Abstract
Acid resistance (AR) in Escherichia coli is defined as the ability to withstand an acid challenge of pH 2.5 or less and is a trait generally restricted to stationary-phase cells. Earlier reports described three AR systems in E. coli. In the present study, the genetics and control of these three systems have been more clearly defined. Expression of the first AR system (designated the oxidative or glucose-repressed AR system) was previously shown to require the alternative sigma factor RpoS. Consistent with glucose repression, this system also proved to be dependent in many situations on the cyclic AMP receptor protein. The second AR system required the addition of arginine during pH 2.5 acid challenge, the structural gene for arginine decarboxylase (adiA), and the regulator cysB, confirming earlier reports. The third AR system required glutamate for protection at pH 2.5, one of two genes encoding glutamate decarboxylase (gadA or gadB), and the gene encoding the putative glutamate:γ-aminobutyric acid antiporter (gadC). Only one of the two glutamate decarboxylases was needed for protection at pH 2.5. However, survival at pH 2 required both glutamate decarboxylase isozymes. Stationary phase and acid pH regulation of the gad genes proved separable. Stationary-phase induction of gadA and gadB required the alternative sigma factor ςS encoded by rpoS. However, acid induction of these enzymes, which was demonstrated to occur in exponential- and stationary-phase cells, proved to be ςS independent. Neither gad gene required the presence of volatile fatty acids for induction. The data also indicate that AR via the amino acid decarboxylase systems requires more than an inducible decarboxylase and antiporter. Another surprising finding was that the ςS-dependent oxidative system, originally thought to be acid induced, actually proved to be induced following entry into stationary phase regardless of the pH. However, an inhibitor produced at pH 8 somehow interferes with the activity of this system, giving the illusion of acid induction. The results also revealed that the AR system affording the most effective protection at pH 2 in complex medium (either Luria-Bertani broth or brain heart infusion broth plus 0.4% glucose) is the glutamate-dependent GAD system. Thus, E. coli possesses three overlapping acid survival systems whose various levels of control and differing requirements for activity ensure that at least one system will be available to protect the stationary-phase cell under naturally occurring acidic environments.
Acid resistance (AR) is perceived to be an important property of Escherichia coli, enabling the organism to survive gastric acidity and volatile fatty acids produced as a result of fermentation in the intestine (8, 9, 12). The ability to resist these acid stresses is believed to be necessary for this organism to colonize and establish a commensal relationship with mammalian hosts. In addition, the low infectious dose associated with enterohemorrhagic E. coli serotype O157:H7 is attributed to its acid-resistant nature (1–4, 18).
Under fasting conditions, the median stomach pH of healthy volunteers is around 2.0 (27). Detailed studies of AR mechanisms in E. coli have exposed three systems that can protect cells against pH 2 to 2.5 (11, 13, 14). The first is a glucose-repressed system induced in Luria-Bertani broth (LB) that is dependent on the alternative sigma factor ςS, encoded by the gene rpoS. The other two clearly defined systems are induced following growth in LB containing 0.4% glucose (LBG) or brain heart infusion broth (BHI) containing 0.4% glucose (BHIG). One system requires glutamic acid during acid challenge to survive pH 2 and is thought to utilize an inducible glutamate decarboxylase, and the other requires arginine and an inducible arginine decarboxylase encoded by adiA. All three systems were identified in stationary-phase cells. How the oxidative system protects cells against acid stress is unknown. However, the two decarboxylase systems are believed to consume protons during the decarboxylation of glutamate or arginine. The end products, γ-aminobutyric acid (GABA, formed from glutamate decarboxylase [GAD]) and agmatine (formed from arginine decarboxylase), are then transported out of the cell in exchange for new substrate. This transport process is catalyzed by specific antiporter systems, GadC for glutamate and an unknown antiporter for arginine. The result is that protons leaking into the cell during acid stress are consumed and excreted from the cell, thereby preventing the internal pH from decreasing to lethal levels. While this appears to be a simple strategy, it is now clear that inducible amino acid-dependent AR requires more than a decarboxylase and an antiporter.
The GAD system encompasses three genes. Two of these genes, gadA and gadB, encode highly homologous glutamate decarboxylase isoforms (25). The third gene, gadC, encodes a putative glutamate:GABA antiporter. The gadB and gadC genes form what appears to be an operon in which gadB is the first gene. In this study, we examined whether one or both decarboxylase genes are expressed, how each is regulated in response to pH and growth conditions, and whether both isoforms contribute to AR. We also discovered that cyclic AMP (cAMP) receptor protein (CRP) and cAMP are required specifically for the oxidative glucose-repressed system, analyzed the apparent acid induction of this system, and performed experiments to determine if there is a fourth system of AR.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was grown at 37°C in minimal E medium (28), minimal E medium containing 0.4% glucose (EG), LBG, or BHIG (the last four are complex media). Antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 25 μg/ml; streptomycin, 100 μg/ml; tetracycline, 20 μg/ml; and chloramphenicol, 30 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| EK181 | (JLS9300) MC4100 rpoS::Tn10 | J. L. Slonczewski |
| EK198 | (GNB7145Km) MC4100 adiA::MudI1734 (Km lac) | G. N. Bennett |
| EK221 | (MDA7K) MC4100 cysB trpB::Tn10 | G. N. Bennett |
| EK227 | E. coli K-12 wild type | AC Matin |
| EK239 | (JLS9391) MC4100 gadC::Tn10 | 11 |
| EK293 | S17-1 λpir | 5 |
| EK345 | (G1297) Δcya::Km Δcrp::Cm | S. Garges |
| EF333 | K12 gadC::Tn10 | EK239 × EK227 |
| EF337 | K12 adiA::MudI1734 (Km lac) gadC::Tn10 | EK198 × EF333 |
| EF362 | K12 rpoS::Tn10 | EK181 × EK227 |
| EF393 | K12 rpsL | EK227, spontaneous Strr |
| EF438 | K12 adiA::pKnock-Ap | |
| EF468 | EK293/pCF305 (gadC internal fragment in pRR10-250v) | This study |
| EF471 | EK293/pCF312 (gadA internal fragment in pRR10-250v) | This study |
| EF490 | K12 rpsL gadC::pRR10 (Ap) | EF393 × EF468 |
| EF491 | K12 gadC::pRR10 (Ap) | EK227 × EF490 |
| EF492 | K12 rpsL gadA::pRR10 (Ap) | EF393 × EF471 |
| EF493 | K12 gadA::pRR10 (Ap) | EK227 × EF492 |
| EF507 | K12 gadB::Km | EK227 × pCF325 |
| EF522 | K12 gadA::pRR10 (Ap) gadB::Km | EF493 × EF507 |
| EF529 | K12 Δcrp::Cm | EK345 × EK227 |
| EF530 | K12 Δcya::Km | EK345 × EK227 |
| EF531 | K12 gadA::pRR10 (Ap) rpoS::Tn10 | EF493 × EF362 |
| EF532 | K12 gadB::Km rpoS::Tn10 | EF507 × EF362 |
| EF547 | K12 gadC::pRR10 (Ap)/pCF348 (gadC+ Tcr) | EF491 × pCF348 |
| EF548 | K12 gadA::pRR10 gadB::Km/pCF348 (gadC+ Tcr) | EF522 × pCF348 |
| EF549 | K12 gadB::Km/pCF348 (gadC+ Tcr) | EF507 × pCF348 |
| Plasmids | ||
| pCR2.1 | Invitrogen | |
| pSK− | Stratagene | |
| pRR10-250v | 10, 19 | |
| pMAK705 | 10 | |
| pHP45ΩKm | 7 | |
| pHP45ΩTet | 7 | |
| pFP256 | pCR2.1 containing gadC+ controlled by lacP | |
| pCF301 | pSK− with internal fragment (oligonucleotides 105 and 106) of gadC | |
| pCF303 | pSK− with internal fragment (oligonucleotides 107 and 109) of gadA | |
| pCF305 | pRR10-250v (EcoRI-HindIII) with gadC (pCF301, EcoRI-HindIII) | |
| pCF312 | pRR10-250v (EcoRI-HindIII) with gadA (pCF303, EcoRI-HindIII) | |
| pCF319 | pCR2.1 with gadBC′ fragment (oligonucleotides 121 and 106) | |
| pCF324 | pMAK705 (XbaI-HindIII) with gadBC′ fragment (XbaI-HindIII) from pCF319 | |
| pCF325 | pCF324 gadBC′::Km | |
| pCF348 | pFP256 with Tet from pHP45ΩTet (gadC+ KmsTcr) |
Genetic and molecular techniques.
Transductions with P1vir, transformations with CaCl2, and conjugations were performed by standard methods (15). General DNA manipulations were carried out as described earlier (23).
AR assays.
Cells were grown overnight in one of several media, including LBG, LB buffered with either 100 mM morpholinepropanesulfonic acid (MOPS; pH 8) or 100 mM morpholinethanesulfonic acid (MES; pH 5.5), BHIG, and EG. The overnight (22-h) stationary-phase cultures were diluted 1:1,000 into prewarmed LB (pH 2.0) or EG (pH 2.5) supplemented where indicated with 1.5 mM glutamate or 0.6 mM arginine. For the glucose-repressed oxidative system, cells were grown overnight in LB-MES (pH 5.5) and diluted in unsupplemented EG (pH 2.5). The glutamate-dependent AR system was tested by growing cells overnight in LBG, which represses the RpoS-dependent oxidative system, and diluting the culture into EG (pH 2.5) supplemented with glutamate. The arginine-dependent AR system was tested by growing cells in BHIG overnight and diluting the culture into EG supplemented with arginine. Viable-cell counts were determined at 0, 2, and 4 h after the acid challenge (13). Controls (acid sensitive) for the oxidative system involved growth overnight in EG (pH 7) followed by dilution to 1 × 106 to 3 × 106 CFU/ml (1:1,000 dilution) in EG (pH 2.5). Acid challenge was carried out for 2 and 4 h at 37°C. Controls (acid sensitive) for the glutamate and arginine systems involved overnight growth in LBG or BHIG, respectively, followed by dilution to 1 × 106 to 3 × 106 CFU/ml in EG (pH 2.5) for 2 h at 37°C. Some experiments involving a challenge by dilution into LB (pH 2.0) were also performed.
Construction of insertion mutants.
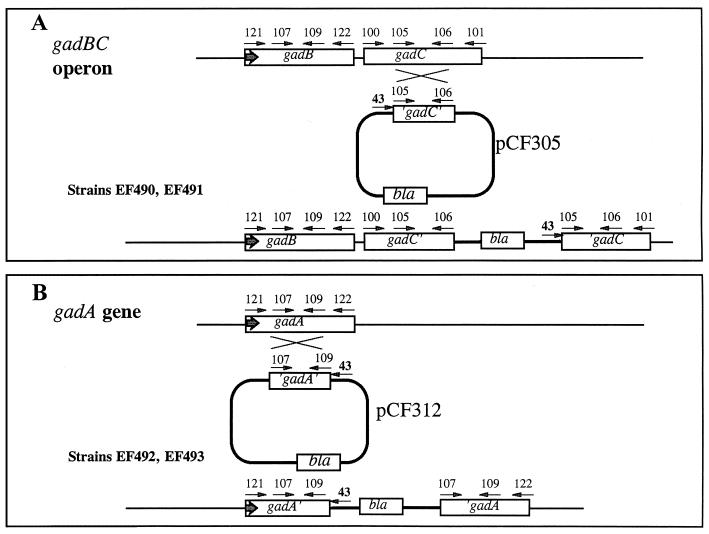
Internal fragments of the gene to be inactivated were amplified with primer pairs 107 plus 109 (gadB or -A) and 105 plus 106 (gadC) (Fig. 1 and Table 2). These fragments were cloned into EcoRV-digested pSK−. The resulting plasmids were digested with EcoRI and HindIII, and the fragments corresponding to the internal portions of the gene were purified and inserted in the vector pRR10-250v (19). The resulting plasmids, pCF305 (pRR10-250v containing an internal fragment of gadC) and pCF312 (pRR10-250v containing an internal fragment of gadA or -B), which replicate only in hosts expressing the π protein (pir), were transferred by conjugation into the nonpermissive host EF393. Since this strain does not possess the pir gene, the only way to obtain stable Ampr Strr exconjugants is by integration of this plasmid into the chromosome via homologous recombination (Fig. 1). These constructs were then transduced via P1 bacteriophage into EK227 (K-12). The proper chromosomal location of the integrated plasmid was confirmed by using PCR with primer pairs 43 plus 121 (gadA) and 43 plus 101 (gadC) (Fig. 1; Table 2). This technique was used to create gadA and gadC mutant strains EF493 and EF491, respectively.
FIG. 1.
Suicide mutagenesis using pRR10-250v vector. (A) Plasmid pCF305 (internal fragment of gadC inserted into pRR10-250v) was integrated into the chromosome of strain EF393 by homologous recombination at the gadC gene. The positions of the different primers used either to generate internal fragments or to verify the final insertion are shown. The sequences of these oligonucleotides are given in Table 2. The integration site was verified by PCR with primer pairs 43 and 101 (gadC), which produced a fragment of the predicted size, 1.4 kb. (B) The procedure was the same as in panel A, except that plasmid pCF312 containing an internal fragment of gadA was used. The integration site was verified by PCR with primer pairs 43 and 121 (gadA), which produced a fragment of the predicted size, 1.2 kb. Insertions that amplify with oligonucleotides 43 and 121 were distinguished as being within gadA or gadB by using oligonucleotides 121 and 106. If the insertion was in gadA, a PCR product was obtained which represented normal gadBC. If the insertion was in gadB, no PCR product was formed. Heavy arrows indicate the direction of transcription.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence | Strand | Location |
|---|---|---|---|
| 43 | GAGCGGATAACAATTTCACACAGG | − | Plasmid, lacZα |
| 100 | AATATGGCTACATCAGTACAGAC | + | Beginning gadC |
| 101 | TTAGTGTTTCTTGTCATTCATCAC | − | End gadC |
| 105 | CGATCTCATTTGGCTATCTGC | + | Internal gadC |
| 106 | CAGTTTCACCCCTTTACCACC | − | Internal gadC |
| 107 | GATCGACAAAGAAGAATATCC | + | Internal gadAB |
| 109 | CGAACGGTGCCAGGAAGCC | − | Internal gadAB |
| 121 | GGAGTTCGAAATGGACCAGAAG | + | Beginning gadAB |
| 122 | AGTTTCGGGTGATCGCTGAG | − | End gadAB |
The gadB::Km strain (EF507) was created by the technique described by Hamilton et al. (10). In this strain an internal fragment of the gadBC operon (from nucleotide 909 of gadB to nucleotide 201 of gadC) was replaced by a kanamycin resistance cassette. A PCR fragment corresponding to the entire gadB gene and a portion of gadC obtained by amplification with primers 121 plus 106 was cloned into pCR2.1. The resulting plasmid, pCF319, was digested with XbaI and HindIII, and the fragment corresponding to gadBC′ was gel purified and cloned into the temperature-sensitive plasmid pMAK705 (10), creating pCF324. This plasmid was cleaved with HincII, removing a fragment from nucleotide 909 of gadB to nucleotide 201 of gadC, which was replaced by a kanamycin resistance cassette from pHP45ΩKm (7). The Km cassette was removed from pHP45ΩKm with HindIII, and blunt ends were formed with Klenow polymerase and ligated to pCF324 missing the HincII fragment. The new plasmid, designated pCF325, was introduced by CaCl2 transformation into wild-type E. coli and plated at 42°C to force integration at gadB. Transformants were grown at 30°C to allow plasmid excision and screened for Kmr (retention of gadB::Km) and Aps (loss of the pMAK vector). Kmr Aps cells were checked by PCR with oligonucleotides 106 and 121 to ensure that proper insertion had taken place.
GAD assay.
Cells were grown in different media to stationary phase, and 1-ml samples were centrifuged and analyzed for GAD activity by resuspending cells in GAD reagent with or without Triton X-100 (3 ml/liter); the former condition measured internal GAD activity, while the absence of Triton X-100 permitted the analysis of glutamate transport via GadC (17). When decarboxylation occurs, the pH of the GAD reagent progressively increases, causing the indicator color to change from yellow to green to blue.
Western blot analysis.
Strains were grown at 37°C as indicated in medium containing the required antibiotics. At an optical density of 600 nm (OD600) of 0.5 (log phase) or 1.8 (stationary phase), cells were collected by centrifugation, resuspended at 1 OD600 unit/ml in 1× loading buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate SDS, 10% glycerol, 2.5% β-mercaptoethanol, 0.1% bromophenol blue). Each protein extract was fractionated on an SDS–10% polyacrylamide gel. After semidry electrophoretic transfer of proteins onto nitrocellulose membranes, the proteins were revealed by using rat primary antibodies (see below), monoclonal anti-rat secondary antibodies coupled to peroxidase (Sigma), and an ECL detection kit (Amersham). The relative amounts of protein were deduced from densitometric analysis of scanned underexposed films.
Preparation of anti-GAD antibody.
Crude E. coli GAD protein was purchased from Sigma (no. G-3757) and purified via two-step fast protein liquid chromatography with an HR10 Mono Q (Pharmacia, Uppsala, Sweden) ion-exchange column followed by size exclusion chromatography on a Superose 12 column (Pharmacia). The protein was dissolved in running buffer A (50 mM Tris-HCl [pH 8.0] and 10% glycerol containing 0.1 mM phenylmethysulfonyl fluoride [Sigma], 2.8 μg of Trasylol [Bayer, Mississauga, Ontario, Canada] per ml, and 0.2 mM pyridoxal phosphate [Sigma]) and was eluted from the ion-exchange column by an NaCl step gradient. Fractions obtained between 0.20 and 0.24 M NaCl were judged to be highly enriched in the 52-kDa GAD protein, as determined by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. These GAD-enriched fractions were concentrated (Centriprep 30; Amicon, Beverly, Mass.) and loaded onto the size exclusion column in buffer A–0.3 M NaCl. GAD-enriched fractions were again identified as above, pooled, and concentrated in a Centricon 30 exchanged with phosphate-buffered saline. The highly purified GAD protein was shown to be enzymatically active in a [14C]glutamate conversion assay. Fast protein liquid chromatography-purified GAD was emulsified in R700 RIBI adjuvant (RIBI Immunochem Research Inc., Hamilton, Mont.), and Sprague Dawley (bios) rats were immunized with 0.1 mg of immunogen. The animals received four such immunizations before the final sera were harvested on day 134, 10 days after the last boost.
RESULTS
Glutamate-dependent AR requires the gadC product.
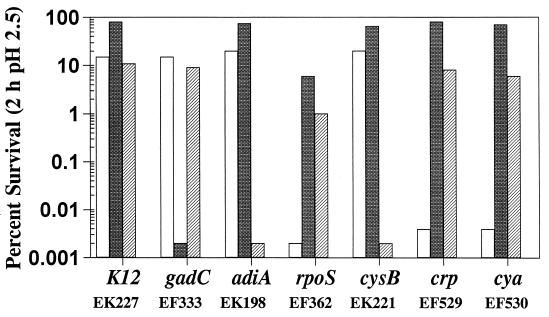
Figure 2 shows the effects of several mutations on the three AR systems. It should be kept in mind that growth in LBG represses the oxidative system, which then allows measurement of the amino acid-dependent AR systems. We were previously able to attribute arginine-dependent AR to arginine decarboxylase (13), the product of the adiA gene (26), and oxidative AR to the alternative sigma factor ςS, encoded by rpoS (14). These results, along with those indicating that CysB specifically controls the arginine-dependent system, are confirmed in Fig. 2. A gadC::Tn10 mutant was originally isolated as being acid sensitive in complex media and was shown to be defective in glutamate-dependent AR (11). We predicted that the gadC insertion would selectively remove glutamate-dependent AR and tested the ability of this mutant to induce the three AR systems. As shown in Fig. 2, the gadC mutant lacked only the glutamate-dependent system. Qualitative assay of GAD activity with permeabilized cells revealed that the gadC mutant possessed considerable GAD activity (data not shown). Thus, the gadC defect affected only the putative antiporter system that exchanges extracellular glutamate for intracellular GABA and did not alter GAD activity proper. However, GadC was required to detect GAD activity in whole-cell GAD assays performed without Triton X-100 to permeabilize the membrane, again consistent with the predicted role of GadC in this system (data not shown).
FIG. 2.
Genetic dissection of AR. Various mutants of E. coli K-12 were tested for the three AR systems. The test for the oxidative system (open bars) involved overnight growth in LB-MES (pH 5.5) followed by dilution to 1 × 106 to 3 × 106 CFU per ml in EG (pH 2.5). Survival (viable counts) was measured after 2 h at 37°C. The glutamate (shaded bars) and arginine (hatched bars) systems required overnight LBG (glutamate system) or BHIG (arginine system) cultures, which were diluted to 1 × 106 to 3 × 106 CFU/ml in EG (pH 2.5) containing 1.5 mM glutamate and 0.6 mM arginine, respectively. Percent survival is calculated as the number of CFU per milliliter remaining after the acid treatment divided by the initial CFU per milliliter at time zero. Initial cell densities ranged from 1 × 106 to 3 × 106 CFU/ml. Experiments were repeated two or three times. Variations were within 50% of the stated value.
Participation of GAD isoforms in glutamate-dependent AR.
A full understanding of how E. coli utilizes GAD for AR requires that potential roles for the two isoforms of GAD be explored. Suicide mutagenesis was used to generate mutations in the gadA, gadB, and gadC genes. The construction of these insertions and their confirmation by PCR analysis is described in Materials and Methods. AR was monitored for the gadA and gadB mutants, and because gadB and gadC form an operon, a gadB mutant containing a gadC+ plasmid (pCF348) was also tested. The gadA (EF493) and the gadB/pCF348 gadC+ (EF549) strains possessed normal glutamate-dependent AR (Table 3), indicating that both GADs can function in AR. The gadB single mutant (EF507) was acid sensitive, as expected, because of a polar effect on gadC. A gadA gadB/pCF348 gadC+ (EF548) mutant, however, exhibited no glutamate-dependent AR due to the lack of both GAD isoforms (Table 3). The results indicate that both gadA and gadB genes are expressed and that either GAD isoform will provide glutamate-dependent AR at pH 2.5.
TABLE 3.
Glutamate AR of the different gad mutants
| Strain | Genotype | Adaptation mediuma | % Survival inb:
|
|
|---|---|---|---|---|
| Glutamate mediuma | Control mediuma | |||
| EK227 | Wild type | LBG | 71 | <0.005 |
| BHIG | 90 | <0.006 | ||
| EF491 | gadC::pRR10 | LBG | <0.008 | <0.005 |
| BHIG | <0.007 | <0.006 | ||
| EF493 | gadA::pRR10 | LBG | 42 | <0.006 |
| BHIG | 66 | <0.006 | ||
| EF507 | gadB::Km | LBG | <0.005 | <0.005 |
| BHIG | <0.006 | <0.007 | ||
| EF549 | gadB::Km/pGadC(Tc) | BHIGc | 52 | <0.004 |
| EF547 | gadC::pRR10/pGadC(Tc) | BHIGc | 30 | <0.004 |
| EF548 | gadA gadB/pGadC(Tc) | BHIGc | <0.005 | <0.005 |
Adaptation was carried out overnight either in LBG or in BHIG at 30°C. Acid challenge was performed by diluting 1:1,000 the overnight culture in EG at pH 2.5 with or without (control) 1.5 mM glutamate.
The percentage of survival was determined by measuring the number of surviving cells after 4 h of acid treatment.
Isopropyl-β-d-thiogalactopyranoside was added at 0.5 mM.
Stationary-phase and acidic pH regulation of GadA and GadB.
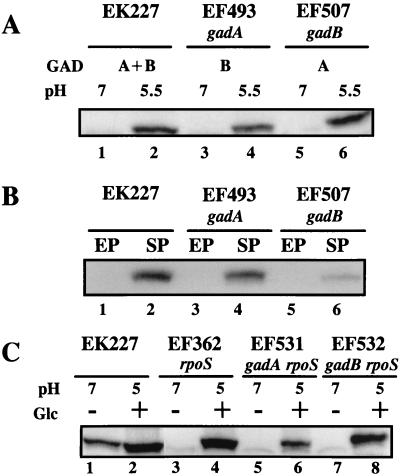
As shown in Table 4, glutamate-dependent AR was absent in exponential-phase cells grown in LBG or in EG (pH 5.5) but was present in stationary-phase cells grown in LBG, minimal EG (pH 5.5), or minimal E medium plus glycerol (pH 5.5). Western blot analysis was used to examine the regulation of GAD synthesis and determine if GAD production correlated with AR. The contributions of GadA and GadB to total GAD levels were determined by using the gadB and gadA mutants, respectively. Exponential-phase cells grown at pH 7 in EG possessed very small amounts of both GadA and GadB (Fig. 3A, lanes 1, 3, and 5). Surprisingly, exponential-phase cells grown in minimal medium (pH 5.5) strongly induced both GadA and GadB, even though these cells could not mount a glutamate-dependent AR (lanes 2, 4, and 6). Thus, some factor in addition to GAD may be required for glutamate-dependent acid resistance. The finding that log-phase cells grown at pH 5.5 possess significant levels of GAD protein but show no GAD activity in the whole-cell qualitative GAD assay (see Materials and Methods) supports this hypothesis.
TABLE 4.
Glutamate-dependent AR in log-phase and stationary-phase cells
| Growth phase | % Survival after adaptation ina:
|
||
|---|---|---|---|
| EG (pH 5.5) | E medium plus glycerol (pH 5.5) | LBG | |
| Exponential | 0.07 | NDb | 0.08 |
| Stationary | 70 | 74 | 76 |
EK227 was grown to exponential phase (OD600 = 0.5) or stationary phase (overnight or for 22 h) in the media indicated. Glycerol was added at 0.4%. Cells were diluted 1:1,000 into EG containing 1.5 mM glutamate (pH 2.5). Survival was measured after 4 h.
ND, not determined.
FIG. 3.
Western blot analysis of GadA versus GadB expression. (A) Exponential-phase cells (OD600 = 0.5) were grown in EG (pH 7 or 5.5). GAD isoforms (A and/or B) present in each strain are indicated. (B) Exponential-phase (OD600 = 0.4) and stationary-phase (18 h) cells were grown in LB-MOPS (pH 8). (C) Cells were grown to early stationary phase (OD600 = 1.8) in unbuffered LB either with or without 0.4% glucose as a means of changing the pH. Final pH without glucose was approximately pH 7. The final pH with glucose was approximately pH 5. Equivalent amounts of protein were loaded in each lane and probed with anti-GAD antibody.
Figure 3B illustrates that stationary-phase induction of GAD occurred even at pH 8 in LB and revealed differential expression of the gad genes. Stationary phase clearly induced GadB to a much higher level than it induced GadA (Fig. 3B, lanes 4 and 6). Very little GAD protein was observed in exponential-phase cells grown at high pH.
Dual control of gad expression by stationary phase and acidic pH was revealed in that stationary-phase cells grown at low pH possessed even higher levels of GAD protein than did stationary phase cells grown at pH 7 (Fig. 3C, lanes 1 and 2). The results in Fig. 3A and B indicate that both genes are induced by low pH and that gadB is more strongly induced by the stationary phase than is gadA.
As expected, the level of glutamate-dependent AR correlated with the amount of GAD in stationary-phase cells, with 80% survival (Fig. 3C, lane 2) and 32% survival (Fig. 3C, lane 1) after acid challenge in EG (pH 2.5) supplemented with glutamate (Fig. 3C, lane 2, has more GAD than Fig. 3C, lane 1). It is interesting that although acid induced GAD in log-phase cells (Fig. 3A, lane 2) to levels similar to what was observed in pH 7 stationary-phase cells (Fig. 3C, lane 1), the log-phase cells were acid sensitive. These results, along with those in Table 4, suggest that glutamate-dependent AR requires adequate levels of GAD as well as the stationary-phase induction of other unknown factors.
Effect of RpoS on glutamate-dependent AR and GAD.
Figure 2 illustrates that an rpoS mutant possessed somewhat reduced glutamate-dependent AR. Since the product of rpoS is an alternative sigma factor that plays an important role in the expression of many stationary-phase-inducible genes, we examined an rpoS mutant for effects on GAD production. Figure 3C indicates that ςS was required for both gadA and gadB expression in pH 7 stationary-phase cells (lanes 1, 3, 5, and 7) but that this alternative sigma factor was not required for expression of either gene during growth at pH 5 in LBG (lanes 2, 4, 6, and 8). This suggests that there are at least two levels of control over gadA and gadBC expression, one in which ςS mediates stationary-phase induction and a second, unknown regulatory circuit that senses the pH. It is interesting that GadB expression was not induced as strongly as GadA by acid pH (Fig. 3A, lane 4 versus 6, respectively). This was confirmed by removing stationary-phase control of gadB and gadA by using an rpoS mutation (Fig. 3C, lanes 6 and 8). In this situation, acid exposure increased GadA production more than it did GadB production. In contrast, GadB was more strongly induced than GadA by stationary phase (Fig. 3B, lanes 4 and 6).
Effects of CRP on AR.
Previous work has shown that the RpoS-dependent oxidative system is repressed by glucose (13). Because of this observation, we investigated whether the expression of this system was also dependent on cAMP and CRP and whether CRP had any effect on the glutamate-dependent system. Figure 2 (bar sets 6 and 7) illustrates that AR via the oxidative system was severely diminished in cya and crp mutants, indicating that both cAMP and CRP are required. The glutamate-dependent system, however, was unaffected. Thus, it appears that rpoS-dependent systems play several roles in AR. Some rpoS-dependent genes collaborate with CRP-dependent genes in the oxidative AR system. In addition, the gad genes themselves are partially regulated by RpoS (see above).
Glutamine inhibits the glucose-repressed, oxidative AR system in a gadC mutant.
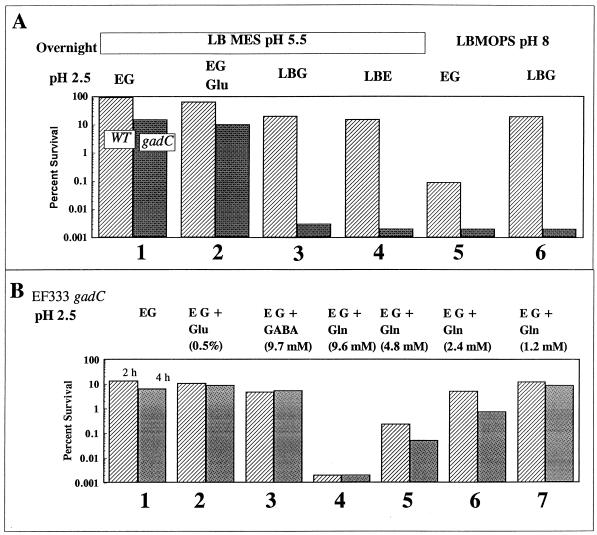
Figure 2 and Fig. 4A (bar sets 1 and 2) reveal that the gadC mutant (shaded bars in Fig. 4A) exhibited a normal oxidative AR when cells were grown in LB-MES (pH 5.5) and acid challenged (pH 2.5) in minimal EG (Fig. 4A, bar set 1). However, the same mutant did not survive when challenged in LBG (pH 2.5), indicating that not only was the glutamate system not functioning but also the oxidative system, which should be present, was not functioning (bar set 3). Adding E salts to LB did not alter this phenotype (bar set 4). After adding various amino acid pools to minimal EG (pH 2.5), we traced the acid-sensitive phenotype to the presence of glutamine in the medium (Fig. 4B, bar set 4). Glutamine had no effect on the AR of a gadC+ cell (data not shown). The minimum concentration of glutamine required to have an effect was 2.4 mM (bar set 6). Glutamate and GABA did not have any effect (bar sets 2 and 3, respectively), suggesting that glutamine might be transported into the cell and converted intracellularly to glutamate and then to GABA via GAD. However, since the GadC transporter was not present, GABA would accumulate intracellularly. It was predicted that GABA accumulation was responsible for the extreme sensitivity to acid in the presence of glutamine. However, a gadA gadB gadC mutant which will not form GABA because it lacks all GAD activity remained acid sensitive in the presence of glutamine (data not shown). Thus, GABA is not the sensitizing molecule.
FIG. 4.
Effect of glutamine on AR. (A) Cells (EK227 [hatched bars] and EF333 [gadC; shaded bars]) were grown overnight in either LB-MES (pH 5.5) or LB-MOPS (pH 8) as indicated and then diluted 1:1,000 into various pH 2.5 media, also as indicated. Survival was measured after 2 h. (B) EF333 (gadC) was grown overnight in LB-MES (pH 5.5) and diluted 1:1,000 into pH 2.5 EG containing various amino acids. Survival was measured at 2 h (hatched bars) and 4 h (shaded bars).
The lack of GadC might also cause excessive accumulation of glutamate, which may be responsible for making the cell acid sensitive. Glutamine transport coupled to deamination could result in glutamate accumulation in a gadC mutant if GadC ordinarily serves as an outlet for excess glutamate. To provide evidence that the glutamine-dependent acid sensitivity of a gadC mutant involved glutamate accumulation rather than the lack of GadC, the acid resistance of a gadA gadB/pCF348 gadC+ mutant (EF548) was tested in EG (pH 2.5) (conditions for oxidative system) with or without glutamine. This mutant lacks GAD but retains GadC antiporter. If glutamate accumulation was involved in sensitization to acid by glutamine, then a lack of GAD, even with GadC present, should also produce glutamine-dependent acid sensitivity. The results confirmed this hypothesis. As with the gadC mutant, the gadA gadB/pgadC+ strain was acid sensitive in the presence of glutamine (data not shown). These results suggest that strategies designed to inhibit GadC antiporter activity or GAD activity in vivo may have a detrimental effect on cell survival in a variety of acidic environments if glutamine is present.
Glutamate is an activator of oxidative (RpoS-dependent) AR.
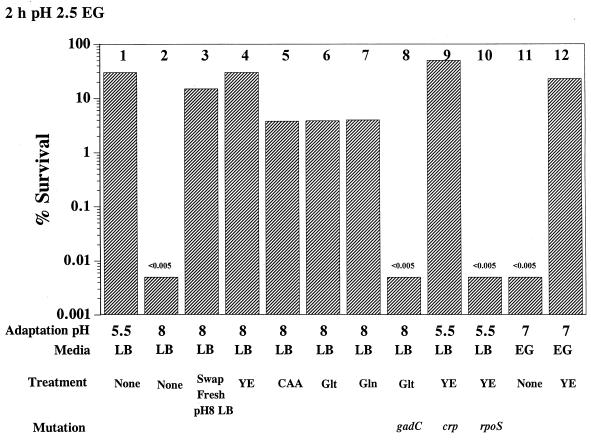
As noted above, the RpoS-dependent, glucose-repressed oxidative AR system is induced when cells are grown to stationary phase in complex media at pH 5.5. The system is not evident in cells grown in LB at pH 8 even though RpoS levels are high in these cells (data not shown). To determine if the pH requirement involves the presence of extracellular signaling molecules or perhaps the degradation of preexisting signaling molecules, a series of medium exchange experiments were designed. The results (Fig. 5, bars 2 and 3) show that cells grown to stationary phase at pH 8 actually possessed the oxidative AR system but could use this system only if they were first resuspended briefly in fresh LB. This suggested two possibilities; either an inhibitory compound was synthesized at pH 8 or a compound required to activate the system was degraded at pH 8 but not at pH 5.5. To address the activator possibility, fresh Casamino Acids and yeast extract were added separately to spent media at pH 8 to see if some compound present in one of these ingredients would rescue oxidative AR in pH 8-grown cells. The brief addition of Casamino Acids or yeast extract to overnight cultures before testing AR at pH 2.5 had a dramatic effect, restoring AR to levels obtained following overnight growth in LB (pH 5.5) (Fig. 5, bar 2 versus bars 4 and 5). The addition of chloramphenicol before the addition of yeast extract did not prevent this phenomenon, indicating that this activator does not trigger new gene expression but somehow activates a preexisting system (data not shown). Through the addition of various amino acid pools, we discovered that either glutamate (5.9 mM) or glutamine (9.6 mM) was the amino acid responsible for activating the system (bars 6 and 7). It is interesting that neither glutamate nor glutamine (not shown) activated the oxidative AR system in a gadC mutant (bar 8), suggesting a link between the glutamate and oxidative AR systems.
FIG. 5.
Demonstration of an activator of oxidative AR in yeast extract. Cells were grown overnight (18 h) in the media and at the pHs indicated. Treatments included transferring cells to fresh LB (pH 8). In addition, Casamino Acids (CAA; 0.5%), yeast extract (YE; 0.5%), glutamate (Glt; 5.9 mM), or glutamine (Gln; 9.6 mM) was added directly to overnight cultures immediately prior to acid challenge (less than 1-min exposure). Acid challenge involved diluting cells to 1 × 106 to 3 × 106 CFU/ml in EG (pH 2.5). The results shown indicate survival for 2 h at pH 2.5. The gadC, crp, and rpoS mutants used were EF491, EF529, and EF362, respectively.
Yeast extract (or glutamate) also restored acid resistance in a crp mutant and in glucose-repressed wild-type cells (Fig. 5, bar 9, and Fig. 6, bar 7). Thus, CRP-dependent genes are not required for the oxidative system if glutamate or glutamine is provided. However, yeast extract did not rescue an rpoS mutant (Fig. 5, bar 10), indicating that glutamate or glutamine is required for the RpoS-dependent subsystem. The activator model also explains why induction of this system seemed to require growth in complex medium. Cells grown to stationary phase in EG at any pH failed to exhibit this system (13). However, the brief addition of yeast extract to stationary-phase cells grown in EG activated the RpoS-dependent system (Fig. 5, bars 11 and 12).
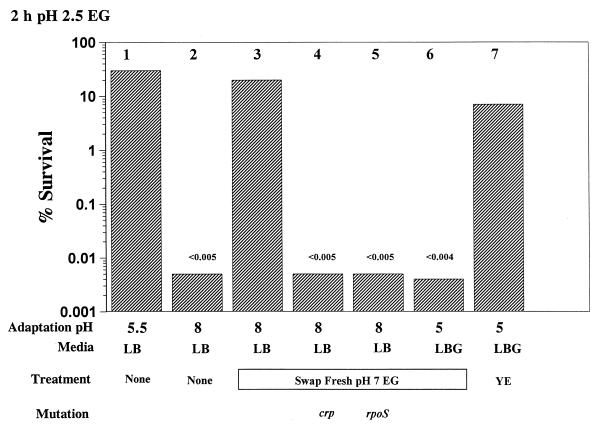
FIG. 6.
Demonstration of an inhibitor of oxidative AR produced at pH 8. Cells were grown overnight (18 h) in the media and at the pHs indicated. Treatments included transferring cells to fresh EG (pH 7). In addition, yeast extract (YE; 0.5%) was added directly to overnight cultures immediately prior to acid challenge (less than 1-min exposure). Acid challenge involved diluting cells to 1 × 106 to 3 × 106 CFU/ml in EG (pH 2.5). Results shown indicate survival for 2 h at pH 2.5.
Growth at pH 8 produces an inhibitor of RpoS-dependent oxidative AR.
Not only would adding yeast extract (glutamate) to pH 8-grown cells activate the RpoS-dependent system, but also it was discovered that transferring pH 8-grown cells to fresh EG (pH 7) activated this system (Fig. 6, bar 3). This suggested that pH 8-grown cells synthesize and secrete an inhibitor of the oxidative AR system. This inhibitor could be removed by washing cells but not by boiling or by treatment with pronase (data not shown). Dilution did not rescue AR in the rpoS or crp mutants (Fig. 6, bars 5 and 4) or in glucose-repressed wild-type cells (Fig. 6, bar 6). Consequently, we cannot determine whether the inhibitor affects the RpoS or CRP subsystems. The data does suggest that since yeast extract (or glutamate) rescued AR of the crp mutant (Fig. 5, bar 9), glutamate does not activate the CRP subsystem. It now seems that the apparent pH control of the oxidative AR system involves the synthesis at pH 8 of an inhibitor.
Is there a fourth AR system exhibited by LBG- or BHIG-grown cells?
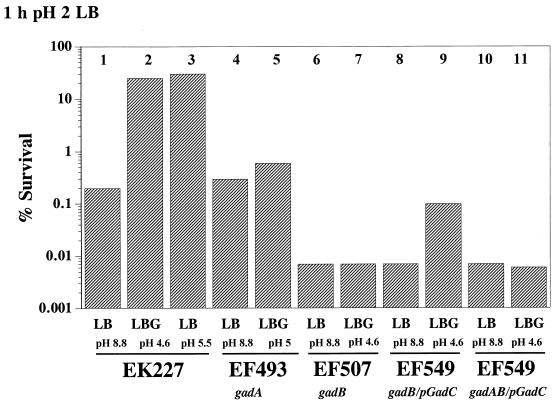
Diez-Gonzalez et al. (6) have provided evidence that LBG-grown commensal strains of E. coli exhibit a more pronounced AR at pH 2.0 than do LB-grown cells. They attributed the difference to the low pH resulting from the fermentative production of volatile fatty acids in LBG. However, of the three AR systems clearly defined in E. coli, the oxidative system is repressed in LBG, the arginine-dependent system is poorly effective at pH 2 (14), and GAD is produced at high levels in both LBG and LB. Therefore, it was not apparent why LBG-grown cells would have dramatically better AR at pH 2.0 than would LB-grown cells. Could a fourth AR system exist that is revealed only at pH 2? To address this question, we repeated the experiment with wild-type and gadA gadB/pCF348 (gadC+) strains of E. coli K-12. The results, shown in Fig. 7 (bars 1 and 2), illustrate that LBG-grown cultures of K-12 survive pH 2 challenge much better than LB-grown cultures do, confirming the previous results (6). The data (Fig. 7, bar 5) further revealed that gadA mutants grown in LBG survived a pH 2 challenge very poorly, indicating that the primary AR system involved in this medium is glutamate dependent. In addition, bars 5 and 9 illustrate that the loss of either GadA or GadB severely diminished the AR at pH 2. The reason why LBG-grown cells have higher AR than do LB-grown cells was suggested by the Western blot results shown in Fig. 3C (lanes 1 and 2). LBG-grown cells contained considerably more GAD than did LB-grown cells, a result consistent with the theory that the difference in AR lies at the level of the GAD proteins produced. The observation that AR at pH 2.5 needed only one of the two GAD genes (Table 3) but AR at pH 2 required both GAD genes (Fig. 7) also helps explain why E. coli has retained both copies of this gene.
FIG. 7.
Effect of gad mutations on AR at pH 2 in LB. Cells were grown to stationary phase in the media indicated (see the legend to Fig. 1). The final pH before acid challenge is also shown. Cells were diluted to approximately 106 CFU/ml in acid challenge media. The results presented indicate survival after 1 h at pH 2 in LB.
Table 5 shows the results of testing the AR at pH 2 of cells grown in BHI or BHIG. We have observed that growth in BHI enhances AR with all three known systems, and we wondered whether a new system might be exposed by using this medium. The data revealed that although a gadC mutant (EF333) grown in BHIG exhibited residual LB AR at pH 2, that resistance was due to arginine decarboxylase, since an adiA gadC mutant (EF337) failed to show any significant AR. Mutant cells lacking GadC that were grown in BHI did not exhibit the glucose-repressed oxidative AR system because of the glutamine-dependent acid sensitivity noted above. These results indicate that there are only three systems of AR in K-12 that are capable of protecting cells at pH 2 and that the most effective system involves GAD. Similar to K-12, a gadA mutant of O157:H7 (ATCC 43895) possessed very little residual AR at pH 2 (data not shown). This suggests that O157:H7 does not possess a unique AR system.
TABLE 5.
Adaptation in BHI and effects of gad and adiA mutations on AR at pH 2
| Strain | Pertinent genotype | % Survival after adaptationa
|
|
|---|---|---|---|
| BHI | BHIG | ||
| EK227 | Wild type | 20 | 65 |
| EK198 | adiA | 70 | 55 |
| EF333 | gadC | 0.003 | 0.8 |
| EF337 | adiA gadC | 0.002 | 0.003 |
Cells were grown to stationary phase in the media indicated. The cells were diluted to 1 × 106 to 3 × 106 CFU/ml in acid challenge media (pH 2 LB). The results presented indicate survival after 1 h.
Are VFAs required to induce AR?
Diez-Gonzalez et al. also suggested that the production of volatile fatty acids (VFAs) in LBG is responsible for inducing AR at pH 2 (6). The implication was that low pH alone will not induce this level of resistance. This hypothesis was tested by growing cells to stationary phase in LB buffered to pH 5.5 rather than in LBG. The results indicate that pH alone, in the absence of VFA production, will induce resistance to pH 2 (Fig. 7, bar 3). Thus, the production of VFAs is not required for AR at pH 2.
DISCUSSION
The AR properties of pathogenic E. coli strains such as O157:H7 contribute to the low infectious dosages of these organisms by allowing small numbers to pass the stomach acidity barrier. Because of this, AR is considered to be an important virulence factor. Our laboratory has shown that commensal and O157 strains of E. coli possess three stationary-phase-dependent AR systems that protect cells under extremely acidic conditions (13, 14). The present results confirm that there are only three discernible systems. The oxidative or glucose-repressed AR system is controlled by the alternative sigma factor ςS, cAMP, and CRP. This RpoS-dependent system is activated by glutamate or glutamine during adaptation, but these amino acids have no effect if they are added during challenge. The arginine-dependent system is under positive control by the CysB protein, while the most effective AR system (GAD) is regulated partly by RpoS, but its main control by acidic pH occurs through an as yet unknown regulator.
Both decarboxylase systems are clearly induced by acidic conditions, although the gadBC and gadA genes are partially induced simply by entry into stationary phase, as is the RpoS-dependent system. Induction of GAD by acidic pH was previously shown (29); however, the present study illustrates that both gadA and gadB, encoding the two isoforms of GAD, are regulated in similar fashions, with ςS controlling stationary-phase induction but not controlling induction. Expression of gadA is affected predominantly by acidic pH whereas expression of gadB is affected primarily by entry into stationary phase. The data also indicate that both GAD enzymes are required for optimal AR at pH 2, since the loss of either one makes cells much more sensitive to pH 2 but has little obvious effect at pH 2.5. A surprising finding was that induction of the GAD enzymes alone is insufficient for glutamate-dependent AR, since log-phase cells grown at pH 5.5 produced large amounts of GadA and GadB but failed to survive pH 2.5. This suggests that additional genes required to survive pH stress must be induced during stationary phase. The participation of other genes was predicted because when one closely examines how the decarboxylase systems should work, the systems have the appearance of futile proton cycles. For example, glutamic acid at pH 2.5 outside the cell is protonated. After transport, it will deprotonate because of the higher intracellular pH. The subsequent consumption of protons by decarboxylation would seem to just compensate for the protons released when glutamate enters the cell, so that there would be no net removal of intracellular protons. Thus, while it is clear that inducible GAD and arginine decarboxylase systems play important roles in E. coli AR, the way in which they actually accomplish AR is unknown.
A GAD-dependent AR system has also been reported in Lactococcus lactis. The system in this organism is controlled by a gene called gadR (24) and is induced by low pH, glutamate, and chloride ions. However, the addition of NaCl to E. coli cultures did not affect the induction of GAD, suggesting a distinct difference in regulation.
New insights were also obtained into the control of the oxidative AR system by pH and complex media. This system ordinarily requires both RpoS- and CRP-dependent gene products and is glucose repressed. It was not clear why induction of this system required growth in complex medium at pH 5.5, since RpoS levels were high in cells grown to stationary phase in LB or minimal medium at pH 5.5 or 8 (data not shown). In addition, neither CRP nor cAMP levels appear to change dramatically under these conditions. The results presented indicate that pH control involves the synthesis of an inhibitor made at pH 8 but not pH 5.5. This inhibitor appears to interfere with the activity rather than with the synthesis of the system. Its influence is removed by washing pH 8 cells prior to acid challenge. The identity of the inhibitor is under investigation.
The complex-medium requirement for inducing the RpoS-dependent oxidative AR system is due to the presence of glutamate and glutamine in yeast extract. Glutamate and glutamine appear to activate a preformed RpoS-dependent system that is produced simply due to entry into stationary phase. Growth at pH 5.5 or 8 will partially consume glutamate and glutamine, making the oxidative system more reliant on CRP, although growth at pH 8 also appears to produce an inhibitor of this system. The fact that glutamate and glutamine can restore AR to a crp mutant or to glucose-repressed cells supports the theory that the RpoS-dependent system can stand alone in protecting cells against pH stress. CRP is not essential for AR. Because CRP dependence can be circumvented by adding excess activator (glutamate) of the RpoS-dependent system but not by removing the inhibitor, it is conceivable that a CRP-dependent pathway may contribute to intracellular glutamate synthesis. In the absence of glucose, this putative pathway might produce enough glutamate to allow the RpoS-dependent AR system to function. Once the oxidative system is induced and active, the way in which it protects cells at pH 2 in minimal media remains a mystery. One could envision intracellular glutamate serving as a counterion for K+ entering the cell due to an RpoS-dependent K+/H+ antiport system, but there is no evidence for this.
Another question raised about RpoS-dependent AR as a result of these studies is why glutamine masks the system in a gadC mutant. It must be more than coincidence that activation of the oxidative system in cells grown at pH 8 requires the addition of glutamate or glutamine and that this activation involves GadC. It has been shown that acidic environments will cause a decrease in intracellular glutamate levels, so that one might predict that the RpoS-dependent AR system may depend upon this (16, 20). However, too much intracellular glutamate might be deleterious. If this is true, excess glutamate might be siphoned from the cell via the GAD system. In this model, exogenous glutamine would be transported into the cell and converted to glutamate via glutaminase. If the GadC antiporter is required to release excess glutamate but is missing, the resulting glutamate accumulation might be too high for AR purposes. Ordinarily, the GadC antiporter is not required for the RpoS-dependent AR system. It is only necessary when glutamine is added at the challenge pH (pH 2.5) or when the CRP subsystem is nonfunctional and extra glutamate is needed from the medium during adaptation. GadC may only provide an overflow for excess internal glutamate or a conduit to build glutamate concentrations if they are not high enough in the cell. It should be noted that although evidence indicating roles for medium components affecting log-phase acid habituation were published previously, those log-phase systems are clearly different from the more efficient stationary-phase systems presented here (21, 22).
One question concerning the role of AR in the virulence of pathogenic E. coli involves how these systems are induced in nature. Recent evidence reported by Diez-Gonzales et al. (6) supports the hypothesis that AR can be induced in E. coli growing in the intestinal tracts of cattle. However, it was suggested that VFAs present in the intestinal contents induced a system that may be unique in protecting cells at pH 2. The results presented here indicate that lowered pH alone (no VFAs) can induce a system suitable to protect cells to pH 2 and that this system is the glutamate-dependent system. It is still possible that VFAs contribute to induction in the intestinal environment by decreasing pH but that the VFAs are not essential.
An earlier report from our laboratory has shown that, once induced, all three AR systems in commensal or O157:H7 strains will persist for at least 1 month under refrigerated conditions (14). Thus, O157 strains with AR systems induced by growth in the gastrointestinal tracts of cattle do not need to grow in contaminated foods prior to ingestion in order to infect at low infectious doses. Based on these findings, we are currently investigating strategies designed to subvert AR in these organisms.
ACKNOWLEDGMENTS
We thank Alexey Atrazhev for help in purifying GAD, as well as M. Spector, M. Moreno, and S. Price for many helpful discussions. Technical assistance by Neelam Ahmad is also gratefully acknowledged.
This work was supported by grant 97-35201-4751 from the U.S. Department of Agriculture.
REFERENCES
- 1.Arnold K W, Kaspar C W. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin M M, Datta A R. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheville A M, Arnold K W, Buchrieser C, Cheng C-M, Kaspar C W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conner D E, Kotrola J S. Growth and survival of Escherichia coli O157:H7 under acidic conditions. Appl Environ Microbiol. 1995;61:382–385. doi: 10.1128/aem.61.1.382-385.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Lorenzo V C, Timmis K M. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 6.Diez-Gonzalez F, Callaway T R, Kizoulis M G, Russell J B. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science. 1998;281:1666–1668. doi: 10.1126/science.281.5383.1666. [DOI] [PubMed] [Google Scholar]
- 7.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 8.Giannella R A, Broitman S A, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut. 1972;13:251–256. doi: 10.1136/gut.13.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giannella R A, Broitman S A, Zamcheck N. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Ann Intern Med. 1973;78:271–276. doi: 10.7326/0003-4819-78-2-271. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersh B M, Farooq F T, Barstad D N, Blankenshorn D L, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewetson J T. The bacteriology of certain parts of the alimentary canal and of the inflammatory processes arising therefrom. Br Med J. 1904;2:1457–1460. doi: 10.1136/bmj.2.2291.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Smith M P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 16.Ogahara T, Ohno M, Takayama M, Igarashi K, Kobayashi H. Accumulation of glutamate by osmotically stressed Escherichia coli is dependent on pH. J Bacteriol. 1995;177:5987–5990. doi: 10.1128/jb.177.20.5987-5990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice E W, Johnson C H, Dunnigan M E, Reasoner D J. Rapid glutamate decarboxylase assay for detection of Escherichia coli. Appl Environ Microbiol. 1993;59:4347–4349. doi: 10.1128/aem.59.12.4347-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 19.Roberts R C, Burioni R, Helinski D R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990;172:6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roe A J, McLaggan D, Davidson I, O’Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowbury R J. An assessment of environmental factors influencing acid tolerance and sensitivity in Escherichia coli, Salmonella spp. and other enterobacteria. Lett Appl Microbiol. 1995;20:333–337. doi: 10.1111/j.1472-765x.1995.tb01314.x. [DOI] [PubMed] [Google Scholar]
- 22.Rowbury R J, Hussain N H, Goodson M. Extracellular proteins and other components as obligate intermediates in the induction of a range of acid tolerance and sensitisation responses in Escherichia coli. FEMS Microbiol Lett. 1998;166:283–288. doi: 10.1111/j.1574-6968.1998.tb13902.x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sanders J W, Leenhouts K, Burghoorn J, Brands J R, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith D K, Kassam T, Singh B, Elliott J F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stim K P, Bennett G N. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdu E, Viani F, Armstrong D, Fraser R, Siegrist H H, Pignatelli B, Idstrom J P, Cederberg C, Blum A L, Fried M. Effect of omeprazole on intragastric bacterial counts, nitrates, nitrites, and N-nitroso compounds. Gut. 1994;35:455–460. doi: 10.1136/gut.35.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 29.Yoshida T, Yamashino T, Ueguchi C, Mizuno T. Expression of the Escherichia coli dimorphic glutamic acid decarboxylases is regulated by the nucleoid protein H-NS. Biosci Biotechnol Biochem. 1993;57:1568–1569. doi: 10.1271/bbb.57.1568. [DOI] [PubMed] [Google Scholar]