTUs detected in association with Phaeocystis globosa colonies and in culture media.
Data were subset to include only OTUs that were present in at least one single-colony or media sample (i.e., not only present in seawater)—reducing the number of unique OTUs from 781 to 341—and the relative abundance of each remaining OTU in every sample is represented by bubble size. Bubbles are colored by P. globosa source strain or inoculating seawater and are grouped by trial (original or recruitment) and sample type (single colony, media, and inoculating seawater). Columns are biological replicates of each sample type and rows are individual OTUs. OTUs are grouped on the y-axis by taxonomic orders, which appear alphabetically.
Impact of microbiomes on Phaeocystis growth when nitrate or B-vitamin deprived
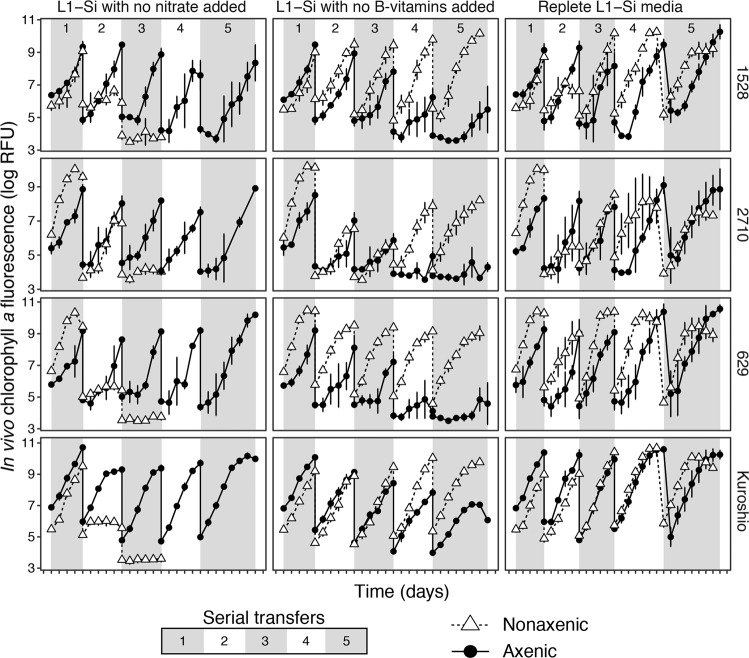
Growth was monitored in axenic and nonaxenic culture replicates of each P. globosa strain in three media formulations: replete L1–Si with no modifications, L1–Si media with nitrate omitted, and L1–Si media with B-vitamin solution omitted (no added source of thiamine, biotin, or cobalamin). Growth was maintained in replete L1–Si media for all strains across the five serial transfers monitored (Fig. 5). In contrast, growth slowed for nonaxenic replicates of all four strains in L1–Si media without nitrate after the second transfer and ceased after the third transfer, but was unaffected in axenic replicates. Conversely, growth slowed for axenic replicates of all four strains in L1–Si media without B-vitamins after the fourth transfer but was unaffected in nonaxenic replicates. These results suggest that when nitrogen is limiting, co-cultured bacteria compete for the limiting resource, whereas when vitamins become limiting, co-cultured bacteria mitigate limitation.
Fig. 5. Growth curves for four strains of Phaeocystis globosa grown in L1–Si media with no nitrate added, L1–Si media with no B-vitamins added, and in replete L1–Si media.
Each set of conditions were established in triplicate—each point is a mean of three biological replicates and error bars are one standard deviation of the mean—and cultures were maintained for five sequential transfers of five (transfers 1–4) to seven days (transfer 5). Serial transfers are denoted by alternating shaded regions on the plots and are numbered in the top panels. Growth was maintained in all replicates across all five transfers in replete L1–Si media; growth slowed after the fourth transfer in axenic replicates in L1–Si media without B-vitamins; growth slowed after the second transfer (and then ceased after the third) in nonaxenic replicates grown in L1–Si media without nitrate.
Discussion
This study shows similarly composed bacterial communities are consistently found in co-culture with Phaeocystis globosa strains that were isolated from different ocean basins across several decades (Figs. 1, 3, 4), thus demonstrating microbiome maintenance without continued access to bacterioplankton in the ocean. Moreover, original communities directly associated with individual colonies were a predictable subset of co-cultured bacteria in culture media, and bacterial communities recruited to colonies converged towards the original composition of colony microbiomes (Fig. 4). The consistency of these communities is evidence Phaeocystis globosa colonies harbor a persistent core microbiome and selective processes drive microbiome assembly. Core constituents of original and recruited colony microbiomes included bacteria belonging to orders Alteromonadales, Burkholderiales, and Rhizobiales (Fig. 4). Of these, only Alteromonadales bacteria are core components of diatom phycosphere communities [20], but Rhizobiales bacteria have been found in association with the diatom Asterionellopsis glacialis [57]. Burkholderiales and Rhizobiales are commonly found in plant rhizospheres [58, 59]. Differences in Phaeocystis phycosphere community composition compared to other phytoplankton reflects the uniqueness of the Phaeocystis colony microenvironment. Furthermore, bacteria in co-culture with P. globosa mitigated nutrient stress when B-vitamins were withheld but accelerated culture collapse when nitrate was withheld (Fig. 5). This implies bacterial counterparts in the Phaeocystis phycosphere play a dual role alternating between mutualism and opportunism with their host [60].
The Phaeocystis globosa microbiome is a stable-state system shaped by selective assembly mechanisms
Bacterial communities in co-culture with long-term Phaeocystis cultures are consistently composed and stable through time. The original media microbiomes were almost identical in P. globosa CCMP1528, CCMP2710, CCMP629, and Kuroshio strains, despite each of these strains being isolated from a different ocean basin during a different decade. Likewise, the original individual colony microbiomes were strikingly similar (Figs. 3, 4). However, despite sharing many core constituents (Fig. 4), overall compositions of original Kuroshio strain colony microbiomes were significantly different from the original colony microbiomes of the other strains (Fig. 3B). Since the Kuroshio strain was isolated most recently and was not sourced from a common culture bank, its different original microbiome suggests time and place of isolation influences the composition of Phaeocystis microbiomes—as reported for other phytoplankton [61]—but time spent in a common culture facility reduces initial variability. Convergence of colonial microbiomes of all four strains, including the Kuroshio strain, after antibiotic treatment and exposure to bacteria from seawater (Figs. 3B, 4) is further evidence that Phaeocystis microbiomes are stable systems shaped by selective, deterministic assembly mechanisms: given access to the same bacteria, all four strains recruit bacterial communities that are not significantly different from each other (F = 1.26, R2 = 0.09, p = 0.117).
Selection of P. globosa microbiome bacteria likely arises from the unique physical and chemical characteristics of colonies. First, the large size of colonies relative to individual Phaeocystis cells and other phytoplankton creates more potential habitat for bacteria and, therefore, increases the diversity that can be supported [62]. Second, the colony skin likely represents a physical barrier limiting access to the inner matrix [63], representing a first step in microbiome selection. Third, the colony matrix is made up of a unique medley of mucopolysaccharides, polysaccharides, and other organic molecules, including amino sugars [30, 31, 35], which would attract and support bacteria with unique enzymatic capabilities to exploit these resources [33]. Finally, additional community filtering may arise from feedback from and between microbiome bacteria, which can produce antibacterial compounds [64] or elicit phytoplankton defense responses [20]. For instance, the Phaeocystis colony matrix has high concentrations of acrylate, which has antimicrobial activity [27, 65]. Together, these Phaeocystis traits lead to consistent microbiome community assembly across individual colonies and across host strains, as was seen in this study. However, the convergence of recruited microbiomes and their differences from original microbiomes also demonstrate some functional redundancy in microbiome bacteria (i.e., phylogenetically distinct bacteria can occupy the same niche) [37].
Core members of original and recruited Phaeocystis globosa colony microbiomes
Core members of the Phaeocystis globosa colony microbiome—bacteria with OTUs detected in almost all original and recruited colony microbiomes—belonged to orders Alteromonadales (family Alteromondacea), Burkholderiales (families Oxalobacteraceae and Comamonabacteraceae), and Rhizobiales (family Rhizobiaceae) (Fig. 4). Of these, only Alteromonadales bacteria are common inhabitants of diatom phycospheres [20]. In a BLAST search [66] against the NCBI nr/nt database (accessed: 10/4/2021 [67]), the three Alteromonadacea OTUs consistently abundant in colony microbiomes shared 99–100% nucleotide identity with sequences from Alteromonas macleodii. A. macleodii—a ubiquitous particle-associated marine copiotroph [68]—is an opportunistic, rather than symbiotic, member of diatom microbiomes [20] that directly competes with diatoms for nitrate [69]. Given how prevalent these bacteria are on marine particles and phytoplankton, it is unsurprising they are also associated with Phaeocystis. However, as with diatoms, we suspect the relationship between Phaeocystis and A. macleodii is more opportunistic than symbiotic in nature.
While bacteria in orders Burkholderiales and Rhizobiales are not commonly found living in association with other phytoplankton studied thus far, they are often associated with plant rhizospheres. The Burkholderiales OTU from the family Oxalobacteraceae that was consistently associated with P. globosa colonies shared 100% sequence identity with bacteria in the genus Herbaspirillum, which have been isolated from rhizospheres of many plants, and from soil and aquatic environments. Herbaspirillum strains have genomic potential to fix nitrogen, reduce nitrite, and produce siderophores [70]. Herbaspirillum strains also produce plant growth hormones—such as gibberellins and indole acetic acid [71]—and have been shown to promote rice, corn, and sugarcane growth [72]. The other core Burkholderiales OTU (family Comamonadaceae) shared 100% sequence identity with sequences deriving from the genus Delftia. Delftia spp. are considered plant growth-promoting rhizobacteria (PGPR), have capacity to produce siderophores [73], and can metabolize aromatic compounds, including benzene [74]. From the order Rhizobiales, one core OTU shared 100% identity with Ochrobactrum pseudogrigonensis sequences. O. pseudogrigonensis is a rhizosphere bacterium that promotes plant growth through induced stress tolerance, e.g., by producing antioxidants [59]. The other core Rhizobiales OTU shared 100% identity with Rhizobium radiobacter. Many R. radiobacter strains are plant pathogens, but nonpathogenic strains promote plant growth and can induce resistance to other pathogens [58]. While further work is needed to fully characterize how members of the core Phaeocystis microbiome influence Phaeocystis growth, our results indicate that, apart from A. macleodii, the core members of the P. globosa colony microbiome are broadly symbiotic bacteria with the potential to promote growth through phytohormones, nitrogen-cycling, or siderophore production.
Impact of microbiomes on Phaeocystis growth under nitrogen and vitamin deprivation
Taken together, consistent and reproducible compositions of P. globosa microbiomes and the growth-promoting potential of most core microbiome members point toward beneficial interactions between Phaeocystis and microbiome bacteria. Interestingly, growth experiments involving axenic and nonaxenic P. globosa strains grown under nitrogen and vitamin deprivation revealed contrasting bacterial effects. Axenic cultures persisted longer than nonaxenic cultures when nitrate was withheld (Fig. 5), demonstrating competition between Phaeocystis and co-cultured bacteria for this important resource. This result aligns with the high relative abundance of A. macleodii in colony microbiomes (Fig. 4), as A. macleodii competes with phytoplankton for nitrate [69]. In a natural setting, these results might indicate microbiome interactions disadvantage Phaeocystis if nitrogen is limiting. However, given that A. macleodii is often also associated with diatoms [20], diatoms may be subject to similar ecological competition with heterotrophs. Indeed, the influence of nitrogen limitation on Phaeocystis and diatom bloom dynamics is not straightforward, and each group has come out on top when nitrogen was limiting in different ecosystems [23, 25, 75].
In contrast to when nitrate was withheld, axenic P. globosa growth was diminished compared to nonaxenic cultures when B-vitamins were withheld (Fig. 5), demonstrating co-cultured bacteria can produce essential B-vitamins (i.e., thiamine, cobalamin; [42]) and stave off growth-diminishing vitamin stress. This is significant because B-vitamins can be limiting in coastal areas where Phaeocystis blooms occur [41, 76, 77]. While several major heterotrophic cobalamin and thiamine synthesizing bacteria—namely Oceanospirillales [7], Rhodospirillales, and Flavobacteriales bacteria [78]—were in our dataset, they were more consistently present and abundant in media microbiomes than in colony microbiomes (Fig. 4). Nonetheless, two Oceanospirillales OTUs were abundant core components of original Kuroshio strain colonial microbiomes, and an Oceanospirillales and a Rhodospirillales OTU were both recruited at relatively high abundance to almost all colonies sampled in the recruitment trial (Fig. 4). The bacteria represented by these OTUs could indeed contribute to vitamin synthesis in Phaeocystis colony microbiomes. However, the nutrient-limitation experiments only included Phaeocystis cultures retaining original microbiomes, where most recognized vitamin-producing bacteria were not tightly associated with colonies. Therefore, it is possible that other colony-associated bacteria are vitamin synthesizers or that vitamin synthesizers were more loosely associated with colonies (i.e., not attached to colony surfaces or inhabiting the colonial matrix). A wide diversity of marine bacteria are capable of vitamin synthesis [78, 79], thus necessitating evaluation of genomic potential and gene expression in Phaeocystis microbiomes to ultimately identify bacterial vitamin sources. Regardless of which microbiome constituents produce or consume limiting nutrients, challenging axenic and nonaxenic P. globosa with different limiting conditions clearly demonstrated interactions with the same total microbiome can be beneficial under one set of conditions and detrimental under another.
Conclusions and future directions
Though interactions between phytoplankton and bacteria occur on small scales, they have large-scale repercussions for primary production and carbon cycling in aquatic ecosystems. A more complete understanding of these interactions that includes diverse phytoplankton taxa will improve our capabilities to model and predict changes to the marine environment. This study showed Phaeocystis globosa has consistent and reproducible core microbiomes co-cultured in media and associated with individual colonies. Members of the core microbiome have potential to produce growth-promoting phytohormones, fix nitrogen, synthesize vitamins, and release siderophores. Our experiments showed co-cultured bacteria provide vitamin resources and drawdown nitrate. Further work including metagenome and metatranscriptome sequencing to assess genomic potential and gene expression in Phaeocystis microbiomes is required to fully characterize mechanisms of interaction and currencies of exchange between Phaeocystis and microbiome bacteria. Importantly, this study was limited to cultured Phaeocystis strains, and additional work is required to determine if the core-microbiomes reported here are also associated with Phaeocystis colonies in natural bloom conditions. Evaluating the composition and function of microbiomes of field-collected Phaeocystis colonies will ultimately illuminate the ecological relevance of Phaeocystis colony microbiomes. Additionally, future work should expand to other bloom-forming Phaeocystis species, determine how bacterial interactions impact Phaeocystis growth under other ecologically-relevant conditions—such as iron and phosphorus limitation—and investigate how increasing sea-surface temperatures associated with global climate change will impact microbiome interactions with Phaeocystis species.
Supplementary information
Acknowledgements
Research was funded by the Marine Biophysics Unit of the Okinawa Institute of Science and Technology (OIST) Graduate University. MMB was supported by an OIST Junior Research Fellowship, a Postdoctoral Scholarship granted by Woods Hole Oceanographic Institution, and a Simons Foundation Postdoctoral Fellowship in Marine Microbiology (award # 874439). We thank the captain and crew of the Okinawa Prefectural Fisheries and Ocean Research Center ship Tonan Maru for collecting seawater from the East China Sea for the culture of the Kuroshio strain of Phaeocystis globosa. We further thank Kazumi Inoha for coordinating seawater sampling on the Tonan Maru. Dawn Moran provided valuable advice for treating phytoplankton cultures with antibiotics. The OIST DNA sequencing section (Onna, Okinawa, Japan) performed the sequencing for this project.
Author contributions
MMB, SM, MAS, and HS conceived and planned the research; MMB performed experiments and analyzed data; All authors participated in discussions interpreting results; MMB wrote the original manuscript and all authors provided comments and approved the final version.
Data availability
All sequencing data generated for this manuscript is available from the NCBI SRA under accession PRJN779092.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Margaret Mars Brisbin, Email: mmarsbrisbin@whoi.edu.
Harriet Alexander, Email: halexander@whoi.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01263-2.
References
- 1.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, et al. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA. 2016;113:3143–51. doi: 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seymour JR, Amin SA, Raina J-B, Stocker R Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat Microbiol. 2017;2:17065. [DOI] [PubMed]
- 3.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 4.Cirri E, Pohnert G. Algae-bacteria interactions that balance the planktonic microbiome. N. Phytol. 2019;223:100–6. doi: 10.1111/nph.15765. [DOI] [PubMed] [Google Scholar]
- 5.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 6.Grant MAA, Kazamia E, Cicuta P, Smith AG. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal-bacterial cocultures. ISME J. 2014;8:1418–27. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO, et al. Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci USA. 2015;112:9938–43. doi: 10.1073/pnas.1501615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suleiman M, Zecher K, Yücel O, Jagmann N, Philipp B. Interkingdom cross-feeding of ammonium from marine methylamine-degrading bacteria to the diatom Phaeodactylum tricornutum. Appl Environ Microbiol. 2016;82:7113–22. doi: 10.1128/AEM.01642-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011;3:331–5. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratnarajah L, Blain S, Boyd PW, Fourquez M, Obernosterer I, Tagliabue A. Resource colimitation drives competition between phytoplankton and bacteria in the Southern Ocean. Geophys Res Lett. 2021;48:e2020GL088369. doi: 10.1029/2020GL088369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Løvdal T, Eichner C, Grossart H-P, Carbonnel V, Chou L, Martin-Jézéquel V, et al. Competition for inorganic and organic forms of nitrogen and phosphorous between phytoplankton and bacteria during an Emiliania huxleyi spring bloom. Biogeosciences. 2008;5:371–83. doi: 10.5194/bg-5-371-2008. [DOI] [Google Scholar]
- 13.Arrigo KR, Robinson DH, Worthen DL, Dunbar RB, DiTullio GR, VanWoert M, et al. Phytoplankton community structure and the drawdown of nutrients and CO2 in the Southern Ocean. Science. 1999;283:365–7. doi: 10.1126/science.283.5400.365. [DOI] [PubMed] [Google Scholar]
- 14.Geider R, La Roche J. Redfield revisited: variability of C:N:P in marine microalgae and its biochemical basis. Eur J Phycol. 2002;37:1–17. doi: 10.1017/S0967026201003456. [DOI] [Google Scholar]
- 15.Smayda TJ. Normal and accelerated sinking of phytoplankton in the sea. Mar Geol. 1971;11:105–22. doi: 10.1016/0025-3227(71)90070-3. [DOI] [Google Scholar]
- 16.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–84. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tréguer P, Bowler C, Moriceau B, Dutkiewicz S, Gehlen M, Aumont O, et al. Influence of diatom diversity on the ocean biological carbon pump. Nat Geosci. 2018;11:27–37. doi: 10.1038/s41561-017-0028-x. [DOI] [Google Scholar]
- 18.Ferrer-González FX, Widner B, Holderman NR, Glushka J, Edison AS, Kujawinski EB, et al. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 2021;15:762–73. doi: 10.1038/s41396-020-00811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mönnich J, Tebben J, Bergemann J, Case R, Wohlrab S, Harder T. Niche-based assembly of bacterial consortia on the diatom Thalassiosira rotula is stable and reproducible. ISME J. 2020;14:1614–25. doi: 10.1038/s41396-020-0631-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibl AA, Isaac A, Ochsenkühn MA, Cárdenas A, Fei C, Behringer G, et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc Natl Acad Sci USA. 2020;117:27445–55. doi: 10.1073/pnas.2012088117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoemann V, Becquevort S, Stefels J, Rousseau V, Lancelot C. Phaeocystis blooms in the global ocean and their controlling mechanisms: a review. J Sea Res. 2005;53:43–66. doi: 10.1016/j.seares.2004.01.008. [DOI] [Google Scholar]
- 22.Peperzak L, Colijn F, Gieskes WWC, Peeters JCH. Development of the diatom-Phaeocystis spring bloom in the Dutch coastal zone of the North Sea: the silicon depletion versus the daily irradiance threshold hypothesis. J Plankton Res. 1998;20:517–37. doi: 10.1093/plankt/20.3.517. [DOI] [Google Scholar]
- 23.Hai D-N, Lam N-N, Dippner JW. Development of Phaeocystis globosa blooms in the upwelling waters of the south central coast of Viet Nam. J Mar Syst. 2010;83:253–61. doi: 10.1016/j.jmarsys.2010.04.015. [DOI] [Google Scholar]
- 24.Wang X, Song H, Wang Y, Chen N. Research on the biology and ecology of the harmful algal bloom species Phaeocystis globosa in China: Progresses in the last 20 years. Harmful Algae. 2021;107:102057. doi: 10.1016/j.hal.2021.102057. [DOI] [PubMed] [Google Scholar]
- 25.Jiang M, Borkman DG, Scott Libby P, Townsend DW, Zhou M. Nutrient input and the competition between Phaeocystis pouchetii and diatoms in Massachusetts Bay spring bloom. J Mar Syst. 2014;134:29–44. doi: 10.1016/j.jmarsys.2014.02.011. [DOI] [Google Scholar]
- 26.Nissen C, Vogt M. Factors controlling the competition between Phaeocystis and diatoms in the Southern Ocean and implications for carbon export fluxes. Biogeosciences. 2021;18:251–83. doi: 10.5194/bg-18-251-2021. [DOI] [Google Scholar]
- 27.Mars Brisbin M, Mitarai S. Differential gene expression supports a resource-intensive, defensive role for colony production in the bloom-forming haptophyte, Phaeocystis globosa. J Eukaryot Microbiol. 2019;66:788–801. doi: 10.1111/jeu.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Meng R, Smith WO, Jr, Doan-Nhu H, Nguyen-Ngoc L, Jiang X. Bacterial composition associated with giant colonies of the harmful algal species Phaeocystis globosa. Front Microbiol. 2021;12:737484. doi: 10.3389/fmicb.2021.737484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delmont TO, Hammar KM, Ducklow HW, Yager PL, Post AF. Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front Microbiol. 2014;5:646. doi: 10.3389/fmicb.2014.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verity PG, Whipple SJ, Nejstgaard JC, Alderkamp A-C. Colony size, cell number, carbon and nitrogen contents of Phaeocystis pouchetii from western Norway. J Plankton Res. 2007;29:359–67. doi: 10.1093/plankt/fbm021. [DOI] [Google Scholar]
- 31.Alderkamp A-C, Buma AGJ, van Rijssel M. The carbohydrates of Phaeocystis and their degradation in the microbial food web. Biogeochemistry. 2007;83:99–118. doi: 10.1007/s10533-007-9078-2. [DOI] [Google Scholar]
- 32.Smriga S, Fernandez VI, Mitchell JG, Stocker R. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci USA. 2016;113:1576–81. doi: 10.1073/pnas.1512307113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mühlenbruch M, Grossart H-P, Eigemann F, Voss M. Mini-review: Phytoplankton-derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ Microbiol. 2018;20:2671–85. doi: 10.1111/1462-2920.14302. [DOI] [PubMed] [Google Scholar]
- 34.Raina J-B, Fernandez V, Lambert B, Stocker R, Seymour JR. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol. 2019;17:284–94. doi: 10.1038/s41579-019-0182-9. [DOI] [PubMed] [Google Scholar]
- 35.Solomon CM, Lessard EJ, Keil RG, Foy MS. Characterization of extracellular polymers of Phaeocystis globosa and P. antarctica. Mar Ecol Prog Ser. 2003;250:81–89. doi: 10.3354/meps250081. [DOI] [Google Scholar]
- 36.Shen P, Qi Y, Wang Y, Huang L. Phaeocystis globosa Scherffel, a harmful microalga, and its production of dimethylsulfoniopropionate. Chin J Oceano Limnol. 2011;29:869–73. doi: 10.1007/s00343-011-0515-7. [DOI] [Google Scholar]
- 37.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–43. doi: 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Bouwman AF, Liu X, Beusen AHW, Van Dingenen R, Dentener F, et al. Harmful algal blooms in chinese coastal waters will persist due to perturbed nutrient ratios. Environ Sci Technol Lett. 2021;8:276–84. doi: 10.1021/acs.estlett.1c00012. [DOI] [Google Scholar]
- 39.Foster RA, Kuypers MMM, Vagner T, Paerl RW, Musat N, Zehr JP. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 2011;5:1484–93. doi: 10.1038/ismej.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helliwell KE. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. N. Phytol. 2017;216:62–68. doi: 10.1111/nph.14669. [DOI] [PubMed] [Google Scholar]
- 41.Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE, et al. Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr. 2007;52:1079–93. doi: 10.4319/lo.2007.52.3.1079. [DOI] [Google Scholar]
- 42.Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci USA. 2010;107:20756–61. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 44.Guillard RRL, Hargraves PE. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–6. doi: 10.2216/i0031-8884-32-3-234.1. [DOI] [Google Scholar]
- 45.Hamilton PB, Lefebvre KE, Bull RD. Single cell PCR amplification of diatoms using fresh and preserved samples. Front Microbiol. 2015;6:1084. doi: 10.3389/fmicb.2015.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.dos Reis MC, Romac S, Le Gall F, Marie D, Frada MJ, Koplovitz G, et al. Exploring the phycosphere of Emiliania huxleyi: from bloom dynamics to microbiome assembly experiments. bioRxiv 2022;02;21:481256. [DOI] [PubMed]
- 47.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Glo FO, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team. R: A language and environment for statistical computing. 2018. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- 52.Mcmurdie PJ, Holmes S phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed]
- 53.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package. R package version. 2019;2:5–4. [Google Scholar]
- 55.Ares Á, Brisbin MM, Sato KN, Martín JP, Iinuma Y, Mitarai S. Extreme storms cause rapid but short-lived shifts in nearshore subtropical bacterial communities. Environ Microbiol. 2020;22:4571–88. doi: 10.1111/1462-2920.15178. [DOI] [PubMed] [Google Scholar]
- 56.Radwan SSA, Al-Mailem DM, Kansour MK. Gelatinizing oil in water and its removal via bacteria inhabiting the gels. Sci Rep. 2017;7:13975. doi: 10.1038/s41598-017-14296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Behringer G, Ochsenkühn MA, Fei C, Fanning J, Koester JA, Amin SA. Bacterial communities of diatoms display strong conservation across strains and time. Front Microbiol. 2018;9:659. doi: 10.3389/fmicb.2018.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glaeser SP, Imani J, Alabid I, Guo H, Kumar N, Kämpfer P, et al. Non-pathogenic Rhizobium radiobacter F4 deploys plant beneficial activity independent of its host Piriformospora indica. ISME J. 2016;10:871–84. doi: 10.1038/ismej.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty U, Chakraborty BN, Dey PL, Chakraborty AP, Sarkar J. Biochemical responses of wheat plants primed with Ochrobactrum pseudogrignonense and subjected to salinity stress. Agric Res. 2019;8:427–40. doi: 10.1007/s40003-018-0394-7. [DOI] [Google Scholar]
- 60.Johnson WM, Alexander H, Bier RL, Miller DR, Muscarella ME, Pitz KJ, et al. Auxotrophic interactions: a stabilizing attribute of aquatic microbial communities? FEMS Microbiol Ecol. 2020;96;11:fiaa115. [DOI] [PMC free article] [PubMed]
- 61.Ajani PA, Kahlke T, Siboni N, Carney R, Murray SA, Seymour JR. The Microbiome of the cosmopolitan diatom Leptocylindrus reveals significant spatial and temporal variability. Front Microbiol. 2018;9:2758. doi: 10.3389/fmicb.2018.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connor EF, McCoy ED. The statistics and biology of the species-area relationship. Am Nat. 1979;113:791–833. doi: 10.1086/283438. [DOI] [Google Scholar]
- 63.Hamm CE, Simson DA, Merkel R, Smetacek V. Colonies of Phaeocystis globosa are protected by a thin but tough skin. Mar Ecol Prog Ser. 1999;187:101–11. doi: 10.3354/meps187101. [DOI] [Google Scholar]
- 64.Geddes BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun. 2019;10:3430. doi: 10.1038/s41467-019-10882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sieburth JM. Acrylic acid, an‘ antibiotic’ principle in Phaeocystis blooms in Antarctic waters. Science. 1960;132:676–7. doi: 10.1126/science.132.3428.676. [DOI] [PubMed] [Google Scholar]
- 66.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2016;44:D67–72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.López-Pérez M, Gonzaga A, Martin-Cuadrado A-B, Onyshchenko O, Ghavidel A, Ghai R, et al. Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep. 2012;2:696. doi: 10.1038/srep00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diner RE, Schwenck SM, McCrow JP, Zheng H, Allen AE. Genetic manipulation of competition for nitrate between heterotrophic bacteria and diatoms. Front Microbiol. 2016;7:880. doi: 10.3389/fmicb.2016.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monteiro RA, Balsanelli E, Wassem R, Marin AM, Brusamarello-Santos LCC, Schmidt MA, et al. Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant Soil. 2012;356:175–96. doi: 10.1007/s11104-012-1125-7. [DOI] [Google Scholar]
- 71.Bastián F, Cohen A, Piccoli P, Luna V, Baraldi R. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998;24:7–11. doi: 10.1023/A:1005964031159. [DOI] [Google Scholar]
- 72.Gyaneshwar P, James EK, Reddy PM. Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium‐tolerant rice varieties. N. Phytol. 2002;154:131–45. doi: 10.1046/j.1469-8137.2002.00371.x. [DOI] [Google Scholar]
- 73.Guo H, Yang Y, Liu K, Xu W, Gao J, Duan H, et al. Comparative genomic analysis of Delftia tsuruhatensis MTQ3 and the identification of functional NRPS genes for siderophore production. Biomed Res Int. 2016;2016:3687619. doi: 10.1155/2016/3687619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vásquez-Piñeros MA, Martínez-Lavanchy PM, Jehmlich N, Pieper DH, Rincón CA, Harms H, et al. Delftia sp. LCW, a strain isolated from a constructed wetland shows novel properties for dimethylphenol isomers degradation. BMC Microbiol. 2018;18:108. doi: 10.1186/s12866-018-1255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riegman R, Noordeloos AAM, Cadée GC. Phaeocystis blooms and eutrophication of the continental coastal zones of the North Sea. Mar Biol. 1992;112:479–84. doi: 10.1007/BF00356293. [DOI] [Google Scholar]
- 76.Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109:14041–5. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gobler CJ, Norman C, Panzeca C, Taylor GT, Sañudo-Wilhelmy SA. Effect of B-vitamins (B1, B12) and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Micro Ecol. 2007;49:181–94. doi: 10.3354/ame01132. [DOI] [Google Scholar]
- 78.Gómez-Consarnau L, Sachdeva R, Gifford SM, Cutter LS, Fuhrman JA, Sañudo-Wilhelmy SA, et al. Mosaic patterns of B-vitamin synthesis and utilization in a natural marine microbial community. Environ Microbiol. 2018;20:2809–23. doi: 10.1111/1462-2920.14133. [DOI] [PubMed] [Google Scholar]
- 79.Bertrand EM, Saito MA, Jeon YJ, Neilan BA. Vitamin B12 biosynthesis gene diversity in the Ross Sea: the identification of a new group of putative polar B12 biosynthesizers. Environ Microbiol. 2011;13:1285–98. doi: 10.1111/j.1462-2920.2011.02428.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated for this manuscript is available from the NCBI SRA under accession PRJN779092.