Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) helicase NSP13 plays a conserved role in the replication of coronaviruses and has been identified as an ideal target for the development of antiviral drugs against SARS-CoV-2. Here, we identify a novel NSP13 helicase inhibitor punicalagin (PUG) through high-throughput screening. Surface plasmon resonance (SPR)-based analysis and molecular docking calculation reveal that PUG directly binds NSP13 on the interface of domains 1A and 2A, with a KD value of 21.6 nM. Further biochemical and structural analyses suggest that PUG inhibits NSP13 on ATP hydrolysis and prevents it binding to DNA substrates. Finally, the antiviral studies show that PUG effectively suppresses the SARS-CoV-2 replication in A549-ACE2 and Vero cells, with EC50 values of 347 nM and 196 nM, respectively. Our work demonstrates the potential application of PUG in the treatment of coronavirus disease 2019 (COVID-19) and identifies an allosteric inhibition mechanism for future drug design targeting the viral helicases.
Keywords: SARS-CoV-2, NSP13, Helicase, Inhibitor, Punicalagin
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has placed a tremendous burden on the global health, economy and social life (Chan et al., 2020b; Chen et al., 2020b; Tan et al., 2020; Wang et al., 2020; Wu et al., 2020; Zhu et al., 2020). SARS-CoV-2 belongs to the Coronaviridae family in the order of Nidovirales. It is an enveloped positive-sense single-stranded RNA virus, and has about 30 kb RNA genome encoding two polyproteins (pp1a and pp1b) which can be proteolytically cleaved into 16 non-structural proteins (NSP1-16), including the RNA-dependent RNA polymerases (RdRp, NSP12), viral proteases (NSP3 and NSP5) and the helicases NSP13 (Chan et al., 2020a). These NSPs together form the machinery for viral replication and transcription (Ziebuhr, 2005).
NSP13 belongs to the helicase superfamily 1B (SF1B), and possesses a nucleotide triphosphate (NTP)-dependent 5ˊ to 3ˊ double-stranded DNA or RNA unwinding activity in vitro, although it mainly functions as an RNA helicase in vivo (Lee et al., 2010; Tanner et al., 2003). Structural studies have shown that NSP13 consists of five domains, an N-terminal zinc binding domain (ZBD), a helical “stalk” domain, a beta-barrel 1B domain, and two canonical RecA-type ATPase domains 1A and 2A that are responsible for nucleotide binding and hydrolysis (Jia et al., 2019; Newman et al., 2021; Yan et al., 2020). Biochemical and biophysical analyses suggest that NSP13 helicase forms a replication-transcription complex (RTC) with holo-RdRp, playing a pivotal role in RTC backtracking for proofreading and template switching during sub-genomic RNA transcription (Chen et al., 2020a). In addition to the helicase activity, NSP13 also has an RNA 5ˊ triphosphatase activity that may play a role in mRNA capping (Ivanov et al., 2004).
Given its conserved and indispensable role in the replication of coronaviruses, NSP13 helicase has been considered as a promising target for the development of antiviral drugs (Frick, 2003; Habtemariam et al., 2020). Despite decades of research efforts to identify chemical inhibitors against NSP13 helicase (Adedeji et al., 2012; Borgio et al., 2020; Newman et al., 2021; Tanner et al., 2005; Ugurel et al., 2020; White et al., 2020; Yu et al., 2012), to date, no NSP13 inhibitors have been approved for SARS-CoV-2, presumably due to limited efficacy. In the present study, we have performed a high throughput screening (HTS) for NSP13 inhibitors using the fluorescence resonance energy transfer (FRET)-based DNA unwinding assay, and identified that the natural product punicalagin (PUG) is an allosteric NSP13 inhibitor. It exhibits potent inhibitory efficacy against the replication of SARS-CoV-2 in Vero cells with an EC50 value of 68.56 nM, demonstrating an application potential in the treatment of COVID-19.
2. Materials and methods
2.1. Compounds and cell lines
All compounds were purchased from TopScience (Shanghai China). Vero cell line was obtained from the National Collection of Authenticated Cell Cultures, China.
2.2. Protein expression and purification
The full-length cDNA of SARS-CoV-2 NSP13 (GenBank accession no. QLL35519.1) was synthesized by GenScript Corporation (Nanjing, China) and subcloned into pET-28a bacterial expression vector with BamH1/EcoR1 restriction sites. The resulting construct was transformed into Rosetta (DE3) competent cells. Cells were cultured in Luria-Bertani (LB) medium at 37 °C until OD600 reached ∼0.6, then protein expression was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 18–24 h at 18 °C. Cells expressing His6-NSP13 were harvested, resuspended with lysis buffer (20 mM HEPES pH 7.2, 500 mM NaCl, 10 mM imidazole, 5% glycerol and 1 mM β-mercaptoethanol) and disrupted by French Pressure (Union Biotech, China). After centrifugation, the supernatant was collected and the His6-NSP13 proteins were pooled through Ni-NTA affinity column (Union-Biotech, China), and eluted with lysis buffer supplemented with 200 mM imidazole. The eluted proteins were dialyzed against SA buffer containing 20 mM HEPES pH 7.2, 100 mM NaCl, 5% glycerol and 1 mM β-mercaptoethanol, and further purified through Source 15S cation exchange column and Superdex 200 10/300 GL size exclusion column (GE Healthcare). The purified proteins in storage buffer (20 mM HEPES pH 7.2, 100 mM NaCl) were aliquoted and stored at −80 °C for future use.
2.3. DNA unwinding assay
The helicase activity of NSP13 was evaluated by both FRET- and gel-based assays, using the 5ˊ-flap DNA as substrates. The FRET-based assay was performed as previously described (Lee et al., 2010). Briefly, the substrate DNA was prepared by annealing the DNA strands 5ˊ-AATG-TCTGACGTAAAGCCTCTAAAATGTCTG-3ˊ-BHQ-2 and CY3-5ˊ-CAGACATTTTAGAGG-3ˊ. NSP13 proteins at various concentrations were mixed with 250 nM substrate DNA and 625 nM single stranded Trap-DNA (5ˊ-CCTCTAAAATGTCTG-3ˊ) in the unwinding buffer (50 mM HEPES pH 7.2, 20 mM NaCl, 4 mM MgCl2 and 0.1 mg/ml BSA), then the reaction was initiated by the addition of 2 mM ATP. The fluorescence intensity was measured at λex/λem = 542 nm/567 nm on a microplate reader (i3X, Molecular Devices). The gel-based DNA unwinding assay was conducted with similar reaction conditions but different substrate DNA and Trap-DNA (Table S1). Reactions were terminated by protease K containing solution. The subsequent reactants were analyzed by 10% polyacrylamide gel electrophoresis (PAGE) and visualized with the ChemDoc™ imaging system (Bio-Rad).
2.4. High throughput screening of NSP13 inhibitors
The high throughput screening of NSP13 inhibitors was performed based on the FRET-based DNA unwinding assay, using a library containing 7920 chemical compounds which were either FDA-approved drugs or bioactive compounds (TopSicence). For the primary screening, each compound at 20 μM was pre-incubated with 50 nM NSP13 at room temperature for 15 min, then the reaction was initiated by the addition of substrate mixture containing 250 nM substrate DNA, 625 nM trap ssDNA and 2 mM ATP. After incubation at 25 °C for 1 h, reactions were stopped and the fluorescence was recorded. Compounds with over 70% inhibitory activity against NSP13 compared to vehicle control were selected for a secondary screen, in which each compound was assayed at two different concentrations (2 μM and 20 μM) with three replicates. Active compounds from secondary screen were subjected to further biochemical and cell-based analyses.
2.5. Luciferase-coupled ATP assay
A luciferase-based luminescence assay was carried out to determine the ATPase activity of NSP13 according to the manufacture's protocols (Promega). Briefly, various concentrations of NSP13 proteins were mixed with 10 μM ATP in kinase buffer containing 50 mM HEPES pH 7.2, 150 mM NaCl, 10 mM MgCl2 and 0.5% Tween 20. After a 30 min-incubation at 30 °C, the reaction was stopped with Kinase-Glo reagent (Promega) and the luminescence intensity that reflected the amount of remaining ATP post reaction was measured on a microplate reader (SpectraMax-L, Molecular Devices). The ATPase activity of NSP13 was calculated based on the amount of ATP consumption.
2.6. Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was carried out to evaluate the effects of chemical inhibitors on the DNA binding affinity of NSP13. Briefly, 125 nM of FAM-labeled DNA (Table S1) was incubated with various concentrations of NSP13 in DNA binding buffer (20 mM HEPES, pH 7.2, 50 mM NaCl, 5 mM MgCl2, 5% glycerol) on ice for 60 min. Then, the protein-DNA mixtures were separated by native PAGE and visualized with the ChemDoc™ imaging system (Bio-Rad).
2.7. Surface plasmon resonance (SPR) assay
SPR-based ligand binding assay was performed to determine the binding affinity between NSP13 protein and its chemical inhibitors according to the manufacturés protocols (Cytiva). Briefly, NSP13 protein was coupled to a CM5 chip following the amine conjugation procedure. Compounds dissolved in DMSO were serially diluted with phosphate buffered saline (PBS) and injected onto the NSP13-coupled and reference channels, with binding and dissociation durations of 60 s and 180 s, respectively. Real-time binding signals were recorded and processed by the GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, USA).
2.8. Molecular docking
The molecular docking analysis for the binding of PUG to SARS-CoV-2 NSP13 was conducted with AutoDock 4.2 package (Morris et al., 2009) as previously described (He et al., 2022). Briefly, the starting structure of NSP13 was obtained from the Protein Data Bank (www.rcsb.org) (PDB ID: 7NNG) (Newman et al., 2021), and the geometry of the ligand was optimized using GAMESS (Schmidt et al., 1993). The docking results of 350 runs were grouped into clusters according to the binding conformations, and the average binding energy was analyzed using the default parameters implemented in AutoDockTools 1.5 (Morris et al., 2009). The dominant cluster containing the most abundant conformations and the lowest binding energy was selected for ligand binding analysis using the LIGPLOT (Wallace et al., 1995) and Pymol (http://www.pymol.org/) programs.
2.9. Cytotoxicity assay
The cytotoxicity of PUG on Vero cells and A549-ACE2 (A549 cells stably expressing ACE2) were evaluated by the Cell Counting Kit-8 (CCK-8, Meilunbio) according to the manufacture's protocols. Briefly, Vero cells were seeded in 96-well plates with 1 × 104 cells/well and further cultured for 18 h. Cells were treated with a series of concentrations of compounds or left untreated in the culture media for 24 h, followed by 4 h incubation in the media supplemented with 10% CCK-8. Viable cells were counted according to the absorbance at 450 nm.
2.10. In vitro evaluation of the antiviral activity of PUG against SARS-CoV-2
The antiviral activity of PUG was analyzed as previously described (Yu et al., 2021). Briefly, Vero or A549-ACE2 cells cultured in 96-well plates were pre-treated with various concentrations of compounds for 1 h, then the SARS-CoV-2 (BetaCoV/Shenzhen/SZTH-003/2020, GISAID No. EPI_ISL_406594) was added at a multiplicity of infection (MOI) of 0.02 and further incubated for 2 h at 37 °C. After incubation, cells were washed and further cultured with fresh media supplemented with indicated concentrations of compounds. Viral RNAs were extracted from the cell culture supernatant at 48 h post infection using the QIAamp RNA Viral Kit (Qiagen, Heiden, Germany), and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using a commercial kit targeting the ORF1b and N gene of SARS-CoV-2 (Mabsky Biotech Co., Ltd.). The resulting cells were collected for indirect immunofluorescence assay (IFA) experiment using the anti-nucleocapsid (N) antibody (Genetex).
To determine the effects of PUG on the viruses from different families, MDCK and A549 cells were infected with influenza A virus (IAV) H1N1 and human adenovirus (HAdV) type 7 (HAdV-7), and treated with various concentrations of PUG similarly as described above, then the viral yields were determined by qRT-PCR assay using commercial kits specific for IAV and HAdVs (Mabsky Biotech Co., Ltd.).
All the antiviral activity assays were carried out at BSL-3 (SARS-CoV-2) or BSL-2 (H1N1 and HAdV-7) facilities of the third People's Hospital of Shenzhen.
2.11. Data statistics
Values of half-maximal inhibitory concentrations (IC50), effective concentration (EC50) and cytotoxic concentration (CC50) were calculated by nonlinear regression using GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, USA). All data are expressed as the mean ± SD of triplicate assays.
3. Results
3.1. Identification of PUG as an NSP13 inhibitors
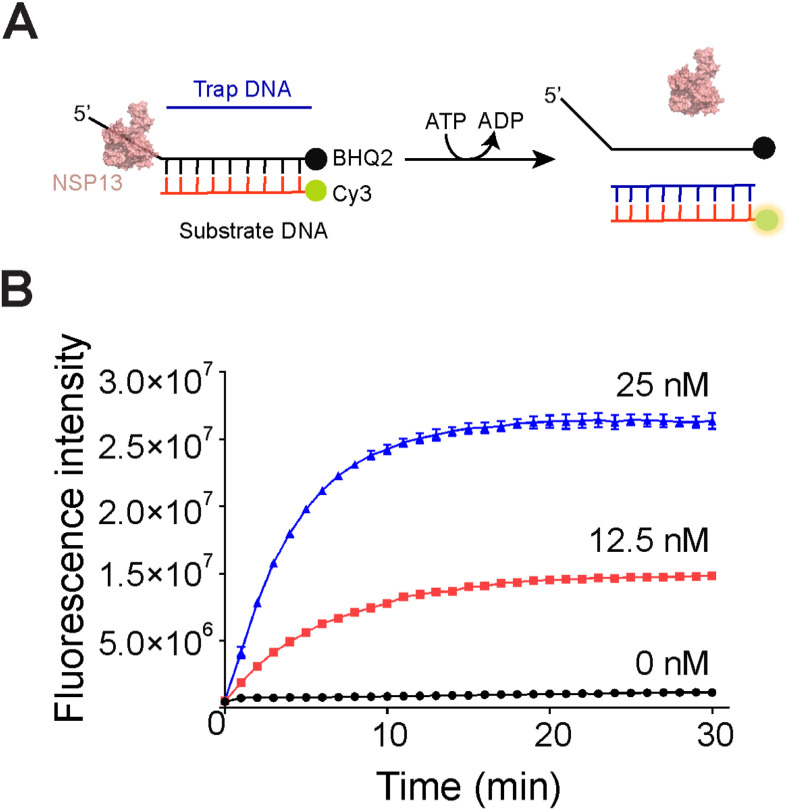
For high throughput screening of NSP13 inhibitors, the full-length NSP13 protein (aa.1–601) was expressed in E.coli cells and purified with a His6-tag at the N-terminus (Fig. S1A). Although NSP13 mainly functions as an RNA helicase in vivo, it possesses a 5ˊ to 3ˊ DNA unwinding activity (Lee et al., 2010). Therefore, to facilitate manipulation, the helicase activity of NSP13 was assayed by using a 5ˊ-flap DNA as substrate. We then tested if the purified recombinant NSP13 protein was active by native polyacrylamide gel analysis. NSP13 protein was incubated with FAM-labeled 5ˊ-flap DNA and a single-stranded Trap-DNA that is complementary to the short strand of 5ˊ-flap DNA (Table S1). In the presence of ATP, NSP13 could unwind the 5ˊ-flap DNA in a dose-dependent manner, resulting in the replacement of flap strand by the Trap-DNA (Fig. S1B). We next extended the DNA unwinding assay to the Cy3/BHQ2 FRET system, for which the two strands of 5ˊ-flap DNA were conjugated with Cy3 fluorescent and BHQ2 quencher probes, respectively. Once unwound, the Cy3-coupled strand would form a stable duplex with the Trap-DNA by replacing the BHQ2-coupled strand, leading to the increase of fluorescence intensity (Fig. 1 A). NSP13 dose-dependently stimulated the fluorescence intensity in an ATP-dependent manner (Fig. 1B). Together, these results thereby suggest that the recombinant NSP13 protein was active in DNA unwinding.
Fig. 1.
Establishment of HTS assay. (A) Schematic representation of FRET-based DNA unwinding assay. NSP13 protein is depicted as 3D structural model. The complete sequences for 5ˊ flap and Trap-DNA are listed in Table S1. (B) Real-time monitoring of DNA unwinding activity of various concentrations of NSP13. Values are means ± SD, n = 3.
We next carried out a high-throughput screening (HTS) of NSP13 inhibitors using the Cy3/BHQ2 FRET assay. In the primary screen with a library containing 7920 compounds, 191 compounds were identified with more than 70% inhibitory activity at 20 μM. These compounds were then subjected to a confirmation screen at two different concentrations (20 μM and 2 μM). As a result, 129 compounds exhibited more than 70% inhibitory activity against NSP13 at 20 μM, including Baicalein (Keum et al., 2013), Myricetin (Yu et al., 2012), Scutellarein (Yu et al., 2012) and Bismuth subsalicylat(Yang et al., 2007) that have previously been reported to have inhibitory effects on NSP13 of either SARS-CoV or SARS-CoV-2. Among those, we picked 11 compounds which exhibited over 85% inhibition at 2 μM for further evaluation of antiviral and cytotoxic activities (Table 1 ). After these procedures (summarized in Fig. S2A), we finally identified a natural product punicalagin, [2,3-hexahydroxydiphenoyl-gallagyl-D-glucose] (PUG, Fig. 2 A), as a novel potent NSP13 inhibitor, with an IC50 value of 427 nM (Fig. 2B). The inhibition potency was further confirmed by the gel-based DNA unwinding assay, in which the helicase activity of NSP13 was nearly abolished by PUG at the concentrations over 1 μM (Fig. 2C).
Table 1.
The half-maximal inhibitory concentrations (IC50) of compounds against NSP13 helicase activity.
| Name | PubChem CID | IC50(μM) |
|---|---|---|
| Rosmanol | 13966122 | 8.93 |
| Katacine | 6708803 | 5.98 |
| UMI-77 | 992586 | 4.38 |
| Zinc Orotate | 108934 | 4.12 |
| MC-Val-Cit-PABC-PNP | 57587849 | 2.45 |
| Silver sulfadiazine | 441244 | 2.11 |
| (−)-Gallocatechin gallate | 199472 | 1.34 |
| Tannic acid | 16129778 | 1.25 |
| Dyngo-4a | 136751773 | 0.63 |
| Rhodiosin | 76959646 | 0.48 |
| Punicalagin | 44584733 | 0.43 |
Fig. 2.
The inhibitory effects of PUG on NSP13. (A) The chemical structure of PUG. (B) Dose-inhibition curve for PUG in the FRET-based DNA unwinding assay. Values are means ± SD, n = 3. (C) Native polyacrylamide gel analysis of NSP13 helicase activity in the presence of increasing concentrations of PUG. Asterisk (*) indicates FAM-labeled DNA strands.
3.2. PUG directly binds NSP13 in the ATPase domain
To detect the direct binding between PUG and NSP13, we performed a surface plasmon resonance (SPR) analysis. PUG displayed fast association and slow dissociation rates, resulting in a KD of 21.62 nM (Fig. 3 A). To explore how PUG might interact with NSP13, we next tried to determine the structure of NPS13 bound to PUG but did not succeed. Alternatively, we then conducted a molecular docking calculation using the published crystal structure of SARS-CoV-2 NSP13 (PDB: 7NNG) (Newman et al., 2021). The results from 350 docking runs were grouped into six clusters according to the binding conformations. Among the six clusters, there was one dominant cluster accounting for approximately 70% of total docking conformations (Fig. S3). Furthermore, this cluster held the lowest average binding energy score as calculated by AutoDock Tools 1.5, indicating a robust structural stability (Fig. S3). Thus, a representative structure of this dominant cluster was selected for further ligand binding analysis. In the docking model, PUG docks onto the interface between domains 1A and 2A of NSP13 and partially occupies the binding site of NTP (Fig. 3B and C). The ellagic acid (EA) skeleton and 4,6-gallagyl groups protrude into the pocket and forms multiple hydrogen bonds with the catalytic residues of NSP13, including G285, T286 and G287 of Walker A, D374 and E375 of Walker B, and R443 (Sensor I). The hexahydroxydiphenoyl-D-glucose group, on the other side, stretches out of the pocket and forms van der Waals contacts with the backbone of α7 on domain 1A. In addition, it also accepts two hydrogen bonds from the carboxylate group of E319 (Fig. 3C). In agreement with these results, point mutation of E319A or E375A caused about 2-fold reduction of the inhibitory potency of PUG (Fig. 3D and E).
Fig. 3.
Direct interaction between PUG and NSP13. (A) SPR measurement of the binding affinity between PUG and NSP13. (B) The binding site of PUG on NSP13 as revealed by molecular docking calculation. NSP13 is shown in surface representation, with each domain labeled and shown in different colors. PUG is shown in stick. The ssRNA/DNA and NTP binding regions are indicated by dashed line and circle, respectively. (C) Detailed interaction between PUG and NSP13. NSP13 is shown in cartoon representation with the same color scheme in (B). The key residues involving in PUG binding are labeled and shown in sticks. Hydrogen bonds are indicated by black dashed lines. (D, E) Effects of E319A (D) and E375A (E) mutations on the inhibitory potency of PUG compared to the wild type (WT). Values are means ± SD, n = 3.
To further investigate the structure-activity relationship (SAR) of PUG, we next tested two structural related compounds, punicalin (PUL) and EA (Fig. 4 A). In both FRET- and gel-based assays (Fig. 4B and C), PUL showed slightly weak inhibitory effect than PUG against the DNA unwinding activity of NSP13 (IC50 ∼ 540 nM). Hence, while the hexahydroxydiphenoyl-D-glucose group plays a role in the binding of PUG to NSP13, the PUL moiety makes the dominant contribution. Moreover, EA was about six times weaker than PUG in NSP13 inhibition (IC50 ∼2.8 μΜ), demonstrating the importance of 4,6-gallagyl groups in the proximity of EA skeleton. Overall, the SAR results are highly consistent with the above docking model.
Fig. 4.
Comparisons of PUG with punicalin (PUL) and ellagic acid (EA) on the DNA unwinding activity of NSP13. (A) Structures of PUL and EA. (B) FRET-based measurement of NSP13 helicase activity in the presence of indicated compounds or DMSO (Veh). Values are means ± SD, n = 3. (C) Native polyacrylamide gel analysis of DNA unwinding by NSP13 in the presence of indicated compounds. Asterisk (*) indicates FAM-labeled DNA strands and pound (#) indicates the fluorescence emitted by the compound EA.
3.3. Mechanism of inhibition for PUG
We next probed the potential mechanism of inhibition for PUG. Because the molecular docking results suggest that PUG partially occupies with the binding site of NTP, we therefore tested if PUG has an effect on the ATPase activity of NSP13 through a luciferase-coupled ATP assay. In agreement with the results of DNA unwinding assays, PUG and PUL showed similar effects on the inhibition of ATP hydrolysis by NSP13, whereas EA showed much weaker inhibition (Fig. 5 A and Fig. S2B). Further enzyme kinetics study revealed that the value of Km increased along with increasing concentrations of PUG, whereas Vmax remained nearly unchanged (Fig. 5B). These results thereby suggest that PUG blocks NTP entry through direct competition with NTP. We next tested if PUG might have any effect on the interaction between NSP13 and its DNA substrate. To this end, a gel mobility-based protein/DNA binding experiment was carried out. As shown in Fig. 5C and D, NSP13 could retard the mobility of FAM-labeled 5ˊ flap DNA, indicating the formation of NSP13/DNA complex. Notably, pre-incubation of NSP13 with PUG, PUL or EA reduced the complex formation in a concentration-dependent manner. Again, PUG and PUL exhibited comparable inhibitory effects on NSP13/DNA binding, whereas EA had a much weaker inhibition. Different to ATP/PUG, there seemed no direct competition between DNA and PUG in NSP13 binding (Fig. 3B). However, a structural comparison between the APO (PDB ID: 7NIO) and AMP-PNP-bound (PDB ID: 7NNO) NSP13 revealed marked conformational changes in PUG-binding site and nucleic substrate-binding motifs (Fig. 5E). Thus, we proposed that PUG binding might keep NSP13 in an open conformation (APO form, in blue color), and allosterically induces conformational changes in the nucleic acid binding region, thereby preventing DNA from binding. Collectively, these results thus suggest that PUG inhibits NSP13 through blocking the binding of both ATP and nucleic acid substrates (Fig. 5F).
Fig. 5.
The inhibition mechanism of PUG against NSP13. (A) Determination of ATP hydrolysis by NSP13 in the presence of indicated compounds using the luciferase-coupled ATP assay. Values are means ± SD, n = 3. (B) The enzyme kinetics analysis of NSP13 in the presence of various concentrations of PUG. Values are means ± SD, n = 3. (C) Effects of various compounds on the DNA binding activity of NPS13 detected by EMSA assay. (D) Quantification of DNA binding in (C). (E) Comparison of the APO (blue, PDB ID: 7NIO) and AMP-PNP-bound (pink, PDB ID: 7NNO) structures of NSP13. Major conformational changes are indicated by red arrows. Dashed line indicates the ssRNA/DNA binding region. (F) Schematic representation summarizing the mechanism of action for NSP13 inhibition by PUG.
3.4. PUG potently inhibits SARS-CoV-2 replication in A549-ACE2 and Vero cells
NSP13 helicases is a key component of the machinery for viral replication and transcription (Ziebuhr, 2005). We investigated the effect of PUG, as a potent NSP13 inhibitor, on the replication of SARS-CoV-2 in A549-ACE2 and Vero cells. First, the CCK-8 cell viability assay showed that PUG had relatively low cytotoxicity, with half-maximal cytotoxic concentration (CC50) of 47 μM and 33 μM for A549-ACE2 (Fig. 6 A) and Vero (Fig. 6B) cells, respectively. For antiviral assay, cells pre-treated PUG were infected with SARS-CoV-2 for 2 h (MOI = 0.02), and further cultured in fresh media supplemented with PUG for 48 h. The SARS-CoV-2 viruses released into culture media were quantified by qRT-PCR analysis, while those retained in the cells were imaged by indirect immunofluorescence assay (IFA) using the anti-nucleocapsid (N) antibody. In the qRT-PCR assay, PUG potently inhibited SARS-CoV-2 replication with half maximal effective concentration (EC50) of 347 nM and 196 nM for A549-ACE2 (Fig. 6C) and Vero (Fig. 6D) cells, respectively. Consistent with the results of qRT-PCR assay, the cellular level of N protein was greatly reduced upon the treatment of PUG in IFA assay (Fig. 6E and F), indicating a suppression of viral protein production. In contrast, PUG did not show detectable antiviral effects against H1N1 and HAdV-7 viruses even at 10 mM (Fig. S4), indicating the specific inhibition of PUG against SARS-CoV-2. Taken together, these results thereby suggest that PUG could potently inhibit SARS-CoV-2 replication in A549-ACE2 and Vero cells.
Fig. 6.
Antiviral activity of PUG in Vero cells. (A, B) Cytotoxicity of PUG to A549-ACE2 (A) and Vero (B) cells measured by CCK-8 assay. Values are means ± SD, n = 3. (C, D) Dose-response curve of viral inhibition by PUG. A549-ACE2 (C) or Vero (D) cells were infected with SARS-COV-2 in the presence of various concentrations of PUG for 48 h, then the viral yields were determined by qRT-PCR assay. Values are means ± SD, n = 3. (E, F) Indirect immunofluorescence analysis of SARS-COV-2 virus in the cells upon the treatment of PUG at the indicated concentrations. A549-ACE2 (E) or Vero (F) Cells were immunostained 48 h post-infection for the SARS-CoV-2 nucleocapsid protein (red) and cell nuclei (blue).
4. Discussion
In this work, we describe a high throughput small-molecule screening targeting the helicase activity of NSP13, and identify PUG as a novel NSP13 inhibitor with potent antiviral activity. Our studies support that NSP13 helicase is a promising target for the development of antiviral drugs to fight against the current COVID-19 pandemic.
We show by molecular docking analysis that PUG directly binds NSP13 on the interface between 1A and 2A domains, and overlays with triphosphate-binding site of NTP, suggesting a direct competition mechanism. This binding model correlates well with our results of ATPase assay and SAR analysis with two structural related compounds, PUL and EA. In addition, we further show that PUG also prevents substrate DNA binding through an allosteric mechanism. Previous structural studies have shown that the 1A and 2A domains undergo large conformational changes in different nucleotide status (Newman et al., 2021). The ATPase domain adopts a closed conformation in AMP-PNP-bound state and a more open conformation in APO state (Newman et al., 2021). Such domain motion has been suggested to drive the movement of NSP13 along DNA/RNA in an inchworm manner (Newman et al., 2021). We note that accompanied by the conformational change of 1A and 2A domains, the PUG-binding pocket shrinks from 10 Å in width (defined by the distance between the Cα atoms of A316 and S539) in open conformation to about 6 Å in closed conformation which is unlikely to accommodate a PUG molecule. Moreover, because the 1A and 2A domains have been shown to interact with the phosphodiester backbone of RNA substrate (Yan et al., 2020), we thus propose that PUG binding may keep NSP13 in open conformation and induce a conformational change in domain 1A, thereby preventing NSP13 from binding to substrate DNA/RNA.
The natural compounds from plants have been widely used for medicinal and beneficial health applications, most of which are secondary metabolites produced by plants to defend against herbivore insects or microbial pathogens, including terpenoids, polyphenols, nitrogen and sulfur-containing compounds (Zaynab et al., 2018). As a kind of polyphenol, PUG is found primarily in the peel of pomegranate and possesses various pharmacological activities, such as anti-inflammatory (Huang et al., 2021; Lee et al., 2008), antioxidant (Moilanen et al., 2016; Seeram et al., 2005), antimicrobial (Rosas-Burgos et al., 2017; Taguri et al., 2004; Xu et al., 2017), anti-tumor (Adaramoye et al., 2017) and antiviral activities(Haidari et al., 2009; Howell and D'Souza, 2013). Here, we demonstrated that PUG could effectively protect human alveolar basal epithelial cells from infection by SARS-CoV-2, and has relatively low cytotoxicity (CC50 ∼ 47 μM). Furthermore, previous animal studies on PUG concluded that PUG is not toxic to rats after high-dose oral administration (6% PUG-containing diet) for 37 days (Cerda et al., 2003). Thus, PUG is a promising therapeutic agent for the treatment of current COVID-19 pandemic. In addition, since NSP13 is the most conserved nonstructural protein within the coronavirus family (Fig. S5), PUG might also have therapeutic potential for other coronaviruses, such as SARS-CoV and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), although it does not inhibit the viruses H1N1 and HAdV-7 from different families.
Recently, PUG has also been reported to have potential anti-SARS-CoV-2 effects. For example, PUG was identified as an allosteric inhibitor of SARS-CoV-2 3CLpro main protease (Du et al., 2021; Khalifa et al., 2020; Tito et al., 2021), which functions to cleave the polyproteins to generate nonstructural proteins. Moreover, PUG was also shown to interfere with the interaction between the virus spike glycoprotein and the cellular receptor ACE2 (Surucic et al., 2021; Tito et al., 2021), and thereby might have potential inhibitory effects on the virus entry into the host cells. Our results showed that PUG directly binds NSP13 with a KD value of 21.6 nM, inhibits the helicase activity of NSP13 (IC50 ∼ 430 nM). Given the high binding affinity between NSP13 and PUG, it appears that NSP13 might be the main target of PUG for its anti-SARS-CoV-2 effects. However, because these PUG-targeting proteins play different roles in the different stages of virus life cycle, we conclude that the potent anti-SARS-CoV-2 efficacy of PUG in the cells may be attributed to the multi-targeting effect.
As a hydrolyzable tannin, PUG can be hydrolyzed into PUL, EA or urolithins in the small intestine by the human gut microbiota (Cerda et al., 2005; Nunez-Sanchez et al., 2014). Thus, it has been proposed that the health benefits of tannin-containing foods intake may be attributed to their metabolites (Cerda et al., 2005; Nunez-Sanchez et al., 2014). Our SAR study suggests that PUL possesses a comparable inhibitory activity as PUG, whereas EA is six times weaker than PUG in the inhibition of NSP13 helicase activity. In light of this, further studies of drug delivery are needed to protect PUG or PUL from gastric degradation, for example, to administrate the drugs intravenously or encapsulate them with gelatin in nanoparticles as previously suggested (Li et al., 2011).
Author contributions
Zhonghui Lin designed and supervised the project. Lian Lu performed drug screen, cytotoxicity assay and all biochemistry experiments. Huiqiao Yao conducted molecular docking calculation under the supervision of Jinyu Li. Yanqun Wang contributed to the design of antiviral experiments. Yang Yang and Yun Peng performed cell-based antiviral assays and interpreted the data. Lian Lu and Zhonghui Lin wrote the manuscript. All authors analyzed the data and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the National Natural Science Foundation of China, 31971222 (Z. L.), 32170936 (Y. Y.) and 22173020 (J. L.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105389.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adaramoye O., Erguen B., Nitzsche B., Hopfner M., Jung K., Rabien A. Punicalagin, a polyphenol from pomegranate fruit, induces growth inhibition and apoptosis in human PC-3 and LNCaP cells. Chem. Biol. Interact. 2017;274:100–106. doi: 10.1016/j.cbi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgio J.F., Alsuwat H.S., Al Otaibi W.M., Ibrahim A.M., Almandil N.B., Al Asoom L.I., Salahuddin M., Kamaraj B., AbdulAzeez S. State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Arch. Med. Sci. 2020;16:508–518. doi: 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda B., Ceron J.J., Tomas-Barberan F.A., Espin J.C. Repeated oral administration of high doses of the pomegranate ellagitannin punicalagin to rats for 37 days is not toxic. J. Agric. Food Chem. 2003;51:3493–3501. doi: 10.1021/jf020842c. [DOI] [PubMed] [Google Scholar]
- Cerda B., Periago P., Espin J.C., Tomas-Barberan F.A. Identification of urolithin a as a metabolite produced by human colon microflora from ellagic acid and related compounds. J. Agric. Food Chem. 2005;53:5571–5576. doi: 10.1021/jf050384i. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573 e1513. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R., Cooper L., Chen Z., Lee H., Rong L., Cui Q. Discovery of chebulagic acid and punicalagin as novel allosteric inhibitors of SARS-CoV-2 3CL(pro) Antivir. Res. 2021;190 doi: 10.1016/j.antiviral.2021.105075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D.N. Helicases as antiviral drug targets. Drug News Perspect. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtemariam S., Nabavi S.F., Banach M., Berindan-Neagoe I., Sarkar K., Sil P.C., Nabavi S.M. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch. Med. Res. 2020;51:733–735. doi: 10.1016/j.arcmed.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidari M., Ali M., Ward Casscells S., 3rd, Madjid M. Pomegranate (Punica granatum) purified polyphenol extract inhibits influenza virus and has a synergistic effect with oseltamivir. Phytomedicine. 2009;16:1127–1136. doi: 10.1016/j.phymed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- He Y., Fu W., Du L., Yao H., Hua Z., Li J., Lin Z. Discovery of a novel Aurora B inhibitor GSK650394 with potent anticancer and anti-aspergillus fumigatus dual efficacies in vitro. J. Enzym. Inhib. Med. Chem. 2022;37:109–117. doi: 10.1080/14756366.2021.1975693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.B., D'Souza D.H. The pomegranate: effects on bacteria and viruses that influence human health. Evid. Based Complement. Alternat. Med. 2013;2013 doi: 10.1155/2013/606212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Wu K., Zeng S., Liu W., Cui T., Chen Z., Lin L., Chen D., Ouyang H. Punicalagin inhibited inflammation and migration of fibroblast-like synoviocytes through NF-kappaB pathway in the experimental study of rheumatoid arthritis. J. Inflamm. Res. 2021;14:1901–1913. doi: 10.2147/JIR.S302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum Y.-S.L., Jin Moo, Yu Mi-Sun, Chin Young-Won, Jeong Yong-Joo. Inhibition of SARS coronavirus helicase by Baicalein. Bull. Kor. Chem. Soc. 2013;34:2. [Google Scholar]
- Khalifa I., Zhu W., Mohammed H.H.H., Dutta K., Li C. Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CL(pro) : an in silico approach with 19 structural different hydrolysable tannins. J. Food Biochem. 2020 doi: 10.1111/jfbc.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.R., Kwon H.M., Park K., Oh S., Jeong Y.J., Kim D.E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.I., Kim B.S., Kim K.S., Lee S., Shin K.S., Lim J.S. Immune-suppressive activity of punicalagin via inhibition of NFAT activation. Biochem. Biophys. Res. Commun. 2008;371:799–803. doi: 10.1016/j.bbrc.2008.04.150. [DOI] [PubMed] [Google Scholar]
- Li Z., Percival S.S., Bonard S., Gu L. Fabrication of nanoparticles using partially purified pomegranate ellagitannins and gelatin and their apoptotic effects. Mol. Nutr. Food Res. 2011;55:1096–1103. doi: 10.1002/mnfr.201000528. [DOI] [PubMed] [Google Scholar]
- Moilanen J., Karonen M., Tahtinen P., Jacquet R., Quideau S., Salminen J.P. Biological activity of ellagitannins: effects as anti-oxidants, pro-oxidants and metal chelators. Phytochemistry. 2016;125:65–72. doi: 10.1016/j.phytochem.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.A., Douangamath A., Yadzani S., Yosaatmadja Y., Aimon A., Brandao-Neto J., Dunnett L., Gorrie-Stone T., Skyner R., Fearon D., Schapira M., von Delft F., Gileadi O. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez-Sanchez M.A., Garcia-Villalba R., Monedero-Saiz T., Garcia-Talavera N.V., Gomez-Sanchez M.B., Sanchez-Alvarez C., Garcia-Albert A.M., Rodriguez-Gil F.J., Ruiz-Marin M., Pastor-Quirante F.A., Martinez-Diaz F., Yanez-Gascon M.J., Gonzalez-Sarrias A., Tomas-Barberan F.A., Espin J.C. Targeted metabolic profiling of pomegranate polyphenols and urolithins in plasma, urine and colon tissues from colorectal cancer patients. Mol. Nutr. Food Res. 2014;58:1199–1211. doi: 10.1002/mnfr.201300931. [DOI] [PubMed] [Google Scholar]
- Rosas-Burgos E.C., Burgos-Hernandez A., Noguera-Artiaga L., Kacaniova M., Hernandez-Garcia F., Cardenas-Lopez J.L., Carbonell-Barrachina A.A. Antimicrobial activity of pomegranate peel extracts as affected by cultivar. J. Sci. Food Agric. 2017;97:802–810. doi: 10.1002/jsfa.7799. [DOI] [PubMed] [Google Scholar]
- Schmidt M.W., Baldridge K.K., Boatz J.A., Elbert S.T., Gordon M.S., Jensen J.H., Koseki S., Matsunaga N., Nguyen K.A., Su S.J., Windus T.L., Dupuis M., Montgomery J.A. General atomic and molecular electronic-structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- Seeram N.P., Adams L.S., Henning S.M., Niu Y., Zhang Y., Nair M.G., Heber D. In vitro antiproliferative, apoptotic and antioxidant activities of punicalagin, ellagic acid and a total pomegranate tannin extract are enhanced in combination with other polyphenols as found in pomegranate juice. J. Nutr. Biochem. 2005;16:360–367. doi: 10.1016/j.jnutbio.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Surucic R., Travar M., Petkovic M., Tubic B., Stojiljkovic M.P., Grabez M., Savikin K., Zdunic G., Skrbic R. Pomegranate peel extract polyphenols attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 Receptor: in silico and in vitro studies. Bioorg. Chem. 2021;114 doi: 10.1016/j.bioorg.2021.105145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguri T., Tanaka T., Kouno I. Antimicrobial activity of 10 different plant polyphenols against bacteria causing food-borne disease. Biol. Pharm. Bull. 2004;27:1965–1969. doi: 10.1248/bpb.27.1965. [DOI] [PubMed] [Google Scholar]
- Tan W., Zhao X., Ma X., Wang W., Niu P., Xu W., Gao G.F., Wu G. vol. 2. China CDC Wkly; 2020. pp. 61–62. (A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases - Wuhan, China 2019-2020). [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Zheng B.J., Zhou J., Watt R.M., Jiang J.Q., Wong K.L., Lin Y.P., Lu L.Y., He M.L., Kung H.F., Kesel A.J., Huang J.D. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem. Biol. 2005;12:303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito A., Colantuono A., Pirone L., Pedone E., Intartaglia D., Giamundo G., Conte I., Vitaglione P., Apone F. Pomegranate peel extract as an inhibitor of SARS-CoV-2 spike binding to human ACE2 receptor (in vitro): a promising Source of novel antiviral drugs. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.638187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel O.M., Mutlu O., Sariyer E., Kocer S., Ugurel E., Inci T.G., Ata O., Turgut-Balik D. Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13) Int. J. Biol. Macromol. 2020;163:1687–1696. doi: 10.1016/j.ijbiomac.2020.09.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.A., Lin W., Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase. J. Phys. Chem. Lett. 2020;11:9144–9151. doi: 10.1021/acs.jpclett.0c02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Shi C., Wu Q., Zheng Z., Liu P., Li G., Peng X., Xia X. Antimicrobial activity of punicalagin against Staphylococcus aureus and its effect on biofilm formation. Foodb. Pathog. Dis. 2017;14:282–287. doi: 10.1089/fpd.2016.2226. [DOI] [PubMed] [Google Scholar]
- Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., Wang Q., Rao Z., Lou Z. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Tanner J.A., Wang Z., Huang J.D., Zheng B.J., Zhu N., Sun H. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem. Commun. 2007:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G., Keum Y.S., Jeong Y.J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Sun G., Sui Y., Li H., Mai Y., Wang G., Zhang N., Bi Y., Gao G.F., Xu P., Jiang L., Yuan C., Yang Y., Huang M. Potent inhibition of severe acute respiratory syndrome coronavirus 2 by photosensitizers compounds. Dyes Pigments. 2021;194 doi: 10.1016/j.dyepig.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaynab M., Fatima M., Abbas S., Sharif Y., Umair M., Zafar M.H., Bahadar K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018;124:198–202. doi: 10.1016/j.micpath.2018.08.034. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.