Abstract
Bifidobacteria are among the first colonizers of the infant gut, and human milk oligosaccharides (HMOs) in breastmilk are instrumental for the formation of a bifidobacteria-rich microbiota. However, little is known about the assembly of bifidobacterial communities. Here, by applying assembly theory to a community of four representative infant-gut associated Bifidobacterium species that employ varied strategies for HMO consumption, we show that arrival order and sugar consumption phenotypes significantly affected community formation. Bifidobacterium bifidum and Bifidobacterium longum subsp. infantis, two avid HMO consumers, dominate through inhibitory priority effects. On the other hand, Bifidobacterium breve, a species with limited HMO-utilization ability, can benefit from facilitative priority effects and dominates by utilizing fucose, an HMO degradant not utilized by the other bifidobacterial species. Analysis of publicly available breastfed infant faecal metagenome data showed that the observed trends for B. breve were consistent with our in vitro data, suggesting that priority effects may have contributed to its dominance. Our study highlights the importance and history dependency of initial community assembly and its implications for the maturation trajectory of the infant gut microbiota.
Subject terms: Community ecology, Microbial ecology, Microbiome
Introduction
Aberrant development of the human gut microbiota during infancy is linked to various health conditions that can manifest themselves throughout adulthood [1]. However, little is known about how the microbial community, following its introduction to the gut, becomes established and structured. Past studies have shown that the postpartum microbial colonization period is critical for gut microbiota development, being influenced by various factors, such as delivery mode (vaginal vs caesarean section) and feeding regime (formula vs breastmilk) [2, 3]. The role played by deterministic factors such as host genetics, which may select for specific taxa through niche-based mechanisms, has previously been examined [4]. However, the impact of certain processes such as assembly history and priority effects, which are stochastic in terms of species colonization order and identity [5], on the structuring of host-associated gut microbial communities has not been fully addressed. Priority effects, i.e., the effect of arrival order of a strain/species in a particular environmental niche on species interactions and community composition [6], present an important mechanism that can lead to the formation of multiple community states, including ecologically important transient stable states [7]. Although the role of species colonization history in structuring the human gut microbiota is increasingly appreciated [8, 9], we know little about whether and how priority effects contribute to the assembly of bifidobacterial communities in the infant gut.
Bifidobacteria are among the first colonizers of the infant gut and commonly represent over 70% of the total gut microbial community during breastfeeding [10–12]. The early colonization by bifidobacteria is at least in part facilitated by their ability to metabolize human milk oligosaccharides (HMOs). HMOs are a group of complex unconjugated glycans that, despite being the third most abundant solid component in breastmilk, provide no (direct) nutritional value to the host as they are resistant to digestion by pancreatic enzymes [13]. Instead, HMOs act as highly selective agents that promote the growth of certain bifidobacteria [14–17], though HMO-utilization strategies differ significantly by species and strain. Furthermore, while breastfeeding has been associated with an overall higher prevalence and abundance of bifidobacteria [18, 19], they are reported to be absent in some breastfed infants [20]. In such infants, bifidobacteria may have been absent during a critical colonization window [21], e.g. establishment of bifidobacteria is reported to be delayed or absent in infants born via caesarean section [10, 22]. Thus, bifidobacteria are used as probiotics to encourage the development of a healthy gut microbiota [23–25]. However, the efficacy of exogenously administered bifidobacteria depends on its relationship with the resident gut microbiota [26], and persistent colonization remains an issue [27, 28]. Therefore, elucidating mechanisms influencing the formation of bifidobacterial communities, in particular the role of assembly history and priority effects, may guide future strategies for microbiota-based therapeutic interventions, as well as explain differential establishment of bifidobacterial communities within the infant gut.
Here, we investigated the influence of assembly history on the compositional structure of infant gut-associated bifidobacterial communities. The four bifidobacterial species used in this study, Bifidobacterium bifidum JCM 1254, Bifidobacterium breve UCC2003, Bifidobacterium longum subspecies longum MCC10007 (B. longum), and Bifidobacterium longum subspecies infantis ATCC 15697 T (B. infantis), employ species-specific HMO assimilation strategies, which were predicted to dictate competitive outcomes. In vitro cultivation experiments in HMO-containing medium showed that assembly history significantly influenced community structure. Our results revealed that despite its limited HMO-assimilation ability, B. breve dominated through priority effects, which were partly underpinned by its ability to utilize fucose, an HMO degradant produced by other species. We also analysed publicly available in vivo faecal metagenome data from a cohort of infants and observed trends consistent with our in vitro findings, particularly for B. breve. Together, our study demonstrates the importance of priority effects on the establishment of bifidobacterial communities.
Results
Genome-based predictions and monoculture characterization of infant gut-associated bifidobacterial strains
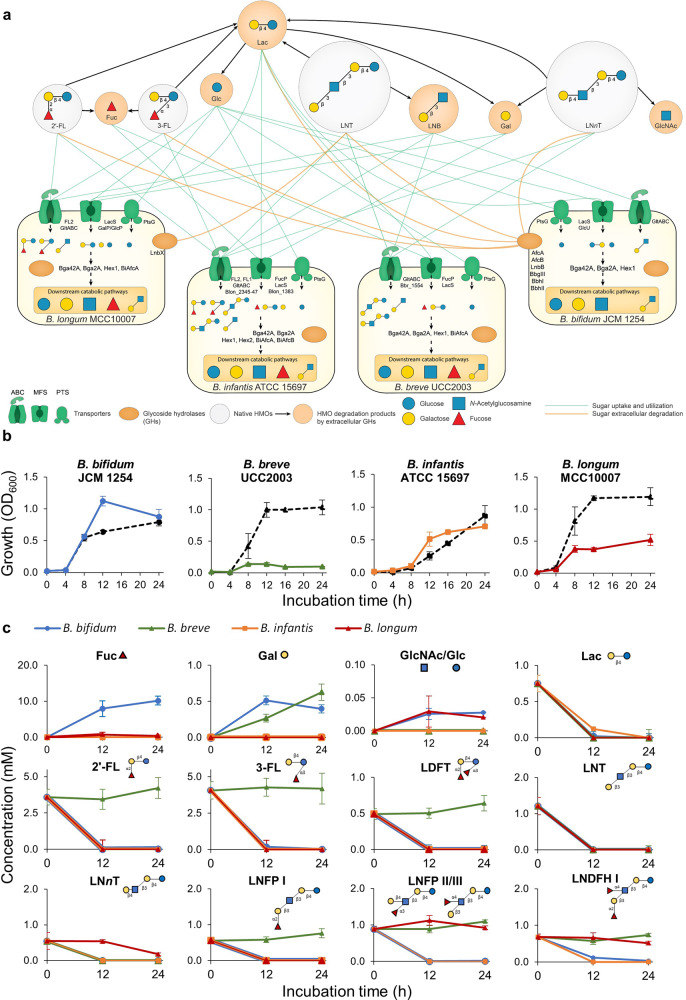
Four infant gut-associated Bifidobacterium strains were used in this study: B. bifidum JCM 1254, B. breve UCC2003, B. infantis ATCC 15697 T, and B. longum MCC10007. The genome of B. bifidum JCM 1254 was de novo sequenced in this study. We combined genomics-based (in silico) metabolic reconstruction with in vitro culturing to assess the HMO utilization capabilities of these strains. Our HMO mixture, purified from pooled breastmilk, included the following oligosaccharides: 2′-fucosyllactose (2′-FL), 3-fucosyllactose (3-FL), lactodifucotetraose (LDFT), lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), lacto-N-fucopentaose (LNFP) I, LNFP II/III, and lacto-N-difucohexaose (LNDFH) I (Supplementary Table 1). Using a subsystem-based approach implemented in the SEED genomic platform [29] we first established the presence/absence of genes encoding transporters and catabolic pathways for HMO constituents/degradation products (oligosaccharides: lacto-N-triose II (LNTri II), lacto-N-biose (LNB), and lactose (Lac); and their monosaccharide components: glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc)) (see Supplementary Table 2). This analysis included repertoires of corresponding glycoside hydrolases (GHs) and transporters involved in HMO utilization in each of the four Bifidobacterium strains (Fig. 1a).
Fig. 1. Genomic and monoculture characterization of infant gut-associated bifidobacteria used in this study.
a Schematic summary of predicted HMO-utilization phenotypes of each bifidobacterial strain. The HMO-utilization phenotypes of each bifidobacterial strain (B. bifidum JCM 1254, B. breve UCC2003, B. infantis ATCC 15697 T, and B. longum MCC10007) were characterized based on genomic predictions. Each strain is shown with its respective transporters and GHs (see Supplementary Table 2). The utilization pathways for four representative HMO molecules (2′-FL, 3-FL, LNT, and LNnT) and their mono- and di-saccharide degradants (Lac, LNB, Fuc, Glc, Gal, GlcNAc) are shown. The green lines indicate sugar uptake and utilization by each strain. The orange lines connect extracellular bacterial enzymes with the substrates. The black arrows connect HMO molecules with their degradants after extracellular degradation. b Monoculture growth curves of each strain grown in HMO-supplemented YCFA medium. Growth of each strain was monitored by measuring OD600 at each time point. All strains were also grown in Lac as a positive control, shown as black dotted lines. Data showing the total OD600 represent averages and error bars represent ± standard deviation of biological quadruplicates. c HMO-utilization profiles of each strain. Culture supernatant was collected at each time point. The remaining sugars in the medium were labeled with 2-AA and analyzed by HPLC (as described in the Materials and Methods section). The consumption of each sugar by each strain is indicated by different colors (blue: B. bifidum, green: B. breve, orange: B. infantis, red: B. longum). Data represent averages and error bars represent ± standard deviation of biological quadruplicates. It should be noted that LNB was not detected at the indicated time points.
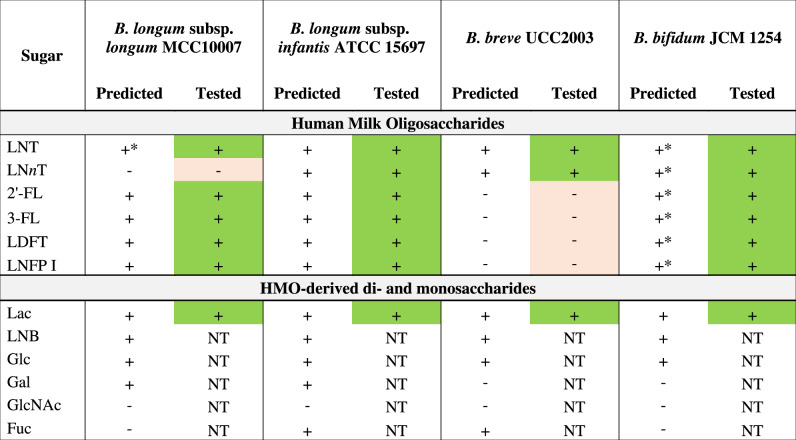
Genomic analysis revealed that the strains used in this study encode varying sets of GHs and transporters for HMO utilization, reflecting different strategies implemented by these Bifidobacterium strains. Overall, B. infantis ATCC 15697 T had the most expansive set of HMO transporters and corresponding intracellular GHs, whereas B. bifidum JCM 1254 had the highest number of extracellular GHs for HMO degradation [30]. While most of the purified HMOs used in our study are fucosylated, there was an apparent absence of the Fuc catabolic pathway in B. bifidum JCM 1254, which is a common trait of this species. B. longum MCC10007, B. infantis ATCC 15697 T, and B. breve UCC2003 possess cytoplasmic Fuc catabolic enzymes, but only B. infantis ATCC 15697 T and B. breve UCC2003 encode the Fuc transporter FucP [31, 32] (Supplementary Table 2). All four strains were predicted to utilize lactose based on the presence of Lac permease LacS [33] and β-1,4-galactosidase Bga2A (GH2) [34] orthologs in all four genomes (Table 1, Supplementary Table 2).
Table 1.
Predicted HMO/HMO degradation product utilization phenotypes of strains in the study.
+ = predicted utilizer (transporter, GH/catabolic enzyme genes are present) +* = predicted degrader (extracellular GH genes are present) - = predicted non-utilizer.
Colours: green = consumes a sugar; light red = does not consume a sugar; NT = not tested.
LNT Lacto-N-tetraose, LNnT Lacto-N-neotetraose, 2′-FL 2′-Fucosyllactose, 3-FL 3-Fucosyllactose, LDFT Lactodifucotetraose, LNFP Lacto-N-fucopentaose, Lac Lactose, LNB Lacto-N-biose I, Glc Glucose, Gal Galactose, GlcNAc N-Acetylglucosamine, Fuc Fucose.
The HMO-consumption behaviour of these strains was evaluated in vitro through monoculture studies in a medium containing 1% (w/v) of either Lac (as the positive control) or a mixture of HMOs purified from pooled breastmilk as the sole carbon source (Fig. 1b), and sugar consumption was monitored using HPLC (Fig. 1c). Consistent with expectations, all strains grew well on Lac. With HMOs, strains predicted to encode the most expansive set of HMO-active GHs and associated HMO transporters, i.e., B. bifidum and B. infantis, showed substantial growth (final OD600 > 0.7). B. longum showed moderate growth (final OD600 > 0.5), while B. breve showed limited growth (final OD600 < 0.3) (Fig. 1b). Overall, carbohydrate consumption profiles were consistent with the phenotypes predicted from genomic data (Table 1): B. bifidum and B. infantis consumed all HMOs, B. longum consumed LNT and fucosylated HMOs (2′-FL, 3-FL, LDFT, and LNFP I), the latter of which is enabled by the presence of the fucosyllactose (FL) transporter (found in approximately 3 % of B. longum strains [16, 26]), and B. breve only utilized LNT and LNnT (Fig. 1c).
Pairwise cultivation of bifidobacterial strains in HMO-supplemented medium
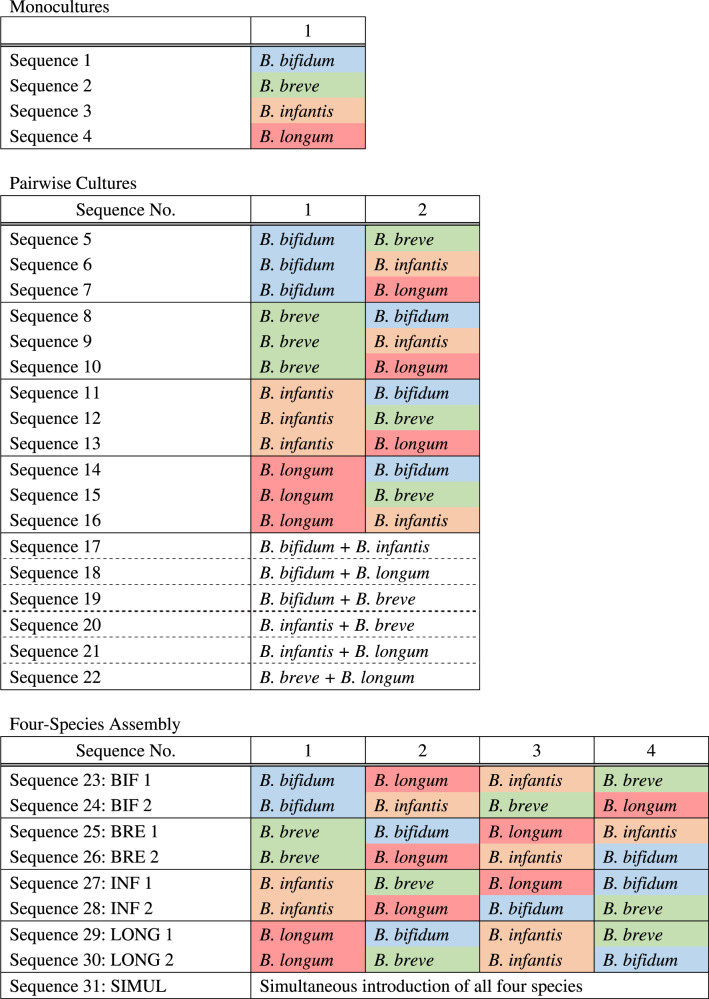
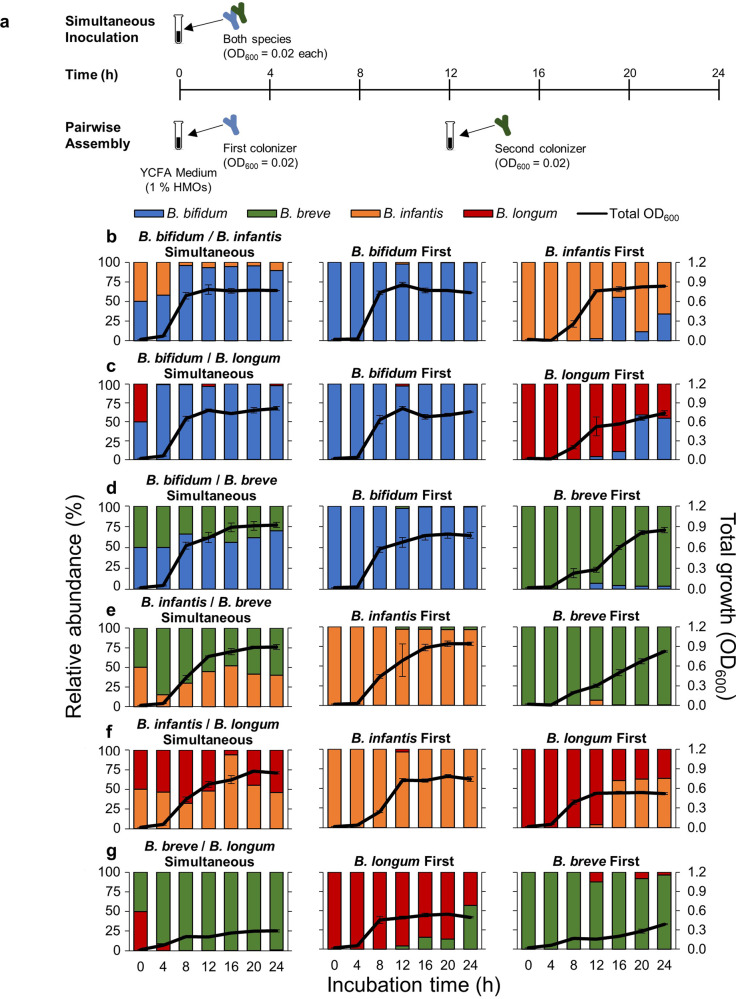
To determine if priority effects influence the outcome of competition among bifidobacterial species, we performed all possible pairwise cultivations (Table 2). Here, we consider any significant effect of arrival order on the absolute and relative abundances of each species as priority effects. Priority effects can be either facilitative, in which the early arriving species positively affects the later arriving species (e.g., changing the types of resources available and enabling later arriving species to colonize), or inhibitory, in which the early arriving species negatively affects the later arriving species (e.g., using up resources and making them unavailable for later arriving species) [6]. For each pair, one species was inoculated into the culture medium first, and the second species was inoculated 12 h later. As a control, both species were also inoculated into the medium simultaneously (Fig. 2a). Experimental duration was 24 h so that all species had at least 12 h in the medium after inoculation (at which growth plateaus during monoculture (Fig. 1b)), and sugars in the medium were depleted in most assembly sequences (Extended Data Figs. 1–4). Pairwise growth curves are summarized in Fig. 2b–g, and a species was considered dominant if its relative abundance was >50 % of the community. Furthermore, we quantified the strength of priority effects using the equation proposed by Vannette and Fukami [35], by comparing a given species’ absolute abundance when it was the first colonizer with its absolute abundance when it was the second, invading species (Supplementary Table 3).
Table 2.
Community assembly sequences used in this experiment.
Fig. 2. Pairwise cultivation of infant gut-associated bifidobacteria.
a Experimental design. Pairwise culturing experiments were performed for all possible combinations. In simultaneous cultures, both species were introduced together into medium supplemented with HMOs at the beginning of the experiment (at 0 h). To test for the effect of assembly history, the first species was inoculated at the beginning of the experiment, and the second species was inoculated 12 h later. b–g Culturing results for each pair. Total OD600 indicated by the black line, was measured at each time point, and the relative abundance of each species was quantified using qPCR. The relative abundance of each strain is indicated by different colors (blue: B. bifidum, green: B. breve, orange: B. infantis, red: B. longum). Relative abundances at 0 h are calculated theoretical values. Data showing the total OD600 represent averages and error bars represent ±standard deviation of biological quadruplicates. The bar graphs showing the relative abundances of each species show averages of biological quadruplicates. b B. bifidum and B. infantis pair. c B. bifidum and B. longum pair. d B. bifidum and B. breve pair. e B. infantis and B. breve pair. f B. infantis and B. longum pair. g B. breve and B. longum pair.
When B. bifidum JCM 1254 was cultivated together with B. infantis ATCC 15697 T, the early arriving species dominated through inhibitory priority effects (Fig. 2b). As B. bifidum and B. infantis are both capable of utilizing all types of HMOs, they both depleted most of the available resources within 12 h when inoculated first (Extended Data Figs. 1, 2). Priority effects were stronger for B. bifidum (Supplementary Table 3), and it was a stronger competitor overall as it dominated in simultaneous culture and also maintained a stable population even when B. infantis was first (Fig. 2b). For the B. bifidum and B. longum pair, the final relative abundance for B. bifidum was greater regardless of arrival order (Fig. 2c). However, priority effects and significant growth were observed for B. longum when it was the first colonizer, possibly because it was able to utilize the HMO-degradants produced by B. bifidum (Fig. 2c, Supplementary Table 3).
For the B. breve and B. bifidum pair, B. bifidum dominated in simultaneous culture and when it was inoculated first (Fig. 2d). When B. breve was first, priority effects and substantial growth were observed, as it dominated and drove B. bifidum to near extinction (Fig. 2d, Supplementary Table 3) despite its limited HMO-consumption ability and low growth in HMO-supplemented medium during monoculture (Fig. 1b). However, B. breve growth remained low until B. bifidum inoculation, upon which B. breve showed biphasic growth (Fig. 2d). A similar pattern was observed for the B. breve and B. infantis pair, in which priority effects were observed for B. breve (Supplementary Table 3) and it outcompeted B. infantis if inoculated first. However, B. breve growth remained low until B. infantis arrival (Fig. 2e), suggesting that the presence of B. bifidum and B. infantis had a facilitative effect on B. breve growth.
In the B. infantis and B. longum pair, B. infantis generally dominated regardless of arrival order (Fig. 2f). However, priority effects were observed for B. longum (Supplementary Table 3) and it was able to maintain a stable population even after B. infantis inoculation (Fig. 2f). As for the B. breve and B. longum pair, the average final total OD600 was low in all assembly sequences (Simultaneous: 0.30, B. longum first: 0.50, B. breve first: 0.37) and priority effects were observed for neither species (Supplementary Table 3), but B. breve dominated in all scenarios by 24 h (Fig. 2g). Furthermore, while HMOs were completely consumed in most pair combinations by the end of the cultivation period, oligosaccharides with longer chains (LNFP II/III and LNDFH I) remained in the spent medium of B. breve and B. longum cultures (Extended Data Figs. 1–4), which was consistent with their predicted HMO assimilation phenotypes.
Four-species assemblages in HMO-supplemented medium
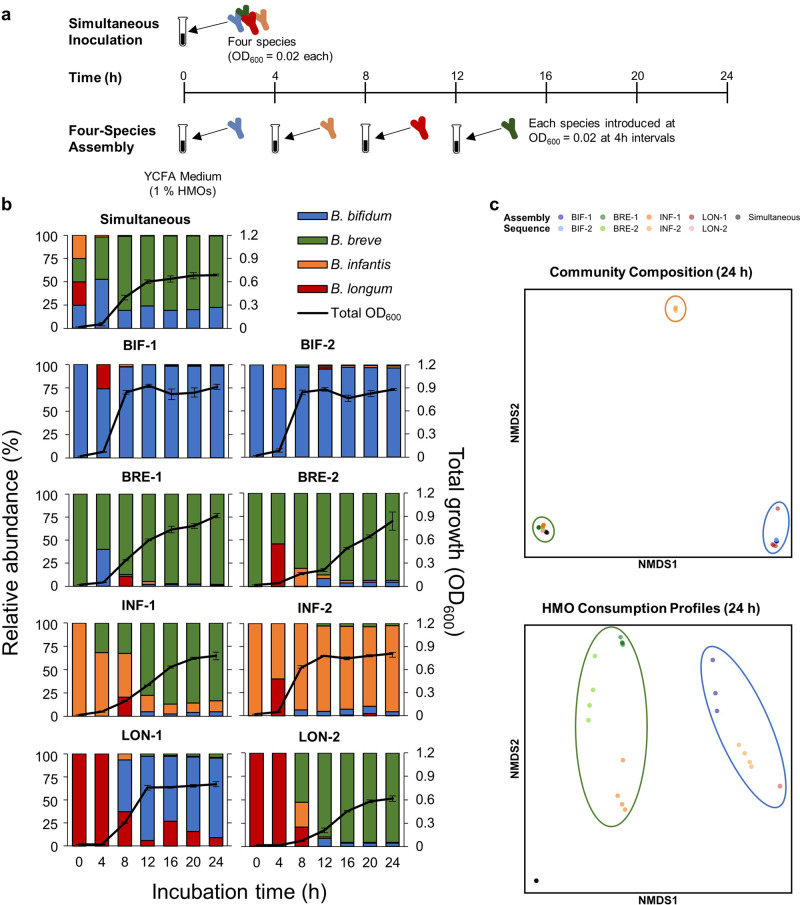
Assembly history was manipulated with all four bifidobacterial strains. As a control, all four species were inoculated simultaneously at the beginning of the experiment (Fig. 3a). Eight assembly sequences were selected so that each species was inoculated first, twice. The second, third, and fourth species were randomly selected (Table 2). Data were collected for 24 h so that all species were in the medium for at least 12 h after inoculation. At 24 h, most HMOs in the medium were depleted, except 3-FL and LNDFH I in the INF-1, BRE-1, and BRE-2 sequences (Extended Data Fig. 5).
Fig. 3. Results of four-species assemblages.
a Experimental design. Culturing experiments were performed with all four species, and assembly order was manipulated. Assembly history sequences are summarized in Table 2. As a control, all four species were introduced into medium supplemented with HMOs at the beginning of the experiment (Simultaneous). b Culturing results. Total OD600, indicated by the black line, was measured at each time point, and the relative abundance of each species was quantified using qPCR. Data represent averages of biological quadruplicates, and error bars indicate ± standard deviation. The relative abundance of each strain is indicated by different colors (blue: B. bifidum, green: B. breve, orange: B. infantis, red: B. longum). c NMDS plots based on the community composition and HMO consumption profiles at 24 h. Each color represents a different assembly sequence. In the top panel, three clusters form based on the final community composition: blue (B. bifidum dominant communities), green (B. breve dominant communities), and orange (B. infantis dominant communities). In the bottom panel, two clusters based on the HMO consumption profiles form: blue (communities with high amounts of fucose remaining) and green (communities with 3-FL and LNDFH I unconsumed) circles.
B. breve (average relative abundance: 77.5%) was the dominant species in simultaneous culture, followed by B. bifidum (average relative abundance: 21.9%) and B. infantis (average relative abundance: 0.6%) at 24 h, while B. longum was below the detection limit and presumed to be extinct within 4 h (Fig. 3b). For communities in which B. bifidum was introduced first (BIF-1 and BIF-2 sequences), B. bifidum was the dominant species with a final OD600 of 0.90 and 0.85, respectively. B. breve also dominated when it was inoculated first, and the community profile and final OD600 were comparable between the two sequences (BRE-1 and BRE-2). However, the growth curves of the BRE-1 and BRE-2 sequences (OD600) were dependent on the identity of the second species. While the average total OD600 reached 0.74 at 12 h when B. bifidum was second (BRE-1), it was 0.28 when B. longum was second (BRE-2). In assembly sequences in which B. infantis was first, B. infantis dominated when B. longum was second (INF-2). However, B. infantis was outcompeted by B. breve in the final community at 24 h when B. breve was second (INF-1). With regards to B. longum, not only was it unable to become the dominant competitor in all assembly sequences, but it was also undetectable by 24 h in all sequences except for LON-1, where B. longum was inoculated first and followed by B. bifidum.
Nonmetric multidimensional scaling (NMDS) based on the final community structure at 24 h revealed three different possible community outcomes: 1) B. breve dominant, 2) B. bifidum dominant, and 3) B. infantis dominant (Fig. 3c). Further analysis with permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis distances revealed that assembly history had a significant influence on final community structure (F8,27 = 64.28, p < 0.001), and the identity of the first species (Partial R2 = 0.91, p < 0.001) and the second species (Partial R2 = 0.04, p < 0.001) determined the final community outcome.
Regression analysis was performed to quantify the strength of priority effects. Calculations were performed to predict final population abundance of each species based on arrival order. A significant negative relationship was found for B. breve (F1,30 = 335.3, p < 0.001, β = −0.29, R2 = 0.92), B. bifidum (F1,30 = 30.94, p < 0.001, β = −0.24, R2 = 0.51), and B. infantis (F1,30 = 6.615, p = 0.015, β = −0.12, R2 = 0.18), indicating that the earlier these three species were introduced into the community, the higher its final abundance. However, no significant relationship was found for B. longum (F1,30 = 1.811, p = 0.177, β = −0.01, R2 = 0.03).
We also examined the HMO-consumption profiles of each assembly sequence (Extended Data Fig. 5). NMDS and PERMANOVA based on the final HMO-consumption profiles at 24 h showed that the amount of LNDFH I, Fuc, 3-FL, and LNFP II/III remaining in the medium significantly contributed to the variation among communities (Fig. 3c, Supplementary Table 4). Two main clusters formed, one of which contained communities dominated by B. bifidum with high amounts of fucose remaining in the spent medium, while the second contained communities dominated by B. breve, in which 3-FL and LNDFH I remained unconsumed (Extended Data Fig. 5).
Metagenomic data mining analysis
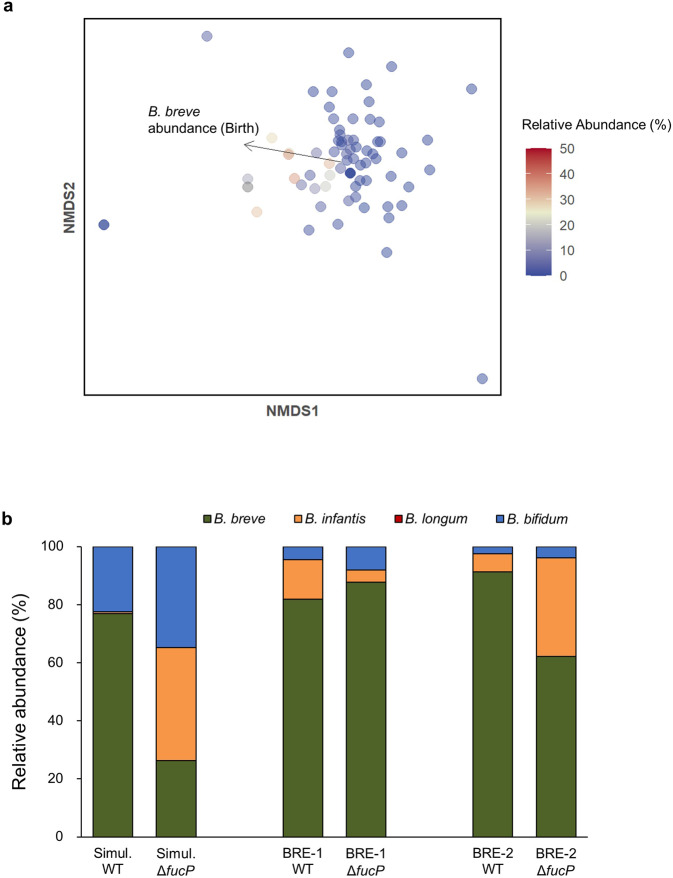
We further analysed bifidobacterial communities in a cohort of European infant-mother pairs from a study performed by Bäckhed et al. [36]. This metagenomic dataset followed the infant gut microbiota at 0, 4, and 12 months of age, as well as the mother’s gut microbiota at the time of delivery. The microbiotas of 73 infants who were at least partially breastfed (exclusively breastfed or mixed fed) at both 0 and 4 months of age were analysed. By 12 months of age, the majority of the infants had been weaned and the average relative abundance of bifidobacteria was 2.46% (Extended Data Fig. 6). PERMANOVA revealed that the relative abundance of B. breve in the infant gut microbiota at birth contributed to the bifidobacterial community structure at 4 months (Partial R2 = 0.09, p = 0.0499; Fig. 4a, Supplementary Table 5). However, such significance was not observed for B. bifidum, the B. longum group, and other species. We also assessed whether the presence of a species at birth is associated with its dominance (>50% of the bifidobacterial community) at 4 months using Fisher’s exact test. Based on this analysis, we found that the presence of B. breve at birth (40 out of 73 infants) was associated with its dominance at 4 months (p = 0.029), while such patterns were not observed for B. bifidum or the B. longum group. However, correlation analyses showed that at birth and 4 months, there was a significant correlation between the abundance of B. bifidum and the total abundance of Bifidobacterium species (Birth: Pearson’s r = 0.33, p = 0.004; 4 months: Pearson’s r = 0.43, p = 0.0002). Additionally, the presence of B. bifidum was positively correlated with B. longum (Pearson’s r = 0.27, p = 0.02) at birth. The stimulatory effect of B. bifidum on the growth of other bifidobacterial species was observed in our in vitro pairwise and four-species assembly cultivation experiments (Figs. 2b, 3c), which is consistent with previous work that reported that the presence of B. bifidum in breastfed infant guts is associated with a higher prevalence of bifidobacteria at genus level [10].
Fig. 4. In silico data mining and in vitro culturing analyses demonstrate that B. breve benefits from priority effects.
a Analysis of in vivo microbiome data. Taxonomic identification was performed using Kraken2 v2, and species abundances were estimated using Bracken v2.6.1. Nonmetric multidimensional scaling (NMDS) ordination plots of bifidobacterial community data from the guts of 4-month old infants who were at least partially breastfed (exclusively breastfed or mixed fed) at birth and at 4 months of age (n = 73) (see Extended Data Fig. 6 and Supplementary Table 5). Each point corresponds to one individual. The color gradient indicates the B. breve abundance within the total gut microbiota at 4 months of age. Statistically significant loadings are indicated as black arrows. b Culturing results using the ΔfucP mutant of B. breve UCC2003. Sequences of 4-species culturing in which B. breve dominated was used, and the experiment was repeated with the wild type (WT) and ΔfucP mutant of B. breve UCC2003. Relative abundance of each species was quantified using qPCR, and each strain is indicated by different colors (blue: B. bifidum, green: B. breve, orange: B. infantis, red: B. longum). Data represent averages of biological triplicates.
Fucose utilization ability of B. breve contributes to its dominance in vitro
B. breve dominated if present in the community early (Figs. 2d, e, g, 3b), despite its limited growth in HMO medium during monoculture (Fig. 1b, c) and the prevalence of genes for utilizing fucosylated HMOs is low (<13 %) among sequenced B. breve strains [26] (Fig. 1b, c). In our in vitro system, we found that Fuc was consumed in B. breve dominant communities, while left unconsumed when other species dominated (Fig. 3c, Extended Data Fig. 5). Furthermore, B. breve UCC2003 and all B. breve strains (46 out of 46 strains whose complete genomes are reported in the NCBI database as of October 27, 2021) possess homologs of the Fuc transporter FucP (Bbr_1742) [31] (Supplementary Table 2). Therefore, we compared wild-type (WT) and ΔfucP strains of B. breve UCC2003 to examine whether its ability to import fucose contributed to its persistence and/or dominance. We selected sequences in which B. breve dominated (Simultaneous, BRE-1, BRE-2; Table 2) and repeated the culturing experiments. Compared to when the WT strain was used, the average relative abundance of B. breve decreased from 76.9% to 26.3% (Δ50.6 %; Student’s two-tailed t-test, p = 0.024) in the simultaneous sequence and from 91.3% to 62.2% (Δ29.1 %; Student’s two-tailed t-test, p = 0.027) in the BRE-2 sequence (Fig. 4b). No significant change was observed in the BRE-1 sequence (Student’s two-tailed t-test, p = 0.08), in which B. bifidum is inoculated after B. breve. Furthermore, analysis of culture supernatant showed that while fucose accumulated in sequences using B. breve ΔfucP, it was depleted in sequences with WT B. breve (Extended Data Fig. 7). These results strongly suggest that the ability to utilize fucose contributed to the competitive ability of B. breve within bifidobacterial communities. This contrasts with B. infantis, which leaves Fuc unconsumed when co-cultured with other species, even though it also possesses FucP (Extended Data Fig. 5b, INF-2).
Discussion
Using infant gut-associated bifidobacterial communities in HMO-supplemented environments, we experimentally controlled the order of species arrival to evaluate the effect of assembly history and priority effects. Four strains that display a variety of species-specific mechanisms for HMO assimilation were used. Based on the predicted phenotypes and monoculture data, we hypothesized that in HMO environments, (1) B. bifidum JCM 1254 and B. infantis ATCC 15697 T would be strong competitors due to their ability to assimilate a variety of HMOs through extracellular and intracellular GHs, respectively, (2) B. longum MCC10007 would be a moderate competitor, as it cannot consume LNnT, but can consume LNT and specific fucosylated sugars such as 2′-FL, 3-FL, LDFT, and LNFP I, and (3) B. breve UCC2003 would be a weak competitor as its HMO-consumption ability is limited to LNT and LNnT (Fig. 1c; Supplementary Table 2). Here, we found that strong competitors like B. bifidum and B. infantis can dominate through inhibitory priority effects in vitro, in which the early arriving species apparently depletes resources for later arriving species. On the other hand, B. breve, despite its limited HMO-utilization ability, benefitted from facilitative effects by utilizing fucose and other degradants provided by competitors like B. bifidum and B infantis. Our results highlight the importance of early arriving species during community assembly, whose mechanisms are influenced by species-specific competitive strategies and sugar consumption phenotypes.
Regarding B. bifidum and B. infantis, inhibitory priority effects were observed in both pairwise and four-species assemblages (except the INF-1 sequence), and communities behaved as would be predicted from genomic data and assembly order. For example, in the B. bifidum and B. infantis pairwise culture, the early-arriving species became dominant (Fig. 2b), and the outcome of competition was dependent on which species arrived and acquired the resources first [6]. The results of the four-species assemblages also showed that assembly history had a significant effect on the final community structure. A negative relationship between arrival order and final abundance is indicative of inhibitory priority effects [37] and regression analyses of 4-species assemblages showed that such negative relationships were observed for B. bifidum and B. infantis. In B. bifidum- and B. infantis-dominant communities, most of the carbohydrates in the medium were depleted soon after inoculation (Extended Data Figs. 1, 2, 5), suggesting that B. bifidum and B. infantis dominated through niche pre-emption. Furthermore, different assembly sequences gave rise to three different community states (Fig. 3c), a predicted consequence of priority effects [6, 38, 39]. PERMANOVA revealed that the identity of the first and second colonizers primarily determined the community outcome, highlighting the significant effect early arriving species have on the final community. For instance, in communities in which B. infantis was inoculated first (INF-1, INF-2), B. infantis dominated when followed by B. longum (INF-2; Fig. 3b). However, it was unexpectedly outcompeted when B. breve was inoculated second (INF-1; Fig. 3b).
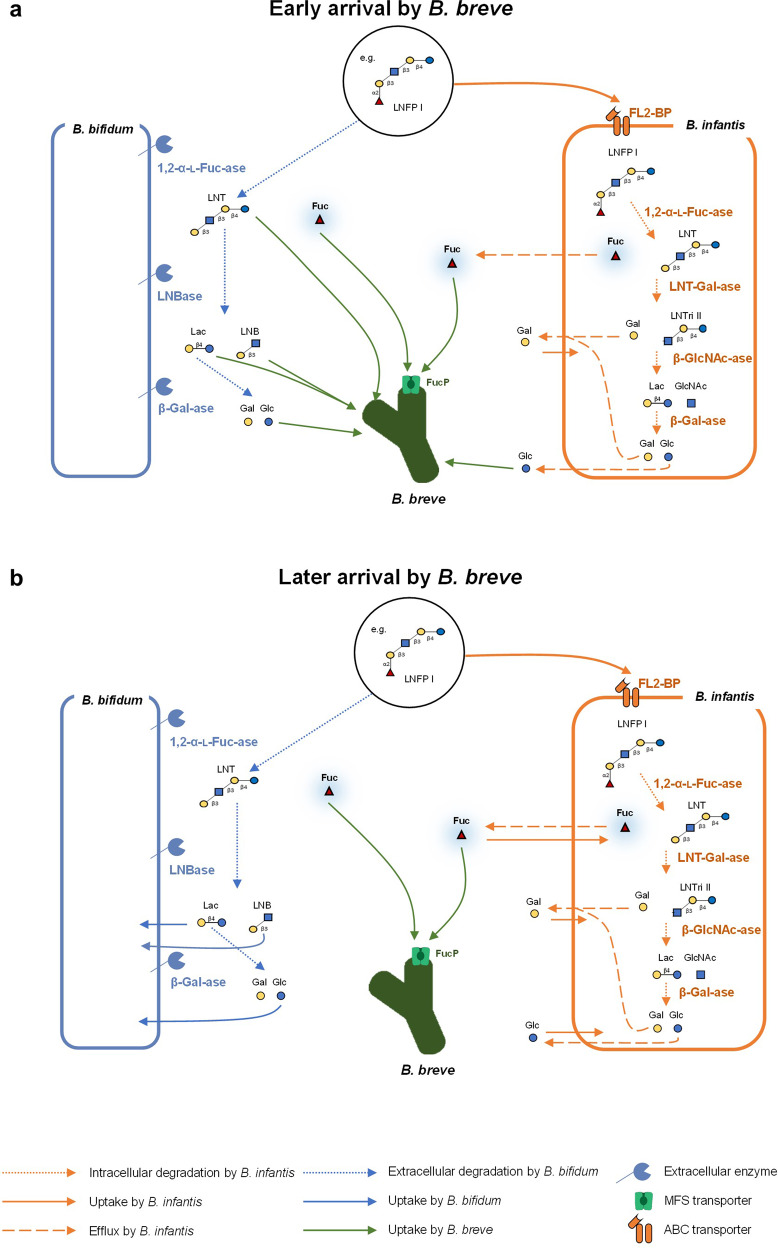
The behaviour of B. breve was not in accordance with our initial expectations and the facilitative effects of other taxa were observed in our in vitro experiments. Despite considerable fitness differences predicted based on HMO-utilization ability, B. breve was able to dominate against stronger competitors (B. bifidum and B. infantis) in both pairwise and 4-species assemblages if inoculated early (Figs. 2d, e, g, 3b). While most of the carbohydrates in the medium in B. bifidum- and B. infantis-dominant communities were depleted, (Extended Data Figs. 1, 2, 5), sugars such as 3-FL and LNDFH I were left unconsumed in B. breve-dominant communities (Extended Data Fig. 5c). The later-arriving strains that can utilize those sugars were unable to outcompete B. breve, suggesting B. breve does not dominate through niche pre-emption. One possible explanation is the differential fucose-utilization phenotypes among infant gut-associated bifidobacteria, which allows B. breve to proliferate through priority effects (Fig. 5). In the INF-1 sequence, in which B. breve is introduced second after B. infantis, fucose is present in the medium at 12 h and decreases as B. breve abundance increases (Fig. 3b, Extended Data Fig. 5b). During pairwise culture, B. breve growth was significantly enhanced when co-cultured with B. bifidum or B. infantis (Fig. 2d, e), both of which liberate fucose and other monosaccharides during HMO metabolization [40]. With B. bifidum, fucose is released through extracellular degradation and left unconsumed [30, 41, 42]. Therefore, niche differentiation may be helping B. breve to maintain a competitive presence when cultured with B. bifidum. On the other hand, B. infantis temporarily excretes fucose and other monosaccharides after intracellular degradation of HMOs, possibly to counteract increasing internal osmotic pressure, and re-imports them later [40]. In co-culture with B. infantis, the timing of arrival becomes crucial — for B. breve to utilize fucose, it must be present in the community after B. infantis temporarily excretes fucose, but before B. infantis re-imports fucose (Fig. 5). Indeed, when the cultivation experiments were performed with ΔfucP B. breve, an isogenic derivative of B. breve UCC2003, the relative abundance of B. breve decreased significantly (Fig. 4b), supporting the fact that fucose (metabolism) enhances the competitive ability of B. breve.
Fig. 5. Predicted HMO-utilization pathways that enable B. breve to benefit from priority effects.
The HMO-utilization pathways, with LNFP I as a representative example, are shown for B. bifidum, B. infantis, and B. breve. Similar mechanisms are expected for other fucosylated HMOs. Activity, enzymes, and transporters attributable to B. bifidum are shown in blue, B. infantis in orange, and B. breve in green. Solid lines indicate assimilation by each species, round-dot lines indicate enzymatic degradation, and dashed lines indicate efflux. B. breve alone cannot assimilate LNFP I. a When B. breve is the prior colonizer, it is ready to utilize degradants as soon as they are made available by either B. bifidum or B. infantis. b When B. breve is the latter colonizer, most of the degradants are consumed before B. breve is introduced. LNT Lacto-N-tetraose; LNFP, Lacto-N-fucopentaose; LNTri II, Lacto-N-triose II, Fuc, Fucose; Glc, Glucose; GlcNAc, N-Acetylglucosamine; Gal, Galactose.
Bifidobacterial communities in the gut of a breastfed infant often fall into one of two groups: B. longum-dominant or B. breve-dominant [10, 43–45]. A variety of factors such as delivery mode, feeding regime, and maternal microbiota can affect the assembly of bifidobacteria in the infant gut, and the ecological mechanisms that lead to B. longum dominance are yet to be determined. With regards to B. breve, only a limited subset of strains can utilize fucosylated HMOs [26], and in general, its HMO-consumption ability is limited to LNT and LNnT. Therefore, factors that contribute to B. breve dominance have not been well understood. Here, an analysis of an in vivo metagenomic dataset revealed that if B. breve was present in the infant gut microbiota at birth, it was more likely to dominate in the community at 4 months of age (Fig. 4a, Extended Data Fig. 6), and our in vitro results suggest that fucose can provide B. breve a competitive advantage. We do note that B. breve, as well as B. infantis (another species that possesses FucP), does not grow well on fucose alone [46]. A study using B. infantis ATCC15697T found that compared to growth on individual sugars, growth significantly increased when the strain was presented with excess fucose in combination with a preferred substrate [47]. Therefore, while fucose alone is not an efficient energy source, it may be serving as a molecule that enhances competitiveness when combined with other sugars. However, continued research is needed to understand the mechanisms by which fucose promotes bifidobacterial growth.
The role of priority effects in the development of the infant gut microbiota is gaining interest, and breastfeeding is a key factor that can significantly influence ecological interactions [48]. Therefore, we focused on bifidobacteria, which are among the first colonizers of the infant gut and utilizers of HMOs found in breastmilk. Here, we show that their sugar-utilization phenotypes determined the outcomes of competition and we have also identified several future research needs. For example, the physiological effects different bifidobacterial species have on infant health has yet to be elucidated. Infant-type bifidobacteria produce indole lactic acid (ILA), an anti-inflammatory molecule that helps regulate intestinal homeostasis in the infant host, and the ability of ILA production is linked with HMO assimilation ability [49, 50]. While the ILA production ability is widespread among infant-type species, only fucose-utilizing species like B. breve also produce 1,2-propanediol, which is utilized by other gut commensals such as Eubacterium hallii to produce propionate [51–55]. Additionally, the presence of B. bifidum is associated with a higher population abundance of Bifidobacterium at the genus level [10]. Therefore, the identity of the dominant bifidobacterial species may affect host health both directly and indirectly through interactions with other taxa, and future work should address the effect of other gut commensals. While Bifidobacterium is often dominant in breastfed infant guts [10], several Bacteroides species are known to utilize HMOs [56]. Bacteroides have been reported to dominate in the absence of bifidobacteria [57], and mutual exclusion may be occurring through the depletion of HMOs. Furthermore, a study comparing infants from different European countries found that while both Russian and Finnish infants were breastfed (199 days and 268 days on average, respectively), Bacteroides remained the dominant taxa in Finnish infants and Bifidobacterium in Russian infants [58], suggesting that once established, community structure is not easily affected by continued dispersal from external sources such as different maternal body sites, home environment, and breastmilk [8, 59–62]. Additionally, a study that administered probiotic B. breve to low birth weight infants suggests that the infant gut microbiota is established within 24 h of birth [63]. In that study, a bifidobacteria-rich microbiota formed earlier when the first dose of B. breve was administered a few hours after birth, as compared to when administered more than 24 h after birth, indicating that the first 24 h of life is a crucial colonization window for gut microbes. To further determine the extent to which priority effects can inform probiotic therapies, additional work that closely mimics the infant gut environment are needed.
As in many other experimental studies, we cannot exclude the possibility that our results are specific to the strain and medium we used, as HMO utilization phenotypes of bifidobacterial species can vary within a species [26]. Additional work utilizing multiple strains from each species, as well as strains isolated from infant faeces could further elucidate the role of priority effects within bifidobacterial communities. However, with regards to B. breve, all known strains possess homologs of FucP and the fucose catabolic pathway, suggesting the possibility that fucose may enhance the competitive ability of other B. breve strains as well. Therefore, our results provide insight into previously unexplained B. breve abundance in breastfed infants [10, 43–45]. Despite certain limitations, we demonstrate the prevalence of priority effects in bifidobacterial communities, particularly for B. breve, and found that early introduction by just a few taxa has the potential to divert the maturational trajectory of the microbial community. Our results suggest that long-term persistence of bifidobacteria and other probiotic strains might be achieved if interventions in the infant gut microbiota are implemented during the early stages of community assembly. As several HMO species are becoming more commercially available in large quantities, additional work using chemostat studies and animal models could provide further insight into how the timing and use of probiotic species can help selectively manipulate the gut microbiota in a way that is both predictable and advantageous to provide disease protection and health-promoting effects for the infant host.
Materials and methods
Chemicals
Fuc, Glc, Gal, 2-AA (anthranilic acid), and sodium cyanoborohydride were purchased from Nacalai Tesque (Kyoto, Japan). GlcNAc and Lac were purchased from Wako Pure Chemical Industries (Osaka, Japan). LNnT and LNFP I were purchased from Dextra Laboratory (Reading, UK). 2′-FL, 3-FL, LDFT, and LNT were obtained from IsoSep (Tullinge, Sweden), or provided as gifts from Glycom A/S (Hørsholm, Denmark). LNFP II, LNFP III, and LNDFH I were purchased from Carbosynth (Berkshire, UK), and isomaltoheptaose was purchased from Seikagaku Kogyo (Tokyo, Japan). LNB was synthesized as described previously [64]. All reagents employed in this study were of analytical grade.
Preparation of oligosaccharides from human milk
Milk samples were collected from healthy Japanese mothers who had not taken any antibiotics for at least 1 month before collection at Nagao Midwife Clinics (Kyoto, Japan). Informed consent was obtained from all mothers. The study was reviewed and approved by the Ethics Committee of Kyoto University (R0046-1) and the University of Shiga Prefecture (71-3) and was performed per the Declaration of Helsinki. HMOs were purified from the collected human milk samples as described previously [40]. During the purification process, sialylated oligosaccharides were eluted near the void fractions as polymeric compounds, and therefore absent from the HMO mixture used in this study. Some amount of Lac, which is a contaminant derived from the HMO purification process, was also detected. The composition of the HMO mixture used in this study was determined by analysis with HPLC and is summarized in Supplementary Table 1.
Bacterial strains and culture conditions in HMO-supplemented medium
Bifidobacterium bifidum JCM 1254 and Bifidobacterium longum subspecies infantis JCM 1222 (ATCC 15697 T) (B. infantis) were obtained from the Japan Collection of Microorganisms (JCM; RIKEN BioResource Center, Japan). Bifidobacterium breve NCIMB 8807 (UCC2003) was obtained from the National Collection of Industrial Food and Marine Bacteria Ltd. (NCIMB; Aberdeen, UK). Bifidobacterium longum subspecies longum MCC10007 (B. longum) was obtained from the Morinaga Culture Collection (MCC, Morinaga Milk Industries, Co. Ltd., Zama, Japan). The ΔfucP mutant derivative of B. breve UCC2003 was constructed in a previous study [31]. All strains used in this study were stored in glycerol stocks and kept frozen at −80 °C until use. All strains were routinely grown in and pre-cultured in Gifu Anaerobic Medium (GAM) broth (Nissui Pharmaceutical, Tokyo, Japan) under anoxic conditions using the AnaeroPack system (Mitsubishi Gas Chemical Co., Tokyo, Japan) at 37 °C. Culturing experiments were performed in YCFA medium, a medium with yeast, casitone, and fatty acids [65], which is known for supporting the growth of the majority of gut commensal species [66]. As a sole carbon source, the medium was supplemented with 1% HMOs or Lac as a positive control (w/v). All cultures were incubated under anoxic conditions at 37 °C and growth was monitored by measuring OD600 at each time point.
Whole genome sequencing
Genomic sequencing for B. bifidum JCM 1254 was performed by first extracting genomic DNA from the strain sub-cultured in de Mann, Rogosa, Sharpe (MRS) medium. The libraries were prepared using a Nextera XT DNA Library Prep Kit (Illumina Inc.) per manufacturer instructions. Paired-end sequencing (29-fold coverage) was conducted on a MiSeq platform (Illumina Inc.) using MiSeq v3 Reagent Kit. Quality trimming of raw reads and de novo genomic assembly were performed in CLC Genomics Workbench (version 8.0) (Qiagen, Valencia, CA, USA) with default settings. Contigs with less than 100 nucleotides were removed. The open reading frame (ORF) predictions and annotations were performed with the DDBJ Fast Annotation and Submission Tool (DFAST) with the default settings [67]. Genomes for B. breve UCC2003 (Accession Number: NC_020517), B. longum MCC10007 (Accession Number: SAMN06368573), and B. infantis ATCC 15697 T (Accession Number: NC_011593) were retrieved from the NCBI GenBank public database (https://www.ncbi.nlm.nih.gov/genbank/).
Bacterial genome annotation, in silico reconstructions, and phenotype predictions
In-depth genomic analysis of Bifidobacterium genomes was performed in the web-based mcSEED (microbial communities SEED) environment, a private clone of the publicly available SEED platform [68]. Genomes of B. longum subsp. longum MCC10007, B. longum subsp. infantis ATCC 15697 T, B. breve UCC2003, and B. bifidum JCM 1254 were first annotated using RAST [68] and then imported into mcSEED. Subsystem-based, context-driven functional assignments of genes combined with manual curation were used to reconstruct and analyse the representation of HMO and HMO constituent utilization pathways in the selected genomes. Data on functional elements (transporters, glycoside hydrolases, downstream catabolic enzymes) involved in bifidobacterial HMO metabolism (Supplementary Table 2) was collected by the following: (1) extensive literature search using PaperBLAST;[69] (2) exporting information from the Carbohydrate Active enZyme (CAZy) [70] and Transporter Classification (TCDB) [71] databases.
Experimental design
To test for priority effects in infant-gut associated bifidobacterial communities, culturing was performed in three different phases: monocultures, pairwise cultures, and four-species assemblages. For pairwise cultures and four-species assemblages, inoculation order was manipulated to examine its effect on community structure. For all culturing experiments, each strain was inoculated with an initial OD600 = 0.02 into 400 μL of YCFA medium containing 1 % HMOs as a sole carbon source, and sampling was performed every 4 h for 24 h. At each sampling time point, OD600 was measured to monitor growth. An aliquot of the culture medium was also collected and centrifuged. The supernatant was collected for sugar concentration analysis. The bacterial pellet was used for DNA extraction, which was performed using the phenol-chloroform method [72].
All cultivation experiments were performed in quadruplicate. Monoculture experiments for each strain were performed to characterize its growth in HMO-supplemented medium, as well as its sugar utilization profiles. Then, pairwise cultures were performed by allowing one species to colonize first and introducing the second species 12 h later, at which the growth curve of the first species plateaus. As a control, both species were inoculated simultaneously (Fig. 1a). All possible pairwise combinations were tested (Table 2; Pairwise). A series of four-species assemblages were also performed, in which 8 assembly sequences were selected so that each species was inoculated first, twice. The second, third, and fourth species were randomly selected. As a control, all four species were inoculated in the community simultaneously (Table 2; Four-Species Assembly). For four-species assemblages, each species was introduced at 4-hour intervals (Fig. 2a).
Sugar concentration analysis
Culture supernatant was collected at each time point, clarified by centrifugation, and stored at −30 °C until use. For analysis with HPLC, the samples were thawed and mixed with isomaltoheptaose (internal standard). The sugars were fluorescence-labelled with 2-AA, and the reaction mixtures were desalted by solid-phase extraction as described previously [40, 73]. HPLC was performed using a Thermo U3000 HPLC system (Thermo Fisher Scientific, Waltham, MA). This was equipped with a TSKgel Amide-80 HR column (4.6 × 250 mm, φ = 5 μm) (Tosoh, Tokyo, Japan) at 65 °C, which was equilibrated with 85% solvent A (acetonitrile)/15% solvent B (100 mM ammonium formate buffer, pH 4.3). The elution was performed using a linear increase of solvent B (from 15 to 85%) over 90 min at a flow rate of 1 mL/min. Using a Waters 2475 Fluorescence Detector (Waters Corp., Milford, MA), the labelled sugars were detected at an excitation wavelength of 350 nm and an emission wavelength of 420 nm. The concentrations of mono- and oligosaccharides remaining in the spent medium were calculated based on the standard curves generated using similarly labelled standard sugars, and the data were normalized using the internal standard. The concentration of Fuc was measured separately with a colorimetric assay using fucose dehydrogenase (FDH) as described previously [74].
Species abundances
From the DNA extracted with bead-beating and the phenol-chloroform method [41, 72], the relative abundance of each species within each sample was quantified using quantitative PCR (qPCR). qPCR was performed with a Thermal Cycler Dice Real-Time System (TaKaRa Bio., Kusatsu, Japan) as described previously [75]. The primer sets used in this study are listed in Supplementary Table 6. Primer specificity was confirmed by agarose gel electrophoresis following PCR using the genomic DNA of each species as templates (Extended Data Fig. 8). Known concentrations of genomic DNA extracted from each species were used as reference curves for species-specific quantification.
Metagenome data mining and taxonomic profiling
Faecal metagenomic data were obtained from SRA (Sequence Read Archive; Accession Number: PRJEB6456) [36]. Data for the 98 infant-mother pairs were available for four different time points: the mother at the time of infant birth, and the infant at 0, 4, and 12 months of age. Of the 98 subjects, infants who were at least partially breastfed (exclusively breastfed or mixed-fed) at 4 months of age were chosen for analysis. Raw reads containing the letter ‘N’ (base pair not identified) were discarded. Reads containing the bacteriophage phiX DNA sequence were identified using Bowtie2 (version 2.3.4.1) [76] with pre-set options and the mapped reads were discarded. Removing of adapters, short reads (<50 bp), as well 3’-end quality trimming (score < 17), was performed using cutadapt (version 2.9) [77]. Next, reads that mapped against the human genome (GRCh 38) using Bowtie2 were also discarded.
The high-quality reads were used for taxonomic classification using Kraken2 version 2.1.2 [78] with standard database, and the abundance of each species was estimated using Bracken version 2.6.1 [79]. We also used METAnnotatorX2 v2.1 [80] with multiple manually curated RefSeq NCBI databases (http://probiogenomics.unipr.it/sw/METAnnotatorX2/METAnnotatorX2_BLASTn_taxonomy_databases.zip) to validate our results (Extended Data Fig. 9, Supplementary Table 7). The predicted taxonomy profiles were normalized based on the microorganisms’ genome sizes. Note that analyses were performed at the species level and did not differentiate between subspecies. Therefore, B. longum and B. infantis populations were both categorized within the B. longum group. If a read was aligned to more than one taxonomic sequence in the database with equal alignment scores, those taxonomies or genes were given a value of 1 divided by the number of hits. Taxonomic ratios were then calculated.
Statistical analysis and quantification of priority effects
Statistical analysis was performed using R ver. 4.0.2 (www.r-project.org). The effect of assembly order on in vitro community composition was analysed with several multivariate techniques. To determine the effect size and significance of each covariate with permutational multivariate analysis of variance (PERMANOVA), the ‘envfit’ function in the package ‘vegan’ [81] was used, and ordination was performed using nonmetric multidimensional scaling (NMDS). The strength of priority effects for pairwise cultures was calculated using the equation proposed by Vannette and Fukami [35]. For four-species assembly, the strength of priority effects was assessed using regression analysis by relating the final population abundance of each species to its arrival order. Negative relationships between population abundance and arrival order were considered to be indicative of inhibitory priority effects [37]. The metagenomic data extracted from public databases, as described above, were also analysed with the ‘envfit’ function and visualized with NMDS as described above. We assessed whether the presence of a species at birth is associated with its dominance (>50% of the bifidobacterial community) at 4 months using Fisher’s exact test.
Supplementary information
Acknowledgements
We thank Director Saeko Nagao of the Nagao Midwife Clinic for collecting breastmilk samples. We also extend our thanks to Glycom A/S for providing LNT, LNnT, 2′-FL, 3-FL, and LDFT. We would also like to thank Dr. Fredrik Bäckhed for helpful information regarding the metadata for the metagenomic dataset. Computations were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics. This study was supported in part by Grants-in-Aid for JSPS Research Fellows (19J14598 to MNO) and JSPS-KAKENHI (21H02116 to TaK). DvS is a member of APC microbiome Ireland which is funded by Science Foundation Ireland through the Irish Government’s National Development Plan (Grant Numbers SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2). The contribution by the SBP team was supported by NIDDK grant to ALO (Grant Number: R01 DK030292).
Author contributions
MNO, LJ, and TaK conceived the project and established the experimental design. AAA, DAR, and ALO were responsible for the bacterial genome annotation, in silico reconstructions, and phenotype predictions. KY and TO were responsible for the whole genome sequencing of B. bifidum JCM 1254. KY, TO, and JzX contributed to the metagenomic data analyses. JH collected the breastmilk samples, and MNO, MK, MS, and TaK were responsible for purifying HMOs from the samples. MNO, AN, AG, ToK, and TU contributed to the sugar concentration analyses. MNO performed the culturing experiments with the wild-type strains. MNO and DvS were responsible for the experiments with the mutant strain. MNO performed the statistical analyses and drafted the manuscript. MNO, LJ, DvS, ALO, MS, and TaK edited the paper. All authors discussed the data and contributed to the completion of the final manuscript.
Data availability
The genome sequence of B. bifidum JCM 1254 was submitted to GenBank (accession numbers: BQJY01000001-BQJY01000062). Source data are provided with this paper. B. longum MCC10007 is available upon reasonable request.
Competing interests
Employment of MNO and MS at Kyoto University is supported by Morinaga Milk Industry Co., Ltd. KY, TO, and JzX are employees of Morinaga Milk Industry Co., Ltd. ALO and DAR are co-founders of PhenoBiome Inc., a company pursuing the development of computational tools for predictive phenotype profiling of microbial communities. The authors declare no other conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miriam N. Ojima, Email: ojima.miriamnozomi.5p@kyoto-u.ac.jp
Takane Katayama, Email: takane@lif.kyoto-u.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01270-3.
References
- 1.Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, et al. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr. 2020;46:1–13. doi: 10.1186/s13052-020-0781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82–343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akagawa S, Tsuji S, Onuma C, Akagawa Y, Yamaguchi T, Yamagishi M, et al. Effect of delivery mode and nutrition on gut microbiota in neonates. Ann Nutr Metab. 2019;74:132–9. doi: 10.1159/000496427. [DOI] [PubMed] [Google Scholar]
- 4.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–5. [DOI] [PubMed]
- 5.Vellend M, Srivastava DS, Anderson KM, Brown CD, Jankowski JE, Kleynhans EJ, et al. Assessing the relative importance of neutral stochasticity in ecological communities. Oikos. 2014;123:1420–30. doi: 10.1111/oik.01493. [DOI] [Google Scholar]
- 6.Fukami T. Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu Rev Ecol Evol Syst. 2015;46:1–23. doi: 10.1146/annurev-ecolsys-110411-160340. [DOI] [Google Scholar]
- 7.Fukami T, Nakajima M. Community assembly: Alternative stable states or alternative transient states? Ecol Lett. 2011;14:973–84. doi: 10.1111/j.1461-0248.2011.01663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debray R, Herbert RA, Jaffe AL, Crits-Christoph A, Power ME, Koskella B. Priority effects in microbiome assembly. Nat Rev Microbiol. 2022;20:109–21. doi: 10.1038/s41579-021-00604-w. [DOI] [PubMed] [Google Scholar]
- 10.Tannock GW, Lawley B, Munro K, Pathmanathan SG, Zhou SJ, Makrides M, et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol. 2013;79:3040–8. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv. 2019;5:eaaw7696. doi: 10.1126/sciadv.aaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–96. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 14.Macrobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macrobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2020;12:71. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci Biotechnol Biochem. 2016;80:621–32. doi: 10.1080/09168451.2015.1132153. [DOI] [PubMed] [Google Scholar]
- 18.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–45. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gore C, Munro K, Lay C, Bibiloni R, Morris J, Woodcock A, et al. Bifidobacterium pseudocatenulatum is associated with atopic eczema: A nested case-control study investigating the fecal microbiota of infants. J Allergy Clin Immunol. 2008;121:135–40. doi: 10.1016/j.jaci.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 20.Lewis ZT, Mills DA. Differential establishment of bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Work Ser. 2017;88:149–59. doi: 10.1159/000455399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tannock GW, Lee PS, Wong KH, Lawley B. Why don’t all infants have bifidobacteria in their stool? Front Microbiol. 2016;7:6–10. doi: 10.3389/fmicb.2016.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyman M, van Houten MA, van Baarle D, Bosch AATM, Man WH, Chu MLJN, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Underwood MA, Kalanetra KM, Bokulich NA, Lewis ZT, Mirmiran M, Tancredi DJ, et al. A comparison of two probiotic strains of bifidobacteria in preterm infants. J Pediatr. 2013;163:1585–91. doi: 10.1016/j.jpeds.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plummer EL, Bulach DM, Murray GL, Jacobs SE, Tabrizi SN, Garland SM. Gut microbiota of preterm infants supplemented with probiotics: sub-study of the ProPrems trial. BMC Microbiol. 2018;18:1–8. doi: 10.1186/s12866-018-1326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitajima H, Sumida Y, Tanaka R, Yuki N, Takayama H, Fujimura M. Early administration of Bifidobacterium breve to preterm infants: Randomised controlled trial. Arch Dis Child Fetal Neonatal Ed. 1997;76:101–7. doi: 10.1136/fn.76.2.F101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojima MN, Yoshida K, Sakanaka M, Jiang L, Odamaki T, Katayama T. Ecological and molecular perspectives on responders and non-responders to probiotics and prebiotics. Curr Opin Biotechnol. 2022;73:108–20. doi: 10.1016/j.copbio.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25:716–29. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 28.Costeloe K, Hardy P, Juszczak E, Wilks M, Millar MR. Bifidobacterium breve BBG-001 in very preterm infants: A randomised controlled phase 3 trial. Lancet. 2016;387:649–60. doi: 10.1016/S0140-6736(15)01027-2. [DOI] [PubMed] [Google Scholar]
- 29.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh T, Ojima MN, Sakanaka M, Ashida H, Gotoh A, Katayama T. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms. 2020;8:481. doi: 10.3390/microorganisms8040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, et al. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:1–14. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins MA, Ryan KS. Generating a fucose permease deletion mutant in Bifidobacterium longum subspecies infantis ATCC 15697. Anaerobe. 2021;68:102320. doi: 10.1016/j.anaerobe.2021.102320. [DOI] [PubMed] [Google Scholar]
- 33.O’Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Micro Biotechnol. 2013;6:67–79. doi: 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, et al. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–8. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 35.Vannette RL, Fukami T. Historical contingency in species interactions: Towards niche-based predictions. Ecol Lett. 2014;17:115–24. doi: 10.1111/ele.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Pu Z, Jiang L. Dispersal among local communities does not reduce historical contingencies during metacommunity assembly. Oikos. 2015;124:1327–36. doi: 10.1111/oik.02079. [DOI] [Google Scholar]
- 38.Chase JM. Community assembly: when should history matter? Oecologia. 2003;136:489–98. doi: 10.1007/s00442-003-1311-7. [DOI] [PubMed] [Google Scholar]
- 39.Schröder A, Persson L, De Roos AM. Direct experimental evidence for alternative stable states: A review. Oikos. 2005;110:3–19. doi: 10.1111/j.0030-1299.2005.13962.x. [DOI] [Google Scholar]
- 40.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep. 2018;8:13958. doi: 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, et al. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–7. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 43.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–41. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 44.Avershina E, Storrø O, Øien T, Johnsen R, Wilson R, Egeland T, et al. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl Environ Microbiol. 2013;79:497–507. doi: 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:20–4. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James K, Bottacini F, Contreras JIS, Vigoureux M, Egan M, Motherway MO, et al. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Rep. 2019;9:1–20. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dedon LR, Özcan E, Rani A, Sela DA. Bifidobacterium infantis metabolizes 2′fucosyllactose-derived and free fucose through a common catabolic pathway resulting in 1,2-propanediol secretion. Front Nutr. 2020;7:1–16. doi: 10.3389/fnut.2020.583397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sprockett D, Martin M, Costello E, Burns A, Holmes S, Gurven M, et al. Microbiota assembly, structure, and dynamics among tsimane horticulturalists of the Bolivian Amazon. Nat Commun. 2019;11:1–14. [DOI] [PMC free article] [PubMed]
- 49.Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. 2021;6:1367–82. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020;88:209–17. doi: 10.1038/s41390-019-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bunesova V, Lacroix C, Schwab C. Fucosyllactose and l-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16:248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-Moyano S, Totten SM, Garrido D, Smilowitz JT, Bruce German J, Lebrilla CB, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–9. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawson MAE, O’Neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–48. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. Trophic interactions of infant bifidobacteria and Eubacterium hallii during l-fucose and fucosyllactose degradation. Front Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engels C, Ruscheweyh HJ, Beerenwinkel N, Lacroix C, Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vatanen T, Kostic AD, D’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–53. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–94. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe. 2019;25:324–335. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 60.Martín R, Langa S, Reviriego C, Jiménez E, Marín ML, Xaus J, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–8. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 61.Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009;75:965–9. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heikkilä MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95:471–8. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Shimizu T, Hosaka A, Kaneko N, Ohtsuka Y, Yamashiro Y. Effects of Bifidobacterium breve supplementation on intestinal flora of low birth weight infants. Pediatr Int. 2004;46:509–15. doi: 10.1111/j.1442-200x.2004.01953.x. [DOI] [PubMed] [Google Scholar]
- 64.Nishimoto M, Kitaoka M. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci Biotechnol Biochem. 2007;71:2101–4. doi: 10.1271/bbb.70320. [DOI] [PubMed] [Google Scholar]
- 65.Duncan SH, Hold GL, Harmsen HJM, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52:2141–6. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 66.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–6. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y. Arita M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci Microbiota, Food Heal. 2016;35:173–84. doi: 10.12938/bmfh.16-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price MN, Arkin AP. PaperBLAST: Text-mining papers for information about homologs. bioRxiv. 2017;2:1–10. doi: 10.1128/mSystems.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016;44:D372–9. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, et al. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–84. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anumula KR. Advances in fluorescence derivatization methods for high-performance liquid chromatographic analysis of glycoprotein carbohydrates. Anal Biochem. 2006;350:1–23. doi: 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 74.Cohenford MA, Abraham A, Abraham J, Dain JA. Colorimetric assay for free and bound l-fucose. Anal Biochem. 1989;177:172–7. doi: 10.1016/0003-2697(89)90035-3. [DOI] [PubMed] [Google Scholar]
- 75.Kato K, Odamaki T, Mitsuyama E, Sugahara H, Xiao JZ, Osawa R. Age-related changes in the composition of gut Bifidobacterium species. Curr Microbiol. 2017;74:987–95. doi: 10.1007/s00284-017-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 78.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;2017:1–17. [Google Scholar]
- 80.Milani C, Lugli GA, Fontana F, Mancabelli L, Alessandri G, Longhi G, et al. METAnnotatorX2: A comprehensive tool for deep and shallow metagenomic data set analyses. mSystems. 2021;6:1–15. doi: 10.1128/mSystems.00583-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence of B. bifidum JCM 1254 was submitted to GenBank (accession numbers: BQJY01000001-BQJY01000062). Source data are provided with this paper. B. longum MCC10007 is available upon reasonable request.