Abstract
VanA and VanB form an oxygenative demethylase that converts vanillate to protocatechuate in microorganisms. Ferulate, an abundant phytochemical, had been shown to be metabolized through a vanillate intermediate in several Pseudomonas isolates, and biochemical evidence had indicated that vanillate also is an intermediate in ferulate catabolism by Acinetobacter. Genetic evidence supporting this conclusion was obtained by characterization of mutant Acinetobacter strains blocked in catabolism of both ferulate and vanillate. Cloned Acinetobacter vanA and vanB were shown to be members of a chromosomal segment remote from a supraoperonic cluster containing other genes required for completion of the catabolism of ferulate and its structural analogs, caffeate and coumarate, through protocatechuate. The nucleotide sequence of DNA containing vanA and vanB demonstrated the presence of genes that, on the basis of nucleotide sequence similarity, appeared to be associated with transport of aromatic compounds, metabolism of such compounds, or iron scavenging. Spontaneous deletion of 100 kb of DNA containing this segment does not impede the growth of cells with simple carbon sources other than vanillate or ferulate. Additional spontaneous mutations blocking vanA and vanB expression were shown to be mediated by IS1236, including insertion of the newly discovered composite transposon Tn5613. On the whole, vanA and vanB appear to be located within a nonessential genetic region that exhibits considerable genetic malleability in Acinetobacter. The overall organization of genes neighboring Acinetobacter vanA and vanB, including a putative transcriptional regulatory gene that is convergently transcribed and overlaps vanB, is conserved in Pseudomonas aeruginosa but has undergone radical rearrangement in other Pseudomonas species.
Vanillate demethylase is a member of a superfamily of reductive dioxygenases with a broad range of activities (47). The enzymes are of interest because they provide model systems for analysis of electron transfer and because they demonstrate the consequences of modular rearrangement of catalytic domains during enzyme evolution (22).
Recent attention has focused upon vanillate as an intermediate in ferulate catabolism by Pseudomonas species (Fig. 1). Nucleotide sequencing of Pseudomonas strain ATCC 19151 genes encoding vanillate demethylase revealed open reading frames designated vanA and vanB; these genes exhibited the high G+C content characteristic of the genus (5). Examination of DNA cloned from Pseudomonas sp. strain HR199 revealed vanA and vanB genes on an EcoRI restriction fragment that also contained an open reading frame encoding vanillin dehydrogenase; the latter open reading frame was in the same operon as an open reading frame encoding a protein with amino acid sequence similarity to enoyl coenzyme A (enoyl-CoA) hydratase (57).
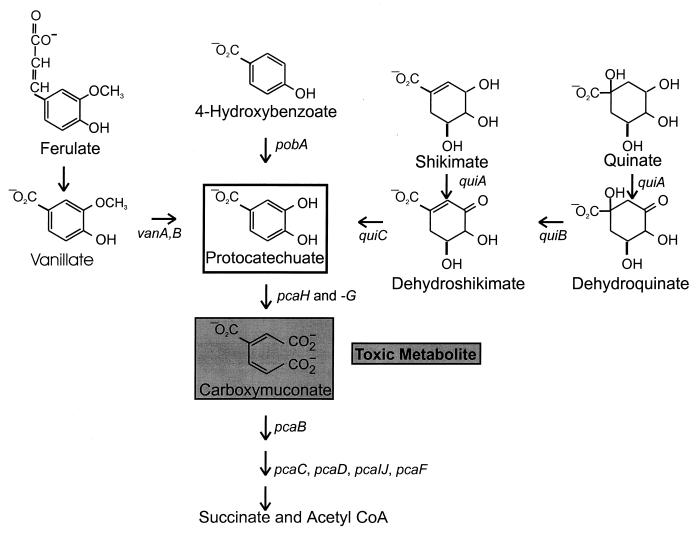
FIG. 1.
Vanillate and many other plant products are metabolized through protocatechuate. The pob, qui, and pca genes are clustered in the Acinetobacter chromosome. The open box indicates protocatechuate, and the shaded box indicates carboxymuconate, which is produced by the action of protocatechuate 3,4-dioxygenase on protocatechuate. Arrows indicate metabolic reactions and are accompanied by the designations of genes encoding the enzymes that catalyze the reactions. Metabolic accumulation of carboxymuconate in strain ADP230, defective in pcaB, prevents the growth of cells in the presence of substrates that can be metabolized to carboxymuconate.
Mutations blocking ferulate catabolism in Pseudomonas putida WCS358 allowed cloning from the organism of two restriction fragments containing DNA necessary for the metabolic pathway (62). The presence of vanA and vanB in one of the clones confirmed the role of these genes in ferulate catabolism. The other clone contained open reading frames for vanillin dehydrogenase and enoyl-CoA hydratase. Analysis of enzymes associated with ferulate metabolism in Pseudomonas fluorescens bv. VAN103 (48) demonstrated an enoyl-CoA hydratase that also acted as a lyase cleaving the hydrated derivative of ferulyl-CoA into vanillin and acetyl-CoA (23). Thus, in at least some Pseudomonas species, ferulate metabolism appears to proceed by thioester formation, hydration, and cleavage, giving rise to vanillin, which is oxidized to vanillate.
Acinetobacter and fluorescent Pseudomonas species are the predominant representatives of the “Pseudomonas” group within the γ subdivision of the proteobacteria as classified by the National Center for Biotechnology Information. Genetic comparison of pathways for aromatic catabolism in the two taxa has revealed some similarities and marked differences. In general, isofunctional enzymes from the two taxa have amino acid sequence identity of roughly 50%. Differences are evident at the DNA level in that the Pseudomonas genes characteristically have a G+C content above 58% (32) whereas the Acinetobacter genes, with a few notable exceptions, have a G+C content below 46% (42). Numerous rearrangements accompanied the divergence of the respective genes, yet transposition did not always lead to their scattering in the chromosome of their hosts. Apparent selection for grouping of genes with related physiological function is evident in the Acinetobacter pca-qui-pob supraoperonic cluster (Fig. 1) in a chromosomal segment containing more than 20 kb of DNA (11, 12, 16, 17, 26, 42). Recent investigations have demonstrated tight linkage to pobA of a gene encoding a CoA-ligase required for catabolism of ferulate and its structural analogs, caffeate and coumarate (61). An objective of the present study was to determine if vanA and vanB were linked to the pca-qui-pob gene cluster.
Earlier investigations demonstrated that vanillate and protocatechuate (Fig. 1) are formed during the metabolism of ferulate by Acinetobacter calcoaceticus DSM586 (10). The conclusion that the vanillate pathway was the sole mechanism for ferulate utilization in Acinetobacter would be supported by the demonstration that mutations blocking vanillate demethylase prevented growth on ferulate. An organism well suited for such genetic analysis is Acinetobacter strain ADP1. This organism, formerly assigned to the species A. calcoaceticus but now recognized to be a representative of a separate taxon within Acinetobacter (3, 4, 15), is highly competent for natural transformation (36, 37). Furthermore, the physiological properties of this strain allow the design of a strategy for selection of mutants blocked in ferulate metabolism (Fig. 1).
Such a selection procedure was first used to survey the properties of spontaneous mutants blocked in pcaH and pcaG, structural genes for protocatechuate 3,4-dioxygenase. Among 94 independently selected strains, 4 contained a newly discovered insertion sequence, IS1236, within pcaH (25). Similar selection for strains blocked in p-hydroxybenzoate metabolism (Fig. 1) yielded strains with defects in either pobA, the structural gene for p-hydroxybenzoate hydroxylase, or pobR, which encodes the transcriptional activator of pobA (11, 13). Spontaneous mutations blocking pobR are caused predominantly by insertion of IS1236 at different locations throughout the gene (24).
In this report we describe the isolation of spontaneous vanA and vanB mutants and demonstrate that the genes for vanillate demethylase are essential for the growth of Acinetobacter with ferulate. A significant fraction of the vanA and vanB mutations were caused by IS1236, which also was shown to contribute to the structure of a newly discovered composite transposon, Tn5613, which can inactivate the van genes. Unlike other known genes associated with protocatechuate metabolism, vanA and vanB occupy a location separate from the pca-qui-pob cluster in the Acinetobacter chromosome.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Properties of bacterial strains used in this investigation are summarized in Table 1, and relevant properties of plasmids are presented in Fig. 2. Escherichia coli cultures were grown at 37°C in Luria-Bertani (LB) medium (60). Acinetobacter cultures were grown at 37°C in basal medium supplemented with carbon sources as indicated in the text.
TABLE 1.
Bacterial strains used in this investigation
| Strain | Parental strain | Genotype | Nature of mutation | Selection | Source or reference |
|---|---|---|---|---|---|
| ADP1 | BD413 | Wild type | 36 | ||
| ADP230 | ADP1 | pcaΔBDK1 | Engineered deletion | 30 | |
| ADP642 | ADP230 | pcaΔBDK1 ΔvanAB643 | Spontaneous deletion | Resistance to ferulate | This investigation |
| ADP643 | ADP642 | ΔvanAB643 | Growth with protocatechuate | This investigation | |
| ADP644 | ADP230 | pcaΔBDK1 vanA645 | Spontaneous insertion | Resistance to ferulate | This investigation |
| ADP645 | ADP644 | vanA645 | Growth with protocatechuate | This investigation | |
| ADP646 | ADP230 | pcaΔBDK1 vanB647::Tn5613 | Spontaneous insertion | Resistance to ferulate | This investigation |
| ADP647 | ADP646 | vanB647::Tn5613 | Growth with protocatechuate | This investigation | |
| ADP648 | ADP230 | pcaΔBDK1 vanB649 | Spontaneous bp substitution | Resistance to ferulate | This investigation |
| ADP649 | ADP648 | vanB649 | Growth with protocatechuate | This investigation | |
| ADP650 | ADP230 | pcaΔBDK1 ΔvanAB651 | Spontaneous deletion | Resistance to ferulate | This investigation |
| ADP651 | ADP650 | ΔvanAB651 | Growth with protocatechuate | This investigation | |
| ADP652 | ADP230 | pcaΔBDK1 ΔvanA653 | Spontaneous 13-bp deletion | Resistance to ferulate | This investigation |
| ADP653 | ADP652 | ΔvanA653 | Growth with protocatechuate | This investigation | |
| ADP654 | ADP230 | pcaΔBDK1 ΔvanAB655 | Spontaneous deletion | Resistance to ferulate | This investigation |
| ADP655 | ADP654 | ΔvanAB655 | Growth with protocatechuate | This investigation | |
| ADP656 | ADP230 | pcaΔBDK1 vanB657 | Spontaneous insertion | Resistance to ferulate | This investigation |
| ADP657 | ADP656 | vanB657 | Growth with protocatechuate | This investigation | |
| ADP672 | ADP230 | pcaΔBDK1 vanA673::Tn10′a | Mini-Tn10 | Resistance to both vanillate and kanamycin | This investigation |
| ADP673 | ADP672 | vanA673::Tn10′a | Growth with protocatechuate; loss of Kanamycin resistance | This investigation | |
| ADP674 | ADP230 | pcaΔBDK1 vanA675::Tn10′avanB675::Tn5613 | Mini-Tn10; spontaneous mutation | Resistance to both vanillate and kanamycin | This investigation |
| ADP675b | ADP672 | vanA675::Tn10′avanB675::Tn5613 | Growth with protocatechuate; resistance to kanamycin | This investigation |
The portion of Tn10 remaining in these strains is the 50-bp segment left by nearly precise excision (58).
This strain retains the mini-Tn10 kanamycin resistance marker, but it is not in the van genes.
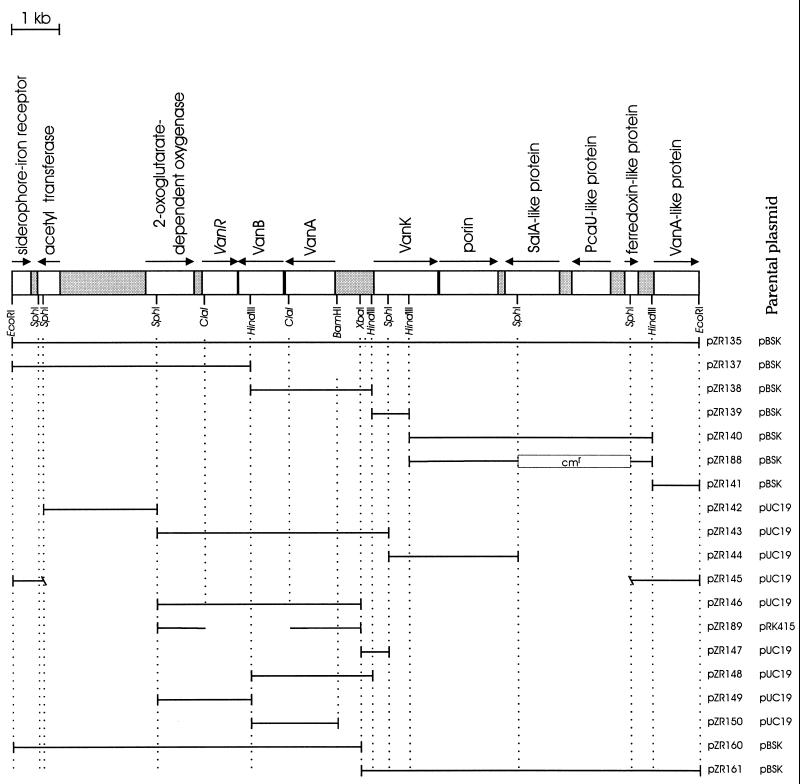
FIG. 2.
Plasmids used in this investigation. The DNA in each depicted insert is represented as a horizontal line extending between the restriction sites that mark the limits of the insert. The vectors pRK415 (39), pUC19 (66), and pBSK (Stratagene) have been described previously. All of the inserts were derived from the 14-kb EcoRI fragment in pZR135. The interruption in the horizontal line in pZR189 indicates the deletion produced by digestion with ClaI before recovery by gap repair of vanB chromosomal DNA in this plasmid. At the top of the figure are listed the functions tentatively assigned to open reading frames (indicated as open rectangles) on the basis of amino acid sequences similar to those of known proteins; the arrows indicate the directions of transcription. It must be emphasized that the some of the tentative functions assigned to proteins were shown to be incorrect as part of this investigation: a knockout mutation blocking the expression of the SalA-like protein did not prevent growth on salicylate, and the VanA-like protein did not complement mutations blocking the expression of vanA. Not depicted here are four additional BamHI sites (GGATCC) that were detected by sequencing the vanA-vanK intergenic region.
Isolation of mutant strains.
Selection for loss of van functions was imposed by demanding growth of strain ADP230, a mutant that accumulates the toxic intermediate carboxymuconate when exposed to vanillate, on plates containing succinate as a growth substrate and either vanillate or ferulate as a source of the toxic metabolite (Fig. 1). After genetic restoration of the ability to metabolize carboxymuconate, natural transformation was used to map the mutations blocking vanillate metabolism (24, 30, 42).
Mutagenesis with mini-Tn10 (34) was achieved by mixing 4 × 107 E. coli SM10λpir(pLOFKm) cells and 107 Acinetobacter strain ADP230 cells on LB plates containing 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG). After 8 h of incubation, the cells were transferred from these plates and spread on plates containing 10 mM succinate, 2 mM vanillate, and 25 mg of kanamycin per ml. Colonies emerging on this medium were screened for resistance to 2 mM ferulate. During maintenance in the absence of kanamycin, the cell line derived from ADP672 lost resistance to the antibiotic, giving rise to ADP673 (Table 1).
Colonies containing spontaneous mutants were selected by spreading about 108 ADP230 cells on plates containing 3 mM ferulate and 10 mM succinate. After single-colony isolation on the same medium, mutant strains were screened for their ability to grow on plates containing 10 mM succinate and 2 mM vanillate. The properties of strains that grew on this medium are summarized in Table 1.
Natural transformation.
Recipient Acinetobacter cells were grown in a shaker overnight in 5-ml cultures with 10 mM succinate, and succinate was added to an additional concentration of 10 mM the next morning. After 30 min of incubation, 200 μl of the culture was mixed with either cell lysate or plasmid DNA on a plate of selective medium. For transformation prior to selection, 200 μl of the overnight culture was added to a tube containing 5 ml of 10 mM succinate and incubated for 3 h, after which the cells were plated on selective medium.
Manipulation of DNA.
Chromosomal DNA and cell lysate were prepared as previously described (25). Plasmid DNA was isolated with the Wizard Miniprep kit from Promega. Standard techniques of molecular biology were used in plasmid and gene manipulations (60).
Screening by replica plating for E. coli clones containing the Acinetobacter vanillate demethylase DNA.
Chromosomal DNA from wild-type Acinetobacter ADP1 was digested with EcoRI (New England Biolabs), and the resulting fragments were ligated to pBSK (previously linearized with EcoRI). The ligated material was introduced into E. coli DH5α by transformation, and the transformants were plated onto LB agar plates containing ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and IPTG. The resulting E. coli colonies were screened for the presence of vanillate demethylase genes by assessment of the ability of plated replicas to transform the mutant ADP675 so that recombinants grew with 2 mM vanillic acid as the sole carbon source (2).
DNA sequencing and sequence analysis.
Sequencing of double-stranded plasmid DNA or PCR templates was performed in an ABI 373 automated sequencer (Perkin-Elmer ABI) with T3, T7, m13 universal, and reverse primers and synthetic oligonucleotides. Subclones, nested deletions, and PCR fragments were made to complete the 14-kb sequence of the insert in pZR135. PCR fragments were used to sequence mutant genes. Homology searches were performed with the gapped BLAST database search program (1), and sequence analysis was performed with DNAMAN (Lynnon Biosoft).
Designed deletions.
Knockout mutations for open reading frames encoding the SalA-like protein and the PcaU-like protein were created by replacing the internal SphI fragment of pZR140 with DNA encoding chloramphenicol resistance in pCAT19 (21). The insert from the resulting plasmid, pZR188 (Fig. 2), was introduced into wild-type Acinetobacter strain ADP1 by transformation followed by selection for chloramphenicol resistance. The resulting mutant, strain ADP1070, was tested for its ability to grow with ferulate, vanillate, or salicylate. To construct a plasmid for recovery of DNA by gap repair (28), the lacZ::Knr cassette from pKOK6 (41) was inserted into the NcoI site of vanB in pZR143. Digestion with XbaI, using one site from the pZR143 polylinker, then yielded a fragment which was ligated into the broad-host-range vector pRK415. Digestion of this plasmid with ClaI released DNA containing all of vanB, and the remaining linearized plasmid was used to recover chromosomal DNA containing vanB647.
p-Toluidine test for metabolic accumulation of protocatechuate.
The color test developed by Parke (55) to screen for protocatechuate accumulation was used to detect formation of the compound from vanillate in E. coli cells containing different recombinant plasmids.
PCR amplifications.
PCR amplifications were performed with 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min. Special conditions were also used with the amplification of ADP647, ADP657, and ADP645 with primers Seq1 to Seq6: the extension time was 5 min, and the annealing temperature was 45°C.
PCR primers.
PCR primers of special importance in efforts to characterize van mutants were located in open reading frames as follows: Seq1, 5′ GTAACTCGGAGAGACTGTACGTC 3′, regulatory protein (Fig. 2); VanB22, 5′ GACGTACAGTCTCTCCGAGTTAC 3′, regulatory protein (Fig. 2); Seq3, 5′ GGGAAGTGTTCAGTCAGGTTGCC 3′, vanB (positions 1772 to 1749); VanB11, 5′ CGCCTATTCTCTCCATGGC 3′, vanB (positions 1655 to 1673); VanB21, 5′ CGTCAATATGTGAGCCTGCG 3′, beginning of vanB (positions 1423 to 1403); VanA2, 5′ CAGGCGTAACTACAATCGACAAC 3′, end of vanA (positions 979 to 1003); Seq5, 5′ CAGCCCACCGCCATAACTCCACTCT 3′, vanA (positions 631 to 606); Seq6, 5′ CAATAAGTGATAAGGAGTCG 3′, upstream of vanA (positions 192 to 214); IS1, 5′ GGCCATTTCTTGGATCTCC 3′, 5′ end of IS1236 (positions 60 to 78); IS2, 5′ CGGCTGGTTAAGCCCAGAAGC 3′, 3′ end of IS1236 (positions 154 to 1174).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 14-kb EcoRI fragment containing Acinetobacter vanA and vanB appears in the GenBank nucleotide sequence database under accession no. AF009672. The GenBank accession number for the complete Tn5613 nucleotide sequence is AF091240, and the GenBank accession number for the 2.7-kb nucleotide sequence of the partial open reading frame carried as an insert in pZR153 is AF011339.
RESULTS
Isolation of mutants unable to metabolize either ferulate or vanillate to protocatechuate.
To test the possibility that vanillate is an intermediate in the catabolism of ferulate by Acinetobacter, strains were selected on the basis of their inability to convert ferulate to carboxymuconate, a toxic intermediate that accumulates and prevents growth in strains containing the pcaΔBDK1 deletion (Fig. 1), and then were screened for potential blocks in their conversion of vanillate to protocatechuate. After exposure to mini-Tn10, ADP230 derivatives that had acquired the transposon were selected by demanding growth in the presence of kanamycin, and colonies that exhibited kanamycin resistance were screened for growth with succinate in the presence of ferulate. Further screening of ferulate-resistant isolates revealed two strains, ADP672 and ADP674, that grew with succinate in the presence of either ferulate or vanillate but failed to grow with succinate in the presence of protocatechuate (Table 1). Replacement of the pcaΔBDK1 deletion in these strains produced the respective transformants ADP673 and ADP675, which grew in the presence of protocatechuate and, as would be expected for strains blocked in vanillate demethylase, failed to grow in the presence of either ferulate or vanillate (Table 1).
As described below, a genetic remnant of mini-Tn10 remained in the van genes of strains ADP673 and ADP675, but the kanamycin marker in these strains was lost and transposed nearby in the chromosome. It therefore was evident that multiple mutations accompanied mini-Tn10 inactivation of the van genes in these organisms, and in order to rule out the possibility that such transpositions-induced mutations affected additional genes required for vanillate catabolism, spontaneous mutants blocked in conversion of vanillate to protocatechuate were selected. Eight spontaneous vanillate-resistant derivatives of strain ADP230 were independently obtained and shown to be unable to grow in the presence of protocatechuate (Table 1). As would be expected if vanillate demethylase were required for ferulate catabolism, the mutant strains grew in the presence of ferulate. Replacement of pcaΔBDK1 in the spontaneous mutants gave rise to strains that grew at the expense of protocatechuate and not at the expense of either vanillate or ferulate (Table 1).
Cloning of DNA fragments containing genes for vanillate demethylase.
Screening of E. coli colonies carrying a library of EcoRI fragments of Acinetobacter DNA revealed four strains, each of which contained a different recombinant plasmid that transformed Acinetobacter strain ADP675 so that it grew on vanillate. Two of the plasmids (pZR135 and pZR136) contained 14-kb inserts in opposite orientations with respect to the plasmid promoter. The other two plasmids (pZR151 and pZR156) contained inserts of 3.3 and 5.5 kb, respectively. As judged by the p-toluidine color test (55), E. coli DH5α(pZR135) produced a substantial amount of protocatechuate from vanillate, whereas E. coli DH5α(pZR136), containing the 14-kb EcoRI insert in reverse orientation with respect to the plasmid lac promoter, formed a slight amount of protocatechuate and E. coli strains containing pZR151 or pZR156 did not form discernible amounts of protocatechuate.
Subcloning and characterization of mutant strains by gene replacement.
Plasmid pZR135 and the five HindIII subclones derived from it (Fig. 2) were tested for their ability to restore wild-type function to the 10 mutant strains blocked in the conversion of vanillate to protocatechuate. The plasmids did not produce a wild-type phenotype when used as donors in transformation for strains ADP643 or ADP655; these strains did yield wild-type recombinants when treated with DNA from the parental strain ADP1. These mutant strains and strain ADP651 did not yield wild-type recombinants when used as the donor in transformation of the other strains blocked in vanillate catabolism. It thus appears that strains ADP643 and ADP655 have undergone deletions extending beyond the limits of the insert in pZR135; the deletion in strain ADP651 eliminates alleles for which van mutations were obtained but falls within the confines of the pZR135 insert. The remaining van mutants exhibited the ability to be transformed to the wild type by pZR135 and by either of two HindIII subclones, pZR138 or pZR137 (Fig. 2). This evidence suggested that the HindIII restriction site between the chromosomal fragments inserted in pZR138 and pZR137 (Fig. 2) lies within the genes required for vanillate catabolism.
Most of the mutant strains that were transformed by one of the HindIII subclones appear to have mutations at separate loci as evidenced by growth of wild-type recombinants after crosses between pairs of the strains. Exceptions to this pattern indicated that some independent mutations may have occurred at identical or nearby loci, and subsequent sequencing of the mutant DNA proved that this was the case. Anomalous results were obtained with strain ADP675, which subsequently was shown to have acquired two separate mutations in the van region.
Nucleotide sequences of different Acinetobacter EcoRI restriction fragments that transform a van-deficient strain so that it grows with vanillate.
Three different EcoRI chromosomal fragments conferred upon strain ADP675 the ability to grow with vanillate. The 5.5-kb insert of pZR156 contained an internal EcoRI site. Sequence analysis of the ends of the pZR156 insert revealed DNA corresponding to benK and benD. The location of restriction sites within the insert confirmed that the cloned genes encoded proteins required for the oxygenative metabolism of benzoate and corresponded to the insert of the previously described plasmids pIB1351 and pIB1352 (49, 50), together with an EcoRI fragment internal to benD. The 3.3-kb insert of pZR151 also contained an internal EcoRI site; a 2.7-kb EcoRI subclone in pZR153 remained able to confer upon ADP675 the ability to grow on vanillate. Sequencing of the 2,755-bp insert in pZR153 revealed that it consisted entirely of a large partial open reading frame beginning with sequences encoding tandem repeats of approximately 100 amino acids, similar to the fibronectin type III repeats in bacterial cellulases (29, 45, 46).
The complete 14,358-bp nucleotide sequence of the pZR135 insert revealed 12 open reading frames and two large DNA segments (one of 1,781 bp and one of 811 bp) with no obvious coding functions (Fig. 2). The open reading frames are of general interest because they are components of a 100-kb DNA fragment that has been selected in wild-type Acinetobacter yet is frequently lost during maintenance of the organism in the laboratory (27).
Of immediate significance to this investigation were the two open reading frames encoding proteins with amino acid sequences closely resembling those determined for VanA and VanB, proteins that form vanillate demethylase in Pseudomonas species (5, 57, 62). The possibility that the Acinetobacter open reading frames assigned vanA and vanB function was enhanced by the fact that DNA from either pZR137 or pZR138 (Fig. 2) replaced mutations in these genes and, as described below, was confirmed by sequencing of mutant genes.
The locations of Acinetobacter vanA and vanB are indicated in Fig. 2. Also indicated is an apparent regulatory gene, tentatively designated vanR, that converges upon vanB; the convergently transcribed genes overlap by 21 bp.
To identify regions that might be associated with transcriptional termination, the 14-kb sequence of the EcoRI fragment was scanned for inverted repetitions containing complementary sequences exceeding 10 bp. The strongest free energy of association, 10.1 kcal/mol, was exhibited by an 11-bp palindrome separated by 7 bp. The palindrome begins 13 bp downstream from the translational terminus of vanB and is followed by the sequence TTTTTT. This potential terminator of the vanA and vanB transcript is of particular interest because it lies well within the convergently transcribed vanR.
The longest palindromic pair proved to be 14 bp long and separated by 401 bp. This inverted repetition and an additional inverted repetition of 12 bp are within the 811-bp vanK-vanA intergenic region; all of the repeated sequences contain the internal palindromic sequence TGGATCCA. Further examination showed that GGATCC, the BamHI restriction site, occurs five times in the 811-bp vanK-vanA intergenic region and only three times in 70,000 bp of known nucleotide sequence from Acinetobacter strain ADP1. A close match to GGATCC is exhibited by the termini of IS1236, which are TGATCC and GGATCA. It is possible that palindromes containing the sequence GGATCC served as a class of preferred targets for IS1236 integration.
Among open reading frames that resemble Acinetobacter vanA is the partial open reading frame at one end of the pZR135 insert. Over 171 aligned residues, the protein encoded by this open reading frame has identity of 33% to Acinetobacter VanA, less than the sequence identity of 72% for Acinetobacter VanA and its closest known homolog, VanA from Pseudomonas sp. strain HR199 (57).
Two neighboring open reading frames in pZR135 closely resemble open reading frames with known functions in aromatic catabolism. One of these, designated the gene for a SalA-like protein in Fig. 2, encodes a protein with amino acid sequence identity of 30% to the known P. putida salicylate monooxygenase (68). The other open reading frame, whose product is designated PcaU-like protein in Fig. 2, encodes a product that resembles Rhodococcus opacus PcaR (30% sequence identity [18]) and Acinetobacter PcaU (27% sequence identity), the transcriptional activator of genes for protocatechuate catabolism (26). The ferredoxin-like protein (Fig. 2) most closely resembles (33% amino acid identity over 100 residues) a Pseudomonas protein, NagAb, believed to be part of the electron transport chain for both salicylate 5-hydroxylase, which converts salicylate to gentisate, and naphthalene dioxygenase (20).
The functions of the salA-like and pcaU-like genes were explored by introduction of a deletion mutation removing 410 bp from the transcriptional terminus of the 813-bp pcaU-like gene and 886 bp from the beginning of the salA-like gene. Strains containing this mutation were unimpaired in their catabolism of salicylate, vanillate, and protocatechuate as judged by growth on plates; therefore, the altered genes appear to be associated with other functions.
Two genes that may form a transcriptional unit in pZR135 appear to be associated with transport. One of these, designated the gene for VanK in Fig. 2, encodes a product with amino acid sequence with 31% identity to Acinetobacter PcaK, a member of a family of proteins known to transport p-hydroxybenzoate and protocatechuate in bacteria (7, 31, 35, 42, 52, 63). The other transport related open reading frame, indicated as a gene with outer membrane protein (OMP)-like function in Fig. 2, encodes a product with 32% sequence identity to its closest known homolog, P. putida PhaK, which is required for growth of the organism on phenylacetate (53). The putative Acinetobacter porin also exhibits close sequence similarity to four gene products assigned OMP functions in Pseudomonas aeruginosa (65, 67); comparison of the products of the Acinetobacter OMP-like gene with the Pseudomonas genes revealed amino acid sequence identities ranging between 25 and 31%.
Upstream of the regulatory gene that overlaps with vanB (Fig. 2) is an ORF encoding a protein with up to 27% identity to several members of the 2-oxoglutarate-dependent oxygenase family in a variety of plants. Because two members are multifunctional gibberellin oxidases that can convert the aldehyde of this plant hormone to its acid (43), this raises the possibility that the Acinetobacter enzyme catalyzes the conversion of vanillin to vanillate. In Pseudomonas sp. strain HR199, this function is performed by an NAD-dependent dehydrogenase and its gene is near vanAB on the chromosome (57).
Transcribed divergently from this oxygenase gene, cloned in pZR135 and separated by over 1,700 bp of DNA without any obvious coding potential, is an ORF for a protein with up to 31% identity (48 of 152 aligned residues) to various yeast hypothetical acetyltransferases. This protein is less closely related to an N-acetyltransferase that provides resistance to the antibiotic streptothricin in diverse bacteria. Lastly, on the end of the pZR135 insert and transcribed in the opposite direction to the putative acetyltransferase gene is the end of a gene encoding a protein with 23 to 33% identity to the corresponding segment of various iron-scavenging outer membrane receptors including the E. coli FhuA ferrichrome receptor, the P. aeruginosa FptA iron-pyochelin receptor, and the Vibrio anguillarum FatA iron-anguibactin receptor. Ligands for the last two receptors are structurally related (9, 56, 64) to other pathogenic bacterial siderophores including yersiniabactin (56) from various Yersinia species and acinetobactin (14, 64) from Acinetobacter baumanii.
Nucleotide sequences of vanA and vanB genes containing spontaneous mutations.
The primers Seq1 and Seq6 were used to amplify the chromosomal DNA containing vanA and vanB from the Acinetobacter chromosome, and internal primers (Seq2, Seq3, and Seq5) were used to sequence mutations in these genes. As expected, the PCR did not yield amplified fragments with the deletion mutations ΔvanAB643, ΔvanAB651, and ΔvanAB655. Sequencing revealed that ΔvanA653 is a 13-bp deletion removing 7 bp of directly repeated nucleotide sequence (Fig. 3), suggesting that the mutation was directed in part by hybridization between slipped DNA strands (Fig. 4).
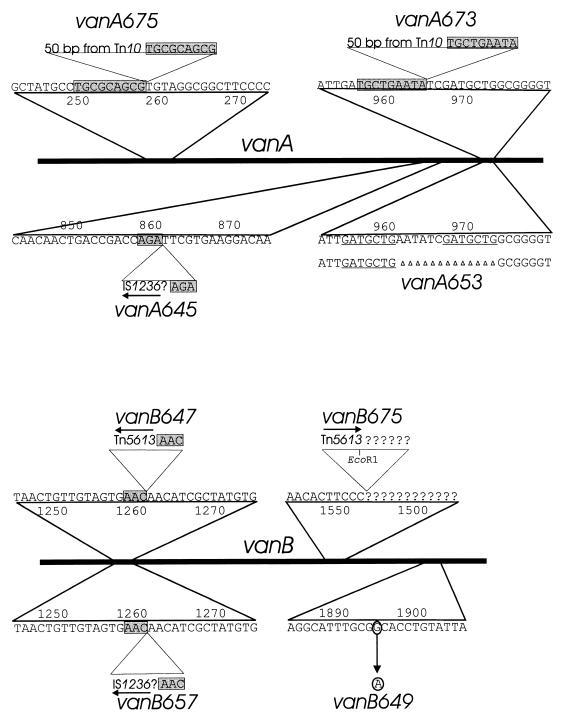
FIG. 3.
Characterized mutations in vanA and vanB. Linear bars represent vanA and vanB; expanded portions of the sequence depict the locations of mutations. Shaded rectangles indicate direct repetitions of chromosomal sequence flanking sites of insertion for mutations caused by insertion sequences or transposons. Horizontal arrows show the direction of transcription of the open reading frames within IS1236. Underlining marks the 7-bp direct chromosomal sequence repetition that appears to have directed the 13-bp vanA653 deletion. Characterization of DNA upstream from vanB674 was made possible by cloning of an EcoRI fragment containing DNA extending into the EcoRI site of Tn5613. Properties of DNA downstream from this site in vanB675 remains unknown.
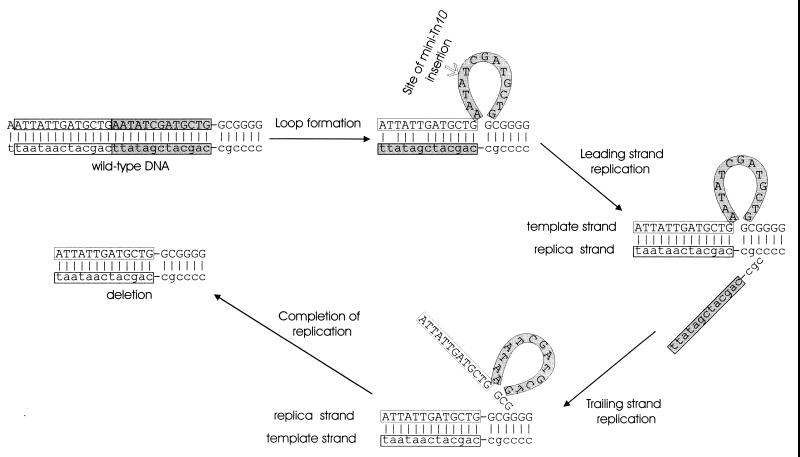
FIG. 4.
Possible contributions of DNA strand slippage to mutations in vanA. The 11-bp deletion in vanA653 removed a 7-bp direct sequence repetition, suggesting that mispairing between slipped strands may have misaligned DNA so that the deletion took place during replication. The single-stranded loop caused by mispairing may have been a target for mini-Tn10 insertion, resulting in vanA675.
Only one of the eight selected spontaneous mutations is a base substitution: the vanB649 mutation contains G-to-A nucleotide substitution causing the amino acid substitution G269D in VanB (Fig. 3). The substituted amino acid is underlined in the peptide sequence CEQGICGTCITR, which is believed to be associated with iron-sulfur binding and is completely conserved in four different Pseudomonas isolates. The glycyl residue is conserved in enzymes as distantly related as pthalate dioxygenase reductase (8), toluenesulfonate methyl-monooxygenase reductase (38), and 3-chlorobenzoate-3,4-dioxygenase reductase (47); therefore, replacement of the flexible neutral amino acid residue by the charged aspartyl residue would be expected to be highly disruptive to the protein structure.
Unexpectedly, chromosomal DNA containing any of the remaining three spontaneous mutations, vanA645, vanB647, or vanB657, could not be amplified by PCR, prompting the hypothesis that these mutations contained long DNA inserts. Since IS1236 is a known cause of insertion mutations in Acinetobacter (24), the primers Is1 and Is2, complementary to nucleotide sequences near the left and right termini, respectively, of IS1236 (24), were paired with primers from vanA and vanB to determine if the insertion sequence could be located within the mutant genes. The results of this analysis and subsequent nucleotide sequencing demonstrated the contribution of IS1236 to all three mutations (Fig. 3).
Insertion of a single copy of the 1.2-kb IS1236, however, could not account for the three negative PCR results. Therefore, gap repair (28) was tried with the two of the mutants by using a plasmid in which digestion with ClaI had removed vanB DNA (Fig. 2). An 8-kb DNA insert containing vanB647 was recovered from the chromosome of ADP647. Sequencing of this fragment revealed a 3-bp direct repeat of vanB DNA (Fig. 3) flanking a previously undescribed 3,552-bp composite transposon designated Tn5613. At the ends of the transposon are identical copies of IS1236 in the same orientation. Between these and in the opposite orientation is a 546-bp open reading frame. The G+C content of the open reading frame is 36%, somewhat less than the 39% G+C content of the entire Tn5613, and the translation product of the open reading frame shows no obvious similarity to any of the amino acid sequences reported in GenBank.
Procedures used to recover vanB647 by gap repair did not yield a product when applied to strains containing vanB657. Sequencing of PCR-amplified DNA flanking IS1236 in vanB657 demonstrated that at least one copy of the IS element is inserted in the same manner as Tn5613 in vanB647 (Fig. 3), but PCR with one primer complementary to the open reading frame within Tn5613 gave no indication of corresponding DNA in vanB657. Furthermore, PCRs with primers designed to amplify between the two IS elements of Tn5613 in strains with vanB657 or vanB645 also failed to yield a product (in contrast to results with the other six spontaneous mutants), suggesting that the transposon was not intact in these two strains. The complete nature of the vanB657 and vanB645 mutations remains a mystery.
Nucleotide sequences of vanA and vanB genes containing mutations caused by mini-Tn10.
Mini-Tn10 proved to be unstable in the two mutants that emerged after selection for the kanamycin resistance marker carried by this transposon. The marker was lost in the cell line that gave rise to the kanamycin-sensitive strain ADP673, which did not grow with vanillate. Mapping of vanA673 in this strain suggested that it was extremely close to the 13-bp deletion van653 (Fig. 3), raising the possibility that the DNA strand slippage giving rise to this mutation influences the behavior of the transposon at the same location (Fig. 4). As indicated in Fig. 3, the departed mini-Tn10 left behind a 50-bp signature sequence of nearly precise excision (58), and this sequence is flanked by 9-bp direct repeats (40) of chromosomal nucleotide sequence (Fig. 3).
Multiple mutations emerged in strain ADP675 after its treatment with mini-Tn10 followed by selection for resistance to the intracellular accumulation of β-carboxymuconate. The mutant strain retained its resistance to kanamycin. The gene conferring this trait was recovered from an EcoRI library from the mutant strain and proved to be in a DNA fragment containing a portion of the van genes. One end of this EcoRI restriction fragment corresponded to the site within a portion of the putative vanA-like gene in pZR135 (Fig. 2), and the other EcoRI site was within Tn5613. Sequence analysis demonstrated that the kanamycin resistance determinant was not in vanA or vanB. Evidence for its temporary insertion was revealed by vanA675, which contains the 50 bp sequence corresponding to nearly precise excision flanked by 9-bp chromosomal repeats (40, 58). Strain ADP675 also contained the portion of Tn5613 extending to its internal EcoRI site in vanB675 (Fig. 3). This was demonstrated by sequencing from Tn5613 into DNA upstream from vanB (Fig. 3). PCR primers downstream from vanB675 did not yield an amplified product with chromosomal DNA from ADP675, and so it seemed likely that a portion of this DNA was deleted downstream from the site of Tn5613 insertion in the mutant strain.
DISCUSSION
vanA and vanB, essential for metabolism of both ferulate and vanillate, are separated from the pca-qui-pob gene cluster in the Acinetobacter chromosome.
At the outset of this investigation, it was not clear whether vanillate was an obligatory intermediate in the utilization of ferulate by Acinetobacter. Essential participation of vanillate as a catabolite is demonstrated by the fact that spontaneous Acinetobacter mutations blocking vanA and vanB prevent growth with ferulate, and this growth property is restored by exposure of the mutant cells to DNA containing the wild-type genes. Such a conclusion could not be drawn on the basis of sequence evidence alone. The 14-kb DNA fragment containing vanA and vanB also contains an open reading frame resembling vanA in sequence but apparently not involved in growth with vanillate. Also within this DNA fragment is a gene that, on the basis of sequence similarity, appears to encode salicylate hydroxylase. However, inactivation of this open reading frame does not prevent growth with salicylate, and so the function of this gene awaits elucidation.
The precise locations of vanA and vanB are unknown, but the genes appear to be lacking in organisms that have undergone the spontaneous deletion eliminating a 100-kb DNA fragment from the Acinetobacter ADP1 chromosome (27). Such strains grow with protocatechuate but not with vanillate and do not reveal DNA fragments corresponding to portions of vanA or vanB on PCR with primers that do produce such fragments after amplification of wild-type chromosomal DNA. The 100-kb deletion was fortuitously discovered during mapping of the Acinetobacter chromosome and occurs in a region that appears to contain multiple copies of IS1236 (24, 27), perhaps reflecting in part the presence of Tn5613. If the van genes do lie within the deleted region, then preferential localized transposition of Tn5613 may explain why this transposon was not detected previously among spontaneous mutations in pob or pca genes. The discovery of Tn5613 adds a new genetic marker that may facilitate the typing of Acinetobacter strains and rapid identification of the strains most likely to be associated with hospital infection (3, 4, 15). The identity of Tn5613 as a transposon associated primarily with Acinetobacter and not with Pseudomonas is affirmed by the G+C content of 39% for the transposon as a whole and 36% for the internal open reading frame.
The 100 kb of spontaneously deleted DNA and by implication the van region are distant from the 20-kb pca-qui-pob gene cluster in the Acinetobacter chromosome (27). This makes vanA and vanB the only genes associated with the metabolic activities summarized in Fig. 1 that are separate from the cluster. It is possible that the locations of vanA and vanB are related to their evident instability. Loss of the genes would cause ferulate to be converted to vanillate, while coumarate and caffeate, which are related to ferulate in both structure and nutritional source, would be metabolized completely through protocatechuate. Thus, the existence of spontaneous van-deficient strains in natural Acinetobacter populations might allow the production of vanillate from ferulate as a chemical signal between plants and bacteria.
Spontaneous mutations caused by Tn5613.
Insertions of IS1236 at different positions in Acinetobacter pobR cause most of the spontaneous mutations in this gene and are accompanied by a target site duplication of 3 bp (24). Insertion of IS1236 into pcaH is accompanied by less precise duplications, and such insertions appear to cause only about 10% of the spontaneous mutations in pcaH and pcaG (25). In this study, the newly discovered transposon Tn5613, which includes two copies of IS1236, was found to be a significant contributor to spontaneous mutation in vanA and vanB and to have generated a 3-bp duplication upon insertion in vanB647 (Fig. 3). The mutations vanA645 and vanB657 appear to be more complex but include at least one copy of IS1236 as well as a 3-bp target site duplication. Insertion of a different transposon in these two strains, composed of IS1236 elements including one from Tn5613, is consistent with the inability to PCR amplify across the mutation together with the inability to detect by PCR an intact Tn5613 element.
Contrasting arrangement in the Pseudomonas chromosome of genes flanking vanAB.
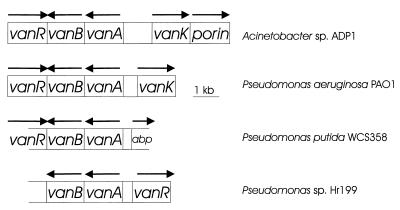
The overlap of the 3′ ends of vanB and the convergently transcribed putative regulatory gene vanR, an arrangement not found previously in ADP1 for genes involved in aromatic catabolism, prompted an analysis of DNA including vanA and vanB in other organisms. DNA for a VanR homolog, a regulatory protein in the GntR family (33), was identified in two pseudomonads, but, unexpectedly, in P. putida WCS358 (62) it was convergently transcribed from vanB, as in ADP1, whereas in Pseudomonas sp. strain HR199 (57) it was divergently transcribed from vanA (Fig. 5). Due either to mutation or sequence data ambiguity, a frameshift is required in both Pseudomonas sequences to maximize the amino acid alignment with their putative ADP1 homolog. Furthermore, the P. putida WCS358 locus corresponding to vanK in ADP1 appears to be occupied by a gene for the periplasmic ATP-binding protein component of an ABC-type transport system (Fig. 5). The VanK amino acid sequence identifies it as a member of the major facilitator superfamily of transport proteins, structurally distinct from ATP-binding cassette transporters (54); therefore, significantly different classes of protein appear to have been called upon during evolution of the transport capacity associated with the vanAB region in different soil bacteria. Based on preliminary sequence information from the Pseudomonas Genome Project, P. aeruginosa PAO1 has a vanAB region similar in organization to that of Acinetobacter sp. strain ADP1 (Fig. 5). The presence of this unusual gene organization in the two genera cannot be attributed to recent horizontal transfer between Pseudomonas and Acinetobacter, because the respective genes each possess the distinctive G+C contents characteristic of their host chromosome.
FIG. 5.
Comparative organization of the vanAB chromosomal region. From top to bottom in the figure, the percent amino acid identities to the corresponding proteins in Acinetobacter sp. strain ADP1, followed in parentheses by the numbers of aligned residues, are 69% (340), 69% (346), and 72% (340) for VanA; 46% (314), 47% (316), and 48% (316) for VanB; 61% (163), 57% (26), and 43% (166) for the regulatory protein in the GntR family (VanR); and 50% (415) for VanK. The region labelled abp could encode a protein with up to 33% identity over 148 aligned residues with a protein in various bacteria thought to be the periplasmic ATP-binding component of an ATP-binding cassette-type transport system (although this region includes a stop codon the potential open reading frame). Similarly, the size of the regulatory gene shown above assumes a frameshift in all three Pseudomonas sequences, due to mutation or sequencing error (and not included in the calculation of amino acid identity for the protein in the bottom two Pseudomonas sequences). The P. aeruginosa genes are present on one contig from the 15 September 1998 release of data from the Pseudomonas Genome Project. Arrows indicate the directions of transcription.
Chromosomal linkage of van genes to a putative iron-scavenging receptor gene.
During mapping of the Acinetobacter chromosome, strain ADP1 lost the ability to grow on vanillate, apparently due to the spontaneous deletion of 100 kb of chromosomal DNA including the van region. The detection of several copies of IS1236 in the chromosomal fragment encompassing the deletion in the parental strain (27) and the spontaneous van mutants with insertions of Tn5613, including vanB675 with a flanking deletion, is consistent with the spontaneous deletion being mediated in ADP1, directly or indirectly, by transposable elements. Given the linkage of vanA and vanB to a putative siderophore receptor gene (Fig. 2), the malleability of this region may be analogous to that described for DNA containing genes for iron metabolism in other bacteria (19). In the plague bacterium Yersinia pestis, for instance, genes for iron acquisition are located within a pathogenicity island in a 102-kb unstable region of the chromosome that is prone to spontaneous deletion apparently by recombination between flanking IS elements (6, 19).
Having siderophore genes flanked by repetitive DNA sequences can confer several advantages to a bacterium. On the one hand, this arrangement may facilitate tandem amplification of the genetic region (59), with the increased gene dosage leading to increased siderophore generation. During pathogenic interactions, this would be to the detriment of the host, but the siderophores of the fluorescent pseudomonads are associated with improved plant growth (51). On the other hand, spontaneous loss of genes can be advantageous to bacteria in certain niches (44): deletion of genes for ferrisiderophore receptors, for instance, would remove an outer membrane protein commonly used as an entry point for phage or the antibiotics of competing microorganisms (51). In Acinetobacter, a frequent additional consequence of deletion of the genetic region containing a putative iron uptake gene would be the loss of vanA and vanB. Such a loss would modify the bacteria into bioreactors capable of converting ferulate to vanillate.
ACKNOWLEDGMENTS
This research was supported by grants from the Army Research Office and the National Science Foundation. A.S. was supported by a postdoctoral fellowship from the Spanish Ministerio de Educacion y Ciencia.
Footnotes
Publication 19 from the Biological Transformation Center in the Yale Biospherics Institute.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averhoff B, Gregg-Jolly L, Elsemore D, Ornston L N. Genetic analysis of supraoperonic clustering by use of natural transformation in Acinetobacter calcoaceticus. J Bacteriol. 1992;174:200–204. doi: 10.1128/jb.174.1.200-204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet P J, Jeanjean S. Differentiation of Acinetobacter calcoaceticus sensu stricto from related Acinetobacter species by electrophoretic polymorphism of malate dehydrogenase, glutamate dehydrogenase and catalase. Res Microbiol. 1995;146:773–785. doi: 10.1016/0923-2508(96)81073-4. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet P J, Jeanjean S, Vieu J F, Dijkshoorn L. Species, biotype, and bacteriophage type determinations compared with cell envelope protein profiles for typing Acinetobacter strains. J Clin Microbiol. 1990;28:170–176. doi: 10.1128/jcm.28.2.170-176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunel F, Davison J. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol. 1988;170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 9.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delneri D, Degrassi G, Rizzo R, Bruschi C V. Degradation of trans-ferulic and p-coumaric acid by Acinetobacter calcoaceticus DSM 586. Biochim Biophys Acta. 1995;1244:363–367. doi: 10.1016/0304-4165(95)00021-3. [DOI] [PubMed] [Google Scholar]
- 11.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiMarco A A, Averhoff B A, Kim E E, Ornston L N. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene. 1993;125:25–33. doi: 10.1016/0378-1119(93)90741-k. [DOI] [PubMed] [Google Scholar]
- 13.DiMarco A A, Ornston L N. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J Bacteriol. 1994;176:4277–4284. doi: 10.1128/jb.176.14.4277-4284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echenique J R, Arienti H, Tolmasky M E, Read R R, Staneloni R J, Crosa J H, Actis L A. Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J Bacteriol. 1992;174:7670–7679. doi: 10.1128/jb.174.23.7670-7679.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrenstein B, Bernards A T, Dijkshoorn L, Gerner-Smidt P, Towner K J, Bouvet P J, Daschner F D, Grundmann H. Acinetobacter species identification by using tRNA spacer fingerprinting. J Clin Microbiol. 1996;34:2414–2420. doi: 10.1128/jcm.34.10.2414-2420.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsemore D A, Ornston L N. The pca-pob supraoperonic cluster of Acinetobacter calcoaceticus contains quiA, the structural gene for quinate-shikimate dehydrogenase. J Bacteriol. 1994;176:7659–7666. doi: 10.1128/jb.176.24.7659-7666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsemore D A, Ornston L N. Unusual ancestry of dehydratases associated with quinate catabolism in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:5971–5978. doi: 10.1128/jb.177.20.5971-5978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulberg D, Lakner S, Golovleva L A, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J Bacteriol. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 20.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522–2530. [DOI] [PMC free article] [PubMed]
- 21.Fuqua W C. An improved chloramphenicol resistance gene cassette for site-directed marker replacement mutagenesis. BioTechniques. 1992;12:223–225. [PubMed] [Google Scholar]
- 22.Gassner G T, Ludwig M L, Gatti D L, Correll C C, Ballou D P. Structure and mechanism of the iron-sulfur flavoprotein phthalate dioxygenase reductase. FASEB J. 1995;9:1411–1418. doi: 10.1096/fasebj.9.14.7589982. [DOI] [PubMed] [Google Scholar]
- 23.Gasson M J, Kitamura Y, McLauchlan W R, Narbad A, Parr A J, Parsons E L H, Payne J, Rhodes M J C, Walton N J. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J Biol Chem. 1998;273:4163–4170. doi: 10.1074/jbc.273.7.4163. [DOI] [PubMed] [Google Scholar]
- 24.Gerischer U, D’Argenio D A, Ornston L N. IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticus chromosome. Microbiology. 1996;142:1825–1831. doi: 10.1099/13500872-142-7-1825. [DOI] [PubMed] [Google Scholar]
- 25.Gerischer U, Ornston L N. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:1336–1347. doi: 10.1128/jb.177.5.1336-1347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerischer U, Segura A, Ornston L N. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 28.Gregg-Jolly L A, Ornston L N. Recovery of DNA from the Acinetobacter calcoaceticus chromosome by gap repair. J Bacteriol. 1990;172:6169–6172. doi: 10.1128/jb.172.10.6169-6172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen C K. Fibronectin type III-like sequences and a new domain type in prokaryotic depolymerases with insoluble substrates. FEBS Lett. 1992;305:91–96. doi: 10.1016/0014-5793(92)80871-d. [DOI] [PubMed] [Google Scholar]
- 30.Hartnett G B, Averhoff B, Ornston L N. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J Bacteriol. 1990;172:6160–6161. doi: 10.1128/jb.172.10.6160-6161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harwood C S, Nichols N N, Kim M K, Ditty J L, Parales R E. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol. 1994;176:6479–6488. doi: 10.1128/jb.176.21.6479-6488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 33.Haydon D J, Guest J R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 34.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwabuchi T, Harayama S. Biochemical and genetic characterization of 2-carboxybenzaldehyde dehydrogenase, an enzyme involved in phenanthrene degradation by Nocardioides sp. strain KP7. J Bacteriol. 1997;179:6488–6494. doi: 10.1128/jb.179.20.6488-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juni E. Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J Bacteriol. 1972;112:917–931. doi: 10.1128/jb.112.2.917-931.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junker F, Kiewitz R, Cook A M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J Bacteriol. 1997;179:919–927. doi: 10.1128/jb.179.3.919-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 40.Kleckner N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979;16:711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- 41.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 42.Kowalchuk G A, Hartnett G B, Benson A, Houghton J E, Ngai K L, Ornston L N. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene. 1994;146:23–30. doi: 10.1016/0378-1119(94)90829-x. [DOI] [PubMed] [Google Scholar]
- 43.Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurelli A T, Fernandez R E, Bloch C A, Rode C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinke A, Gilkes N R, Kilburn D G, Miller R C, Jr, Warren R A. Multiple domains in endoglucanase B (CenB) from Cellulomonas fimi: functions and relatedness to domains in other polypeptides. J Bacteriol. 1991;173:7126–7135. doi: 10.1128/jb.173.22.7126-7135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meinke A, Gilkes N R, Kwan E, Kilburn D G, Warren R A, Miller R C., Jr Cellobiohydrolase A (CbhA) from the cellulolytic bacterium Cellulomonas fimi is a β-1,4-exocellobiohydrolase analogous to Trichoderma reesei CBH II. Mol Microbiol. 1994;12:413–422. doi: 10.1111/j.1365-2958.1994.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 47.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 48.Narbad A, Gasson M J. Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology. 1998;144:1397–1405. doi: 10.1099/00221287-144-5-1397. [DOI] [PubMed] [Google Scholar]
- 49.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neidle E L, Shapiro M K, Ornston L N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J Bacteriol. 1987;169:5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neilands J B. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 52.Nichols N N, Harwood C S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivera E R, Minambres B, Garcia B, Muniz C, Moreno M A, Ferrandez A, Diaz E, Garcia J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parke D. Application of p-toluidine in chromogenic detection of catechol-protocatechuate diphenolic intermediates in catabolism of aromatic compounds. Appl Env Microbiol. 1992;58:2694–2697. doi: 10.1128/aem.58.8.2694-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Priefert H, Rabenhorst J, Steinbuchel A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J Bacteriol. 1997;179:2595–2607. doi: 10.1128/jb.179.8.2595-2607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross D G, Swan J, Kleckner N. Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. Cell. 1979;16:733–738. doi: 10.1016/0092-8674(79)90089-8. [DOI] [PubMed] [Google Scholar]
- 59.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhart J L I F C, Low K B, Magasanik B, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 2256–2276. [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 61.Smith M A, Huang G, Young D M, Ornston L N. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. The Acinetobacter pca-qui-pob supraoperonic cluster extends to a ligase required for catabolism of coumarate, caffeate and ferulate, abstr. K-148; p. 350. [Google Scholar]
- 62.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 63.Williams P A, Shaw L E. mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J Bacteriol. 1997;179:5935–5942. doi: 10.1128/jb.179.18.5935-5942.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto S, Okujo N, Sakakibara Y. Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch Microbiol. 1994;162:249–254. doi: 10.1007/BF00301846. [DOI] [PubMed] [Google Scholar]
- 65.Yamano Y, Nishikawa T, Komatsu Y. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1. Mol Microbiol. 1993;8:993–1004. doi: 10.1111/j.1365-2958.1993.tb01643.x. [DOI] [PubMed] [Google Scholar]
- 66.Yanisch-Perron G, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 67.Yoneyama H, Yoshihara E, Nakae T. Nucleotide sequence of the protein D2 gene of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1791–1793. doi: 10.1128/aac.36.8.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.You I S, Ghosal D, Gunsalus I C. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3′-flanking region. Biochemistry. 1991;30:1635–1641. doi: 10.1021/bi00220a028. [DOI] [PubMed] [Google Scholar]