Abstract
Inter-individual differences in gut microbiota composition are hypothesized to generate variation in host fitness—a premise for the evolution of host–gut microbe symbioses. However, recent evidence suggests that gut microbial communities are highly dynamic, challenging the notion that individuals harbour unique gut microbial phenotypes. Leveraging a long-term dataset of wild meerkats, we reconcile these concepts by demonstrating that the relative importance of identity for shaping gut microbiota phenotypes depends on the temporal scale. Across meerkat lifespan, year-to-year variation overshadowed the effects of identity and social group in predicting gut microbiota composition, with identity explaining on average less than 2% of variation. However, identity was the strongest predictor of microbial phenotypes over short sampling intervals (less than two months), predicting on average 20% of variation. The effect of identity was also dependent on meerkat age, with the gut microbiota becoming more individualized and stable as meerkats aged. Nevertheless, while the predictive power of identity was negligible after two months, gut microbiota composition remained weakly individualized compared to that of other meerkats for up to 1 year. These findings illuminate the degree to which individualized gut microbial signatures can be expected, with important implications for the time frames over which gut microbial phenotypes may mediate host physiology, behaviour and fitness in natural populations.
Keywords: temporal dynamics, host–microbiota interactions, gut microbiome, meerkats, repeatability, intraclass correlation coefficient
1. Introduction
Inter-individual differences in gut microbiota composition can lead to variation in host health [1], pathogen susceptibility [2–4] and measures of fitness such as survival [5,6]. Although the mechanisms underpinning these relationships remain poorly understood, one possibility is that hosts maintain individualized and stable microbial symbionts that are disproportionally important for mediating long-term physiological and behavioural phenotypes [7]. However, there is increasing evidence that gut microbial communities are highly dynamic [8–10], and the role of individual identity in shaping longitudinal dynamics remains puzzling [11]. This uncertainty hinders efforts to unequivocally link gut microbiota communities to host phenotypes, to understand the temporal scales over which microbe-mediated selection may act, and to decipher how phylosymbioticg relationships between hosts and microbes evolve and persist [12–14].
The individuality and stability of gut microbial communities provide rich information on the ecological and evolutionary processes that maintain their structure and function. Individuality refers to the extent a trait varies between individuals, while stability measures trait variation over time within an individual. For example, strong individual signatures in the gut microbiota may be expected where transmission between hosts is limited, or where personalized gut microbiotas are both advantageous to fitness and under host genetic control. By contrast, individualized microbial signatures are expected to be weak if transmission between hosts is frequent, or in ecological contexts where dynamic (i.e. unstable) gut microbiotas confer a higher fitness advantage than individualized and stable microbiotas [15].
The individuality and stability of gut microbiotas across different host species remains unclear, in part due to the sparsity of longitudinal data. In humans from industrialized countries, the gut microbiota is generally rather individualized, even over many years [16–18]. Yet, this long-term individuality conceals a highly dynamic community that is revealed by daily sampling [10], suggesting that while human gut microbial communities are highly dynamic over the short term, they nevertheless maintain some level of individuality. This long-term individuality is potentially due to modern human lifestyles buffering the gut microbiota from environmental effects or because most studied subjects do not share the same environment, which may accentuate individual differences. Longitudinal studies of wild non-human primates also report highly dynamic gut microbiotas [8,19,20], but recent evidence suggests that individualized responses to changing environments limit the formation of individualized microbial compositions over long time scales [21]. These findings suggest that microbial dynamics, rather than composition per se, are individualized.
In this study, we gathered longitudinal information on the individuality and stability of the gut microbiota using 965 samples collected from 157 wild meerkats (Suricata suricatta; mean number of samples per ID = 6, min. = 3, max. = 14) belonging to 22 social groups, sampled between 1997 and 2019 (electronic supplementary material, figure S1). Meerkats are small insectivorous mongooses living in social groups of two to fifty individuals in the arid regions of southern Africa. Social groups are largely made up of related individuals, and while dispersal and immigration between groups occurs frequently, most individuals remain in the same social group for life. The population under study is part of the Kalahari Research Project, which has monitored tagged individuals since 1993 [22]. Here we analyse 16S gut microbiota data described previously [9], and which was generated using an internal standard to quantify 16S copy number. Previous research demonstrated that the gut microbiota of this population differs from that of previously studied primates in that it undergoes strong diurnal oscillations, yet relatively weak seasonal changes [9], potentially generating particularly high microbial turnover rates on a daily basis. In this study, we only include individuals that were sampled at least three times, therefore we analyse 965 samples out of 1109 reported in [9].
We measured the individuality of the gut microbiota in two ways: firstly by estimating its repeatability, which is defined as the proportion of total phenotypic variation that is attributed to individuals, and is also referred to as the intraclass correlation coefficient (ICC) when applied other sources of variation [23,24]. We additionally measured individuality by estimating beta dissimilarity between pairs of samples, which differs from repeatability by being a relative rather than an absolute measure (i.e. how individualized is a meerkat's gut microbiota compared to that of other meerkats). We measured microbial community stability by estimating taxa turnover between consecutive sampling events.
Applying these definitions, our aims were to (a) quantify repeatability for a range of single-taxon and community phenotypes, and to compare repeatability to the effects of social group membership and year of sample collection; (b) identify microbial phylogenies that are most likely to demonstrate long-term repeatability; investigate how (c) sampling interval and (d) age affect the relative influence of repeatability compared to the effects of social group and year of sample collection; and (e) examine whether changes to repeatability are determined by shifts in overall community stability. Lastly, (f), we apply pairwise beta dissimilarity measures to test whether gut microbial communities remain individualized compared to other meerkats as a function of sampling interval.
2. Results
(a) . Weak repeatability and strong inter-year variation across phenotypes
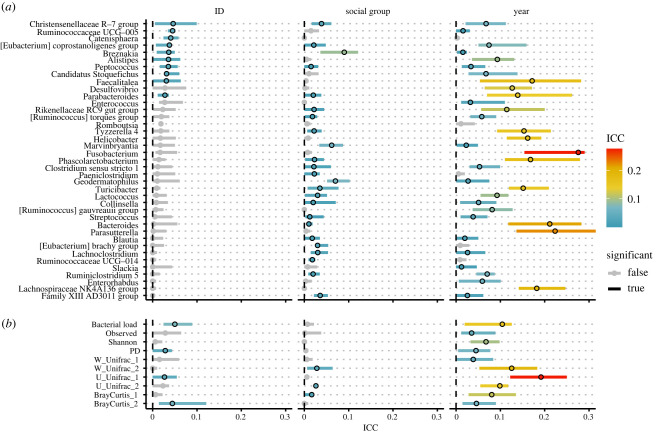
We calculated the ICC for meerkat identity (repeatability), social group membership and year of sample collection for 39 collapsed genera that were detected in 50% of samples, which together accounted for 82% relative abundance (electronic supplementary material, table S1). Year of sample collection had the strongest effects across genera (87% significant; mean ICC = 0.081) with social group membership (62% significant; mean ICC = 0.02) and identity (26% significant; mean ICC = 0.016) having weaker effects (figure 1a). The genera Christensenellaceae (R-7 group) and Ruminococcaceae (UCG-005) were most likely to be characterized by inter-individual variation. Year of sample collection was also the most important predictor of bacterial load, alpha diversity measures, and the first axis of variation of beta diversity ordinations, and was particularly associated with suites of rarer, non-core taxa (captured by unweighted UniFrac; figure 1b). Both individual identity and social group were largely unimportant for explaining gut microbiota alpha and beta diversity across the 20-year study period (figure 1b).
Figure 1.
ICC measures and 95% confidence intervals for meerkat identity (repeatability), social group membership and year of sample collection on (a) the abundances of 39 genera that were detected in at least 50% of samples, and (b) community phenotypes, including bacterial load, three measures of alpha diversity (observed ASV richness, Shannon diversity and Faith's phylogenetic diversity), and the first axis of variation extracted from ordinations based on three beta diversity distances (weighted UniFrac, unweighted UniFrac and Bray–Curtis). Colours are scaled by their relative effect size (ICC), and greyed out if they are not significant. PD = Faiths phylogenetic alpha diversity. (Online version in colour.)
We next modelled taxa abundances at the amplicon sequence variant (ASV) level, including 121 ASVs that were detected in over 30% of samples and which together also accounted for 79% of relative abundance (electronic supplementary material, table S2). Year of sample collection again had the strongest effects across ASVs (87% significant, mean ICC = 0.09 ± 0.06 s.d), with social group membership (60% significant, mean ICC = 0.04 ± 0.03 s.d.) and identity (39% significant, mean ICC = 0.05 ± 0.02 s.d.) having weaker effects (electronic supplementary material, figure S2). Weak correlations existed between the effects of identity, year and social group, with ASVs that tended to be characterized by identity, also tending to be characterized by social group membership (Pearson's r = 0.27, p = 0.003), while taxa characterized by inter-group variation tending to be buffered from year effects (Pearson's r = −0.26, p = 0.006; electronic supplementary material, figure S3). The most individually repeatable ASVs belonged to the genera Blautia and Bacteroides. However, these genera were not significantly repeatable within individuals at the genus level, suggesting that identity effects often act at higher taxonomic resolutions than genus level.
We tested whether the weak influence of identity was dependent on model structure by excluding social group membership and year from models. Excluding social group and year of sample collection considerably inflated the contributions of identity, with the majority of ASVs becoming significantly associated with identity (electronic supplementary material, figure S4).
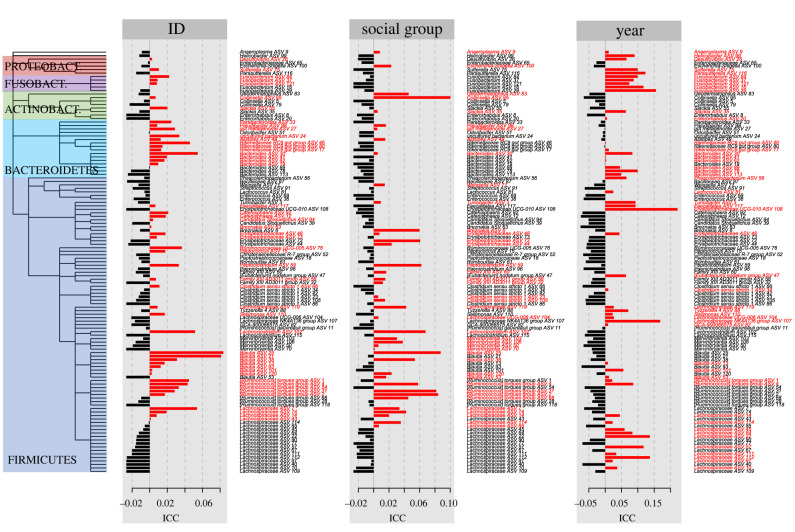
(b) . Specific phylogenetic lineages are more likely to be repeatable
To test whether patterns in ICC were centred around particular phylogenetic branches, we estimated the phylogenetic signal in associations with identity, social group membership and year. We found localized phylogenetic signals for individual identity (Moran's I = 0.00728, p = 0.014), social group (Moran's I = 0.00711, p = 0.018) and year-to-year variation (Moran's I = 0.02, p = 0.003; figure 2). Individual identity was predominantly associated with members of the phylum Bacteroidetes, in particular Rikenellaceae, Alistipes, and some Bacteroides members, as well as some specific Firmicutes genera, including Blautia, and Ruminoccocus torques group. By contrast, social group had wide-spread effects across members of Firmicutes, with particularly notable associations with cellulose-degrading Marvinbryantia, potentially reflecting different levels of plant consumption among social groups. Year effects characterized members of the phyla Fusobacterium and Proteobacteria, some members of Bacteroides, and members of Lachnospiraceae. The most abundant genus, Clostridium sensu stricto 1, which undergoes strong diurnal oscillations [9], demonstrated no phylogenetic signal in association with any variables.
Figure 2.
Phylogenetic signal in ICC for individual identity (repeatability), social group membership and year of sample collection across 121 ASVs with over 30% prevalence in the overall sample. ASVs for which ICC is higher than average are coloured in red. The major phyla are indicated. (Online version in colour.)
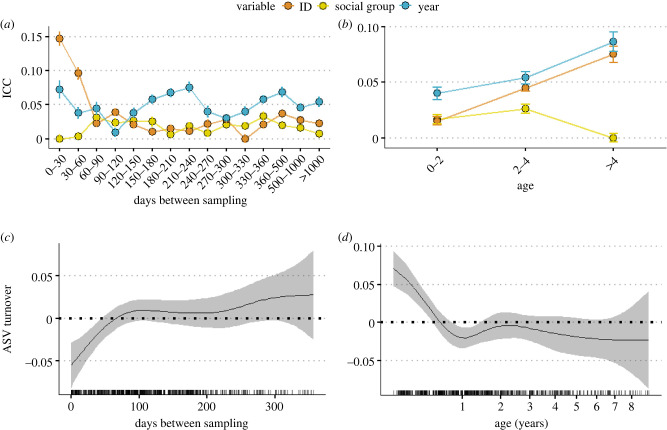
(c) . Repeatability is strong at short time intervals but weakens over time
While identity had weak effects over the whole study period, we hypothesized that it may be more important over shorter time frames. To test this, we examined whether sampling interval affects ICC estimates for identity, social group membership and year for each of the 121 ASVs analysed above. Therefore, we generated subsets of data that reflected a range of sampling intervals, from pairs of samples collected less than a month apart, to pairs collected over 1000 days apart and repeated models for each sampling interval. As predicted, individual identity was more important than social group and year when longitudinal samples were taken within two months of each other, but the importance of identity decreased rapidly as sampling interval increased (figure 3a). When samples were longitudinally sampled within the same month, mean and median repeatability were 0.2 and 0.15, respectively, with some ASVs having a repeatability as high as 0.39 (electronic supplementary material, figure S5). When including only samples that were taken over two months apart, year of sample collection became the most important predictor of ASV abundances (figure 3a). Social group had, on average, weak effects (median ICC < 0.05) across all time frames.
Figure 3.
Temporal trends in ICC (a,b) and ASV turnover between sampling events (c,d) of meerkat gut microbiomes. (a,b) Median ICC (and standard error) of individual identity (repeatability), social group membership and year of sample collection from models predicting the abundances of 121 ASVs when samples are categorized by (a) time intervals between sampling from the same individual; and (b) different meerkat age categories. (c,d) Temporal predictors of ASV turnover between consecutive sampling events from the same individual, extracted from a GAMM, showing the association between ASV turnover and (c) the number of days between samples; and (d) the age of the meerkat at the point of the first sample. (Online version in colour.)
(d) . Repeatability increases with age
We tested whether ICC was also influenced by meerkat age by categorizing samples by meerkat age (young, less than 2 years; adult, 2–4 years; old, greater than 4 years). The effect of identity did indeed increase with age, being lowest in young meerkats (median = 0.015, 10% significantly repeatable), higher in adults (median = 0.05, 37% significantly repeatable) and highest in older meerkats (median = 0.08, 35% significantly repeatable; figure 3b). Interestingly, the effect of year also increased with age, possibly due to older meerkats being sampled over many years (figure 3b). By contrast, the effect of social group decreased in older meerkats (figure 3b), potentially suggesting that as gut microbial communities become more individualized with age, microbial communities are buffered from group effects, such as horizontal transmission from group members.
(e) . Changes in repeatability are reflected by shifts in community stability
Low long-term repeatability at the ASV level may be driven by high community turnover, in which case we would predict that the timeframes associated with low repeatability would also be associated with higher ASV turnover, and vice versa. To test this association, we measured ASV turnover (the proportion of ASV appearing or disappearing) between consecutive samples taken from the same individual, and tested whether ASV turnover was predicted by the amount of time elapsed between samples and meerkat age.
Mean community turnover between sampling events was very high (approx. 80% of ASVs appeared or disappeared between sampling events). Turnover increased with the amount of time elapsed between samples (figure 4c), yet also was dependent on meerkat age, with turnover being higher in meerkats under 1 year of age (figure 4d; electronic supplementary material, table S3).
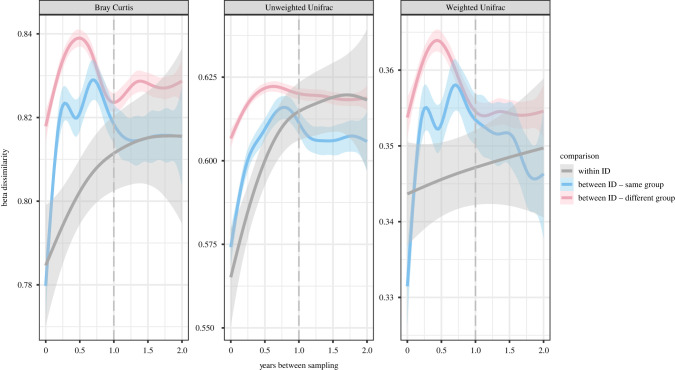
Figure 4.
Beta dissimilarity of meerkat gut microbial communities as a function of sampling interval for samples collected up to 2 years apart, coloured by comparison type (grey, within individual; blue, between individuals from the same social group; and red, between individuals from different social groups) and based on (a) Bray–Curtis; (b) unweighted UniFrac, and (c) weighted UniFrac distances. The dashed grey line indicates the 1 year mark. Lower beta dissimilarity indicates higher similarity in community composition. (Online version in colour.)
(f) . Despite low repeatability, gut microbiota composition remains marginally individualized when compared with other meerkats
Although the effect of identity on gut microbiota composition is negligible after two months, this does not preclude that an individual's gut microbiota remains more similar to itself than to gut microbiotas of other meerkats. We tested this hypothesis by estimating beta similarity between pairwise samples and modelling similarity as a function of time interval between sampling, for samples collected up to 2 years apart. Comparisons were categorized as comparing either samples of the same individual, samples of different individuals within the same social group, or different individuals of different social groups.
We found that samples taken from the same individual did tend to be more similar than to samples taken from other individuals, and match more closely with gut microbiotas from members of the same social group, independent of the distance metric applied (figure 4a–c). However, these effects had low explanatory power, suggesting substantial variation (GAMMs: Bray–Curtis: p < 0.001, R2 = 0.002; unweighted UniFrac: p < 0.001, R2 = 0.002; weighted UniFrac: p < 0.001, R2 = 0.001). Notably, patterns in inter-individual beta dissimilarity over time were nonlinear (figure 4a–c), with gut microbiotas of different individuals beginning to converge again after 1 year, hinting at seasonal effects over the first year.
3. Discussion
Longitudinal studies of wild populations are scarce but invaluable to dissect short- and long-term evolutionary and ecological dynamics. For gut microbiota, long-term data allows us to tease apart how much variation is explained by identity, social group membership and yearly shifts in a way that is not possible from cross-sectional studies conducted over short time scales. We leveraged an extensively sampled and well-studied wild meerkat population (greater than 20 years) to show that, over long time intervals, year of sample collection had stronger effects on the abundances of most common ASVs than identity or social group membership, and in addition is more influential in shaping overall alpha and beta diversity. However, the contribution of identity is considerably higher over shorter time intervals, associated with specific phylogenetic groups of taxa, and increases with meerkat age. Increased repeatability with age is underpinned by an increase in overall microbial community stability, and not due to increased stability of particular taxa. Lastly, we found that while the effect of identity was negligible after two months, meerkat gut microbiotas remained weakly individualized compared with that of other individuals for up to 1 year. These findings suggest that the downstream physiological effects of individualized gut microbiotas are likely to act over the scale of weeks or months, rather than years.
Our findings of weak long-term contributions of individual identity to gut microbial composition, an exponential decay in microbiota similarity within individuals over time, and increases in taxon repeatability with age, all align closely with those found in a decade long study of baboons [8,25], suggesting that such dynamics may be consistent across host species. These patterns in longitudinal dynamics may resolve the conflicting reports of the major drivers of gut microbiota dynamics across different species, with different findings being due to variable sampling intervals and designs rather than inherent differences among species. Nevertheless, this does not preclude the possibility of differences in gut microbiota stability and associated drivers between host species. For example, individual host traits such as age, sex and social dominance rank generate individualized microbial signatures that are stable over short time frames in baboons [25], yet these traits do not have a strong stabilizing effect in meerkats [9].
We distinguished between taxa whose temporal variation is characterized by inter-individual, inter-group or inter-year variation. Pinpointing taxa associated with inter-individual variation is important for being able to link the gut microbiota to immutable host traits encoded by genetics and responsible for host fitness; yet, this is highly challenging when different sources of variation are nested in structure. In humans, members of the phylum Bacteroidetes tend to be characterized by strong inter-individual variation [17], and this also appears to be the case in meerkats, with the Bacteroidetes genera Alistipes, Rikenellaceae and Bacteroides all tending to be associated with meerkat identity. Some specific lineages of Firmicutes were also associated with identity, including Blautia, Ruminoccocus torques group, and Christensenellaceae R-7 group. Many of these genera have been found to be significantly heritable in mammals [8], and therefore may be more likely to be associated with host traits such as genotype, physiology or fitness. Together, these lines of evidence suggest that future studies of mammalian host–gut microbe interactions may benefit from focusing on these lineages as a potential mediator of host health and fitness.
We found that inter-year variation was mostly associated with members of the phyla Fusobacterium and Proteobacteria, as well Bacteroides (phylum Bacteroidetes) and Lachnospiraceae (Phylum Firmicutes). Fusobacterium and Proteobacteria tend to make up only a small component of mammalian gut microbial communities, with some exceptions including bats [26] and some primates [27], yet there is evidence that they are often highly abundant in diseased individuals suffering with a dysbiotic gut microbial community [28–30]. The exact mechanism that causes this year-to-year variation remains unclear, given that the amount of rainfall is not an important predictor of gut microbiota composition in this population [9]. However, longer term climatic conditions, such as drought, are known to affect population health in this system. Climate extremes have long-term effects on reproduction [31,32], mortality [33] and tuberculosis prevalence [34,35], and together these may produce signals of dysbiosis in the gut microbiota.
We found that social group membership played a secondary role to year effects in explaining gut microbial variation, but was nevertheless associated with a wider suite of taxa than meerkat identity. Indeed, excluding social group from models incorrectly inflated the importance of individual identity for explaining gut microbiota composition. In addition, gut microbial beta diversity tended to be more similar to other social group members than to non-group members for up to a year (figure 4). It is unclear whether these group effects are based on shared responses to environment, ample opportunity for microbial transmission between individuals, or reflect the higher genetic relatedness of group members. This underscores the importance of decomposing the often nested effects of identity, social group membership, and long-term conditions when synthesizing the relationship between gut microbial phenotype and both host genotype and fitness.
Taken together, our findings paint a picture of a highly dynamic gut microbial community, whereby individuals can perhaps be distinguished if sampling intervals are short, and idiosyncratic responses leading to weakly individualized gut microbial signatures for up to a year. While our findings do provide some support for individualized dynamics of gut microbiotas, they also call into question how close a match between host genotype and microbial phenotype can be expected. In contrast to stochastically fluctuating taxa, individualized microbes are expected to be heritable [8], and associated with a host's genotype and evolutionary lineage [12,14]. As such, individuality and repeatability form the conceptual basis of linking commensal microbiota with, for instance, host immunogenetics [36]. However, increasing evidence suggests that interactions between host genetics and the gut microbiota may vary across life, with MHC-gut microbiota associations dependent on age in bats [37], and microbiota heritability shifting with age and season in baboons [8]. As such, associations between host genotype and the gut microbiota may be very strong under certain ecological contexts, yet much weaker when averaged across seasons and demographic groups.
In conclusion, our findings demonstrate that the dynamics of specific microbial lineages are differentially driven by identity, social group membership or inter-year variation, with implications for the mechanisms by which host–gut microbe symbioses function and evolve. These results inform how to study gut microbiota dynamics of natural populations in the future, and the time frames over which gut microbial phenotypes may mediate host physiology, behaviour and fitness.
4. Material and methods
(a) . Study population and sample collection
The study population inhabits the Kalahari Desert region in South Africa (−26.96S, 21.83E). Individuals from this population are individually marked and have been monitored three to five times a week since 1993 by the Kalahari Meerkat Project [22]. Faecal samples have been collected across the entire study period from almost all monitored individuals. For this study, we analysed a subset of the samples included in [9], excluding any individuals that had three samples or less. We therefore included a total of 965 samples collected from 157 wild meerkats (mean samples per ID = 7.5, min. = 4, max. = 14) belonging to 22 social groups, sampled between 1997 and 2019 (electronic supplementary material, figure S1). Faecal samples were collected from the ground immediately after a meerkat was observed defecating, and were stored next to an icepack and frozen at −20°C within 6 to 8 h. For long-term storage, samples were then either frozen at −80°C (before 2008) or freeze-dried (after 2008). Effects of storage were minimal and are investigated in Risely et al. [9]. Fifteen soil samples were also collected from the same area, which were used to remove soil contaminants and were treated identically to faecal samples.
(b) . DNA extraction with internal standard, 16S rRNA amplification and sequencing
Before DNA extraction, NAP buffer was added to all samples [38]. A subsample of 0.6 ± 0.05 µg (wet) was taken, and 3 µl of ZymoBIOMICS Spike-in Control I (high microbial load) was added to each subsample prior to DNA extraction. This internal standard consists of cells belonging to Imtechella halotolerans and Allobacillus halotoleranss, two species which are rarely found in gut microbiota communities. An internal standard allows us to quantify ratios of absolute abundance by adding a known number of cells to each sample by which to normalize microbiota counts after sequencing. This method measures 16S copy number rather than absolute abundance, but has shown to accurately reflect variation in absolute abundances when care is taken to standardize faecal sample mass [39–42]. We have shown previously with this dataset that sample identity accounts for 90% of variation in estimated bacterial load, while 10% is technical variation [9].
The bacterial genomic DNA was extracted using the NucleoSpin 96 Soil kit (Macherey-Nagel) following the manufacturer's instructions, and the hypervariable V4 region of the 16S rRNA gene was amplified using the primer pair 515 F (5-GTGCCAGCMGCCGCGGTAA-3) and 806 R (5-GGACTACHVGGGTWTCTAAT-3). We used the Fluidigm Access Array for Illumina Sequencing Systems for indexing and adding Illumina adaptor sequences. After purification (NucleoMag NGS Clean-up and Size Select, Macherey-Nagel) and quantification (QuantiFlour dsDNA Systemt, Promega) of barcoded samples, the normalized pooled sample library was sequenced as paired-end run on Illumina MiSeq platform (2 × 250 bp) at the Institute of Evolutionary Ecology and Conservation Genomics, Ulm University. Samples were sequenced across four Illumina runs (MiSeq Reagent Kit v2, 500-cycles). Extraction and PCR negative controls were included on all runs.
(c) . Microbiome bioinformatics and normalization
All sequence reads were processed using QIIME2 version 2020.2 [43]. Sequences were merged, quality filtered and chimaeras were removed using the DADA2 pipeline [44] to generate 34 248 amplicon sequence variants (ASVs) with an average of 34 050 reads per sample [44,45]. Primers were trimmed and reads were truncated at 244 (forward) and 235 (reverse) base pairs, based on the visualization of quality plots. ASVs were assigned a taxonomy using SILVA version 132 [46]. A tree was built using QIIME2's fragment insertion method [47]. ASVs were filtered if they were not bacteria, not assigned to a phylum (as these are assumed to be spurious), or if they were classified as mitochondria or chloroplasts. These filtering steps discarded 8% of AVS and 1.8% of reads, with the vast majority of these belonging to mitochondria and chloroplasts. We used the function decontam::isContaminant [48] using the ‘prevalence' method to identify soil microbes using 15 sand samples as a reference, and to remove them from the dataset. We then divided taxa counts per sample by Allobacillus halotolerans abundance per sample to quantify ratios of absolute abundance across samples (described in [9]). Both Allobacillus and Imtechella were then removed, and all further analysis were conducted on normalized reads. Because some samples had very high relative abundances of spike-in, we only retained samples for which read depth of the true microbiome (minus the internal reference) was over 5000. After processing and the removal of soil samples, 26 122 ASVs remained in the final dataset, with an average of 27 080 reads per sample.
(d) . Sample metadata
Detailed analysis of the biological and environmental factors that are associated with meerkat gut microbiotas was conducted in [9]. Here, our aim was to quantify the contributions of identity, social group and year whil controlling for important sources of variation identified in that study. The most important predictors of taxa abundances identified were time of day, meerkat age, season, as well as sequencing depth, sequencing run and storage. We therefore included these variables in all models (described below). We measured time of day in reference to sunrise because this is more biologically meaningful than time of day. We calculated sunrise times per day using suncalc::getSunlightTimes [49]. We categorized season into wet (October to April) and dry (May to September).
(e) . Statistical analysis
We quantified the contributions of individual identity, social group membership, and year for predicting the abundances of 39 genera that were detected in over 50% of samples, 121 single-taxon phenotypes at the ASV level that were detected in at least 30% of samples, and seven community phenotypes that represented measures of bacterial load, alpha diversity and beta diversity. We lowered the threshold to 30% at the ASV level because only few ASVs reach high prevalences, while many genera do. Therefore, a 50% threshold for genera level and 30% threshold of ASV level both capture approximately 80% of sequence reads. Altering this threshold does not affect reported ICC values, but rather shifts which taxa are presented. We also estimated ICC for seven community phenotype metrics, including bacterial load (16S copy number estimated using the internal standard), observed ASV richness, Shannon diversity, Faith's phylogenetic diversity, and the first axis of variation from a multidimensional scaling (MDS) ordination using weighted UniFrac, unweighted UniFrac, and Bray–Curtis. We chose MDS over non-multidimensional scaling (NMDS) methods because NMDS models did not converge on our data, even with prevalence filtering. These measures were calculated in R using phyloseq::estimate_richness() [50], metagMisc::phyloseq_phylo_div() (https://github.com/vmikk/metagMisc), and phyloseq::ordinate().
We estimated the adjusted ICC of all three variables when included as random effects in a generalized additive mixed model (GAMM), fitted using the mgcv package [51], controlling for time of day, meerkat age, and sequencing depth as nonlinear factors, and season (wet/dry), sample storage method and sequencing run as fixed factors. We accounted for temporal autocorrelation by including an autocorrelation term in the model, nested by year. Smoothed terms were fitted using cubic (cr) splines.
ICC and 95% confidence intervals of the three random effects were calculated using the R function rptGam::rptgam (https://github.com/elipickh/rptGam). We chose a GAMM approach with a Gaussian distribution because both changes in microbiota abundances across the day and with sequencing depth are nonlinear, and ICC and 95% confidence intervals become increasingly challenging to estimate with other distributions such as Poisson. Reliable approaches for estimating ICC from models with negative binomial error distributions and/or zero inflation parameters are not yet available. GAMMs are also more likely to converge than linear models when data has high levels of nestedness, as it does here. Nevertheless, using a Gaussian approach may not always be appropriate if taxa counts are zero-inflated. We therefore also estimated ICC applying various modelling approaches, including linear models with square root transformed ASV counts modelled with a Poisson distribution, and applying both frequentist and Bayesian methods to linear models and comparing ICC estimates to GAMMs. All methods returned highly correlated estimates for ICC (electronic supplementary material, figure S6), suggesting that estimates are robust to different modelling approaches.
(i) . Phylogenetic signal of microbial phenotypes
We tested for phylogenetic signal in ICC using the functions phylosignal::lipamoran and phylosignal::phyloSignal [52], applying Moran's I index as a measure of the correlation between ICC and bacterial phylogenetic structure.
(ii) . ICC changes with sampling interval and age
To test how ICC changes with sampling interval, we generated subsets of data based on how far apart samples were taken for the same individual, and reran all models for each ASV and each sampling category. To do this, we made pairwise comparisons for every sample collected from the same individual, and calculated sampling interval for each comparison. For sampling interval, we generated X categories (0–30 days, 30–60 days, 60–90 days and so forth). Mean sample size per category was 200, with a minimum sample size of 70, and samples could be included in more than one category. We then reran identical GAM models as described above per ASV and per sampling interval category.
We applied a similar approach to test how ICC changes with meerkat age, but instead of creating subsets of data based on sampling intervals, we subsetted samples based on meerkat age at the time of sampling (young: less than 2 years, n = 346; adult: 2–4 years, n = 465; old: greater than 4 years, n = 154). GAM models were then rerun on the 121 ASVs, using identical parameters as described above, on each age subset.
(iii) . Community stability measured by ASV turnover
We estimates ASV turnover between consecutive samples collected from the same individual using the function codyn::turnover [53]. We then modelled changes to turnover using a GAMM with a Gaussian distribution and the number of days between samples and the age of the meerkat at the first sample as fixed effects, while accounting for meerkat ID, social group membership and year as random effects. Note that because samples tended to be taken regularly from individuals across the sampling interval, meerkat age at the first sample and meerkat age at the second sample were highly correlated (Pearson's r = 0.97, p < 0.0001; electronic supplementary material, figure S7a), therefore both variables had almost identical effects when included in the model. In addition, days between samples was not correlated with age at second sample (Pearson's r = 0.0003, p = 0.9; electronic supplementary material, figure S7b), therefore estimates were not bias by co-correlation between explanatory variables. Removing or adding variables had no effect on the model estimates, indicating that results are robust to changes in model structure.
(iv) . Changes to beta dissimilarity within and between individuals with sampling interval
We calculated pairwise beta dissimilarity for all samples collected within two years of each other (n = 127 885 comparisons), applying Bray–Curtis, unweighted UniFrac, and weighted UniFrac. We limited comparisons to 2 years apart because meerkats are mostly rather short-lived, and within-individual comparisons over 2 years apart were comparatively rare. We assigned pairwise comparisons into three categories: based on whether the samples were taken from the same individual, a different individual in the same group or a different individual in a different group. We then statistically tested the associations applying GAMMs, with beta dissimilarity as the response variable, and sampling interval as the smoothed variable, with smoothing factors fitted separately by comparison type.
Acknowledgements
We are grateful to the Kalahari Research Trust and the Kalahari Meerkat Project for access to facilities and habituated animals in the Kuruman River Reserve, South Africa. This paper has relied on records of individual identities and/or life histories maintained by the Kalahari Meerkat Project and collected by scientists and volunteers. We thank the Northern Cape Conservation Service for permission to conduct fieldwork, and the South African Weather Service (SAWS) for providing weather data. We thank Ben Danzer for facilitating with sample collation and storage and Ulrike Stehle for contributing to laboratory work.
Ethics
The South African Northern Cape Department of Environment and Nature Conservation gave permission to conduct the fieldwork research (FAUNA 1020/2016).
Data accessibility
All sequences and processed data used in this study are available to download at Zenodo [54]. Sequences are additionally stored under NCBI BioProject PRJNA764180. R code can be downloaded at https://github.com/Riselya/Microbiome-repeatability.
Electronic supplementary material is available online [55].
Authors' contributions
A.R.: conceptualization, formal analysis, funding acquisition, investigation, methodology, visualization, writing—original draft, writing—review and editing; D.W.S.: writing—original draft, writing—review and editing; N.M.-K.: writing—review and editing; K.W.: investigation, methodology; T.H.C-B..: data curation, funding acquisition, resources; M.B.M.: data curation, funding acquisition, resources, writing—review and editing; S.S.: conceptualization, funding acquisition, resources, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This study was supported by German Research Foundation DFG SO 428/15-1 (S.S.); European Research Council 294494 (T.H.C.-B.); European Research Council 742808 (T.H.C.-B.); Human Frontier Science; Program RGP0051/2017 (T.H.C.-B.); University of Zurich (M.B.M.); MAVA Foundation KRP 16026 (M.B.M. and T.H.C.-B.).
References
- 1.Gupta VK, Kim M, Bakshi U, Cunningham KY, Davis JM, Lazaridis KN, Nelson H, Chia N, Sung J. 2020. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 11, 1-16. ( 10.1038/s41467-019-13993-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosshart SP, et al. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015-1028. ( 10.1016/j.cell.2017.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung JM, Budischak SA, Hansen C, Bowcutt R, Neill R, Shellman M, Graham AL. 2018. Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol. 16, e2004108. ( 10.1371/journal.pbio.2004108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alavi S, Mitchell JD, Cho JY, Liu R, Macbeth JC, Hsiao A. 2020. Interpersonal gut microbiome variation drives susceptibility and resistance to cholera infection. Cell. 181, 1533-1546. ( 10.1016/j.cell.2020.05.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilmanski T, et al. 2021. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 3, 274-286. ( 10.1038/s42255-021-00348-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worsley SF, Davies CS, Mannarelli M-E, Hutchings MI, Komdeur J, Burke T, Dugdale HL, Richardson DS. 2021. Gut microbiome composition, not alpha diversity, is associated with survival in a natural vertebrate population. Anim. Microb. 3, 1-18. ( 10.1186/s42523-021-00149-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson GL, Cooke AC, Johnson CN, Quinn JL. 2018. The gut microbiome as a driver of individual variation in cognition and functional behaviour. Phil. Trans. R. Soc. B 373, 20170286. ( 10.1098/rstb.2017.0286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grieneisen L, et al. 2021. Gut microbiome heritability is nearly universal but environmentally contingent. Science 373, 181-186. ( 10.1126/science.aba5483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Risely A, Wilhelm K, Clutton-Brock T, Manser MB, Sommer S. 2021. Diurnal oscillations in gut bacterial load and composition eclipse seasonal and lifetime dynamics in wild meerkats. Nat. Commun. 12, 1-12. ( 10.1038/s41467-021-26298-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandeputte D, De Commer L, Tito RY, Kathagen G, Sabino J, Vermeire S, Faust K, Raes J. 2021. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 12, 1-13. ( 10.1038/s41467-021-27098-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat. Med. 24, 392-400. ( 10.1038/nm.4517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moeller AH, Sanders JG. 2020. Roles of the gut microbiota in the adaptive evolution of mammalian species. Phil. Trans. R. Soc. B 375, 20190597. ( 10.1098/rstb.2019.0597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groussin M, Mazel F, Alm EJ. 2020. Co-evolution and co-speciation of host-gut bacteria systems. Cell Host Microbe. 28, 12-22. ( 10.1016/j.chom.2020.06.013) [DOI] [PubMed] [Google Scholar]
- 14.Mallott EK, Amato KR. 2021. Host specificity of the gut microbiome. Nat. Rev. Microbiol. 19, 639-653. ( 10.1038/s41579-021-00562-3) [DOI] [PubMed] [Google Scholar]
- 15.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689-699. ( 10.1016/j.tree.2016.06.008) [DOI] [PubMed] [Google Scholar]
- 16.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJ, Huttenhower C. 2015. Identifying personal microbiomes using metagenomic codes. Proc. Natl Acad. Sci. USA. 112, E2930-E2938. ( 10.1073/pnas.1423854112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Price J, et al. 2017. Strains, functions and dynamics in the expanded human microbiome project. Nature 550, 61-66. ( 10.1038/nature23889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinson JN, Pinkham NV, Peters GW, Cho H, Heng J, Rauch M, Broadaway SC, Walk ST. 2019. Rethinking gut microbiome residency and the Enterobacteriaceae in healthy human adults. ISME J. 13, 2306-2318. ( 10.1038/s41396-019-0435-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren T, Grieneisen LE, Alberts SC, Archie EA, Wu M. 2015. Development, diet and dynamism: longitudinal and cross-sectional predictors of gut microbial communities in wild baboons. Environ. Microbiol. 18, 1312-1325. ( 10.1111/1462-2920.12852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Springer A, Fichtel C, Al-Ghalith GA, Koch F, Amato KR, Clayton JB, Knights D, Kappeler PM. 2017. Patterns of seasonality and group membership characterize the gut microbiota in a longitudinal study of wild Verreaux's sifakas (Propithecus verreauxi). Ecol. Evol. 7, 5732-5745. ( 10.1002/ece3.3148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Björk JR, et al. 2022. Synchrony and idiosyncrasy in the gut microbiome of wild baboons. Nat. Ecol. Evol. 6, 955-964. ( 10.1038/s41559-022-01773-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clutton-Brock TH, Manser M. 2016. Meerkats: cooperative breeding in the Kalahari. Cooperative Breeding in Vertebrates 294, 317. [Google Scholar]
- 23.Nakagawa S, Schielzeth H. 2010. Repeatability for Gaussian and non-Gaussian data: a practical guide for biologists. Biol. Rev. 85, 935-956. ( 10.1111/j.1469-185x.2010.00141.x) [DOI] [PubMed] [Google Scholar]
- 24.Stoffel MA, Nakagawa S, Schielzeth H. 2017. rptR: Repeatability estimation and variance decomposition by generalized linear mixed-effects models. Methods Ecol. Evol. 8, 1639-1644. ( 10.1111/2041-210X.12797) [DOI] [Google Scholar]
- 25.Bjork JR, et al. 2021. Synchrony and idiosyncrasy in the gut microbiome of wild primates. bioRxiv.
- 26.Song SJ, et al. 2020. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio. 11, e02901-19. ( 10.1128/mBio.02901-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallott EK, Amato KR. 2018. The microbial reproductive ecology of white-faced capuchins (Cebus capucinus). Am. J. Primatol. 80, e22896. ( 10.1002/ajp.22896) [DOI] [PubMed] [Google Scholar]
- 28.Shin N-R, Whon TW, Bae J-W. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496-503. ( 10.1016/j.tibtech.2015.06.011) [DOI] [PubMed] [Google Scholar]
- 29.Amitay EL, et al. 2017. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 38, 781-788. ( 10.1093/carcin/bgx053) [DOI] [PubMed] [Google Scholar]
- 30.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. 2017. Proteobacteria: a common factor in human diseases. BioMed Res. Int. 2017, 9351507. ( 10.1155/2017/9351507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodge SJ, Manica A, Flower TP, Clutton-Brock TH. 2008. Determinants of reproductive success in dominant female meerkats. J. Anim. Ecol. 77, 92-102. ( 10.1111/j.1365-2656.2007.01318.x) [DOI] [PubMed] [Google Scholar]
- 32.Bateman AW, Ozgul A, Nielsen JF, Coulson T, Clutton-Brock TH. 2013. Social structure mediates environmental effects on group size in an obligate cooperative breeder, Suricata suricatta. Ecology 94, 587-597. ( 10.1890/11-2122.1) [DOI] [PubMed] [Google Scholar]
- 33.Clutton-Brock TH, Maccoll A, Chadwick P, Gaynor D, Kansky R, Skinner JD. 1999. Reproduction and survival of suricates (Suricata suricatta) in the southern Kalahari. Afr. J. Ecol. 37, 69-80. ( 10.1046/j.1365-2028.1999.00160.x) [DOI] [Google Scholar]
- 34.Patterson S, Drewe JA, Pfeiffer DU, Clutton-Brock TH. 2017. Social and environmental factors affect tuberculosis related mortality in wild meerkats. J. Anim. Ecol. 86, 442-450. ( 10.1111/1365-2656.12649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paniw M, Duncan C, Groenewoud F, Drewe JA, Manser M, Ozgul A, Clutton-Brock T. 2022. Higher temperature extremes exacerbate negative disease effects in a social mammal. Nat. Clim. Change 12, 284-289. ( 10.1038/s41558-022-01284-x) [DOI] [Google Scholar]
- 36.Montero BK, Schwensow N, Gillingham MA, Ratovonamana YR, Rakotondranary SJ, Corman V, Drosten C, Ganzhorn JU, Sommer S. 2021. Evidence of MHC class I and II influencing viral and helminth infection via the microbiome in a non-human primate. PLoS Pathog. 17, e1009675. ( 10.1371/journal.ppat.1009675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleischer R, Schmid DW, Uddin W, Brändel SD, Rasche A, Corman VM, Drosten C, Tschapka M, Sommer S. 2022. Interaction between MHC diversity and constitution, gut microbiota and Astrovirus infections in a neotropical bat. Mol. Ecol. 31, 3342-3359. ( 10.1111/mec.16491) [DOI] [PubMed] [Google Scholar]
- 38.Menke S, Gillingham MA, Wilhelm K, Sommer S. 2017. Home-made cost effective preservation buffer is a better alternative to commercial preservation methods for microbiome research. Front. Microbiol. 8, 102. ( 10.3389/fmicb.2017.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stämmler F, Gläsner J, Hiergeist A, Holler E, Weber D, Oefner PJ, Gessner A, Spang R. 2016. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 4, 28. ( 10.1186/s40168-016-0175-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardwick SA, Deveson IW, Mercer TR. 2017. Reference standards for next-generation sequencing. Nat. Rev. Genet. 18, 473. ( 10.1038/nrg.2017.44) [DOI] [PubMed] [Google Scholar]
- 41.Tourlousse DM, Yoshiike S, Ohashi A, Matsukura S, Noda N, Sekiguchi Y. 2017. Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing. Nucleic Acids Res. 45, e23. ( 10.1093/nar/gkw984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y, Gifford S, Ducklow H, Schofield O, Cassar N. 2019. Towards quantitative microbiome community profiling using internal standards. Appl. Environ. Microbiol. 85, e02634-18. ( 10.1128/AEM.02634-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolyen E, et al. 2018. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints. See https://peerj.com/preprints/27295.
- 44.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581-583. ( 10.1038/nmeth.3869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639-2643. ( 10.1038/ismej.2017.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188-7196. ( 10.1093/nar/gkm864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janssen S, et al. 2018. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. Msystems. 3, e00021-18. ( 10.1128/mSystems.00021-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 1-14. ( 10.1186/s40168-018-0605-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agafonkin V, Thieurmel B. 2017. suncalc: Compute sun position, sunlight phases, moon position, and lunar phase. R package version 0.3.
- 50.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. ( 10.1371/journal.pone.0061217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood SN. 2017. Generalized additive models: an introduction with R. Boca Raton, FL: CRC press. [Google Scholar]
- 52.Keck F, Rimet F, Bouchez A, Franc A. 2016. phylosignal: an R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 6, 2774-2780. ( 10.1002/ece3.2051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallett LM, Jones SK, MacDonald AAM, Jones MB, Flynn DF, Ripplinger J, Slaughter P, Gries C, Collins SL. 2016. codyn: An r package of community dynamics metrics. Methods Ecol. Evol. 7, 1146-1151. ( 10.1111/2041-210X.12569) [DOI] [Google Scholar]
- 54.Risely A, Wilhelm K, Clutton-Brock TH, Manser M, Sommer S. 2021. Data and code for: Diurnal oscillations in gut bacterial load and composition eclipse seasonal and lifetime dynamics in wild meerkats, Suricata suricatta. Version 1.0. Zenodo. ( 10.5281/zenodo.5337076) [DOI] [PMC free article] [PubMed]
- 55.Risely A, Schmid DW, Müller-Klein N, Wilhelm K, Clutton-Brock TH, Manser M, Sommer S. 2022. Gut microbiota individuality is contingent on temporal scale and age in wild meerkats. Figshare. ( 10.6084/m9.figshare.c.6135648) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Risely A, Wilhelm K, Clutton-Brock TH, Manser M, Sommer S. 2021. Data and code for: Diurnal oscillations in gut bacterial load and composition eclipse seasonal and lifetime dynamics in wild meerkats, Suricata suricatta. Version 1.0. Zenodo. ( 10.5281/zenodo.5337076) [DOI] [PMC free article] [PubMed]
- Risely A, Schmid DW, Müller-Klein N, Wilhelm K, Clutton-Brock TH, Manser M, Sommer S. 2022. Gut microbiota individuality is contingent on temporal scale and age in wild meerkats. Figshare. ( 10.6084/m9.figshare.c.6135648) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All sequences and processed data used in this study are available to download at Zenodo [54]. Sequences are additionally stored under NCBI BioProject PRJNA764180. R code can be downloaded at https://github.com/Riselya/Microbiome-repeatability.
Electronic supplementary material is available online [55].