Abstract
Salmonella typhi is the only species of Salmonella which grows exclusively in humans, in whom it causes enteric typhoid fever. Strains of S. typhi show very little variation in electrophoretic types, restriction fragment length polymorphisms, cell envelope proteins, and intervening sequences, but the same strains are very heterogeneous for ribotypes which are detected with the restriction endonuclease PstI. In addition, the genome of S. typhi has been proven to undergo genomic rearrangement due to homologous recombination between the seven copies of rrn genes. The relationship between ribotype heterogeneity and genomic rearrangement was investigated. Strains of S. typhi which belong to 23 different genome types were analyzed by ribotyping. A limited number of ribotypes were found within the same genome type group; e.g., most strains of genome type 3 belonged to only two different ribotypes, which result from recombination between rrnH and rrnG operons. Different genome type groups normally have different ribotypes. The size and identity of the PstI fragment containing each of the seven different rrn operons from S. typhi Ty2 were determined, and from these data, one can infer how genomic rearrangement forms new ribotypes. It is postulated that genomic rearrangement, rather than mutation, is largely responsible for producing the ribotype heterogeneity in S. typhi.
Salmonella typhi grows only in humans, in whom it causes typhoid enteric fever. Independent S. typhi strains from different geographic regions are phenotypically homogeneous. Reeves et al. (21) showed that 26 strains of S. typhi tested by multilocus enzyme electrophoresis had an identical electrophoretic type, leading to the conclusion that S. typhi strains are a single clone. Selander et al. (25) also found S. typhi more homogeneous in electrophoretic type than other species of Salmonella, although they identified two electrophoretic types, Tp1 and Tp2. Data on restriction fragment length polymorphisms from digestion with EcoRI and PstI showed conserved banding patterns for all 22 S. typhi strains studied (5). The cell envelope protein profiles for a series of outer membrane and inner membrane proteins for 32 S. typhi strains showed only very minor differences (6). Each of 15 S. typhi strains had intervening sequences in all seven rrl genes for rRNA, and all those tested had identical sequences (18). All these data show a high degree of homogeneity of S. typhi strains.
Although the above data indicate homogeneity, ribotyping studies of S. typhi by Altwegg et al. (1), Nastasi et al. (19), and Pang et al. (20) found a large number of ribotypes (RTs) among different strains of S. typhi, whereas other Salmonella spp. are relatively homogeneous in RTs. Bacteriophage typing with Vi phage is the most common method used to demonstrate epidemiological associations of S. typhi strains (2), but RT data have also been very valuable for further subdivision of the different phage types (1, 19, 20). The objective of this study was to determine the basis for heterogeneity of RTs in S. typhi.
RTs are determined by probing a Southern blot of a restriction digest of the genome with ribosome sequences; thus, the RT of a strain is a specific pattern of band sizes, each band containing rRNA sequences. In enteric bacteria such as Salmonella (11) and Escherichia coli (4), which have seven rrn operons, seven fragments containing rrn operons are expected if digestion is performed with an enzyme such as PstI, which does not digest within the rrn operon. Each fragment is composed of two arms (the distance from the left end, or 16S end, of the rrn operon to the nearest PstI site, and the distance from the right end, or 5S end, of the rrn operon to the nearest PstI site) plus the rrn operon itself, which is 6 kb; thus, all seven bands in PstI digests that hybridize to the probe are 6 kb or larger. An RT is defined as a specific set of lengths of the seven fragments containing the seven rrn operons. (RTs for enzymes which cut within the rrn operon should have 14 fragments representing the two arms from each operon plus internal rrn fragments if any.) Changes in the fragment lengths with resultant changes in RT can result from (i) point mutations in the genome, leading to gain or loss of restriction sites in one of the two arms of the restriction fragment carrying the rrn operon (nucleotide sequences within the rrn operon are highly conserved), or (ii) chromosomal rearrangements which affect the genome within the fragments carrying the rrn operons.
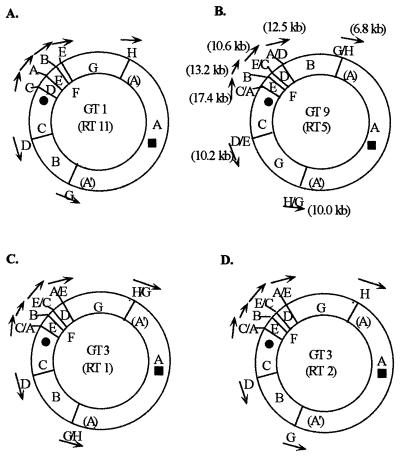
The structure and the order of genes on the chromosomes of different enteric bacteria are usually strongly conserved (9, 23); the genetic and physical maps of S. typhimurium LT2 (11), E. coli K-12 (4), S. enteritidis, and S. paratyphi B (10) are very similar. All fragments from digestion by the endonuclease I-CeuI (which cuts only in rrn operons [13, 17]) are in the order ABCDEFG, as illustrated for genome type 1 (GT1) in Fig. 1A. Within each species, the genomic order of these fragments is also conserved, as exemplified by strains of S. typhimurium (13). However, the genome of S. typhi is frequently rearranged by recombination between rrn operons; by using partial digestion by I-CeuI, 21 different orders were detected among 127 wild-type strains examined (14, 16). These different orders of I-CeuI fragments are defined as GTs and are shown in detail in reference 16. They are illustrated by GT9 (Fig. 1B) and GT3 (Fig. 1C and D), in which the order of fragments is changed. We postulated that homologous recombination between rrn operons results in translocations and inversions; for example, in S. typhi Ty2, which is GT9 (Fig. 1B), linkages of genes are changed so that fragment I-CeuI-B is now linked to the A end of I-CeuI-A while fragment G is linked to the A′ end (a reversal of the order in GT1 [Fig. 1A]); this is postulated to be due to recombination between rrnH and rrnG in GT1, resulting in rrnG/H and rrnH/G in strain GT9 (Ty2) (12). Homologous recombination between rrn operons had previously been shown to occur in S. typhimurium LT2 (3) and E. coli K-12 (7) at frequencies as high as 10−4 per cell and to result in the formation of rearrangements of the types we have observed in S. typhi.
FIG. 1.
Genomic rearrangements of the I-CeuI fragments of GT1, GT3, and GT9 in S. typhi (12, 14). The RTs of each GT are also shown. The letters for each I-CeuI fragment are within or adjacent to the circles, and each junction between the fragments is the endonuclease cleavage site of I-CeuI (11). The letters outside the circle indicates the rrn genes (in GT1) and inferred rrn recombinants (in GT3 and GT9). The solid circle in I-CeuI-C denotes the origin of replication (oriC), and the square in I-CeuI-A denotes the termination of replication (TER). Arrows beside the rrn operons indicate the orientation from rrs (for 16S rRNA) to rrf (for 5S rRNA). The order of these I-CeuI fragments on the chromosome of strains of S. typhi was determined by the partial-digestion method from data reported previously (12, 14, 16). (A) GT1 is the same as the order in S. typhimurium LT2 and E. coli K-12. The positions of the rrn operons are shown. (B) GT9 is the GT found in Ty2, which has been commonly used as a wild-type strain (12). (C and D) GT3 is the most common GT found among S. typhi strains; it has two dominant ribotypes, RT1 and RT2.
Homologous recombination between two rrn operons is expected to change the RT determined from a PstI digest; the two “arms” of each of the two PstI fragments will be interchanged, resulting in two new fragment lengths, but the other five PstI fragments should remain unaltered. In this study, we tested the hypothesis that recombination between rrn operons, rather than point mutation in PstI restriction sites, is responsible for the formation of new ribotypes in S. typhi. We found that recombination is the major basis for new RT formation, although some role for mutation cannot be excluded. The sizes of the PstI fragments of each of the seven rrn operons of S. typhi Ty2 are identified, and recombination between rrnH and rrnG is shown to be common.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
A total of 127 S. typhi strains were obtained from different sources: Laboratory Centre for Disease Control, Ottawa, Canada; Centers for Disease Control and Prevention, Atlanta, Ga.; Provincial Laboratory of Alberta, Calgary, Canada; Tikki Pang (University of Malaya, Kuala Lumpur, Malaysia); Robert Selander (Pennsylvania State University); and Bruce Stocker (Stanford University). They were identified as S. typhi based on biochemical and antigenic characterizations, which were determined by the laboratories of origin and confirmed by the Laboratory Centre for Disease Control, Ottawa, Canada. Genomic analysis of these strains has been previously reported (16). The strains were grown on Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, 3.5 ml of 1 M NaOH); solid medium also contained 1.5% agar. The minimal medium used is a modified Davis medium (24). Tetracycline was used at 20 μg/ml. Strains were maintained in 15% glycerol at −70°C, and a single colony was isolated prior to use.
Enzymes and chemicals.
Endonucleases were from New England Biolabs (PstI, AvrII = BlnI, I-CeuI, SpeI), and Boehringer Mannheim (XbaI). Other chemicals, such as agarose, were from GIBCO BRL.
Preparation, digestion, and separation of genomic DNA and Southern blotting.
Preparation of high-molecular-weight genomic DNA, endonuclease cleavage of DNA embedded in agarose blocks, and separation by pulsed-field gel electrophoresis were as reported previously (10, 12).
PstI-restricted chromosomal DNA fragments of different strains of S. typhi were separated by conventional gel electrophoresis in Tris-borate-EDTA buffer for 20 h at 100 V. After electrophoresis, the gel was washed in 0.25 N HCl, then in 0.5 N NaOH–1.5 M NaCl, and finally in 1 M ammonium acetate–0.02 N NaOH. Separated DNA fragments in the gel were then transferred to a positively charged nylon membrane (Boehringer Mannheim) by Southern blotting.
The Southern blotting was done as follows. The gel and the positively charged nylon membrane were sandwiched between filter papers soaked in a buffer solution of 1 M ammonium acetate–0.02 N NaOH and blotted for 18 to 24 h. Then the membrane was dried at 80°C for 2 h to fix the DNA onto the membrane.
Preparation of the probe for ribotyping.
The membrane was probed with plasmid pT711 containing the E. coli rrnB operon with the 16S and 23S rRNA and 5S tRNA gene sequences (25); this plasmid was present in strain SGSC2266, an E. coli K-12 strain. SGSC2266 was grown overnight on a plate containing Luria-Bertani agar plus 100 μg of ampicillin per ml. Then the cells were scraped up and the plasmid DNA was isolated by the alkali lysis method as described by Sambrook et al. (22).
Digoxigenin DNA labeling and detection of RTs.
The digoxigenin DNA labeling and detection methods were performed as described by the manufacturer (Boehringer Mannheim). The RTs were detected by exposing a film to the membrane.
Identification of the rrn operon(s) in specific RT bands.
The XbaI–I-CeuI–BlnI–SpeI genome map of S. typhi Ty2 has been developed previously (12). The first three endonucleases cut within the rrn operons of Ty2, while SpeI cuts outside the rrn operons; therefore, some restriction fragments contain a full copy of a rrn operon, while others contain only part of an rrn operon. The genome of Ty2 was digested with XbaI, I-CeuI, BlnI, or SpeI, and the restricted fragments were then separated by pulsed-field gel electrophoresis (12). Restricted fragments which include full or partial rrn operons were then isolated by being excised from the gel. These fragments were then redigested with PstI and probed for rrn operons.
RESULTS
RTs of S. typhi strains.
All 127 strains of S. typhi reported previously (16) and briefly described in Materials and Methods were tested for RT following digestion by PstI; they were separated into 31 different RTs (Table 1), according to data of the type reported in Fig. 2A.
TABLE 1.
RTs detected following PstI digestion among the 127 strains of S. typhi belonging to different GTs
| GT | Total no. of strains in GT | RT | No. of strains with specific RT |
|---|---|---|---|
| 1 | 2 | 11 | 2 |
| 2 | 22 | 6 | 16 |
| 7 | 2 | ||
| 8 | 1 | ||
| 9 | 2 | ||
| 10 | 1 | ||
| 3 | 57 | 1 | 37 |
| 2 | 18 | ||
| 3 | 1 | ||
| 4 | 1 | ||
| 4 | 5 | 2 | 1 |
| 3 | 2 | ||
| 12 | 1 | ||
| 13 | 1 | ||
| 5 | 2 | 14 | 2 |
| 6 | 8 | 9 | 1 |
| 15 | 7 | ||
| 7 | 3 | 13 | 2 |
| 16 | 1 | ||
| 8 | 2 | 6 | 2 |
| 9 | 5 | 5 | 5 |
| 11 | 2 | 17 | 2 |
| 13 | 2 | 18 | 1 |
| 19 | 1 | ||
| 14 | 2 | 20 | 1 |
| 21 | 1 | ||
| 16 | 1 | 22 | 1 |
| 17 | 1 | 23 | 1 |
| 18 | 2 | 24 | 1 |
| 25 | 1 | ||
| 19 | 4 | 23 | 4 |
| 22 | 2 | 26 | 2 |
| 23 | 2 | 16 | 1 |
| 27 | 1 | ||
| 24 | 2 | 28 | 1 |
| 29 | 1 | ||
| 25 | 1 | 30 | 1 |
| 26 | 1 | 31 | 1 |
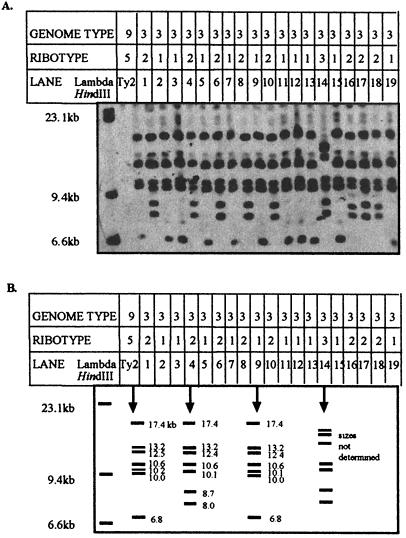
FIG. 2.
(A) Chemiluminescent detection of RTs in some of the S. typhi strains in GT3. The DNA of the strains was first digested by the endonuclease PstI, which cuts outside the rrn genes, and then separated by conventional gel electrophoresis, Southern blotted, and probed. The standard sizes of lambda HindIII and the RT of Ty2 (RT5) are shown for comparison. Lanes 1 to 19 contain strains from GT3. The corresponding RTs are shown for all strains. Seven bands (some appears as doublets) are detected in each RT. Within GT3, the strains show two dominant ribotypes (RT1 and RT2), and one strain shows RT3. Genomic DNA of the following strains are in the indicated lanes: 1, 26T4; 2, 26T7; 3, 26T11; 4, 26T15; 5, 26T18; 6, 26T22; 7, 26T23; 8, 26T29; 9, 26T30; 10, 26T33; 11, 26T34; 12, 26T35; 13, 26T36; 14, 26T41; 15, 26T42; 16, 26T43; 17, 26T45; 18, 26T47; 19, 25T37. (B) Proposed fragment sizes observed in panel A. The sizes of lambda HindIII are 23.1, 9.4, and 6.6 kb. The fragment sizes for strain Ty2 (RT5, GT9), for a single strain of RT2 and GT3 (lane 4) and a single strain of RT1 and GT3 (lane 9) are shown, as well as the fragments of the one strain of RT3 of GT3 (lane 14).
If changes in RTs are due to genomic rearrangements only and not to mutations, strains with the same genomic arrangement should have the same RT. We therefore examined genomic DNA of the 57 strains of S. typhi which are of GT3; a sample of 19 of these is shown in Fig. 2A, lanes 1 to 19. These 19 strains fell into three classes for RT; most strains were RT1 or RT2, while 1 was RT3. The cartoon in Fig. 2B shows the seven fragments of a representative strain of RT1 from lane 9, with predicted sizes in kilobases; a strain of RT2 from lane 4 and a strain of RT3 from lane 14 are also illustrated. The RT of strain Ty2 (GT9) is also shown for comparison. Among the whole set of 57 strains of GT3, 27 were of RT1, 18 were of RT2, 1 was of RT3, and 1 was of RT4 (only some of the data are shown). These data indicate that within a specific GT there is a limited number of different RTs; the sections below explain the basis for the occurrence of the different RTs.
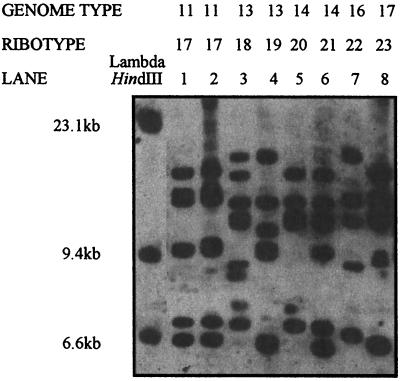
The RTs of eight strains representing five different GTs are shown in Fig. 3. All these strains have different RTs, except for the two strains of GT11, both of which are RT17. The RTs of all 127 strains, which belong to 21 different GTs, are summarized in Table 1. These data show that strains with different GTs almost always have different RTs.
FIG. 3.
Chemiluminescent detection of RTs in S. typhi strains which belong to different GTs. The genomic DNA of these strains was treated as described in the legend to Fig. 2. The standard sizes of lambda HindIII are shown. Lanes: 1 and 2, 26T49 and T189 (both GT11); 3 and 4, 382-82 and SA4865 (both GT13); 5 and 6, 26T38 and R16B7 (both GT14); 7, 26T9 (GT16); 8, In4 (GT17). The corresponding RTs are shown for all strains.
Identification of the specific rrn operon associated with each fragment in the RT.
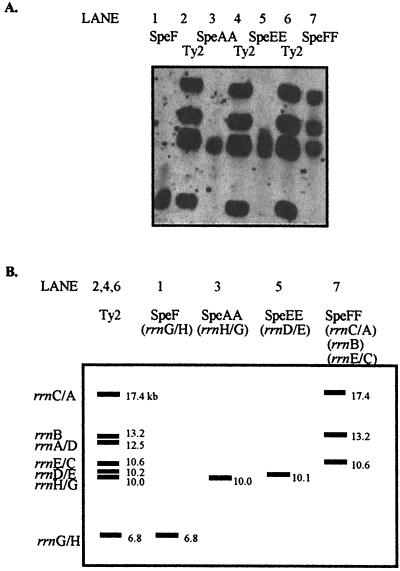
If the formation of new RTs is due to homologous recombination between different rrn operons alone, strains which are defined as having the same GT by the method of partial I-CeuI digestion used by Liu and Sanderson (16) should all have similar RTs. However, they may not be identical, because the partial I-CeuI analysis method cannot detect inversions between rrnC and rrnD on either end of fragment I-CeuI-C or between rrnG and rrnH on either end of I-CeuI-A (Fig. 1A) (see also references 14 and 16). Thus, inversions of these types might occur within GT3. We therefore determined the specific rrn operon associated with each RT fragment in S. typhi Ty2, to see if the fragments which vary within a GT are the ones which we would predict. This was done by performing pulsed-field gel electrophoresis and then excising agarose blocks containing DNA which contains individual fragments that carry known rrn operons of S. typhi Ty2, as described in Materials and Methods; these fragments were then digested with PstI, electrophoresed, and probed. Representative data are given in Fig. 4A, and interpretations are provided in Fig. 4B. For example, lane 1 contains DNA of fragment SpeI F, known from earlier studies (12) to carry rrnG/H; this yields a single band of 6.8 kb, and so the 6.8-kb band carries rrnG/H. Lane 3 contains SpeI-AA, known to carry rrnH/G; this yields a single band of 10.0 kb, and so the 10.0-kb band carries rrnH/G. These two fragments from strain Ty2, of 6.8 and 10 kb, are also present in strains of RT1 in GT3; however, they are missing from RT2, where they are replaced by fragments of 8.0 and 8.7 kb, while all other fragments remain unaltered between RT1 and RT2. Our conclusion is that strains of RT1 carry the inversion of fragment I-CeuI-A which is present in Ty2, while strains of RT2 have the “normal” orientation of this fragment, which is present in S. typhimurium and most other enteric bacteria; these two orientations are illustrated in Fig. 1C and D. Thus the only difference in RT fragments between RT1 (37 strains) and RT2 (18 strains) can be explained by an inversion between rrnG and rrnH; no mutations in PstI sites needs to be invoked to explain the RTs of all these strains. RT3 (Fig. 2, lane 14) and RT4, however, cannot be thus explained, and mutation might be invoked to explain some of the fragments observed.
FIG. 4.
(A) Chemiluminescent detection of rrn operons from S. typhi Ty2 following isolation of individual genomic fragments from pulsed-field gel electrophoresis, digestion by PstI, separation by conventional gel electrophoresis, Southern blotting, and probing. Lanes: 2, 4, and 6, whole genomic DNA of Ty2; 1, rrnGH from the SpeI-F fragment (see Fig. 1 in reference 12); 3, rrnHG from SpeI-AA; 5, rrnDE from SpeI-EE; 7, rrnCA, rrnB, and rrnEC from SpeI-FF. The identity of the individual rrn operon can be determined from the genomic cleavage map of Ty2 as reported previously (12). (B) Interpretation of the above results; the sizes of the bands are shown.
Fragments carrying each of the seven rrn operons of S. typhi Ty2, either alone or (in some cases) in combination, were isolated, digested with PstI, and probed (Table 2). In some cases, either the left or right arm of the PstI fragment and part of the rrn operon were included because the rrn operon was digested by the enzyme used. The sizes of the PstI fragment including each rrn operon, derived from these data, are illustrated in Fig. 1B for strain Ty2.
TABLE 2.
The restriction fragments containing specific rrn operons isolated from S. typhi Ty2 following digestion with SpeI, XbaI, and BlnIa
| rrn operon | I-CeuI junction fragment | Restriction fragment | Fragment size (kb) |
|---|---|---|---|
| rrnGH | BA | BlnI-C | 6.8 |
| rrnGH | BA | SpeI-F | 6.8 |
| rrnHG | GA′ | SpeI-AA | 10.0 |
| rrnDE | CG | SpeI-EE | 10.1 |
| rrnCA (left arm) | C | BlnI-TU | 9.9 |
| rrnCA (left arm) | C | XbaI-DD | 9.9 |
| rrnCA (right arm) | E | BlnI-V | 6.5 |
| rrnB | EF | XbaI-EE | 13.2 |
| rrnCA (right arm) | E | 6.5 | |
| rrnEC (left arm) | F | 6.0 | |
| rrnCA | CE | SpeI-FF | 17.4 |
| rrnB | EF | 13.2 | |
| rrnEC | FD | 10.6 | |
| rrnB | EF | BlnI-X | 13.2 |
| rrnEC (left arm) | F | 6.0 | |
| rrnEC (right arm) | D | BlnI-Y | 5.8 |
| rrnAD | DB | BlnI-Z | 12.5 |
| rrnAD | DB | SpeI-GG | 12.5 |
DISCUSSION
Homologous recombination between the rrn operons results in rearrangements of the DNA fragments between these rrn operons, causing the formation of duplications, deletions, transpositions, and inversions, as illustrated in Fig. 4 of reference (16). These rearrangements can also produce new RTs, since they bring together different PstI fragment lengths. Such RTs, in principle, can also result from mutations in the PstI target sites. However, we conclude that the diversity of RTs results primarily from genomic rearrangements rather than from mutations in the PstI sites, based on the following data.
(i) Among the 57 strains which belong to GT3, 37 were RT1, 18 were RT2, and only 1 was RT3 and 1 was RT4. We concluded that RT1 and RT2 differ only in fragments which we have identified to be due to the postulated recombination between rrnH and rrnG. Recombination between these rrn operons in wild-type strains has been observed before; they are recombined in S. typhi Ty2 (12) and in S. paratyphi A (15). Recombination can occur only between rrn operons with the same polarity; thus, inversions cannot occur within the rrn operons in the half of the chromosome in which the rrn operons are oriented in the same direction, but they can occur between the two halves. The GT detected by partial I-CeuI digestion will detect most changes of order of the I-CeuI fragment but will not detect inversions of I-CeuI-A (due to recombination between rrnH and rrnG) or inversions of I-CeuI-C (due to recombinations between rrnD and rrnC). Thus, the RTs of 55 of the 57 strains of GT3 can be explained as being due to recombination between rrnH and rrnG; there is no need to invoke mutation in PstI sites to explain the occurrence of these strains. However, there may be a minor role for mutation in the PstI sites in producing new RTs among strains in GT3; the sizes of fragments in one strain of RT3 and one of RT4 could not be explained by recombination alone.
(ii) A further indication that new RTs result from the genomic rearrangements which produce new GTs is the observation that strains with different GTs almost always show different RTs (Fig. 3; Table 1).
Researchers working on ribotyping in S. typhi have focused mainly on discriminating among different strains; they assumed that point mutations lead to RT changes (19) or did not discuss the genetic basis (1, 20). Karaolis et al. (8), investigating ribotyping in Vibrio spp., assumed that point mutations lead to RT changes and used the frequency of RT changes to calculate the frequency of overall genomic point mutation.
The method of partial I-CeuI digestion will detect genomic rearrangements due to recombination between parts of the rrn operons and will reveal the order of fragments, thus determining the “rrn skeleton” of the genome, i.e., the number of rrn operons, and the lengths of the DNA intervals between each of these operons. This is a very efficient method for detecting changes in the genome, either rearrangements of the existing DNA or addition or deletion of DNA (indels). However, this method will not detect inversions of fragments I-CeuI-C or I-CeuI-A in Salmonella (or equivalent changes in other genomes). For example, strains of GT3, resulting from homologous recombination between rrnH and rrnG to produce rrnHG and rrnGH, as shown in Fig. 1C and D, cannot be distinguished by the partial I-CeuI digestion method. Ribotyping, on the other hand, will detect new RTs which result either from recombination between any of the rrn operons (without the limits that apply to the I-CeuI partial-digestion method) or from mutation in the endonuclease target sites. For simply revealing distinct types, ribotyping is superior to genome typing because it is somewhat more discriminating, since it can distinguish between strains with inversions in I-CeuI fragments A and C (Fig. 1 and 2) and can also detect mutations in the PstI sites. However, ribotyping alone will not determine the basis for the new RTs unless the analysis is coupled with partial I-CeuI digestion, as in the analysis we present here.
ACKNOWLEDGMENTS
This work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada and by grant RO1AI34829 from the National Institute of Allergy and Infections Diseases.
REFERENCES
- 1.Altwegg M, Hickman-Brenner F W, Farmer J J., III Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi strains. J Infect Dis. 1989;160:145–149. doi: 10.1093/infdis/160.1.145. [DOI] [PubMed] [Google Scholar]
- 2.Anderson E S, Felix A. The Vi type-determining phages carried by Salmonella typhi. J Gen Microbiol. 1953;9:65–88. doi: 10.1099/00221287-9-1-65. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R P, Roth J R. Spontaneous tandem genetic duplication in Salmonella typhimurium arise by unequal recombination between ribosomal RNA (rrn) cistrons. Proc Natl Acad Sci USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 5.Faundez G, Aron L, Cabello F C. Chromosomal DNA, iron-transport systems, outer membrane proteins, and enterotoxin (heat labile) production in Salmonella typhi. J Clin Microbiol. 1990;28:894–897. doi: 10.1128/jcm.28.5.894-897.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco A, Gonzalez C, Levine O S, Lagos R, Hall R H, Hoffman S L, Asril Moechtar M, Gotuzzo E, Levine M M, Hone D M, Morris J G., Jr Further consideration of the clonal nature of Salmonella typhi: evaluation of molecular and clinical characteristics of strains from Indonesia and Peru. J Clin Microbiol. 1989;30:2187–2190. doi: 10.1128/jcm.30.8.2187-2190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill C W, Gray J A. Effects of chromosomal inversion on cell fitness in Escherichia coli K12. Genetics. 1988;119:771–778. doi: 10.1093/genetics/119.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaolis D K R, Lan R, Reeves P R. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J Bacteriol. 1994;176:6199–6206. doi: 10.1128/jb.176.20.6199-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S-L, Hessel A, Cheng H Y M, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella paratyphi B. J Bacteriol. 1994;176:1014–1024. doi: 10.1128/jb.176.4.1014-1024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S-L, Hessel A, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling and pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S-L, Sanderson K E. Genomic cleavage map of Salmonella typhi Ty2. J Bacteriol. 1995;177:5099–5107. doi: 10.1128/jb.177.17.5099-5107.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S-L, Sanderson K E. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S-L, Sanderson K E. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci USA. 1995;92:1018–1022. doi: 10.1073/pnas.92.4.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S-L, Sanderson K E. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J Bacteriol. 1995;177:6585–6592. doi: 10.1128/jb.177.22.6585-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S-L, Sanderson K E. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall P, Lemieux C. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene. 1991;104:241–245. doi: 10.1016/0378-1119(91)90256-b. [DOI] [PubMed] [Google Scholar]
- 18.Mattatall N R, Daines D A, Liu S-L, Sanderson K E. Salmonella typhi contains identical intervening sequences in all seven rrl genes. J Bacteriol. 1996;178:5323–5326. doi: 10.1128/jb.178.17.5323-5326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nastasi A, Mammina C, Villafrate M R. rDNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi infections. Epidemiol Infect. 1991;107:565–576. doi: 10.1017/s0950268800049268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang T, Altwegg M, Martinetti G, Koh C L, Puthucheary S. Genetic variation among Malaysian isolates of Salmonella typhi as detected by ribosomal RNA gene restriction patterns. Microbiol Immunol. 1992;36:539–543. doi: 10.1111/j.1348-0421.1992.tb02053.x. [DOI] [PubMed] [Google Scholar]
- 21.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., III Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sanderson K E. Genetic relatedness in the family Enterobacteriaceae. Annu Rev Microbiol. 1976;30:327–349. doi: 10.1146/annurev.mi.30.100176.001551. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson K E, Stocker B A. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J Bacteriol. 1981;146:535–541. doi: 10.1128/jb.146.2.535-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selander R K, Beltran P, Smith N H, Helmuth R, Rubin F A, Kopecko D J, Ferris K, Tall B D, Cravioto A, Musser J M. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]