Abstract
Organisms living on the seafloor are subject to encrustations by a wide variety of animals, plants and microbes. Sea urchins, however, thwart this covering. Despite having a sophisticated immune system, there is no clear molecular mechanism that allows sea urchins to remain free of epibiotic microorganisms. Here, we test the hypothesis that pigmentation biosynthesis in sea urchin spines influences their interactions with microbes in vivo using CRISPR/Cas9. We report three primary findings. First, the microbiome of sea urchin spines is species-specific and much of this community is lost in captivity. Second, different colour morphs associate with bacterial communities that are similar in taxonomic composition, diversity and evenness. Lastly, loss of the pigmentation biosynthesis genes polyketide synthase and flavin-dependent monooxygenase induces a shift in which bacterial taxa colonize sea urchin spines. Therefore, our results are consistent with the hypothesis that host pigmentation biosynthesis can, but may not always, influence the microbiome in sea urchin spines.
Keywords: polyketide pigments, polyketide synthase, flavin-dependent monooxygenase, echinoderm, microbial communities, host–microbe symbiosis
1. Introduction
The shallows of most temperate and tropical seas contain diverse encrustation of invertebrates, macroalgae and biofilms. Sea urchins, on the other hand, are free of this biofouling: their calcium carbonate spines are stiff and capable of supporting epibiotic organisms, yet they do not. A closer look at their surface reveals tri-jawed claspers—pedicellaria—that can capture prey and remove parasites, detritus and other large eukaryotes (e.g. algae) [1], but these appendages do not appear to remove epibiotic microorganisms. Like many other animals, sea urchins have a sophisticated innate immune system that contains a variety of cell types involved in immune surveillance [2–4]. However, it is not clear that these cells are embedded within the calcium carbonate spines nor are responsible for preventing other organisms from colonizing their surface.
What is also striking about these animals is their coloration; spines of the approximately 950 extant sea urchin species contain polyketide-based echinochromes and spinochromes that produce a wide range of colours. Sea urchins do not use their echinochromes or spinochromes for recognition, dimorphic signalling or camouflage [5]. Instead, in vitro evidence suggest that echinochromes and spinochromes that were isolated from adult spines and tests have a number of biological activities, including antimicrobial properties (e.g. degranulation in the presence of bacterial pathogens) [6–9]. Antimicrobial compounds generally serve a dual role in host–microbe interactions: they protect the host against pathogenic invasions while also influencing which microbes colonize and become part of the host-associated microbial community [10,11]. This raises the question as to whether the molecules involved in pigmentation biosynthesis also play a role in host–microbe interactions.
Sea urchin pigments are genetically determined and are made of a base polyketide with side-chain hydroxyls that can either be substituted for modifications and/or be used in free radical scavenging [12,13]. This base polyketide is made by the enzyme polyketide synthase (PKS) with modifications derived by members of the flavin-dependent monooxygenase (Fmo3) family (electronic supplementary material, figure S1) [14,15]. To test whether sea urchin spines have bioactive pigments that is involved in host–microbe interactions, we performed three sets of field-based collections and experiments [16–18]. First, we collected the spines of several sea urchin species as well as three colour morphs of Lytechinus variegatus. Second, we transferred L. variegatus into captivity to compare to field specimens. Lastly, we used genome editing by CRISPR/Cas9 to remove PKS and Fmo3 from the sea urchin Hemicentrotus pulcherrimus to test whether pigment biosynthesis influences the microbiota of sea urchin spines in vivo [15,19–21].
2. Methods
(a) . Sample collection
To establish a baseline for the spine-associate bacterial community, we tested whether the bacterial communities associated with sea urchin spines are species-specific and of the stability of this community when urchins are transferred to captivity. These comparisons were performed through two separate collections. First, we collected Asthenosoma ijimai (n = 3), Diadema clarki (n = 11), H. pulcherrimus (n = 11) and Pseudocentrotus depressus (n = 5) from the coastal waters of Tateyama, Japan (34°59′10.9″ N, 139°48′54.4″ E). Second, we collected L. variegatus (n = 57) from Florida, USA (Biscayne Bay; 25°39′17.0″ N, 80°10′28.7″ W) and compared these to individuals held in captivity (n = 18) at Brown University (RI, USA). Moreover, unlike the species in our first collection, L. variegatus has several colour morphs and, thus, our second collection included individuals that were predominantly green (n = 7), red (n = 9) or white (n = 6). Spines from all individuals were collected using sterile scissors and forceps, and tissues were placed in sterile 2 ml microfuge tubes filled with ethanol and stored at −20°C. All tools were cleaned with 70% alcohol between individuals.
(b) . Characterization of spine pigments
The diversity of echinochromes and spinochromes was characterized for the three dominant colour morphs (green, red and white) of L. variegatus as well as the purple sea urchin Strongylocentrotus purpuratus. We used high-performance liquid chromatography (LC-MS; 1260 series, Agilent Technologies) coupled with a 6530 Accurate-Mass Q-TOF (Agilent Technologies) and operated in negative (ESI-) electrospray ionization mode. Vials containing pigment were kept at −20 °C prior to LC-MS. Reversed phase column Waters XTerra MS C18, 3.5 µm 2.1 × 50 mm column was used at 40 °C with a sample volume injected of 8 µl and flow rate of 0.3 µl min−1. The HPLC mobile phases consisted of: A = 0.1% formic acid in water and B = acetonitrile. Using an initial volume of 8 µl, the linear gradient elution used the following time program: 0 min 5% B, linear to 95% B at 9.5 min, hold at 95% for 2 min, back to 5% B at 14 min and equilibrate for 8 min. The ESI source conditions were gas temperature 300 °C, drying gas 11 l min−1, nebulizer 35 psig, VCap voltage 3500 V, fragmentor 175 V and skimmer 65 V. The instrument was tuned using an Agilent calibration tuning mix for mass calibration of the Q-TOF instrument. The reference solution provided (reference masses m/z 112.9856 and m/z 1033.9881 for ESI (−)) was used to correct small mass drift during acquisition. Data were collected in both centroid and profile formats. Data were analysed with Agilent MassHunter Qualitative Analysis (v. B.06.00).
(c) . Pigment manipulation via CRISPR/Cas9 guide RNAs
To determine whether the presence of echinochromes and spinochromes influence the microbiota in sea urchin spines, we identified the coding sequences for the pigmentation genes PKS and Fmo3 using the genomic database for H. pulcherrimus (HpBase; http://cell-innovation.nig.ac.jp/Hpul/ [22]). We then designed guide RNAs (gRNAs) for manipulating H. pulcherrimus using CRISPRscan (www.crisprscan.org) and synthesized these according to Moreno-Mateos et al. [23]. A mixture of two gRNAs (200 ng µl−1) and Cas9 mRNA (500 ng µl−1) was injected into freshly fertilized H. pulcherrimus eggs [19,21,24]. Albino larvae were collected at 3 days post-fertilization and young adults were biopsied after 1 year of culture at the Tateyama Marine Laboratory (Tateyama, Japan) for genetic analysis [21]. Spines were then collected after 18 months using the same protocol that is described above.
(d) . DNA extraction and sequencing
Total DNA was extracted from sea urchin spines and DNA kit blanks (n = 10) using a modified protocol for the DNeasy Blood & Tissue Mini Kit (Qiagen). Specifically, spines were submerged in an enzymatic lysis buffer (10 mM Tris (pH 8), 1 mM EDTA and 1% TX-100 plus 10 mg of lysozyme) that was incubated at 37°C for 20 min and then bead-beaten by vortex for 5 min. We then discarded the porous calcium carbonate structure and followed the manufacture's protocol thereafter [25]. DNA was quantified using a Qubit (Life Technologies) and diluted to 5 ng µl−1 using RNase/DNase-free water. Bacterial sequences were then amplified using primers for the V3/V4 regions of the 16S rRNA gene (electronic supplementary material, table S1) [26]. Products were purified using the Axygen AxyPrep Mag PCR Clean-up Kit (Axygen Scientific), indexed using the Nextera XT Index Kit V2 (Illumina Inc.) and then purified again. At each clean-up step, fluorometric quantitation was performed using a Qubit, and libraries were validated using a Bioanalyzer High Sensitivity DNA chip (Agilent Technologies). Illumina MiSeq sequencing (v3, 2 × 300 bp paired-end reads) was performed at the University of North Carolina at Charlotte (Charlotte, USA).
(e) . Bacterial community analysis
Raw reads and quality information were imported into QIIME 2 (v. 2021.2) [27], where forward and reverse reads were paired using VSEARCH [28], filtered by quality score, and denoized using Deblur [29]. QIIME 2-generated ‘features' were analysed as amplicon sequence variants (ASVs) [30] and were assigned taxonomy using SILVA (v.138) [31]. Sequences matching Archaea or present in the DNA kit blanks were discarded. The data table was then subdivided for each set of samples and rarified for each individual comparison: 2183 sequences for species (electronic supplementary material, figure S2), 389 sequences for captivity (electronic supplementary material, figure S3), 3582 sequences for coloration (electronic supplementary material, figure S4) and 3168 sequences for the gene knockouts (electronic supplementary material, figure S5). Samples below each threshold were discarded.
Unweighted and weighted UniFrac [32] values were calculated for each sample in QIIME 2 and compared using principal coordinate analyses to test whether community membership and composition differed between samples that were selected a priori to test differences between host species, spine coloration, environment and gene knockouts. Results from these analyses were then visualized in Prism (v. 9.0.0) and stylized using Adobe Illustrator (v. 24.0.1). Separate permutational multivariate analysis of variances (PERMANOVA) were used to compare the relatedness of these bacterial communities between host species, spine coloration, environment and gene knockouts, and this was followed by pairwise comparisons.
Multiple measures of α diversity (i.e. total ASVs, phylogenetic distance, McIntosh evenness and McIntosh dominance) were then calculated for each sample in QIIME 2. We used an analysis of variance (ANOVA) and a Tukey's post hoc test to compare community diversity between species, coloration and gene knockout, and a t-test for the captivity comparison was performed using Prism. The bacterial groups as well as the number of shared and specific ASVs were summarized. Lastly, we used an analysis of composition of microbiomes (ANCOM) to identify differentially abundant ASVs based on the underlying structure of the community [33].
Our bioinformatic pipeline used to convert raw reads to ASVs for visualization is presented in detail in the supplemental code, and raw sequence reads have been deposited in the Dryad Digital Repository: https://doi.org/10.5061/dryad.v15dv41xk [34].
(f) . Sequence alignment and phylogeny
We used the basic local alignment search tool (BLAST; [35]) to verify the taxonomy of a H. pulcherrimus ASV-of-interest, which suggested that this was an ‘uncultured Arcobacter sp.' instead of an ‘uncultured Halarcobacter sp.' We then identified all other Arcobacter species using the Prokaryotic names with Standing in Nomenclature database (bacterio.net) and downloaded the full 16S rRNA sequences from the National Center for Biotechnology Information (NCBI). These 32 sequences were also compared with the five most closely related sequences from BLAST, three Campylobacter spp., and Dissulfuribacter thermophilus for an outgroup (see electronic supplementary material, table S2 for GenBank accession numbers). All 42 sequences were imported, aligned and trimmed using the Molecular Evolutionary Genetics Analysis software (v. 11.0.9) [36], and this relationship was inferred using maximum likelihood with the optimized DNA substitution model (K2 + G, as determined by BIC criteria) and 1000 bootstrap replicates.
3. Results
(a) . Specificity in the spine microbiome
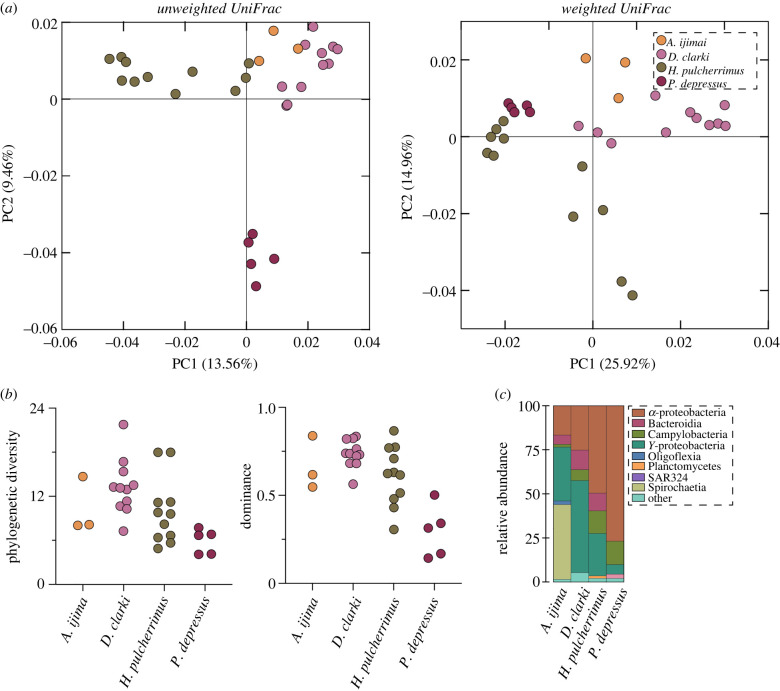
The membership (unweighted UniFrac) and composition (weighted UniFrac) of the spine microbiome differed between the four sea urchin species from Japan (PERMANOVA, p < 0.001 for both; figure 1; electronic supplementary material, table S3). This trend was consistently observed in each pairwise comparison between these species (p < 0.01 for all; figure 1; electronic supplementary material, table S3). The structure (a combined assessment of taxonomic richness (total ASVs and phylogenetic diversity) and taxonomic evenness (dominance and evenness)) of these communities also differed between sea urchin species (ANOVA; observed ASVs: p = 0.002, phylogenetic diversity: p = 0.013, dominance: p < 0.0001, evenness: p < 0.0001; figure 1; electronic supplementary material, figure S2 and tables S4, S5). This difference was largely due to P. depressus, which harboured a community that was dominated by relatively few bacteria taxa (figure 1; electronic supplementary material, figure S2 and tables S4, S5). Despite compositional and structural variation in the spine microbiome, these communities were largely represented by α-proteobacteria (42.1% on average; from 16.7% to 76.8%), γ-proteobacteria (28.1% on average; from 5.5% to 52.3%) and Spirochaetia (10.6%; from 0% to 42.4%) (figure 1; electronic supplementary material, table S6).
Figure 1.
Specificity in the spine microbiome between sea urchin species. (a) Community similarity of the spine bacterial communities of seven sea urchin species, as estimated by unweighted UniFrac for membership (left) and weighted UniFrac for composition (right). (b) The corresponding diversity estimates via phylogenetic diversity (left) and dominance (right) as well as (c) phylum-level profiles of the bacterial communities. (Online version in colour.)
(b) . Captivity restructures the spine microbiome
The membership and composition of the spine microbiome of L. variegatus differed significantly between individuals in the field and in the laboratory (PERMANOVA, p < 0.001 for both; electronic supplementary material, figure S3 and table S3). The structure of these bacterial communities also differed, whereby the bacterial community of laboratory individuals was more diverse and taxonomically dominant (t-test for observed ASVs, phylogenetic diversity, dominance, evenness: all p < 0.0001; electronic supplementary material, figure S3 and table S3). Only 9.6% of all ASVs found in the spines of wild L. variegatus were retained when cultured in the laboratory (electronic supplementary material, figure S3). Individuals from the field were primarily composed of γ-proteobacteria (70.6% on average; from 12.6% to 99.5%) and Spirochaetia (17.8% on average; from 0% to 65.3%), while laboratory individuals associated with α-proteobacteria (16.9% on average; from 2.1% to 37.8%) and γ-proteobacteria (43.4% on average; from 25.7% to 79.9%) (electronic supplementary material, figure S3 and table S7).
(c) . Consistency in the spine microbiome between colour morphs
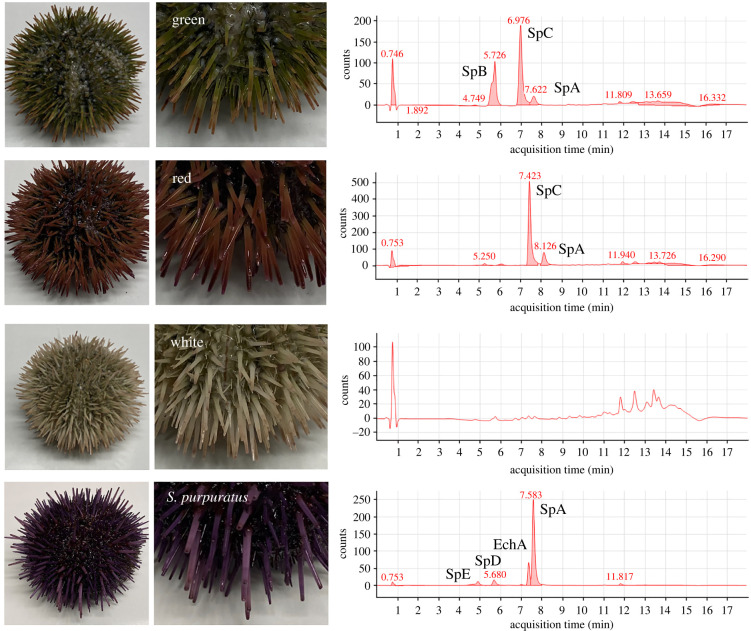
Spines of L. variegatus include spinochromes A, B and C and the combination of these produced the different colour morph (figure 2). All three spinochromes were present in the green colour morph, while only spinochrome A and C were present in the red colour morph. No spinochromes were detectable in the white colour morph (figure 2). For comparison between species, spines of the purple urchin S. purpuratus had spinochromes A, D and E as well as echinochrome A (figure 2).
Figure 2.
Colour morphs and their pigmentation chemistry. Aboral photographs (left) and high-performance liquid chromatography profiles of spine pigments from green (first row), red (second row) and white (third row) colour morphs of Lytechinus variegatus, as well as from Strongylocentrotus purpuratus (fourth row) (Sp, spinochrome; Ech, echinochromes). (Online version in colour.)
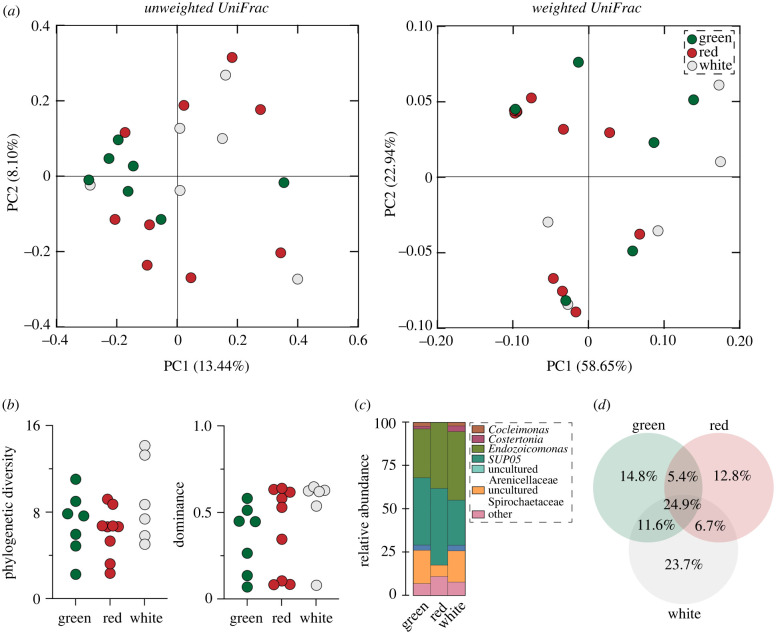
The bacterial communities associated with spines of the green, red and white colour morphs for L. variegatus were similar in membership and composition (PERMANOVA, unweighted UniFrac, p = 0.471; PERMANOVA, weighted UniFrac, p = 0.382; figure 3; electronic supplementary material, table S3). Moreover, the structure of these bacterial communities was also consistent between these colour morphs (ANOVA for each, observed ASVs: p = 0.283, phylogenetic diversity: p = 0.196, dominance: p = 0.397, evenness: p = 0.398; figure 3; electronic supplementary material, figure S4, and tables S4 and S5). The bacterial communities associated with these colour morphs primarily included Endozoicomonas (35.6% on average; from 29.0% to 39.6%), SUP05 (37.1% on average; from 24.9% to 46.1%) and an uncultured Spirochaetaceae (14.7% on average; from 6.8% to 19.8%). The green, red and white colour morphs shared 24.9% of their ASVs while any two colour morphs shared between 5.4% and 11.6% of their ASVs. Moreover, 14.8%, 12.8% and 23.7% of these ASVs were unique to the green, red and white colour morph, respectively (figure 3; electronic supplementary material, table S8).
Figure 3.
Consistency in the spine microbiome between colour morphs of Lytechinus variegatus. (a) Similarity in the membership (unweighted UniFrac) and composition (weighted UniFrac) of the bacterial communities associated with the spines of each colour morph. (b) Estimates of α diversity for each colour morph, as measured by Faith's phylogenetic diversity (left) and McIntosh dominance (right). (c) Genus-level profiles of these bacterial communities. (d) ASV similarity between the green, red and white colour morphs. (Online version in colour.)
(d) . Pigmentation biosynthesis influences the spine microbiome
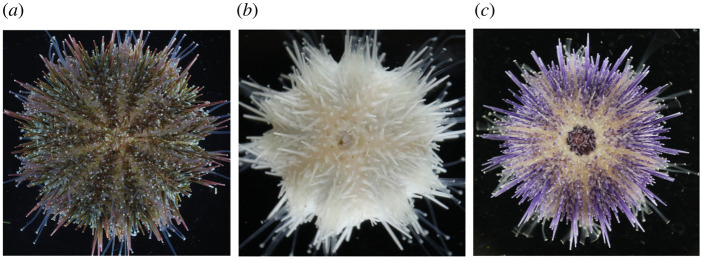
To empirically test whether the presence of pigmentation products influences which microbes reside in sea urchin spines, we targeted two genes that synthesize the non-variable pigmentation in H. pulcherrimus, a sea urchin species that has a long history in developmental biology, a relatively short generation time, and where genome editing using CRISPR/Cas9 has been established [22,37]. Genes targeted for this test were: PKS—that iteratively constructs the base polyketide—and Fmo3—that appears to modify the pigment base (electronic supplementary material, figure S1). As a result of these targeted gene removal, the spines (and all other pigmented structures) were albino for animals where PKS was knocked out and a distinct shift in pigmentation to a pastel purple occurred for animals where Fmo3 was knocked out (figure 4) [21]. No other phenotypic differences were observed.
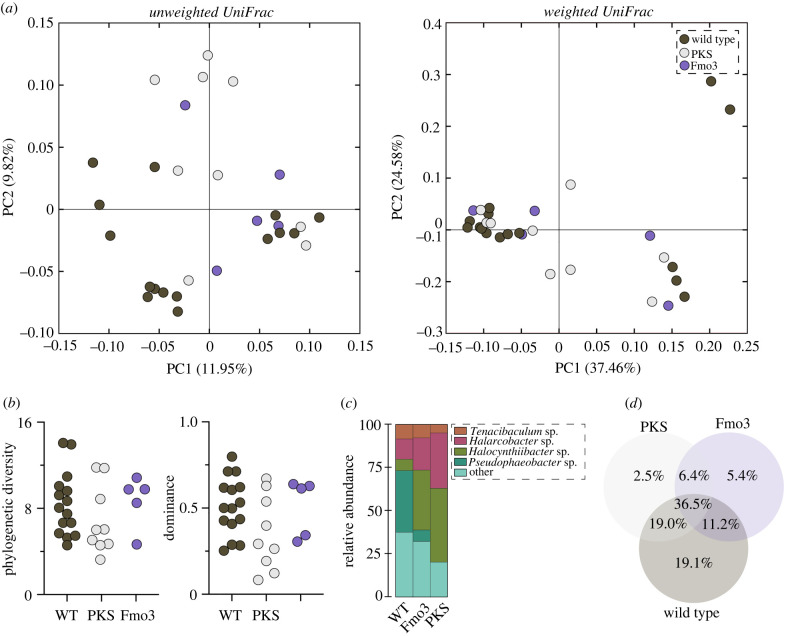
Figure 4.
Pigmentation phenotypes. Phenotypes of (a) the wild-type, (b) polyketide synthase knockout (PKS) and (c) flavin-dependent monooxygenase knockout (Fmo3) in the sea urchin Hemicentrotus pulcherrimus that were established by CRISPR/Cas9 mutagenesis. (Online version in colour.)
The membership, but not the composition, of the spine microbiome differed significantly between the wild-type, PKS knockout and Fmo3 knockout animals (PERMANOVA, unweighted UniFrac: p = 0.004; weighted UniFrac: p = 0.583; figure 5; electronic supplementary material, table S3). Moreover, based on pairwise comparisons, the membership of the PKS (p = 0.011) and Fmo3 (p = 0.035) knockouts was significantly different from the wild-type while being statistically indistinguishable from each other (p = 0.395) (figure 5; electronic supplementary material, table S3). The structure of these bacterial communities did not differ between treatments (ANOVA for each, observed ASVs: p = 0.263, phylogenetic diversity: p = 0.401, dominance: p = 0.124, evenness: p = 0. 125; figure 5; electronic supplementary material, figure S5 and table S3), and the taxonomic representation primarily included Halocynthiibacter sp. (20.9%), Pseudophaeobacter arcticus (28.0%) and Tenacibaculum sp. (21.2%) (figure 5; electronic supplementary material, table S9).
Figure 5.
Pigment composition influences the spine microbiome. (a) Community similarity of the spine bacterial communities of wild-type, PKS knockout and Fmo3 knockout in the sea urchin Hemicentrotus pulcherrimus, as estimated by unweighted UniFrac for membership (left) and weighted UniFrac for composition (right). (b) The corresponding diversity estimates via phylogenetic diversity (left) and dominance (right), as well as (c) phylum-level profiles of the bacterial communities and (d) ASV similarity between treatments. (Online version in colour.)
At the ASV level, the wild-type, PKS knockout and Fmo3 knockout shared 36.5% of their taxa, while any two treatments shared between 6.4% and 19.0%. Moreover, 5.4% and 2.5% of these ASVs were unique to the Fmo3 and PKS knockouts, while 19.1% were unique to the wild-type (figure 5). Interestingly, only one ASVs was differentially abundant relative to the microbiome composition between the wild-type and knockouts (ANCOM, p < 0.05). Comparison of the sequence for this ASV to the NCBI database, as well as to similar sequences, suggest that it is an Arcobacter (electronic supplementary material, figure S6). The relative abundance of this ASV increased 2.31× on average and to as high as 93.0% in one animal where PKS was knocked out (electronic supplementary material, table S9).
4. Discussion
Sea urchin spines are free of epibionts despite being subjected to diverse encrustations on the seafloor and having no clear antimicrobial mechanism. A series of comparisons between the bacterial communities associated with sea urchin spines suggests three primary findings. First, the bacterial communities of the spines are species-specific and that much of this community is lost upon being transferred into captivity. Second, colour morphs of a single sea urchin species have similar bacterial communities in composition and structure. Third, by removing two pigmentation biosynthesis genes from the host genome, we support the hypothesis that these molecules can influence microbiome membership in vivo. Thus, our results suggest that pigmentation biosynthesis could be a factor that influences how sea urchins interact with their microbial community.
Microbiome assembly depends on the host's evolutionary history and the environment [38–41]. One of these two factors is often more influential [39,40], but which host gene(s) or environmental parameter(s) influences these interactions often remains elusive (but see [42,43]). Furthermore, recent efforts have increased the availability of hologenomes to enable the underlying genetic factors of these evolutionarily ancient partnerships to be determined [41,44–46]. Our multi-species and multi-environment comparisons is consistent with this expectation and suggests that both factors underly community assembly of the spine microbiome in sea urchins. By editing the host genome and observing taxonomically convergent shifts in the spine microbiome, we have identified two such host factors in vivo that influence which microbes associates with sea urchin spines. This is the only instance that we are aware of where host genome editing via CRISPR/Cas9 resulted in a change in the microbiome [41].
The experimental removal of host genes involved in host–microbe interactions can disrupt symbiotic homeostasis and, as a result, can cause a microbe to shift from mutualism or neutrality towards parasitism [47,48]. Disrupting pigmentation biosynthesis (by knocking out PKS or Fmo3) is hypothesized to be one example of this, whereby a shift in microbiome membership afforded an opportunistic bacterium the ecological situation to alter its symbiotic relationship with the host. This is consistent with the behaviour of Arcobacter, which are known to transition along the mutualism-parasitism continuum and, specifically, towards pathogenicity in albino organisms [48–51]. Anecdotally, in a previous test, an entire batch of several month old PKS-null individuals died within 2 weeks of each other while wild-type siblings (that were in the same flow-through seawater tanks) remained healthy [21]. Decreased survival has been reported in another sea urchin species (Temnopleurus reevesii) whose PKS gene had also been knocked out [20]. We speculate that the significant increase in Arcobacter and mortality in albino, but not wild-type, animals of two different species with the same gene targeting that yielded the same outcome is not a coincidence, suggesting a mechanism for future experiments.
If pigmentation biosynthesis influences which bacterial lineages that sea urchin associate with, then one expectation would be that different colour morphs would also associate with district bacterial communities [52]. This expectation was not met by L. variegatus, despite a similar pattern having been observed in S. intermedius [52]. We provide four hypotheses for why this may be the case. First, pigmentation biosynthesis could have a general—as opposed to a fine-tune—effect on which bacterial lineages colonize the spines and the regulation of their interactions. Therefore, any disruption to the pigmentation biosynthesis pathway would affect which bacterial lineages associate with the sea urchin. Second, other bioactive molecules derived from pigmentation biosynthesis may also influence host–microbe interactions. This may explain why the white colour morph that are devoid of spinochromes had a bacterial community that was similar to the green and red colour morphs. One may, thus, speculate that the white colour morph and albino knockout may not be analogous. Third, the spectrum of influence that pigmentation has in how sea urchins interact with microbial symbionts may be wide-ranging between host species. Lastly, pigmentation may play a minor role or no role in shaping how sea urchins interact with microbial symbionts, even if we observed significant shifts in the microbiome like those in these experiments.
Taken together, the data presented here suggest that pigmentation biosynthesis influences the microbiome of sea urchin spines in vivo and that these pigments may be central to interaction of hosts and their bacterial communities. Each of the potential explanations described above will require refined isolation of distinct populations and members of the microbiome, to define their growth conditions, and mechanisms of interactions. Nevertheless, the genetic manipulation of the sea urchin H. pulcherrimus provides a proof-of-concept for the utility of CRISPR/Cas9 in studying host–microbe interactions and, specifically, for determining which host genetic factors are involved in partnerships with microbial symbionts [45].
Acknowledgements
We thank Karen Lopez and Daniel Janies (UNC Charlotte, USA) for sequencing resources and technical assistance with sequencing. We thank Scott and Kate from Scotts Services (Miami, USA) for essential work in surveying, collecting and assisting in specimen preparation.
Contributor Information
Gary M. Wessel, Email: rhet@brown.edu.
Tyler J. Carrier, Email: tcarrier@geomar.de.
Data accessibility
Amplicon data, processed data tables and the code used here are available on the Dryad Digital Repository: https://doi.org/10.5061/dryad.v15dv41xk [34].
The data are provided in electronic supplementary material [53].
Authors' contributions
G.M.W.: conceptualization, funding acquisition, investigation, methodology, resources, writing—original draft, writing—review and editing; M.K.: methodology, project administration; A.M.R.: funding acquisition, methodology, project administration, resources, software, supervision; T.J.C.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
G.M.W. was supported by US National Institutes of Health (NIH 1R35GM140897) and the US National Science Foundation (NSF IOS-1923445); A.M.R. was supported by funds from the Bioinformatics Research Center at UNC Charlotte (NC, USA); and T.J.C. was supported by a post-doctoral fellowship from the Alexander von Humboldt Foundation, the CRC 1182 (‘Origin and Function of Metaorganisms'; Deutsche Forschungsgemeinschaft, Project-ID 261376515) and the GEOMAR Helmholtz Centre for Ocean Research.
References
- 1.Hyman L. 1955. The invertebrates. New York, NY: McGraw-Hill. [Google Scholar]
- 2.Hibino T, et al. 2006. The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349-365. ( 10.1016/j.ydbio.2006.08.065) [DOI] [PubMed] [Google Scholar]
- 3.Rast J, Smith L, Loza-Coll M, Hibino T, Litman G. 2006. Genomic insights into the immune system of the sea urchin. Science 314, 952-956. ( 10.1126/science.1134301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith L, et al. 2018. Echinodermata: the complex immune system in echinoderms. In Advances in comparative immunology (ed. Cooper E), pp. 409-501. Berlin, Germany: Springer. [Google Scholar]
- 5.O'Hara T, Byrne M. 2017. Australian echinoderms: biology, ecology and evolution. Victoria, Australia: CSIRO Publishing. [Google Scholar]
- 6.Buckley K, Ho E, Hibino T, Schrankel C, Schuh N, Wang G, Rast J. 2017. IL17 factors are early regulators in the gut epithelium during inflammatory response to Vibrio in the sea urchin larva. eLife 6, e23481. ( 10.7554/eLife.23481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho E, Buckley K, Schrankel C, Schuh N, Hibino T, Solek C, Bae K, Wang G, Rast J. 2016. Perturbation of gut bacteria induces a coordinated cellular immune response in the purple sea urchin larva. Immunol. Cell Biol. 94, 861-874. ( 10.1038/icb.2016.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasseur L, Hennebert E, Fievez L, Caulier G, Bureau F, Tafforeau L, Flammang P, Gerbaux P, Eeckhaut I. 2017. The roles of spinochromes in four shallow water tropical sea urchins and their potential as bioactive pharmacological agents. Mar. Drugs 15, 179. ( 10.3390/md15060179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadek S, Hassanein S, Mohamed A, Soliman A, Fahmy S. 2021. Echinochrome pigment extracted from sea urchin suppress the bacterial activity, inflammation, nociception, and oxidative stress resulted in the inhibition of renal injury in septic rats. J. Food Biochem. 46, e13729. ( 10.1111/jfbc.13729) [DOI] [PubMed] [Google Scholar]
- 10.Franzenburg S, Walter J, Kunzel S, Wang J, Baines J, Bosch T, Fraune S. 2013. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl Acad. Sci. USA 110, E3730-E3738. ( 10.1073/pnas.1304960110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosch T, Zasloff M. 2021. Antimicrobial peptides—or how our ancestors learned to control the microbiome. mBio 12, e01847-21. ( 10.1128/mBio.01847-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise M. 2011. Phenotypic and genetic diversity in the sea urchin Lytechinus variegatus. Durham, NC: Duke University Press. [Google Scholar]
- 13.Perillo M, Oulhen N, Foster S, Spurrell M, Calestani C, Wessel G. 2020. Regulation of dynamic pigment cell states at single-cell resolution. eLife 9, e60388. ( 10.7554/eLife.60388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke R. 2022. Pigment cells: Paragons of cellular development. In Gene regulatory mechanisms in development and evolution: insights from echinoderms (ed. Ettensohn C), pp. 149-182. Amsterdam, The Netharlands: Elsevier. [DOI] [PubMed] [Google Scholar]
- 15.Calestani C, Wessel G. 2018. These colors don't run: regulation of pigment—biosynthesis in echinoderms. In Marine organisms as model systems in biology and medicine (eds Kloc M, Kubiak J), pp. 515-525. Berlin, Germany: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Ridder C, Foret T. 2001. Non-parasitic symbioses between echinoderms. In Echinoderm studies (eds Lawrence J, Jangoux M). Abingdon, UK: Routledge. [Google Scholar]
- 17.Foret T, Lawrence J. 2001. Variation in abundance of subcuticular bacteria in Florida echinoderms. Symbiosis 31, 309-322. [Google Scholar]
- 18.Kelly M, Barker M, McKenzie J, Powell J. 1995. The incidence and morphology of subcuticlar bacteria in the echinderm fauna of New Zealand. Biol. Bull. 189, 91-105. ( 10.2307/1542459) [DOI] [PubMed] [Google Scholar]
- 19.Oulhen N, Wessel G. 2016. Albinism as a visual, In Vivo guide for CRISPR/Cas9 functionality in the sea urchin embryo. Mol. Reprod. Dev. 83, 1046-1047. ( 10.1002/mrd.22757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaguchi S, Yaguchi J, Suzuki H, Kinjo S, Kiyomoto M, Ikeo K, Yamamoto T. 2020. Establishment of homozygous knock-out sea urchins. Curr. Biol. 30, R427-R429. ( 10.1016/j.cub.2020.03.057) [DOI] [PubMed] [Google Scholar]
- 21.Wessel G, Kiyomoto M, Shen T-L, Yajima M. 2020. Genetic manipulation of the pigment pathway in a sea urchin reveals distinct lineage commitment prior to metamorphosis in the bilateral to radial body plan transition. Sci. Rep. 10, 1973. ( 10.1038/s41598-020-58584-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinjo S, Kiyomoto M, Yamamoto T, Ikeo K, Yaguchi S. 2018. HpBase: a genome database of a sea urchin, Hemicentrotus pulcherrimus. Dev. Growth Differ. 60, 174-182. ( 10.1111/dgd.12429) [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Mateos M, Vejnar C, Beaudoin J, Fernandez J, Mis E, Khokha M, Giraldez A. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982-988. ( 10.1038/nmeth.3543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shevidi S, Uchida A, Schudrowitz N, Wessel G, Yajima M. 2017. Single nucleotide editing without DNA cleavage using CRISPR/Cas9-deaminase in the sea urchin embryo. Dev. Dyn. 246, 1036-1046. ( 10.1002/dvdy.24586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ketchum R, Smith E, Vaughan G, Phippen B, McParland D, Al-Mansoori N, Carrier T, Burt J, Reitzel A. 2018. DNA extraction method plays a significant role when defining bacterial community composition in the marine invertebrate Echinometra mathaei. Front. Mar. Sci. 5, 255. ( 10.3389/fmars.2018.00255) [DOI] [Google Scholar]
- 26.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. ( 10.1093/nar/gks808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolyen E, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852-857. ( 10.1038/s41587-019-0209-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. ( 10.7717/peerj.2584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir A, et al. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639-2643. ( 10.1038/ismej.2017.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590-596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228-8235. ( 10.1128/AEM.71.12.8228-8235.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal S, Van Treuren W, White R, Eggesbø M, Knight R, Peddada S. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Micro. Ecol. Health Dis. 26, 27663. ( 10.3402/mehd.v26.27663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessel G, Kiyomoto M, Reitzel A, Carrier T. 2022. Data from: Sea urchin pigmentation influences the microbiome. Dryad Digital Repository. ( 10.5061/dryad.v15dv41xk) [DOI] [PMC free article] [PubMed]
- 35.Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, Yamamoto T. 2010. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15, 875-885. [DOI] [PubMed] [Google Scholar]
- 38.Carrier T, Lessios H, Reitzel A. 2020. Eggs of echinoids separated by the Isthmus of Panama harbor divergent microbiota. Mar. Ecol. Prog. Ser. 648, 169-177. ( 10.3354/meps13424) [DOI] [Google Scholar]
- 39.Thomas T, et al. 2016. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 11870. ( 10.1038/ncomms11870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinstein S, Martínez-Mota R, Stapleton T, Klure D, Greenhalgh R, Orr T, Dale C, Kohl K, Dearing M. 2021. Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl Acad. Sci. USA 118, e2108787118. ( 10.1073/pnas.2108787118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu E, Davenport E. 2022. Host genetics determinants of the microbiome across animals: from Caenorhabditis elegans to cattle. Annu. Rev. Anim. Biosci. 10, 203-226. ( 10.1146/annurev-animal-020420-032054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ketchum R, Smith E, Vaughan G, McParland D, Al-Mansoori N, Burt J, Reitzel A. 2021. Unraveling the predictive role of temperature in the gut microbiota of the sea urchin Echinometra sp. EZ across spatial and temporal gradients. Mol. Ecol. 30, 3869-3881. ( 10.1111/mec.15990) [DOI] [PubMed] [Google Scholar]
- 43.Kurilshikov A, et al. 2021. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156-165. ( 10.1038/s41588-020-00763-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna V, et al. 2021. The Aquatic Symbiosis Genomics Project: probing the evolution of symbiosis across the tree of life. Wellcome Open Res. 6, 254. ( 10.12688/wellcomeopenres.17222.1) [DOI] [Google Scholar]
- 45.Douglas A. 2019. Simple animal models for microbiome research. Nat. Rev. Microbiol. 17, 764-775. ( 10.1038/s41579-019-0242-1) [DOI] [PubMed] [Google Scholar]
- 46.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229-3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McFall-Ngai M. 2007. Care for the community. Nature 445, 153. ( 10.1038/445153a) [DOI] [PubMed] [Google Scholar]
- 48.Hentschel U, Steinert M, Hacker J. 2000. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8, 226-231. ( 10.1016/S0966-842X(00)01758-3) [DOI] [PubMed] [Google Scholar]
- 49.Drew G, Stevens E, King K. 2021. Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 19, 623-638. ( 10.1038/s41579-021-00550-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira S, Queiroz J, Oleastro M, Domingues F. 2016. Insights in the pathogenesis and resistance of Arcobacter: a review. Crit. Rev. Microbiol. 42, 364-383. ( 10.3109/1040841X.2014.954523) [DOI] [PubMed] [Google Scholar]
- 51.Hamann E, et al. 2016. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534, 254-258. ( 10.1038/nature18297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balakirev E, Pavlyuchkov V, Ayala F. 2008. DNA variation and symbiotic associations in phenotypically diverse sea urchin Strongylocentrotus intermedius. Proc. Natl Acad. Sci. USA 105, 16 218-16 223. ( 10.1073/pnas.0807860105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wessel G, Kiyomoto M, Reitzel A, Carrier T. 2022. Data from: Pigmentation biosynthesis influences the microbiome in sea urchins. Figshare. ( 10.6084/m9.figshare.c.6135664) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wessel G, Kiyomoto M, Reitzel A, Carrier T. 2022. Data from: Sea urchin pigmentation influences the microbiome. Dryad Digital Repository. ( 10.5061/dryad.v15dv41xk) [DOI] [PMC free article] [PubMed]
- Wessel G, Kiyomoto M, Reitzel A, Carrier T. 2022. Data from: Pigmentation biosynthesis influences the microbiome in sea urchins. Figshare. ( 10.6084/m9.figshare.c.6135664) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Amplicon data, processed data tables and the code used here are available on the Dryad Digital Repository: https://doi.org/10.5061/dryad.v15dv41xk [34].
The data are provided in electronic supplementary material [53].