Abstract
Muscle satellite cells (SCs) are responsible for muscle homeostasis and regeneration and lncRNAs play important roles in regulating SC activities. Here, in this study, we identify PAM (Pax7 Associated Muscle lncRNA) that is induced in activated/proliferating SCs upon injury to promote SC proliferation as myoblast cells. PAM is generated from a myoblast‐specific super‐enhancer (SE); as a seRNA it binds with a number of target genomic loci predominantly in trans. Further studies demonstrate that it interacts with Ddx5 to tether PAM SE to its inter‐chromosomal targets Timp2 and Vim to activate the gene expression. Lastly, we show that PAM expression is increased in aging SCs, which leads to enhanced inter‐chromosomal interaction and target genes upregulation. Altogether, our findings identify PAM as a previously unknown lncRNA that regulates both SC proliferation and aging through its trans gene regulatory activity.
Keywords: Ddx5, muscle aging, muscle satellite cell, PAM, seRNA
Muscle satellite cells (SCs) are indispensable for muscle regeneration and lncRNAs play important roles in regulating SC activities. Here, we characterize the function of lncRNA PAM in SC. We show that PAM promotes SCs proliferation by binding with Ddx5 to facilitate the chromatin interaction between PAM residing super enhancer (SE) and target loci, Timp2 and Vim. In aging mice, the activity of PAM SE is elevated to enhance the transcription of Timp2, which potentially modulates extracellular matrix (ECM) components in aging muscle.
Abbreviations
- 3C
chromatin conformation capture
- 4C‐seq
chromatin conformation capture sequencing
- ASC
activated satellite cell
- BET
bromodomain and extraterminal protein family
- ChIP‐seq
chromatin immunoprecipitation sequencing
- ChIRP‐seq
chromatin isolation by RNA purification sequencing
- DNA
deoxyribonucleic acid
- eRNA
enhancer RNA
- FISC
freshly isolated satellite cell
- GO
gene ontology
- JQ1
(S)‐Tert‐butyl2‐(4‐(4‐chlorophenyl)‐2,3,9‐trimethyl‐6H‐thieno[3,2‐f][1,2,4]triazolo[4,3‐a][1,4]diazepin‐6‐yl)acetate
- lncRNA
long non‐coding RNA
- mRNA
messenger RNA
- PAM
Pax7 associated muscle lncRNA
- QSC
quiescent satellite cell
- RNA
ribonucleic acid
- rRNA
ribosomal RNA
- RT‐qPCR
real time quantitative polymerase chain reaction
- SC
satellite cell
- SE
super‐enhancer
- seRNA
super‐enhancer RNA
- siRNA
small interfering RNA
1. INTRODUCTION
Skeletal muscle tissue homeostasis and regeneration relies on muscle stem cells, also known as muscle satellite cells (SCs). These cells reside in a niche between the muscle fiber sarcolemma and the basal lamina surrounding the myofiber and are uniquely marked by transcription factor paired box 7 (Pax7). SCs normally lie in a quiescent state, upon activation by injury or disease, the cells quickly activate and express the master myogenic regulator, MyoD, then re‐enter cell cycle and proliferate as myoblasts, subsequently differentiate and fuse to form myotubes (Wang et al., 2014). A subset of SCs undergo self‐renewal and return to quiescence, thus restoring the stem cell pool. Deregulated SC activity contributes to the development of many muscle‐associated diseases. For example, sarcopenia, a highly prevalent elderly disorder condition characterized by declined muscle mass and deficient muscle strength and function, is linked to a progressive reduction in the regenerative capacity of the SCs. It is thus imperative to understand the way SCs contribute to muscle regeneration, and their potential to cell‐based therapies. At cellular level, every phase of SC activity is tightly orchestrated by many molecules and signaling pathways both intrinsically from the cell and extrinsically from the niche; the elucidation of factors and molecular regulatory mechanisms governing SC function thus is of extreme importance, being the first step toward successful use of these cells in therapeutic strategies for muscle diseases.
It has become increasingly clear that long non‐coding RNAs (lncRNAs) are important players regulating SC regenerative activities (Li et al., 2018). For example, SAM promotes myoblast proliferation through stabilizing Sugt1 to facilitate kinetochore assembly (Li et al., 2020); Linc‐YY1 promotes myogenic differentiation and muscle regeneration through interaction with YY1 (Zhou et al., 2015). In another example, we found that in myoblast cells master transcription factor MyoD induces the expression of lncRNAs from super enhancers (SEs), so‐called seRNAs, which in turn regulate target gene expression in cis through interacting with hnRNPL (Zhao et al., 2019). In fact, the functional synergism between enhancer‐generated eRNAs and their associated enhancer activity in regulating target promoter expression is well established. There are myriad of mechanisms that eRNAs or lncRNAs cooperate with protein, DNA or RNA partners to regulate transcription of target genes either in cis and in trans. For example, it is known that specific eRNA can interact with CBP or BRD4 within topologically associating domain (TAD) in a localized manner (Rahnamoun et al., 2018). Similarly, eRNA Sphk1 evicts CTCF, that insulates between enhancer and promoter, thus activating proto‐oncogene SPHK1 expression in cis (Blank‐Giwojna et al., 2019). Some eRNAs or lncRNAs, on the contrary, play a dual molecular function both in cis and in trans, for example, lincRNA‐p21 acts in trans by recruiting heterogeneous nuclear ribonucleoprotein K (hnRNPK) to the target promoter (Huarte et al., 2010). Interestingly, in a separate study, lincRNA‐p21 can also transcriptionally activate Cdkn1a in cis (Dimitrova et al., 2014). Another well‐demonstrated example of trans acting eRNA is FIRRE, that interacts with hnRNPU via a conserved Repeating RNA Domain (RRD) to mediate nuclear localization during hematopoiesis (Hacisuleyman et al., 2014; Lewandowski et al., 2019). Also, distal regulatory region of MyoD transcribed DRR eRNA that interacts with cohesin and transcriptionally activates Myogenin in trans (Tsai et al., 2018). Altogether, these findings demonstrate the diversified modes of action of eRNAs or lncRNAs in regulating target genes, which needs to be more exhaustively investigated.
Here, in this study, we identify PAM, an seRNA that regulates SC proliferation. Expression of PAM is evidently upregulated during SC activation/proliferation; consistently, knockdown of PAM in vitro hinders SC proliferation. High throughput identification of PAM interactome reveals that it regulates tissue inhibitor of metalloproteinases 2 (Timp2) locus in trans through binding and recruiting Ddx5 protein; loss of PAM or Ddx5 results in reduction of chromatin interaction between PAM SE and Timp2 loci. Furthermore, PAM SE activity and chromatin connectivity with Timp2 is elevated in aging SCs; in vivo inhibition of the SE activity by JQ1 reduces Timp2 expression. Altogether, our findings have identified PAM as a seRNA regulator of SC proliferation through its trans regulation of Timp2 synergistically with Ddx5.
2. RESULTS
2.1. lncRNAs profiling identifies PAM as a seRNA promoting SC proliferation
To gain global insights into the catalog of lncRNAs in the course of SC lineage progression, we analyzed our recently generated RNA‐seq datasets (Zhao et al., 2021) from freshly isolated satellite cells (FISC) which are believed to be early activating by the isolation process, FISCs cultured for 24, 48, or 72 hours (h) to become activated (ASC24h), proliferating (ASC48h) and differentiating (ASC72h) (Figure 1a). We also included the published RNA‐seq dataset from quiescent satellite cells (QSCs) which were in situ fixed in ice‐cold 0.5% paraformaldehyde before cell dissociation to preserve their quiescence (Machado et al., 2017). The analysis revealed transcriptomic signatures that were consistent with the SC lineage progression timing (Zhao et al., 2021). A principal component analysis (PCA) also showed that QSC and FISC had similar transcriptomic signatures while ASC time points were more similar to each other (Figure S1a). To identify lncRNAs differentially expressed during the lineage progression, we found a total of 143 lncRNAs were dynamically expressed in QSC, FISC, and ASCs (Figure 1b). To further identify lncRNA candidates that may play a role in promoting SC activation/proliferation, we found a total of 40 lncRNAs were upregulated in ASC24h vs FISC (Figure 1c and Table S1) and 48 in ASC48h vs FISC (Figure S1b and Table S1). Among these differentially expressed lncRNAs, 21 were upregulated in both ASC24h and ASC48h (Figure S1c and Table S1). Gm12603 was among the highest induced ones; it was barely detectable in QSC and FISC but highly induced in activating (17.9‐fold) and proliferating myoblasts (39.6‐fold) (Figure 1d,e). The Gm12603 expression calculated from the RNA‐seq data was also confirmed by RT‐qPCR using in vitro cultured SCs (Figure 1f). Indeed, it was induced when SCs were activated after culturing for 24 h (1856‐fold), the expression remained high at 48 h (746‐fold) but dropped at 96 h when cells differentiated (Figure 1f). The above finding was also validated in vivo by examining Gm12603 expression in acute injury induced regenerating muscles (Chen et al., 2021). Cardiotoxin (CTX) was injected into tibialis anterior (TA) muscle in young adult (2–4 months old) at Day 0, and SCs were sorted out at 1, 2, 3, 4, and 6 days post‐injury (dpi) for RT‐qPCR (Figure 1g). As expected Gm12603 expression was induced at 2 dpi and peaked at 3 dpi (Figure 1h) concomitant with the SC activation/proliferation phase in vivo (Shcherbina et al., 2020). The in vivo expression dynamics of Gm12603 was also confirmed by analyzing publicly available RNA‐seq data generated from SCs collected in regenerating muscles (Figure 1i,j, S1d). Altogether, the above data demonstrate a possible role of lncRNA Gm12603 in promoting SC activation/proliferation. Interestingly, we also found it was bound by Pax7 at its promoter region via analyzing the publicly available Pax7 ChIP‐seq data in myoblasts (Soleimani et al., 2012), further confirming its functional relevancy in SCs. We thus re‐named it Pax7 associated muscle (PAM) lncRNA.
FIGURE 1.
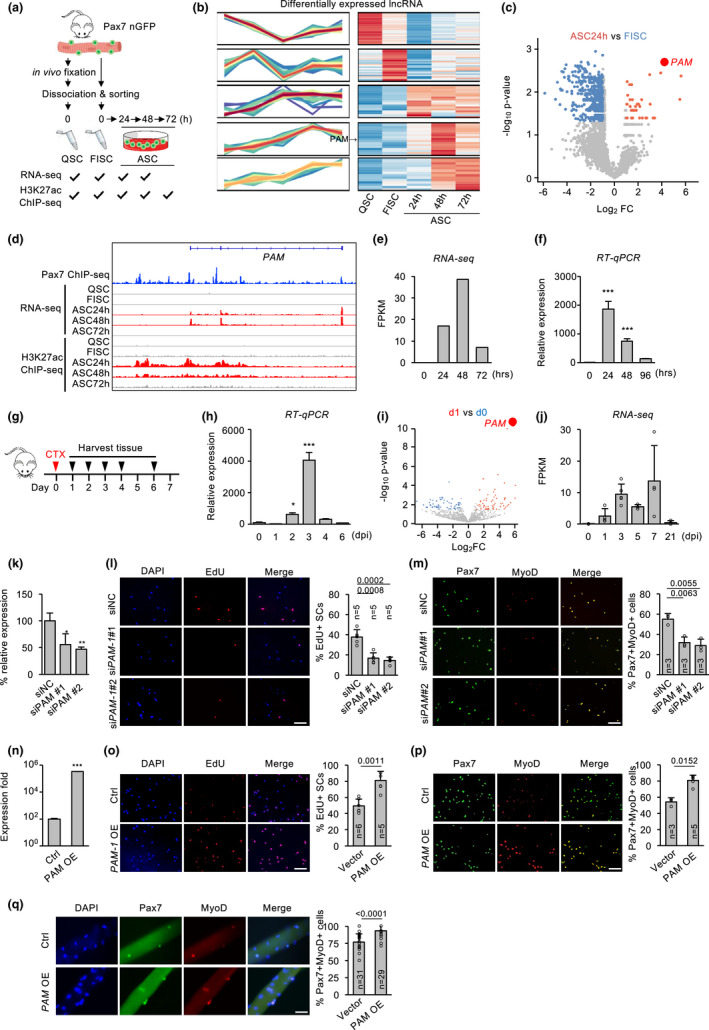
lncRNAs profiling identifies PAM as a seRNA promoting SC proliferation. (a) Transcriptomic and epigenomic discovery of lncRNAs in quiescence SCs (QSCs), freshly isolated SCs (FISCs), or activated SCs (ASCs) in vitro cultured for 24, 48, and 72 hours (h), respectively. All SCs were isolated from muscles of Tg:Pax7‐nGFP mice. (b) Heatmap showing time‐series analysis of differentially expressed lncRNAs during SC lineage progression. PAM was highly expressed in ASCs. (c) Volcano plots showing differentially expressed lncRNAs in in vitro activated ASC24h vs FISC. PAM is indicated in red dot. (d) Genome browser tracks showing temporal expression of PAM by RNA‐seq; histone mark H3K27ac and Pax7 ChIP‐seq tracks are also shown. (e) Bar chart showing PAM expression dynamics (FPKM) from RNA‐seq data. (f) FISCs were cultured for the designated time and PAM expression was detected. (g) Schematic illustration of in acute muscle injury‐induced regeneration. CTX was injected into TA muscles of Tg:Pax7‐nGFP mice and FISCs were collected at the designated days post‐injury (dpi). (h) RT‐qPCR detection of PAM in the above FISCs. (i) Publicly obtained RNA‐seq data from SCs collected at 0, 1, 3, 5, 7, or 21 dpi were analyzed. Volcano plots showing differentially expressed lncRNAs in SCs from Day 1 vs. Day 0 post‐injury. (j) Bar chart showing PAM expression in FPKM from the above data. (k) ASCs were transfected with either control (siNC) or PAM siRNA (siPAM#1 or siPAM#2) and PAM knockdown was confirmed by RT‐qPCR 48 h post‐transfection. (l) EdU incorporation assay was performed in the above cells and the percentage of EdU+ cells were quantified. (m) Pax7+/MyoD+ immunostaining was performed in the above cells and double positively stained cells were quantified. (n) ASCs were transfected with a PAM expression plasmid and PAM overexpression was confirmed 48 h post‐transfection (o,p) EdU incorporation assay (o) or Pax7+/MyoD+ immunostaining (p) were performed in the above cells. The percentage of EdU+ or Pax7+/MyoD+ cells were quantified. (q) Overexpression of PAM in freshly isolated myofibers increased the percentage of Pax7+/MyoD+ cells 48 h post‐transfection. The percentage of EdU+ and Pax7+/MyoD+ cells were quantified. The total number of biologically independent samples are indicated in (l,m, o,q). (data represent the mean ± SD. p‐value was calculated by two‐tailed unpaired t test [*p < 0.05, **p < 0.01, ***p < 0.001]. Scale bars: 100 μm)
PAM is a lncRNA located on chromosome 4: in the intervening region of Interferon alpha (Ifna) family and Cyclin‐dependent kinase 2 inhibitor (Cdkn2a and Cdkn2b) protein‐coding genes. It is also known as Wincr1, a Wnt activated lncRNA in mouse dermal fibroblast affecting extracellular matrix composition via collagen accumulation in dermal fibrosis (Mullin et al., 2017). To dissect its function in SCs, we next cloned its sequence from C2C12 myoblast cell line by Rapid Amplification of cDNA ends (RACE); it was 718 bp long with three exons. More interestingly, we found that PAM is generated from a SE region defined using our published H3K27ac ChIP‐seq datasets (Figure 1d) (He et al., 2021). Concomitant with the expression pattern of PAM, high level of H3K27ac ChIP‐seq signals were observed in ASC24h and ASC 48 h but not in FISC (Figure 1d).
The above findings suggested that PAM may function as a seRNA to promote SC activation/proliferation. To test the potential function of PAM in myoblast proliferation, we knocked down PAM in FISCs from young adult (2–4 months old) with two separate siRNA oligos and performed EdU incorporation after 2 days; a 21 and 23% decrease of EdU+ cells were observed upon PAM knockdown (Figure 1k,l); similarly, by staining for Pax7 and MyoD 48 h after transfection, Pax7+/MyoD+ cells were also significantly reduced by 22% and 25% (Figure 1m). Gain‐of‐function assay was also performed by overexpressing a PAM plasmid in FISCs and increased proliferation was observed by both EdU+ (32% increase) and Pax7+/MyoD+ (26% increase) cells (Figure 1n,p). Altogether, the above results from cultured cells suggested PAM indeed promotes SC proliferation. Similar conclusion was also obtained using in SCs associated with isolated and cultured single muscle fibers; overexpressing PAM led to a 26.66% increase of Pax7+/MyoD+ cells (Figure 1q).
2.2. PAM interacts with inter‐chromosomal loci to modulate target expression
To further elucidate the regulatory mechanism of PAM in SC proliferation, we sought to identify the subcellular localization pattern of PAM as lncRNA function is largely determined by its cellular localization (Yao et al., 2019) and seRNAs are known to be localized in both nucleus and cytoplasm of muscle cells (Zhao et al., 2019). Cellular fractionation was performed using ASC (Figure 2a) and PAM was found to localize largely in nuclear fraction (70.21%); as controls, lncRNAs Xist and Malat1 were predominately nuclear‐localized (93.61% and 87.37%, respectively) while Gapdh mRNAs were enriched in the cytoplasm (89.66%). Similar phenomena were also found when performing cellular fractionation in C2C12 myoblast (Figure 2b). The above finding was further validated by RNA Fluorescence in situ hybridization (FISH) using PAM antisense probe, which also revealed PAM transcript was predominantly enriched in the nucleus of ASC24h but barely detectable in FISC or ASC72h (Figure 2c). Lastly, Subcellular localization of PAM was also confirmed using sucrose gradient centrifugation on C2C12 myoblast lysate which separated protein complex based on their size. Using cohesin loading factor NIPBL as positive control for nuclear fraction, we also found PAM mainly enriched in the nucleus of myoblasts (Figure 2d). Taken together, our results show PAM is a nuclear enriched seRNA.
FIGURE 2.
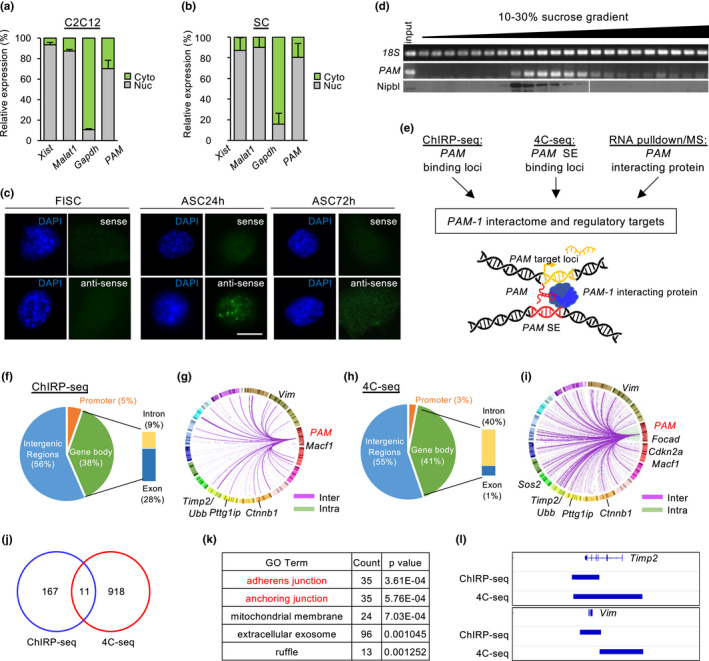
PAM is a nuclear‐retained lncRNA, forming cis and trans chromosomal interactions (a,b). Cellular fractionation of (a) C2C12 cell line and (b) ASC showed PAM was enriched in nucleus not cytosolic fraction. Xist and Malat1 were positive control for nuclear fraction, Gapdh was positive control for cytosolic fraction. (c) Fluorescence in situ hybridization (FISH) in FISC, ASC24h and ASC72h using PAM antisense (AS) probe showed nuclear localization of PAM, sense probe (S) was used as negative control. Scale bar: 10 μm (d) Cellular fractionation using sucrose gradient ultracentrifugation showed PAM was co‐localized in fractions containing nuclear protein Nipbl. (e) Experimental workflow to discover PAM interactome and regulatory targets. For details and controls, see experimental procedures. (f) Pie chart showing distribution of PAM seRNA interacting chromatin across the genome in ChIRP‐seq. (g) Circos plot showing genes associated to PAM seRNA interacting chromatin, each line in the plot represents an interaction, line colors represent inter‐chromosomal (purple) or intra‐chromosomal (green) interactions. Chromosome numbers were colored and arranged in clockwise direction. Top‐ranked genes were named in the figure. (h) Pie chart showing distribution of PAM SE interacting chromatin across the genome in 4C‐seq. (i) Circos plot showing genes associated to PAM SE interacting chromatin, each line in the plot represents an interaction, line colors represent inter‐chromosomal (purple) or intra‐chromosomal (green) interactions. Chromosome numbers were colored and arranged in clockwise direction. Top‐ranked genes were named in the figure. (j) Venn diagram showing overlapping loci between ChIRP‐seq of PAM seRNA and 4C‐seq of PAM SE. (k) Table showing top‐ranked gene ontology (GO) terms of genes associated with overlapping loci from ChIRP‐seq of PAM seRNA and 4C‐seq of PAM SE. (l) Genome browser tracks showing Timp2 and Vim as examples of PAM inter‐chromosomal targets. ChIRP‐seq tracks indicate regions of broad peaks enriched with PAM seRNA binding. 4C‐seq tracks indicate fragments cut by two restriction enzymes and showing significant chromatin interaction with PAM SE
To elucidate the functional mechanism of PAM as a nuclear seRNA, we reasoned it could function to regulate target gene transcription as an integral component of PAM residing SE. Considering SE and seRNA are known to work synergistically by tethering the active transcriptional machinery to target loci, we decided to identify PAM seRNA and PAM SE interacting loci on the genome, respectively (Figure 2e). We first performed PAM Chromatin Isolation by RNA Purification (ChIRP‐seq) in ASCs to identify its binding target loci. As a result, we found that PAM seRNA interacted with 178 DNA regions across the genome which were mainly distributed in intergenic (56%) and gene body regions (36%) (Figure 2f); and these regions were associated with 1199 genes that were enriched for GO terms such as “fiber association,” “actin filament organization,” and “regulation of cell shape” et al. (Figure S2a and Table S2). Strikingly, PAM was dominantly associated with inter‐ but not intra‐chromosomal regions (Figure 2g). Only 5 out of the 178 regions were found on chromosome 4 and not adjacent to PAM gene locus (Table S2). The above findings suggested PAM seRNA may predominantly target gene loci in trans. Next, we set out to identify PAM SE target genes by performing circular chromosome conformation capture (4C‐seq) using PAM SE region as a bait to query genome‐wide chromatin interactions. Among 929 interacting targets, 74% (687) were in trans while only 26% (242) were in cis on chromosome 4 (Figure 2h,i and Table S3).
To further define target loci co‐regulated by PAM and its SE region, we next integrated the targets from the above ChIRP‐seq and 4C‐seq and identified 11 common loci in both datasets (Figure 2j); one of the loci was located 33 Mb downstream of PAM on chromosome 4, while others were all located in other chromosomes (Figure S2b), suggesting PAM and PAM SE may regulate genes in trans together. These 11 loci were associated with 152 genes, which were enriched for GO terms such as “adherens junction,” “anchoring junction” (Figure 2k). Timp2 and Vimentin (Vim) genes were among the top‐ranked (Figure 2l). Timp2 plays a dual role in mediating extracellular matrix by mediating matrix metalloproteinase (MMP) activation and inhibition via interaction with MMP‐14 and MMP‐2 (Young et al., 2004). Overexpression of Timp2 in C2C12 myoblasts was known to delay myogenic differentiation and arrest C2C12 in proliferating state (Lluri & Jaworski, 2005). Vim is an intermediate filament protein to modulate cell shape and motility in myoblast, which is also considered as a reliable marker for regenerating muscle tissue (Bornemann & Schmalbruch, 1992).
2.3. PAM seRNA and PAM residing SE synergistically activate Timp2 and Vim transcription in myoblast
The above findings raised an intriguing possibility that PAM seRNA and PAM SE interact with the inter‐chromosomal target loci Timp2 and Vim to activate their transcription (Figure 2e). Consistent with the notion, we found that PAM, Timp2, and Vim expressions were upregulated during SC activation/proliferation (Figure S2c). In addition, knocking down PAM seRNA using siRNA oligo decreased the expression levels of Timp2 and Vim in both ASCs and C2C12 myoblasts (Figure 3a,b); removing the PAM SE region using CRISPR/cas9 in C2C12 (Figure S2d) yielded similar molecular phenotype (Figure 3c). To test the possibility that PAM functions to tether PAM SE to the target loci, we performed 3C qPCR assay in ASCs and C2C12 myoblasts; in line with the 4C‐seq result (Figure 2h), PAM locus indeed displayed evident interaction with Timp2 and Vim promoters (Figure 3d,e); Furthermore, PAM siRNA oligo mediated knockdown significantly reduced the interaction with Timp2 and Vim (Figure 3d,e and S2e). Consistently, PAM SE knockout also yielded reduction in chromatin interactions with Timp2 and Vim (Figure 3f and S2e). Lastly, knockdown of PAM of PAM SE also led to subtle reduction in H3K27ac signals on Timp2 and Vim in C2C12 myoblasts (Figure 3g and S2f), which was more evidently shown in PAM knockout (Figure 3h). Altogether, our results demonstrate that PAM seRNA and PAM SE can indeed interact with inter‐chromosomal target loci Timp2 and Vim to modulate their expression.
FIGURE 3.
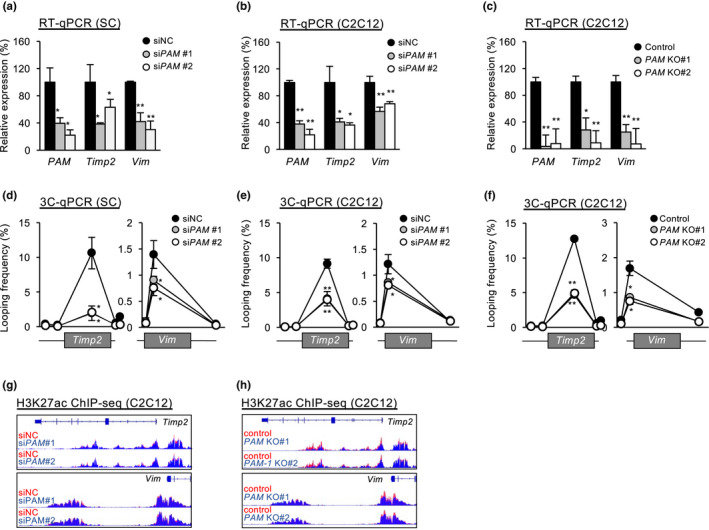
PAM regulates extracellular matrix‐associated genes Timp2 and Vim. (a,b) ASCs (a) or C2C12 (b) were transfected with either control (siNC) or PAM siRNA (siPAM#1 or siPAM#2) oligos, the expression levels of PAM, Timp2 and Vim were detected 48 hours post‐transfection. (c) RT‐qPCR detection of PAM, Timp2 and Vim from C2C12 with or without PAM knockout. (d) ASCs (d) or C2C12 (e) were transfected with either control (siNC) or PAM siRNA (siPAM#1 or siPAM#2) oligos and chromatin conformation capture assay (3C‐qPCR) was performed to detect the chromatin interaction between PAM SE with Timp2 or Vim locus (f) the above 3C‐qPCR was performed in C2C12 cells with PAM knockout. (g,h) Genome browser tracks showing both knockdown (g) and knockout (h) of PAM led to reduction in H3K27ac signal intensity on Timp2 and Vim loci. Data information: Data represent the mean ± SD. p‐value was calculated by two‐tailed unpaired t‐test (*p < 0.05, **p < 0.01)
2.4. PAM regulates inter‐chromosomal targets via associating with Ddx5
To further explore molecular mechanism on how PAM regulates its inter‐chromosomal targets Timp2 and Vim, we performed RNA pulldown followed by mass spectrometry (MS) to identify its protein interactome (Figure 4a). Biotinylated sense probe of PAM was used in the RNA pulldown in vitro, while biotinylated antisense probe was used as negative control for non‐specific protein binding. Among 134 potential protein partners retrieved by the sense probe with at least 5 unique peptide counts (Figure 4b), two known RNA binding proteins (RBPs), DEAD‐Box Helicase 5 (Ddx5) and DEAD‐Box Helicase 17 (Ddx17) were highly ranked showing 22 and 14 unique peptide counts, respectively (Figure 4b). These helicase proteins are commonly found to bind together in multiple cell types to carry out a myriad of molecular functions such as transcription regulation, rRNA processing, mRNA decay, and splicing (Giraud et al., 2018). To validate the above result, we performed RNA pulldown followed by Western blot. PAM seRNA but not GFP control transcripts retrieved an evident amount of Ddx5 and Ddx17 from C2C12 myoblasts, with Ddx5 showing a stronger binding with PAM seRNA (Figure 4c). To test if Ddx5 regulates target expression in cooperation with PAM seRNA, we first performed Ddx5 ChIP‐seq which revealed an evident binding of Ddx5 on Timp2 and Vim loci (Figure 4d), suggesting PAM and Ddx5 are both tethered to the target loci. Consistent with their possible functional synergism, knockdown of Ddx5 in ASC or C2C12 led to down‐regulation of Timp2 and Vim (Figure 4e,f). Furthermore, the above knockdown also reduced the interaction between PAM SE and the promoters of Timp2 and Vim (Figure 4g,h), suggesting Ddx5 indeed promotes the inter‐chromosomal interaction together with PAM. Lastly, knockdown of Ddx5 reduced ASC proliferation as revealed by reduction in both EdU+ (Figure 4i) and Pax7+/MyoD+ cells (Figure 4j). Taken together, our findings demonstrate PAM seRNA and Ddx5 function synergistically in orchestrating the inter‐chromosomal interactions between the PAM SE with the two target loci, Timp2 and Vim, consequently promoting transcriptional activity.
FIGURE 4.
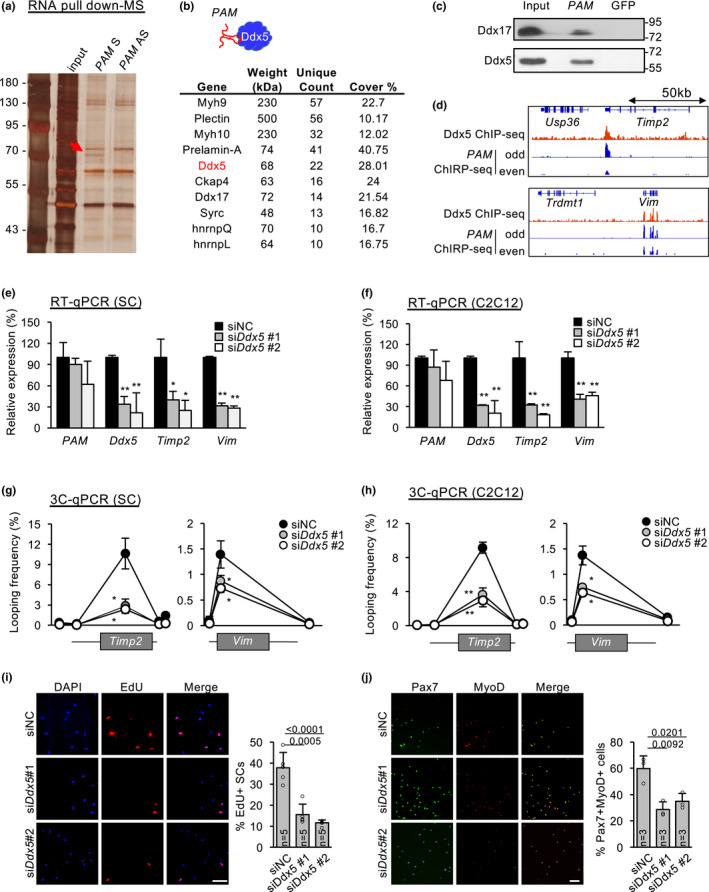
PAM regulates inter‐chromosomal targets via association with Ddx5. (a) RNA pulldown using PAM sense (S) or antisense (AS) experiment followed by SDS‐PAGE. AS was used as negative control. Red arrow indicates enrichment of a protein band at 70 kDa specifically found in pulldown using PAM sense probe. (b) Mass spectrometry (MS) result of the above band showing a list of potential protein binding partners of PAM. (c) RNA pulldown followed by Western blotting of the above identified two candidate protein partners (Ddx5 and Ddx17) of PAM‐ transcript. (d) Genome browser tracks showing Ddx5 ChIP‐seq and PAM ChIRP‐seq on Timp2 or Vim locus. (e,f) ASCs (e) or C2C12 (f) were transfected with either control (siNC) or Ddx5 siRNA (siDdx5#1 or siDdx5#2) oligos, PAM, Ddx5, Timp2 and Vim expression levels were detected by RT‐qPCR 48 h post‐transfection. (g,h) ASCs or C2C12 were transfected with either control (siNC) or Ddx5 siRNA (siDdx5#1 or siDdx5#2) oligos, 3C qPCR was performed to detect chromatin interaction between Timp2 or Vim promoter with PAM locus. (i,j) Knockdown of Ddx5 expression in vitro in cultured ASCs for 48 hours led to reduction in EdU+ (i) or Pax7+/MyoD+ (j) SCs. The percentage of EdU+ and Pax7 + Myod+ cells were quantified. The total number of biologically independent samples are indicated in (i,j). Data information: Data represent the mean ± SD. p‐value was calculated by two‐tailed unpaired t test (*p < 0.05, **p < 0.01). Scale bar: 100 μm
2.5. PAM upregulation in aging SCs drives its target gene activation
Lastly, to test the possible involvement of PAM in SC aging, we isolated SCs from mice of various ages (2, 16, or 24 months) and cultured for 48 h to obtain proliferating ASC. By performing H3K27Ac ChIP‐qPCR, we found the activity of PAM SE was indeed increased by 109% in ASC from 16 month as compared with 2‐month‐old mice, and continued to increase at 24 month (Figure 5a). The expression of PAM and Timp2 also showed an increase (308% and 681%) in 20 vs 2‐month ASC. Interestingly, Vim expression remained unchanged (Figure 5b); the lack of response in Vim could be due to the weak interaction between Vim locus and PAM SE (Figure 3d). In addition, we also obtained RNA‐seq data from injured 2‐ or 24‐month‐old muscles (Shcherbina et al., 2020) and found PAM was increased endogenously in ASC isolated 1 dpi from 24 vs 2‐month‐old mice (Figure S3a). Knockdown of PAM in ASCs from 20‐month‐old muscle also led to reduced proliferation (Figure 5c and S3b). The above data thus suggest that both PAM SE activity and PAM expression is elevated in aging SCs and may contribute to the upregulated Timp2 expression. Consistent to the notion, the interaction between PAM locus and the promoter of Timp2 were found to increase by 46.75% in 20 vs 2‐month ASC (Figure 5d). Lastly, to further confirm the synergistic function of PAM and Ddx5 in increasing Timp2 expression in aging SCs, we found that knockdown of PAM or Ddx5 in ASC in 2‐, 20‐ or 30‐month‐old mice led to down‐regulation of Timp2 (Figure 5e).
FIGURE 5.
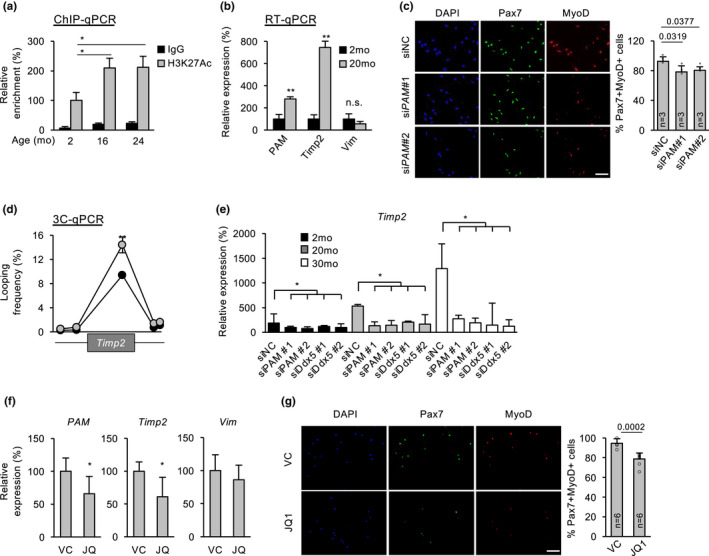
PAM increase in aging SCs drives its target gene upregulation. (a) H3K27ac ChIP‐qPCR showing increase in enrichment on PAM SE in ASCs isolated from aging (16 and 24 months) vs young (2 months) old mice. (b) RT‐qPCR showed upregulation of PAM and target genes Timp2 but not Vim in ASCs from 20 vs 2 month old mice. (c) Knockdown of PAM in aging ASC from 20 months old mice reduced the number of Pax7+/MyoD+ proliferating ASC. (d) 3C‐qPCR assay showed increase in interaction between PAM locus and Timp2 promoter in the above aging ASCs. (e) RT‐qPCR showing the expression dynamics of Timp2 in ASCs from 2, 20 and 30 months old mice. Knockdown of PAM or Ddx5 using siRNA oligo in ASCs from 20 or 30 months old mice showed down‐regulation of Timp2. (f,g) in vivo treatment of JQ1, a Brd4 inhibitor, in 10 month old mice caused down‐regulated expression of PAM and Timp2 but not Vim in FISCs (f), and reduced number of Pax7+/MyoD+ (g) in aging SC. The total number of biologically independent samples are indicated in (c&g). Data information: Data represent the mean ± SD. p‐value was calculated by two‐tailed unpaired t test (*p < 0.05, **p < 0.01). Scale bar: 100 μm
Recently, we have reported the use of BET family of bromodomain protein binding inhibitor JQ1 to down‐regulate enhancer activity in aging mouse muscle tissue (Zhou et al., 2019). Expectedly, we found that intraperitoneal injection of JQ1 in 10‐month‐old mice led to a down‐regulation of PAM seRNA and Timp2 expression in FISCs, but not Vim (Figure 5f). Consistently, we found ASC proliferation was reduced by JQ1 treatment (16% in Pax7 + MyoD+ cells) (Figure 5g). Altogether, our data reinforce the notion that PAM SE activation causes Timp2 upregulation during SC aging.
3. DISCUSSION
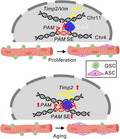
Myogenesis is a complex process that relies on tightly regulated and finely tuned transcriptional regulatory mechanisms. Previous studies have discovered a myriad of lncRNAs that are dynamically regulated during myogenesis (Alessio et al., 2019). Yet, their functional mechanisms in SCs remain largely unexplored. Here, in this study, we identified PAM, a seRNA that binds with Ddx5 protein to synergistically tether the SE to its inter‐chromosomal target loci Timp2 and Vim. Furthermore, we showed that deregulation of PAM in aging SCs drives the target gene deregulation (Figure 6).
FIGURE 6.
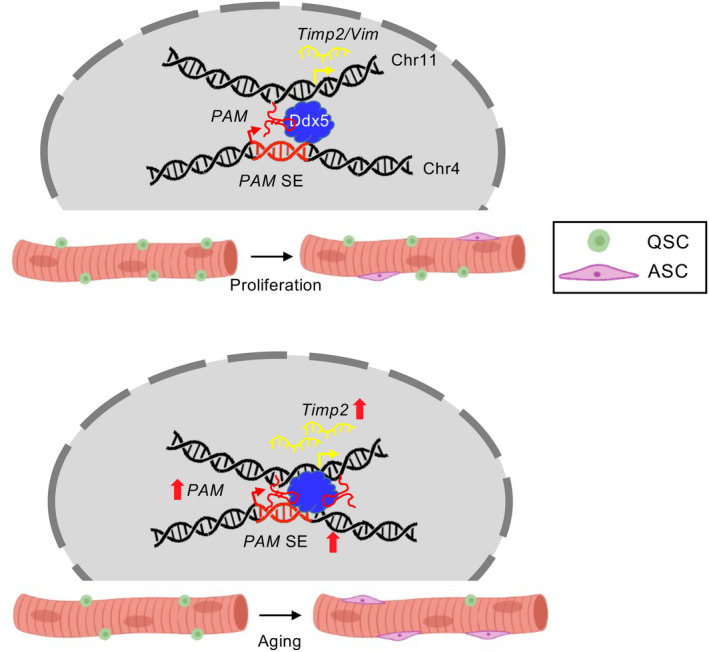
Schematic model showing functional role of seRNA PAM in SC proliferation and skeletal muscle aging. PAM regulates SCs proliferation by binding with Ddx5 to facilitate the chromatin interaction between PAM SE and target loci, Timp2 and Vim during SC proliferation. In aging mice, the activity of PAM SE was elevated, thereby enhancing the transcription of Timp2, which potentially modulates extracellular matrix components in skeletal muscle
Through transcriptomic profiling, PAM was identified as highly induced in activated and proliferating SCs. Loss‐ and gain‐of‐function studies in SCs indeed pinpointed it as a promoting factor for SC proliferation. Its nuclear localization is consistent with its nature of being a seRNA. As part of the SE regulatory machinery, PAM and its residing SE together promote the expression of their target loci, Timp2 and Vim, both encoding extracellular matrix proteins. Increased extracellular matrix (ECM) proteins expressions are essential to SC activation (Zhang et al., 2021). The regulation of ECM composition was conventionally believed to be mediated by surrounding fibroblasts, fibro/adipogenic progenitors, myofibers, and basal lamina. In addition to receiving signals from ECM, emerging evidence demonstrates SCs also contribute to ECM compositions through secretion of matrix metalloproteases and urokinase plasminogen activator (Guerin & Holland, 1995). For example, transcriptome profiling of freshly isolated SCs revealed that cell adhesion and ECM genes, such as Timp and integrin, were differentially expressed when compared with freshly isolated SCs from dystrophic mdx mice (Pallafacchina et al., 2010). This correlation was further demonstrated by systemic delivery of MMP inhibitor, AM409, which impairs SC activation (Pallafacchina et al., 2010). Therefore, upregulation of ECM gene expression mediates the promoting function of PAM during SC proliferation. Furthermore, we showed that PAM upregulation contributes to ECM increase in aging SCs. It is known that the regenerative potential of SCs declines during aging, which is in concurrent with its fibrogenic conversion and muscle fibrosis (Yamakawa et al., 2020). Using publicly available single‐cell RNA‐seq analysis, ECM and SC niche are contributed by multiple cell types, while Timp2 is expressing in SCs, macrophages, mesenchymal stem cells, and endothelial cells (Tabula Muris, 2020) (Figure S3c). Our findings thus provide a potential way to partially restore ECM in aging SCs by down‐regulating PAM expression in a cell‐autonomous manner. While our data suggest an elevated PAM expression in aging SCs, it remains to be determined whether this is due to faster activation of aged SCs or not. In the future, more efforts will be needed to elucidate the potential roles of lncRNAs in aging SCs. For example, a recent study on integrated transcriptome analysis of aging human skeletal muscle revealed a group of differentially expressed lncRNAs, and overexpression of lncRNA PRKG1‐AS1 could increase cell viability and reduce apoptosis in human skeletal myoblast (Zheng et al., 2021).
Mechanistically, our data highlight the important role of PAM seRNA to regulate inter‐chromosomal targets through tethering its residing SE to the target loci. eRNAs or seRNAs are commonly known to regulate enhancer‐promoter interactions as an integrated component of SE activating machinery (Zhao et al., 2019); it is believed that cis regulation of the target loci through intra‐chromosomal interactions is a more prevalent mode compared with trans regulation via inter‐chromosomal interactions (Sartorelli & Lauberth, 2020). However, trans activating eRNAs do exist to translocate to distal chromosomal regions beyond its neighboring loci. For example, MyoD distal enhancer generates DRR eRNA that transcriptionally regulates Myogenin expression in trans via cohesin recruitment (Tsai et al., 2018). Similarly, in human prostate cancer, an adjacent eRNA of kallikrein‐related peptidase 3 (KLK3) can regulate target genes expression in trans (Hsieh et al., 2014). Our findings from integrating ChIRP‐seq and 4C‐seq demonstrate that PAM mainly acts in trans to exert its regulatory function in SCs; thus, provide additional evidence to support the trans regulatory mechanism by eRNAs.
Another important discovery from our study stems from the identification of a direct physical interaction between PAM and Ddx5. Ddx5 and PAM synergistically facilitate the SE‐target interaction; knockdown of Ddx5 impaired the interaction. Although classically known as a RNA helicase controlling mRNA splicing, recent studies demonstrated Ddx5 interacts with a myriad of lncRNAs, and uses the lncRNAs as a scaffold to bring in specific transcriptional machinery or chromatin architectural protein in context‐dependent manner (Giraud et al., 2018). It was also demonstrated that Ddx5 interacts with lncRNA mrhl to mediate cell proliferation in mouse spermatogonia cells (Arun et al., 2012). Ddx5 and Ddx17 (p68 and p72) bind with lncRNA SRA in regulating skeletal muscle differentiation (Caretti et al., 2006). With foundation laid by these studies, we further demonstrated the functional role of Ddx5 in mediating chromatin interactions via its interaction with PAM, underscoring the prevalence of lncRNAs and RNA helicase interaction and broadening the mechanisms through which lncRNAs regulate gene expression.
4. EXPERIMENTAL PROCEDURES
4.1. Mice
Tg:Pax7‐nGFP mouse strain (Sambasivan et al., 2009) was kindly provided by Dr. Shahragim Tajbakhsh. All animal handling procedures and protocols were approved by the Animal Experimentation Ethics Committee (AEEC) at the Chinese University of Hong Kong.
4.2. JQ1 treatment
JQ1 treatment was performed as described previously (Zhou et al., 2019). C57/BL6 mice were caged in groups of five and maintained at controlled temperature (20 ± 1°), humidity (55 ± 10%), and illumination with 12 h light/ 12 h dark cycle. Food and water were provided ad libitum. All procedures involving animal care or treatments were approved by the Animal Ethics Committee (AEEC) at Chinese University of Hong Kong (CUHK) on the protection of animals used for scientific purposes. To investigate the effect of JQ1 on aging skeletal muscle, daily intraperitoneal injection of JQ1 at 50 mg/kg was performed on 10‐month‐old mouse for 14 days (Zhou et al., 2019), with DMSO as control. Then, TA muscle tissues were extracted from mice, and SCs were isolated from mouse skeletal muscle by fluorescence‐activated cell sorting (FACS) using BD FACSAria Fusion cell sorter (BD Biosciences) with cell surface marker Sca1−/CD31−/CD45−/Vcam+ (Chen et al., 2019).
4.3. Cells
Mouse C2C12 myoblast cell (CRL‐1772) was obtained from American Type Culture Collection (ATCC) and cultured in DMEM medium with 10% fetal bovine serum, 1% penicillin/streptomycin at 37°C in 5% CO2. Oligonucleoties of siRNA against mouse PAM and scrambled control were obtained from Ribobio Technologies (Guangzhou, China). siRNAs were transfected at 100 nM into C2C12 using Lipofectamine 2000 (Life Technologies). The sequences of oligonucleotides using for siRNA knockdown were listed in Table S4.
4.4. Satellite cell isolation and culture
Hindlimb skeletal muscles from Tg:Pax7‐nGFP mice were dissected and minced, followed by digestion with Collagenase II (LS004177, Worthington, 1000 units/ml) for 90 min at 37°C in water bath shaker. Digested muscles were then washed in washing medium (Ham's F‐10 medium (N6635, Sigma) containing 10% heat‐inactivated horse serum (Gibco, 26,050,088) with 1% penicillin/ streptomycin, followed by incubating in digestion medium with Collagenase II (100 units/ml) and Dispase (1.1 unit/ml, Gibco, 17,105–041) for additional 30 min. Suspensions were then passed through 20G syringe needle to release myofiber‐associated SCs. Mononuclear cells were filtered with a 40 μm cell strainer, followed by cell sorting using BD FACSAria Fusion Cell Sorter (BD Biosciences). BD FACSDiva (BD Biosciences, version 8.0.1) software was used to manage machine startup, data acquisition and analysis of flow cytometry data. Culture dishes were coated with poly‐D‐lysine (Sigma, P0899) and Matrigel (BD Bioscience, 356,234). FACS isolated SCs were seeded in coated culture dish and cultured in Ham's F10 medium with 10% heat‐inactivated horse serum, 5 ng/ml FGF‐Basic (AA 10–155) (Gibco, PHG0026), or cultured in differentiation medium (Ham's F10 medium with 2% horse serum and 1% penicillin/ streptomycin).
4.5. EdU incorporation assay
EdU incorporation assay was performed as described previously (Li et al., 2020). EdU was added to cultured MuSCs for 4 hours before fixation, followed by fixation in 4% paraformaldehyde (PFA) for 15 min and stained according to the EdU staining protocols provided by manufacturer (Thermo Fisher Scientific, C10086).
4.6. Single myofibers isolation and culture
Extensor digitorum longus (EDL) muscles were dissected and digested in collagenase II (800 units/ml) in DMEM medium at 37°C for 75 min. Single myofibers were released by gentle trituration with Ham's F‐10 medium with heat‐inactivated horse serum and 1% penicillin/ streptomycin, then cultured in this medium for the follow‐up experiments.
4.7. Genomic editing by CRISPR‐Cas9 in C2C12 cells
To delete PAM exon1, target‐specific guide RNAs (gRNAs) were designed using CRISPR design tool (http://crispr.mit.edu), followed by cloning into BbsI digested px330 plasmid (Addgene, 42,230). To perform genomic deletion, a pair of gRNAs containing plasmids were co‐transfected into C2C12 cells with screening plasmid pSIREN‐RetroQ (Clontech) using Lipofectamine 2000. Cells were selected with 2.5 μg of puromycin for 3 days at 48 h post‐transfection. Cells were diluted to 1 cell per well in 96 well plate. Individual colonies were PCR validated. Sequences of gRNAs and genotyping primers were listed in Table S4.
4.8. Plasmids
Full‐length cDNA of Gm12603 (PAM) was cloned into pcDNA3.1 vector using HindIII and KpnI restriction enzymes digestion site. Primer sequences were listed in Table S4.
4.9. RT‐qPCR
RNAs were extracted using Trizol (Life Technologies), followed by reverse transcription using SuperScript III Reverse Transcriptase (Life Technologies). PCRs were performed with SYBR green (Life Technologies). PCR products were analyzed by LC480 II system (Roche). Primers used are listed in Table S4.
4.10. RNA pulldown
RNA pulldown was performed as described previously (Li et al., 2020). PAM DNA constructs were first linearized by single restriction enzyme digestion (NotI and XhoI for antisense and sense transcription respectively). Biotinylated transcripts were generated by these digested constructed by in vitro transcription using Biotin RNA labeling Mix (Roche) and MAXIscript T7/T3 In Vitro Transcription Kit (Ambion). Transcribed RNAs were denatured at 90°C for 2 min, then cooling on ice for 5 min, followed by addition of RNA structure buffer (Ambion) and refolding at room temperature for 20 min. Nuclear proteins from C2C12 cells were collected by resuspending cell pellet in nuclear isolation buffer (40 mM Tris–HCl pH 7.5, 1.28 M sucrose, 20 mM MgCl2, 4% Triton X‐100 and 1x protease inhibitor). Nuclei were collected by centrifugation at 3000 g and 4°C for 10 min. Supernatant was removed, and nuclear pellet was resuspended in 1 ml RIP buffer (25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5 mM DTT, 0.5% NP‐40, 1 mM PMSF, 1x RNase inhibitor, and 1x protease inhibitor), followed by homogenization for 10 cycles (15 s on/off) using Ika homogenizer (Ika‐Werke Instruments, Germany). Nuclear envelops and debris were removed by centrifugation at 16,200 g for 10 min. For RNA pulldown assay, 1 mg of nuclear extracts were incubated with 3 μg of refolded RNA on rotator at room temperature for 1 h. 30 μl of prewashed Dynabeads M‐280 Streptavidin were added to each reaction with incubate on rotator at room temperature for additional 1 h. Streptavidin beads were collected using a magnetic rack, and beads were washed with 1 ml RIP buffer for 5 times. Proteins were eluted by adding Western blot loading buffer and incubated at 95°C for 5 min, followed by removal of beads using magnetic rack. RNA pulldown samples were analyzed by SDS‐PAGE followed by silver staining and LC–MS/MS with Q Exactive and Easy‐nLC 1000 system (Thermo Fisher). Peptides were identified using MASCOT.
4.11. RNA fluorescence in situ hybridization (FISH)
RNA FISH was performed as described previously (Chen et al., 2017). Cells were fixed with 4% formaldehyde in PBS for 15 min at room temperature, followed by permeabilization with 0.5% Triton X‐100, 2 mM VRC (NEB) on ice, and two times 2x SSC wash for 10 min each. Probes were first amplified with PCR using PAM expression plasmid in RNA pulldown experiment. PCR products were then precipitated by ethanol, nick‐translated and labelled with Green d‐UTP (Abbott) and nick translation kit (Abbott). For each FISH experiment, 200 μg of probe and 20 μg of yeast tRNA were lyophilized and redissolved in 10 μl formamide (Ambion), followed by denaturation at 100°C for 10 min and chilled immediately on ice. Denatured probes were mixed with hybridization buffer at 1:1 ratio. 20 μl of hybridization mix was added onto fixed cells, followed by putting coverslip on it and incubated at 37°C for 16 h in a humidified chamber. Cells were then washed twice in 2x SSC, 50% formamide; thrice in 2x SSC; and once in 1x SSC for 5 min each in 42°C. Cells were mounted by coverslip with ProLong Gold Antifade Reagent with DAPI (Invitrogen). Fluorescence images were taken in Olympus microscope FV10000 and FV10‐ASW software (version 01.07.02.02, Olympus).
4.12. Cellular fractionation
Cellular fractionation was performed as described previously (Chen et al., 2017). C2C12 cell pellet from 1 × 106 cells was lysed with lysis buffer (140 mM NaCl, 50 mM Tris–HCl pH 8.0, 1.5 mM MgCl2, 0.5% NP‐40, and 2 mM Vanadyl Ribonucleoside Complex) for 5 min at 4°C, followed by centrifugation at 4°C 300 g for 2 min. The supernatant after centrifugation was considered as cytoplasmic fraction and stored in −20°C for storage, while the pellet was resuspended in 175 μl resuspension buffer (500 mM NaCl, 50 mM Tris–HCl pH 8.0, 1.5 mM MgCl2, 0.5% NP‐40, 2 mM Vanadyl Ribonucleoside Complex) and incubate at 4°C for 5 min. Nuclear‐insoluble fraction in the resuspended pellet was removed by centrifugation at 4°C and 16,000 g for 2 min. RNA was extracted from cytoplasmic and nuclear soluble fraction by Trizol (Life Technologies).
4.13. Sucrose gradient
Sucrose gradient was performed as described previously (Peng et al., 2017). C2C12 cells were lysed in cell lysis buffer (50 mM Tris–HCl pH 7.6, 1 mM EDTA, 1% Triton X‐100, 10% glycerol, 1 mM DTT, 1 mM PMSF, 1x RNase inhibitor, and 1x protease inhibitor). 500 μl of whole‐cell lysate was added to 13.5 ml of 10%–30% sucrose gradient, followed by centrifugation at 38000 RPM at 4°C for 16 h. The centrifuged lysate was fractionated in 500 μl portion. To avoid cross‐contamination, only odd‐numbered fractions were obtained. Protein samples were resolved in SDS‐PAGE, followed by Western blotting. RNA samples were subjected to RT‐PCR of PAM and 18S, followed by 2% agarose gel electrophoresis.
4.14. Chromatin immunoprecipitation using sequencing (ChIP‐seq) and ChIP‐qPCR
ChIP assays were performed as previously described (So et al., 2017). C2C12 cells were crosslinked with 1% formaldehyde at room temperature for 10 min, followed by quenching with 0.125 M glycine for 10 min. Chromatin was fragmented using S220 sonicator (Covaris), followed by incubation with 5 μg of antibodies and 50 μl Dynabeads Protein G magnetic beads (Life Technologies) at 4°C on rotator for overnight. Anti‐histone H3‐K27 acetylation (Abcam, ab4729), anti Ddx5 (Abcam, ab21696) and normal rabbit IgG (Santa Cruz Biotechnology, sc‐2027) were used in ChIP assay. Beads were washed with 1 ml RIPA buffer for 5 times, followed by decrosslinking at 65°C for 16 h and DNA extraction with phenol/chloroform. Immunoprecipitated DNA was resuspended in 50 μl of water. 200 ng of immunoprecipitated DNA was used as starting material for NEBNext® Ultra II DNA Library Preparation kit for Illumina (NEB) according to manufacturer's guideline. DNA libraries were sequenced in Illumina NextSeq 550 platform. ChIP‐qPCRs were performed with 1 μl of ChIP DNA as templated with SYBR Green Master Mix (Life Technologies), quantitative PCRs were performed on Roche LC480 system (Roche). Primers used are listed in Table S4.
4.15. Chromatin isolation by RNA purification using sequencing (ChIRP‐seq)
Biotin labelled probes targeting PAM lncRNA were designed by ChIRP Designer (LGC Biosearch Technologies) and listed in Table S4. Cells were rinsed in PBS, trypsinized, washed once with complete DMEM followed by resuspension in PBS. 10 million of ASCs were collected per ChIRP experiment for separated odd and even probe pools. Cell pellets were crosslinked with 1% Glutaraldehyde in 40 ml PBS on rotator for 10 min at room temperature, followed by quenching the crosslinking reaction with 2 ml 1.25 M Glycine for 5 min and resuspend in 1 ml chilled PBS. Cell pellets were collected at 2000RCF for 5 minutes at 4°C, followed by removing PBS, snap‐frozen with liquid nitrogen and stored at −80°C. Cell pellets were lysed and sonicated according to our standard ChIP‐seq protocol (So et al., 2017), then aliquoted into two 1 ml samples. Before ChIRP experiment, DNA were extracted for quality control with size ranging from 100‐500 bp. For ChIRP experiment, 10 μl of lysate were saved for DNA input. 1 ml of sonicated lysate was mixed with 2 ml of hybridization buffer (750 mM NaCl, 50 mM Tris–HCl pH 7, 1 mM EDTA, 1% SDS, 15% Formamide, 1x protease inhibitor and 1x RNase inhibitor). 100 pmol of odd and even ChIRP probes were added separately to the hybridization mixture and incubate at 37°C for 4 h with rotation. After the hybridization was completed, 100 μl of streptavidin magnetic C1 beads (Life Technologies, 65,001) were washed thrice with hybridization buffer and added to each ChIRP reaction for extra 30 minutes incubation at 37°C with rotation. After the hybridization completed, 1 ml of wash buffer (2X SSC, 0.5% SDS and 1x protease inhibitor) was used to wash the beads for 5 times using magnetic stand. Input control and PAM‐bound DNA was eluted with ChIP elution buffer for each pair of ChIRP reactions using standard elution protocol as ChIP (So et al., 2017). For ChIRP‐seq, DNA libraries were prepared as previous described in ChIP‐seq protocol (So et al., 2017). Raw reads were uniquely mapped to mm9 reference genome using Bowtie2 (Langmead & Salzberg, 2012). Peaks were called by using MACS2 (Zhang et al., 2008).
4.16. 4C‐seq and 3C qPCR
3C experiments were performed as previously described using restriction enzyme BglII to digest fixed chromatins (Peng et al., 2017). Primers for PAM bait region and target regions were listed in Table S4. First round of restriction enzyme digestion in 4C‐seq was the same as 3C qPCR. 4C experiment was then continued with TatI restriction enzyme digestion, incubated overnight at 37°C and circularized using T4 DNA ligase. Gradient range of annealing temperature (55–65°C) were used to determine the optimum annealing temperature for inverse PCR. Primer sequences for inverse PCR were listed in Table S4. PCR products were subject to standard sequencing library preparation as ChIP‐seq and ChIRP‐seq. Sequencing reads with 5'end matching the forward inverse PCR primer sequence were selected and trimmed, remaining sequences containing TatI sites were mapped to mm9 assembly using Bowtie2 (Langmead & Salzberg, 2012) and the interaction regions are identified by fourSig (Williams et al., 2014).
4.17. RNA‐seq
Total RNAs were extracted using Trizol, followed by poly(A) selection (Ambion, 61,006) and library preparation using NEBNext Ultra II RNA Library Preparation Kit (NEB). Barcoded libraries were pooled at 10 pM and sequenced on Illumina HiSeq 1500 platform.
4.18. Statistical analysis
Statistical analysis of experimental data was calculated by the Student's t‐test, whereas * p < 0.05, ** p < 0.01, *** p < 0.001 and n.s. means not significant (p > =0.05).
AUTHOR CONTRIBUTIONS
K.K.H.S., H.S., and H.W. designed the experiments; K.K.H.S., Y.H., and S.Z. conducted the experiments; L.H. provided support on CRISPR/cas9 experiments; Y.L. provided support on RNA pulldown experiments; X.C. and Y.Q. provided support on cellular fractionation; X.C. provided support on RNA FISH; Y.H. analyzed the sequencing data; S.Z. contributed to ex vivo muscle fiber culture; M.H.S. provided resources for molecular experiments; K.K.H.S. and H.W. wrote the paper.
FUNDING INFORMATION
This work was supported by Collaborative Research Fund (CRF) from the Research Grants Council (RGC), University Grants Committee of the Hong Kong Special Administrative Region, China (Project Code: C6018‐19GF to H.W.); General Research Funds (GRF) from the Research Grants Council (RGC), University Grants Committee of the Hong Kong Special Administrative Region, China (Project Codes: 14106521, 14100620, 14115319, and 14106518 to H.W.; 14120420, 14116918, 14120619 and 14103522 to H.S.); Health and Medical Research Fund (HMRF) from Health Bureau of the Hong Kong Special Administrative Region, China (Project Code: 08190626 to H.W.); the research funds from Health@InnoHK program launched by Innovation Technology Commission, the Government of the Hong Kong Special Administrative Region, China; the National Natural Science Foundation of China (NSFC) to H.W. (Project codes: 82172436 and 31871304), NSFC/RGC Joint Research Scheme from the Research Grants Council, University Grants Committee of the Hong Kong Special Administrative Region to H.S. (Project code: N_CUHK 413/18); Theme‐based Research Scheme (TRS) from the Research Grants Council, University Grants Committee of the Hong Kong Special Administrative Region, China (Project code: T13‐602/21‐N); Area of Excellence Scheme (AoE) from the Research Grants Council, University Grants Committee of the Hong Kong Special Administrative Region, China (Project code: AoE/M‐402/20).
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
Supporting information
Figure S1
Figure S2
Figure S3
Table S1
Table S2
Table S3
Table S4
ACKNOWLEDGMENTS
We thank Prof. Zhenguo Wu, Prof. Danny CY Leung, and Prof. Tom HT Cheung for their kind suggestions on skeletal muscle satellite cells and epigenomics. Graphical abstract and Figure 6 were created with biorender.com with publication license AJ2412GJVI.
So, K. K. H. , Huang, Y. , Zhang, S. , Qiao, Y. , He, L. , Li, Y. , Chen, X. , Sham, M. H. , Sun, H. , & Wang, H. (2022). seRNA PAM controls skeletal muscle satellite cell proliferation and aging through trans regulation of Timp2 expression synergistically with Ddx5. Aging Cell, 21, e13673. 10.1111/acel.13673
Funding information
Focused Innovations Scheme: Scheme B, the Chinese University of Hong Kong, Grant/Award Number: 1907307; General Research Funds (GRF), Research Grants Council, University Grants Committee of the HKSAR, Grant/Award Number: 14100018, 14106117, 14115319, 14116918, 14120619 and 14133016; National Natural Science Foundation of China, Grant/Award Number: 31871304; NSFC/RGC Joint Research Scheme, Research Grants Council, University Grants Committee of the HKSAR, Grant/Award Number: N_CUHK 413/18
Karl Kam Hei So and Yile Huang these authors contribute equally to this paper as first authors.
Contributor Information
Hao Sun, Email: haosun@cuhk.edu.hk.
Huating Wang, Email: huating.wang@cuhk.edu.hk.
DATA AVAILABILITY STATEMENT
H3K27ac ChIP‐seq, Ddx5 ChIP‐seq, PAM ChIRP‐seq, and PAM 4C‐seq using in this study have been deposited in Gene Expression Omnibus database under the accession code (GSE180073). The data that support the findings of this study are available in Gene Expression Omnibus database, reference number (GSE121589, GSE132042, GSE134529, GSE189842).
REFERENCES
- Alessio, E. , Buson, L. , Chemello, F. , Peggion, C. , Grespi, F. , Martini, P. , Massimino, M. L. , Pacchioni, B. , Millino, C. , Romualdi, C. , Bertoli, A. , Scorrano, L. , Lanfranchi, G. , & Cagnin, S. (2019). Single cell analysis reveals the involvement of the long non‐coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Research, 47(4), 1653–1670. 10.1093/nar/gkz007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun, G. , Akhade, V. S. , Donakonda, S. , & Rao, M. R. (2012). Mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Molecular and Cellular Biology, 32(15), 3140–3152. 10.1128/MCB.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank‐Giwojna, A. , Postepska‐Igielska, A. , & Grummt, I. (2019). lncRNA KHPS1 activates a poised enhancer by triplex‐dependent recruitment of epigenomic regulators. Cell Reports, 26(11), 2904–2915 e2904. 10.1016/j.celrep.2019.02.059 [DOI] [PubMed] [Google Scholar]
- Bornemann, A. , & Schmalbruch, H. (1992). Desmin and vimentin in regenerating muscles. Muscle & Nerve, 15(1), 14–20. 10.1002/mus.880150104 [DOI] [PubMed] [Google Scholar]
- Caretti, G. , Schiltz, R. L. , Dilworth, F. J. , Di Padova, M. , Zhao, P. , Ogryzko, V. , Fuller‐Pace, F. V. , Hollman, E. P. , Tapscott, S. J. , & Sartorelli, V. (2006). The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Developmental Cell, 11(4), 547–560. 10.1016/j.devcel.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Chen, F. , Zhou, J. , Li, Y. , Zhao, Y. , Yuan, J. , Cao, Y. , Wang, L. , Zhang, Z. , Zhang, B. , Wang, C. C. , Cheung, T. H. , Wu, Z. , Wong, C. C. , Sun, H. , & Wang, H. (2019). YY1 regulates skeletal muscle regeneration through controlling metabolic reprogramming of satellite cells. The EMBO Journal, 38(10), e99727. 10.15252/embj.201899727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , He, L. , Zhao, Y. , Li, Y. , Zhang, S. , Sun, K. , So, K. , Chen, F. , Zhou, L. , Lu, L. , Wang, L. , Zhu, X. , Bao, X. , Esteban, M. A. , Nakagawa, S. , Prasanth, K. V. , Wu, Z. , Sun, H. , & Wang, H. (2017). Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov, 3, 17002. 10.1038/celldisc.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Yuan, J. , Xue, G. , Campanario, S. , Wang, D. , Wang, W. , Mou, X. , Liew, S. W. , Umar, M. I. , Isern, J. , Zhao, Y. , He, L. , Li, Y. , Mann, C. J. , Yu, X. , Wang, L. , Perdiguero, E. , Chen, W. , Xue, Y. , … Wang, H. (2021). Translational control by DHX36 binding to 5'UTR G‐quadruplex is essential for muscle stem‐cell regenerative functions. Nature Communications, 12(1), 5043. 10.1038/s41467-021-25170-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, N. , Zamudio, J. R. , Jong, R. M. , Soukup, D. , Resnick, R. , Sarma, K. , Ward, A. J. , Raj, A. , Lee, J. T. , Sharp, P. A. , & Jacks, T. (2014). LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Molecular Cell, 54(5), 777–790. 10.1016/j.molcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud, G. , Terrone, S. , & Bourgeois, C. F. (2018). Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Reports, 51(12), 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin, C. W. , & Holland, P. C. (1995). Synthesis and secretion of matrix‐degrading metalloproteases by human skeletal muscle satellite cells. Developmental Dynamics, 202(1), 91–99. 10.1002/aja.1002020109 [DOI] [PubMed] [Google Scholar]
- Hacisuleyman, E. , Goff, L. A. , Trapnell, C. , Williams, A. , Henao‐Mejia, J. , Sun, L. , McClanahan, P. , Hendrickson, D. G. , Sauvageau, M. , Kelley, D. R. , Morse, M. , Engreitz, J. , Lander, E. S. , Guttman, M. , Lodish, H. F. , Flavell, R. , Raj, A. , & Rinn, J. L. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nature Structural & Molecular Biology, 21(2), 198–206. 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Ding, Y. , Zhao, Y. , So, K. K. , Peng, X. L. , Li, Y. , Yuan, J. , He, Z. , Chen, X. , Sun, H. , & Wang, H. (2021). CRISPR/Cas9/AAV9‐mediated in vivo editing identifies MYC regulation of 3D genome in skeletal muscle stem cell. Stem Cell Reports, 16(10), 2442–2458. 10.1016/j.stemcr.2021.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, C. L. , Fei, T. , Chen, Y. , Li, T. , Gao, Y. , Wang, X. , Sun, T. , Sweeney, C. J. , Lee, G. S. , Chen, S. , Balk, S. P. , Liu, X. S. , Brown, M. , & Kantoff, P. W. (2014). Enhancer RNAs participate in androgen receptor‐driven looping that selectively enhances gene activation. Proceedings of the National Academy of Sciences of the United States of America, 111(20), 7319–7324. 10.1073/pnas.1324151111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte, M. , Guttman, M. , Feldser, D. , Garber, M. , Koziol, M. J. , Kenzelmann‐Broz, D. , Khalil, A. M. , Zuk, O. , Amit, I. , Rabani, M. , Attardi, L. D. , Regev, A. , Lander, E. S. , Jacks, T. , & Rinn, J. L. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell, 142(3), 409–419. 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski, J. P. , Lee, J. C. , Hwang, T. , Sunwoo, H. , Goldstein, J. M. , Groff, A. F. , Chang, N. P. , Mallard, W. , Williams, A. , Henao‐Meija, J. , Flavell, R. A. , Lee, J. T. , Gerhardinger, C. , Wagers, A. J. , & Rinn, J. L. (2019). The Firre locus produces a trans‐acting RNA molecule that functions in hematopoiesis. Nature Communications, 10(1), 5137. 10.1038/s41467-019-12970-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Chen, X. , Sun, H. , & Wang, H. (2018). Long non‐coding RNAs in the regulation of skeletal myogenesis and muscle diseases. Cancer Letters, 417, 58–64. 10.1016/j.canlet.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Yuan, J. , Chen, F. , Zhang, S. , Zhao, Y. , Chen, X. , Lu, L. , Zhou, L. , Chu, C. Y. , Sun, H. , & Wang, H. (2020). Long noncoding RNA SAM promotes myoblast proliferation through stabilizing Sugt1 and facilitating kinetochore assembly. Nature Communications, 11(1), 2725. 10.1038/s41467-020-16553-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluri, G. , & Jaworski, D. M. (2005). Regulation of TIMP‐2, MT1‐MMP, and MMP‐2 expression during C2C12 differentiation. Muscle & Nerve, 32(4), 492–499. 10.1002/mus.20383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, L. , Esteves de Lima, J. , Fabre, O. , Proux, C. , Legendre, R. , Szegedi, A. , Varet, H. , Ingerslev, L. R. , Barres, R. , Relaix, F. , & Mourikis, P. (2017). In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Reports, 21(7), 1982–1993. 10.1016/j.celrep.2017.10.080 [DOI] [PubMed] [Google Scholar]
- Mullin, N. K. , Mallipeddi, N. V. , Hamburg‐Shields, E. , Ibarra, B. , Khalil, A. M. , & Atit, R. P. (2017). Wnt/beta‐catenin signaling pathway regulates specific lncRNAs that impact dermal fibroblasts and skin fibrosis. Frontiers in Genetics, 8, 183. 10.3389/fgene.2017.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina, G. , Francois, S. , Regnault, B. , Czarny, B. , Dive, V. , Cumano, A. , Montarras, D. , & Buckingham, M. (2010). An adult tissue‐specific stem cell in its niche: A gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Research, 4(2), 77–91. 10.1016/j.scr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Peng, X. L. , So, K. K. , He, L. , Zhao, Y. , Zhou, J. , Li, Y. , Yao, M. , Xu, B. , Zhang, S. , Yao, H. , Hu, P. , Sun, H. , & Wang, H. (2017). MyoD‐ and FoxO3‐mediated hotspot interaction orchestrates super‐enhancer activity during myogenic differentiation. Nucleic Acids Research, 45(15), 8785–8805. 10.1093/nar/gkx488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamoun, H. , Lee, J. , Sun, Z. , Lu, H. , Ramsey, K. M. , Komives, E. A. , & Lauberth, S. M. (2018). RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nature Structural & Molecular Biology, 25(8), 687–697. 10.1038/s41594-018-0102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan, R. , Gayraud‐Morel, B. , Dumas, G. , Cimper, C. , Paisant, S. , Kelly, R. G. , & Tajbakhsh, S. (2009). Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Developmental Cell, 16(6), 810–821. 10.1016/j.devcel.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Sartorelli, V. , & Lauberth, S. M. (2020). Enhancer RNAs are an important regulatory layer of the epigenome. Nature Structural & Molecular Biology, 27(6), 521–528. 10.1038/s41594-020-0446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbina, A. , Larouche, J. , Fraczek, P. , Yang, B. A. , Brown, L. A. , Markworth, J. F. , Chung, C. H. , Khaliq, M. , de Silva, K., Choi, J. J. , Fallahi‐Sichani, M. , Chandrasekaran, S. , Jang, Y. C. , Brooks, S. V. , & Aguilar, C. A. (2020). Dissecting murine muscle stem cell aging through regeneration using integrative genomic analysis. Cell Reports, 32(4), 107964. 10.1016/j.celrep.2020.107964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So, K. K. , Peng, X. L. , Sun, H. , & Wang, H. (2017). Whole genome chromatin IP‐sequencing (ChIP‐seq) in skeletal muscle cells. Methods in Molecular Biology, 1668, 15–25. 10.1007/978-1-4939-7283-8_2 [DOI] [PubMed] [Google Scholar]
- Soleimani, V. D. , Punch, V. G. , Kawabe, Y. , Jones, A. E. , Palidwor, G. A. , Porter, C. J. , Cross, J. W. , Carvajal, J. J. , Kockx, C. E. , van IJcken, W. F. , Perkins, T. J. , Rigby, P. W. , Grosveld, F. , & Rudnicki, M. A. (2012). Transcriptional dominance of Pax7 in adult myogenesis is due to high‐affinity recognition of homeodomain motifs. Developmental Cell, 22(6), 1208–1220. 10.1016/j.devcel.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris, C. (2020). A single‐cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature, 583(7817), 590–595. 10.1038/s41586-020-2496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, P. F. , Dell'Orso, S. , Rodriguez, J. , Vivanco, K. O. , Ko, K. D. , Jiang, K. , Juan, A. H. , Sarshad, A. A. , Vian, L. , Tran, M. , Wangsa, D. , Wang, A. H. , Perovanovic, J. , Anastasakis, D. , Ralston, E. , Ried, T. , Sun, H. W. , Hafner, M. , Larson, D. R. , & Sartorelli, V. (2018). A muscle‐specific enhancer RNA mediates Cohesin recruitment and regulates transcription in trans. Molecular Cell, 71(1), 129–141 e128. 10.1016/j.molcel.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. X. , Dumont, N. A. , & Rudnicki, M. A. (2014). Muscle stem cells at a glance. Journal of Cell Science, 127(Pt 21), 4543–4548. 10.1242/jcs.151209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. L., Jr. , Starmer, J. , Mugford, J. W. , Calabrese, J. M. , Mieczkowski, P. , Yee, D. , & Magnuson, T. (2014). fourSig: A method for determining chromosomal interactions in 4C‐seq data. Nucleic Acids Research, 42(8), e68. 10.1093/nar/gku156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, H. , Kusumoto, D. , Hashimoto, H. , & Yuasa, S. (2020). Stem cell aging in skeletal muscle regeneration and disease. International Journal of Molecular Sciences, 21(5), 1830. 10.3390/ijms21051830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. W. , Wang, Y. , & Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nature Cell Biology, 21(5), 542–551. 10.1038/s41556-019-0311-8 [DOI] [PubMed] [Google Scholar]
- Young, D. A. , Gavrilov, S. , Pennington, C. J. , Nuttall, R. K. , Edwards, D. R. , Kitsis, R. N. , & Clark, I. M. (2004). Expression of metalloproteinases and inhibitors in the differentiation of P19CL6 cells into cardiac myocytes. Biochemical and Biophysical Research Communications, 322(3), 759–765. 10.1016/j.bbrc.2004.07.178 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Liu, Y. , & Zhang, H. (2021). Extracellular matrix: An important regulator of cell functions and skeletal muscle development. Cell & Bioscience, 11(1), 65. 10.1186/s13578-021-00579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, T. , Meyer, C. A. , Eeckhoute, J. , Johnson, D. S. , Bernstein, B. E. , Nusbaum, C. , Myers, R. M. , Brown, M. , Li, W. , & Liu, X. S. (2008). Model‐based analysis of ChIP‐seq (MACS). Genome Biology, 9(9), R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Ding, Y. , He, L. , Li, Y. , Chen, X. , Sun, H. , & Wang, H. (2021). Multiscale 3D genome reorganization during skeletal muscle stem cell lineage progression and muscle aging. bioRxiv, 12(20), 473464. 10.1101/2021.12.20.473464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhou, J. , He, L. , Li, Y. , Yuan, J. , Sun, K. , Chen, X. , Bao, X. , Esteban, M. A. , Sun, H. , & Wang, H. (2019). MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nature Communications, 10(1), 5787. 10.1038/s41467-019-13598-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Liu, T. , Li, Q. , & Li, J. (2021). Integrated analysis of long non‐coding RNAs (lncRNAs) and mRNA expression profiles identifies lncRNA PRKG1‐AS1 playing important roles in skeletal muscle aging. Aging (Albany NY), 13(11), 15044–15060. 10.18632/aging.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , So, K. K. , Li, Y. , Li, Y. , Yuan, J. , Ding, Y. , Chen, F. , Huang, Y. , Liu, J. , Lee, W. , Li, G. , Ju, Z. , Sun, H. , & Wang, H. (2019). Elevated H3K27ac in aged skeletal muscle leads to increase in extracellular matrix and fibrogenic conversion of muscle satellite cells. Aging Cell, 18(5), e12996. 10.1111/acel.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Sun, K. , Zhao, Y. , Zhang, S. , Wang, X. , Li, Y. , Lu, L. , Chen, X. , Chen, F. , Bao, X. , Zhu, X. , Wang, L. , Tang, L. Y. , Esteban, M. A. , Wang, C. C. , Jauch, R. , Sun, H. , & Wang, H. (2015). Linc‐YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nature Communications, 6, 10026. 10.1038/ncomms10026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Table S1
Table S2
Table S3
Table S4
Data Availability Statement
H3K27ac ChIP‐seq, Ddx5 ChIP‐seq, PAM ChIRP‐seq, and PAM 4C‐seq using in this study have been deposited in Gene Expression Omnibus database under the accession code (GSE180073). The data that support the findings of this study are available in Gene Expression Omnibus database, reference number (GSE121589, GSE132042, GSE134529, GSE189842).


