Abstract
Many RNA-binding proteins (RBPs) that assemble into membraneless organelles have a common architecture including disordered prion-like domain (PLD) and folded RNA-binding domain (RBD). An enrichment of PLD within the condensed phase gives rise to formation, on longer time scales, of amyloid-like fibrils (aging). In this study, we employ coarse-grained Langevin dynamics simulations to explore the physical basis for the structural diversity in condensed phases of multi-domain RBPs. We discovered a highly cooperative first-order transition between disordered structures and an ordered phase whereby chains of PLD organize in fibrils with high nematic orientational order. An interplay between homodomain (PLD-PLD) and heterodomain (PLD-RBD) interactions results in variety of structures with distinct spatial architectures. Interestingly, the different structural phases also exhibit vastly different intracluster dynamics of proteins, with diffusion coefficients 5 times (disordered structures) to 50 times (ordered structures) lower than that of the dilute phase. Cooperativity of this liquid-solid transition makes fibril formation highly malleable to mutations or post-translational modifications. Our results provide a mechanistic understanding of how multi-domain RBPs could form assemblies with distinct structural and material properties.
Significance
Assembly of proteins and nucleic acids into dense, liquid-like pockets is associated with several key functions including stress response, gene regulation, DNA repair, and RNA processing. Several RNA-binding proteins such as FUS are known to form liquid-like condensates that progressively harden into more dynamically solid-like structures, a phenomenon that is accelerated by disease mutations. In this study, we discover the mechanistic origins of this transition and show that small mutational or post-translational modifications could result in sharp disorder-order transitions that could characterize accelerated liquid-solid transition in disease mutants.
Introduction
Spatial segregation of biomolecules is a vital mechanism by which cellular machinery and biochemical reactions become spatiotemporally localized (1,2). While classical examples of such spatiotemporal segregation involve physical compartments bound by a membrane, more recent evidence points toward the presence of biomolecule-rich compartments that form and disassemble in response to external cues (3, 4, 5). These structures, known as membraneless organelles (MLOs), have been associated with diverse cellular functions, ranging from cell division and ribosome synthesis to stress response (1,3,6, 7, 8). Despite the diversity in function, the physical mechanisms resulting in their formation show remarkable similarity. The hallmark of these structures is the ability to assemble into droplet-like assemblies via a mechanism called liquid-liquid phase separation (LLPS) (8, 9, 10).
A key class of membraneless assemblies is ribonucleoprotein granules (RNPs), which are a heterogeneous mix of RNA-binding proteins (RBPs) and RNA (11, 12, 13). Proteins localized within RNP granules are often RBPs rich in low-complexity domains enriched in a select few amino acids (3,6,13,14). The low-complexity domains, or prion-like domains (PLDs), typically display a high degree of intrinsic disorder (3,6,13,14). Intrinsically disordered regions (IDRs) have been previously reported to be key drivers of phase separation in RBPs into liquid-like droplets. The interaction of RBPs with nucleic acids is mediated by the folded domains localized within the RNA-binding domain (RBD) of the proteins: the RNA-recognition motif (RRM) and the zinc finger domain (ZnF) (13). The multi-domain architecture of these proteins also results in multiple modes of intermolecular interactions (PLD-PLD and PLD-RBD) stabilizing the droplet phase (13). In their seminal work involving the FUS family of proteins, Wang et al. demonstrated that phase separation could be driven by hydrophobic interactions involving the PLD or through pi-pi and pi-cation interactions between the PLD and the RBD (14). A switch in the predominant mode of intermolecular interaction (PLD-PLD versus PLD-RBD) not only results in altered threshold concentrations for phase separation but also structures with different material or dynamic properties (13). The PLD of FUS is known to form ordered amyloid-fibrillar structures (15). On the other hand, while full-length FUS proteins phase separate into spherical droplets with liquid-like properties initially (16), they are known to progressively reorganize and assume more filamentous geometries, which are dynamically solid-like (17). This liquid-solid transition gains further importance in disease biology where mutations to the FUS low-complexity domain are known to accelerate the process (18). Several experimental studies have probed the ability of FUS, FUS PLD, and FUS variants to spontaneously self-assemble into liquid-like droplets in vitro (13,15,18, 19, 20). Structural elucidation of FUS assemblies using solution and solid-state NMR studies have revealed that the FUS PLD can assume a disordered configuration (in liquid-like FUS droplets) (16) or an ordered solid-like amyloid cross- fold in FUS hydrogels (15). The wide range of structural and dynamic phases accessed by FUS makes it a useful model protein to study structural (and dynamic) transitions in multi-domain RBP assemblies.
Biomolecular simulations of varying resolutions have previously been employed to study the equilibrium (19,21) and dynamic properties (droplet surface tension, shear viscosities) (19) of FUS droplets. Simulations have also been employed to identify the key intermolecular interactions stabilizing the condensed phase in FUS droplets (22). A recent computational study by Garaizar et al. demonstrates how an increase in strength of PLD-PLD interactions could result in spatially heterogeneous condensates (23). While the thermodynamics of FUS phase separation has been extensively characterized using computational models, the factors influencing the morphology of the assemblies is yet to be systematically explored. Furthermore, while most of the earlier computational models can capture the spherical droplet phase in FUS, the “fibril-like” ordered state has not been captured in simulations.
In this study, we employ a coarse-grained representation of the full-length FUS protein to understand the physical mechanisms underlying emergence of order in multi-domain protein assemblies such as those constituted by RBPs. Using this coarse-grained representation, we explore how the variation of homodomain (PLD-PLD) and heterodomain (PLD-RBD) interaction strengths within a narrow, biologically relevant regime (<1 kT) could give rise to self-assembled structures with diverse morphologies. We scan a broad range of conditions to establish a relationship between interaction networks, phosphorylation, intracluster density, and structural order within the condensed phase. Our simulations show that biomolecular assemblies could transition from a disordered to an ordered phase by switching the predominant mode of interaction stabilizing the condensed phase. More generally we observe the emergence of an ordered phase in a first-order-like phase transition whereby loss of entropy due to local ordering of individual polymer chains in fibrils is compensated by favorable interaction energy in a more dense, ordered phase. Our simulations also show that the disorder-order structural transitions are also accompanied by dramatically altered intracluster dynamics of polymers, making them biologically relevant.
Model
FUS proteins contain two domains critical for their phase separation into condensates: a low-complexity PLD and an RBD. The PLD and the RGG region (arginine-, glycine-rich region) of the RBD exhibit a high degree of disorder (24). Earlier studies have established the ability of the full-length FUS proteins (13) as well as the PLDs to form liquid-like droplets (13) at different saturation concentrations.
Coarse-graining resolution
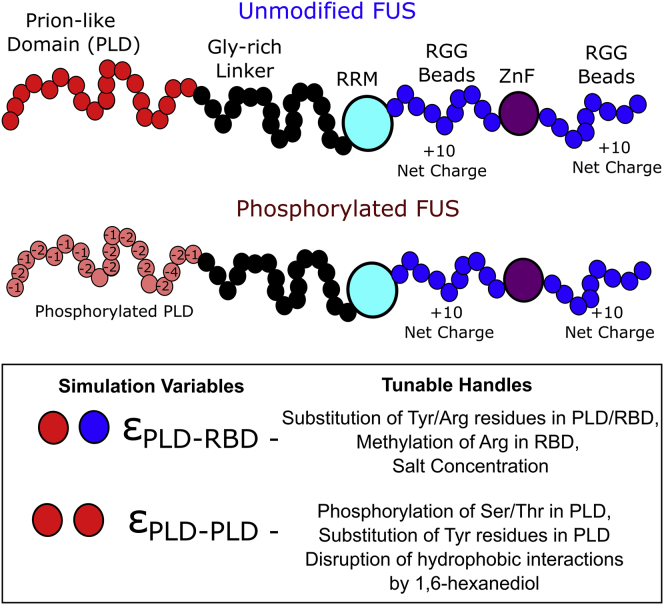
In this work, we employ a coarse-grained model for a model scaffold protein, FUS, whereby we coarse-grain stretches of 8 amino acids on the PLD into an effective interaction site (Fig. 1). In effect, the 160-residue-long PLD of FUS is modeled through 20 effective interacting beads. The RGG region is represented at a resolution of 5 amino acids per bead, with 20 effective RGG interaction beads making up the coarse-grained FUS chain. The choice of different ratios of residues per bead was to ensure that all regions in the coarse-grained polymer chains have a comparable contour length of 20 beads each, thereby ensuring that the orientational ordering is not due to a difference in contour lengths. The slight difference in ratio of amino acids/beads allows us to study ordering due to enthalpic effects and rule out effects due to differences in local rigidity. The charges of the PLD and the RBD beads were fixed based on the net charge of an amino acid stretch coarse-grained by the bead (see Table S1 for the FUS sequence). The net charge on the RBD is positive owing to the high density of arginine residues in the RGG. To model phosphorylated PLD (Fig. 1, phosphorylated FUS), we assume 31 potential phosphosites (25,26) (Table S2). For simulations involving RNA chains, we employed semiflexible polymer chains mimicking 200-residue-long, unstructured single-stranded, homopolymer RNA. Each bead of the coarse-grained RNA chain represents a single RNA base and is negatively charged.
Figure 1.
The model. Coarse-grained representation of unmodified and fully phosphorylated variants of FUS. Red beads represent 8-amino-acid long coarse-grained PLD stretches. Blue beads represent 5-amino-acid long stretches from the RGG region of FUS. The glycine-rich linkers are shown in black. Two folded domains, the RNA-recognition motif (RRM) and the zinc finger domain, are modeled as large idealized beads. The two key simulation variables, εPLD-PLD and εPLD-RBD, and their physical changes in this study are shown in the box. The blue beads in the upper figure are, for simplicity, referred to as RBD beads in the paper. In Tables S1 and S2, we provide the FUS protein sequence and the location of phosphorylation sites used for assigning charges to the coarse-grained beads of the PLD and RGG. Note that this schematic is for representative purposes and not drawn to scale.
Size of coarse-grained beads
The folded stretches of the protein, RRM and the ZnF motif, are modeled as large effective interaction domains. The size (radius) of the RGG and PLD beads was set to 6 Å while the folded RRM and ZnF motifs were of 8 Å. For bead size calculations, we assume that the 6- to 8-amino-acid stretches within the PLD and RGG behave like random coils. According to the Flory theory, polymer chains scale by Naaϒσaa. For random coil regions of the protein (PLD region, linker region, and RGG region of RBD), the scaling exponent of ϒ = 2/5 was used while the compactness of the folded ZnF and RRM domains was modeled with a ϒ of 0.3. We assume a σaa of 4.5 Å for each amino acid. Based on these values, we obtain a bead radius of about 6 Å (per 6–8 residues) for disordered regions of the protein and ∼8 Å for the folded domains. In Table S3, we provide details of the different model parameters chosen for the study. The RBD and the PLD are connected by a flexible linker that does not participate in interactions, mimicking the glycine-rich region in FUS. The glycine-rich linker (∼100 residues long) is modeled with 20 beads in our study. Each effective interaction site harbors a charge based on the corresponding region of the full-length protein sequence and the phosphorylation state of the protein. The persistence length () of the PLD and RGG regions was set to 4 residues while that of the glycine linker was set to 1 residue to model greater flexibility of the glycine-rich region.
Rationale behind choice of key simulation variables
The RRM and the ZnF regions of the protein are folded domains which are primarily involved in FUS-RNA interactions (27,28). These regions have not been reported to play a role in the self-assembly of FUS. The low-complexity PLD and the RGG regions of the protein, on the other hand, have been established to be the primary drivers of LLPS in experimental studies involving FUS (17,31). Therefore, in this study we primarily focus on the interplay between PLD-RBD and PLD-PLD interactions, assuming that the RRM and ZnF domains do not play a key role in self-assembly. For this reason we use two interaction variables, and , as effective interaction parameters (see materials and methods) that capture the physicochemical properties of the two FUS domains (PLD and RBD) as well as solvent conditions. Physically, any change in solvent conditions or in the protein sequence would manifest itself via a change in either (or both) these interaction parameters (tunable handles in Fig. 1). Therefore, for the rest of this paper we present phase diagrams wherein we use and as our key sequence and environment-dependent phase parameters.
Materials and methods
Coarse-grained models for proteins and peptides, at varying resolutions, have been used to study the relationship between biomolecular structure and self-assembly (14,21,29). Owing to the fewer degrees of freedom and higher computational tractability, these coarse-grained models are efficient tools to probe structural transitions in self-assembling polymers (30). To study the diversity in self-assembled states in response to varying PLD-PLD and PLD-RBD interaction strengths, we performed Langevin dynamics simulations. To simulate self-assembly, we perform simulations with 100 coarse-grained FUS chains (61 beads each) in a cubic box with periodic boundary conditions. Previous computational studies with FUS have established that dynamic and equilibrium quantities converge at system sizes of 50 and above, suggesting that a system size of 100 monomers is large enough to avoid finite size effects (19,21). The coarse-grained FUS polymer beads interact via the following potential functions. The neighboring beads in a polymer chain are bonded via the harmonic potential with energy
| (1) |
where and correspond to the positions of the and beads, respectively, and is the resting bond length. denotes the spring constant and has units of kT/ and was set to 5 kT/. This interaction ensures the connectivity between the adjacent beads of the same polymer chain.
To model semiflexible polymers, neighboring bonds in a polymer chain interact via a bending potential
| (2) |
where refers to the angle between and bond. Here, is the bending stiffness with energy unit kT. The value of was chosen such that the persistence length (Lp = ) of the polymer chain was ∼4 FUS PLD/RGG/Linker beads. It must therefore be noted that the contour length of the PLD (LPLD) and the RBD (LRBD) regions in the coarse-grained polymer are five times that of the persistence length of the polymer. The contour length of the full-length coarse-grained FUS is 15 times that of the persistence length, indicating the semiflexible nature of the modeled polymer chains. The persistence length of RNA chains was set at 8 beads, with a contour length of 200 for the RNA chains.
Interactions between any pair of nonbonded beads were modeled using the Lennard-Jones (LJ) potential,
| (3) |
for all rc. Here, refers to the interaction cutoff beyond which LJ interactions are neglected. The cutoff for LJ calculations were set to 2.5 times . The values of and corresponding to different pairwise interactions and bead types can be found in Table S3.
To model electrostatic interactions, we use the standard Debye-Huckel electrostatic potential, which has the form
| (4) |
and are the charges of the two interacting beads. Each RNA bead carries a single negative charge in our simulations. As previously demonstrated by Dignon et al. (21,31), the Debye-Huckel potential can robustly capture features of phase-separating proteins such as TDP43 and FUS. We therefore use the Debye-Huckel potential with a screening length of 1 nm (corresponding to an ionic strength of 100 mM) in our simulations to mimic the screening of electrostatic interactions in a cellular environment.
The attractive part of the LJ potential, , is tuned to vary the strength of the interaction. In this study, the strength of interaction between PLD beads is referred to as while PLD and the disordered regions of RBD (RGG beads) interact with a strength . The pairwise interaction strengths for all bead types—PLD, RBD (referring to the disordered parts of RBD), linker, RRM, and ZnF domains—are listed in Table S3. We used the LAMMPS molecular dynamics package (32) to perform the simulations, whereby the simulator solves Newton’s equations with viscous force, and a Langevin thermostat ensuring an NVT ensemble with temperature K. An integration timestep () of 20 fs was used for the simulations.
Metadynamics
To enable efficient sampling of the self-assembly landscape and ensure the formation of a single large cluster, we employed metadynamics (33). In this method, the evolution of the self-assembling system is influenced by a history-dependent biasing potential. Energy Gaussians are deposited during the course of a trajectory along the free-energy space for a chosen reaction coordinate that effectively describes the process of self-assembly. The deposited potentials are then used to reconstruct the free-energy profiles along this chosen reaction coordinate (33). To drive self-assembly and observe coalescence of protein-rich clusters into a single large cluster, we used the radius of gyration of the peptide system, as the reaction coordinate. is the radius of gyration of an imaginary polymer that links together the center of masses of the individual polymer chains in the simulation. Large values of correspond to a fully mixed system, whereas smaller values indicate a self-assembled state (Figs. S1 and S2). The systems were simulated until the free-energy profiles showed convergence.
Simulation protocol
We first run metadynamics simulations (with the as the collective variable) to compute the free-energy profiles and also ensure that the coarse-grained chains coalesce into one large cluster at simulation timescales. This single large, coalesced structure corresponding to the free-energy minima ( at ∼50 Å in Fig. S1) was then used as a starting configuration for conventional Langevin dynamics trajectories (with no biasing potential) of at least 10 μs or until the simulations reached convergence. In Figs. S3 and S4, we provide proof of principle for convergence of our simulations. To ensure convergence, we also start our simulations with different initial configurations—fully mixed disordered state, fibril-like core state—and then allow these structures to evolve under different regimes of εPLD-PLD and εPLD-RBD. The computed order parameters were found to be independent of the starting configuration, thereby establishing convergence (Fig. S4). These unbiased 10-μs-long simulation trajectories were used to compute statistical quantities (orientational order, interprotein contacts). These unbiased trajectories were also used to compute intracluster dynamics and autocorrelation functions. We run 10 independent Langevin dynamics trajectories (Fig. S3).
Orientational order parameter
To characterize structural order in the FUS assemblies we employ the nematic order parameter, which is commonly used in liquid-crystal literature to quantify the extent of alignment of molecules along a reference axis (34, 35, 36). We first define the long axis of each of the “N” FUS chains within the self-assembly by a unit vector , which connects the two terminal beads of the PLD domain. We compute the extent of alignment in each local region around the “N” polymer chains using the local nematic order parameter S:
| (5) |
where the index i ranges from 1 to N, the total number of polymer chains in the system.
in the above equation is a unit vector that corresponds to the direction of local alignment for a set of Nneigh polymer chains (including the ith chain) that define the local region around any polymer chain “i.” To define a local region around each polymer chain, any chain that lies within an interaction radius of 2.5 is considered to be in physical contact. Therefore, using Eq. 5 we get “N” such scalar order parameter values, each corresponding to a local region defined by every polymer chain in the system. Next, we average over all local order parameters:
| (6) |
In our study, we use <S> (computed using Eq. 6) to define the extent of local ordering within a self-assembled structure. In the rest of this article, we use S instead of <S> to refer to the mean orientational order in the system. This order parameter S ranges from 0 to 1. indicates weak orientational order while suggests high local orientational order.
To obtain the director vector required to compute the orientational order parameter using Eq. 5, we define a second-order tensor, , which reflects nematic ordering for each local region corresponding to the “N” FUS chains in the system:
| (7) |
where , are component vectors ( x, y, or z) corresponding to each of the chains (including the ith chain) with which the ith chain is physically in contact. The index refers to the identity of each chain in the system. We therefore have such independent local order tensors (), each one corresponding to the region defined by every FUS chain and its corresponding Nneigh neighbors. From Eq. 7, we obtain by computing the eigenvector corresponding to the largest eigenvalue of the tensor . We therefore have N such eigenvectors, , each defining the direction of alignment in the region around the ith chain. These director vectors are used to compute local orientational order Si using Eq. 5.
Results
Disorder-order transition is an outcome of hetero- to homodomain interaction network switching
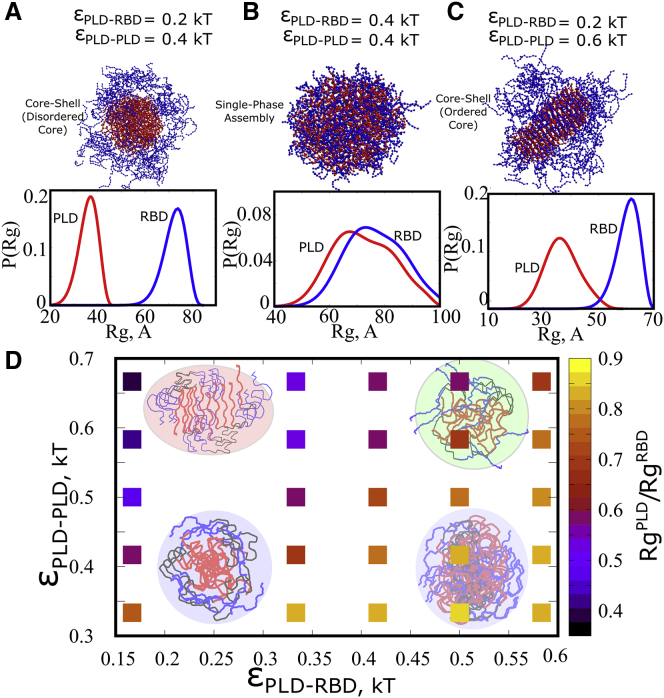
FUS is known to spontaneously phase separate in vitro and also act as a scaffold protein in RNA-protein condensates in the cell (6,16). Previous studies have shown that FUS droplets are stabilized by two primary modes of interactions: PLD-PLD interactions and PLD-RBD interactions. Here, we systematically vary the strength of these two interactions— and —to study how it influences the nature of the FUS condensate.
To drive the self-assembly of the proteins into a single, large, condensed phase at simulation timescales, we employ metadynamics simulations. We use the radius of gyration of the system () of 100 protein chains as a collective variable to define the state of the system. The free-energy profiles were computed along this reaction coordinate, with minima around small values of indicating a fully assembled equilibrium state. In Fig. S1, we demonstrate the free-energy profiles for four such scenarios, with varying strengths of heterodomain () and homodomain () interactions. The free-energy profiles suggest that, for the protein concentration chosen for the study (200 μM) the self-assembled state is favored even for relatively weak interaction strengths ( of 0.15 kT and of 0.3 kT).
Upon establishing that the condensed phase indeed corresponds to an equilibrium state, we then proceed to characterize the interaction network of the condensed phase for a broad range of interaction parameters. In Fig. S5, we plot the number of intermolecular contacts per chain as a function of strengths of homotypic interactions between amino acids in the PLD domain and heterotypic interactions between PLD-RBD domains, and , respectively. For weak interaction strengths, the interaction network within the FUS cluster is sparse (Qinterchain ≤ 100 intermolecular contacts per chain in Fig. S5). For strong PLD-PLD interactions, we observe a dense intermolecular contact network (>100 contacts per chain in Fig. S5) suggesting a different intracluster interaction network in this regime.
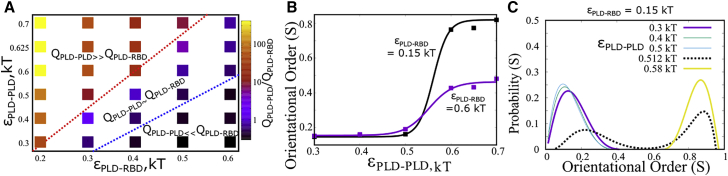
Upon establishing variation in the extent of intermolecular connectivity within the droplet upon tuning the interaction parameters, we probe the nature of interactions that make up these intermolecular contacts. In Fig. 2 A, we plot the ratio of PLD-PLD to PLD-RBD intermolecular contacts to elucidate the dominant intermolecular interactions that stabilize the condensed phase. For weak and strong the FUS clusters are stabilized by homotypic interactions between PLD domains from different chains. On the other hand, in the limit of weak and strong , heterodomain interactions dominate. In a narrow range of interaction strengths, the contact ratios 1( = ). In this limit, the intracluster environment exhibits both homo- and heterodomain contacts with equal likelihood.
Figure 2.
Structural transitions at different regimes of εPLD-PLD and εPLD-RBD. (A) Switching of predominant interaction network stabilizing the condensate, as a function of varying εPLD-PLD and εPLD-RBD. and refer to the intermolecular contacts per chain between PLD-PLD and PLD-RBD domains, respectively. (B) Local orientational order as a function of εPLD-PLD for different strengths of εPLD-RBD. The solid curves are shown as a guide to the eye. (C) Distribution of the local orientational order parameter (S) for different values of εPLD-PLD. The distributions were computed across 100 different orientational local order parameter values and 50 different structures at equilibrium.
Tuning the strength of homo- and heterodomain interactions in the thermally relevant regime results in structures that are stabilized by distinct modes of interaction (Figs. 2 A and S5). Wang et al. have previously established that a hierarchy of interactions—homo- and heterodomain—can drive phase separation of FUS into protein-rich droplets (13). We, therefore, probe whether a switch in the primary mode of intermolecular interactions upon varying the interaction strengths could result in altered structural properties of the assembly. To characterize the morphology of the assemblies, we employ the nematic orientational order parameter , which defines the extent of local orientational order for the PLDs within a cluster. This order parameter can take any value from 0 to 1. indicates a high degree of orientational order. On the other hand, suggests an entirely disordered system. Using this order parameter, we capture the extent of orientational order as a function of varying interaction parameters and . In Fig. 2 B, we plot as a function of increasing PLD-PLD interactions for two different strengths of PLD-RBD interactions. Interestingly, an increase in results in a first order-like, disorder-to-order transition for weak . A further affirmation of the discontinuous nature of this transition could be ascertained from the evolution of the distribution of S for increasing values of (Fig. 2 C). For weak homodomain interactions ( ≤ 0.5 kT in Fig. 3 B), we see the distributions peaking at low values of S (∼0.2), suggesting that the assemblies never access the ordered states in this interaction regime. Interestingly, an increase in from 0.2 to 0.5 kT results in a negligible shift in distribution. However, a slight increase in homodomain interaction strength from 0.5 to 0.512 kT results in a shift from a unimodal distribution (fully disordered) to a bimodal distribution with peaks corresponding to both the ordered and disordered states. Such a bimodality is a typical signature of discontinuity. A further increase in interaction strength to 0.6 kT results in a unimodal distribution with the assemblies populating the ordered state alone. For stronger interaction, the S versus curve saturates at lower values of the order parameter, owing to a competition between homodomain and heterodomain interactions.
Figure 3.
Structural phases. Phase diagram describing the various structural phases in response to varying strengths of homodomain (εPLD-PLD) and heterodomain (εPLD-RBD) interactions. Structures are described as ordered, disordered, and partially ordered based on the mean value of the nematic orientational order parameter S. Assemblies with S > 0.6 are labeled as ordered structures while those with S < 0.4 are labeled as disordered structures. PLD residues are shown in red and RBD regions are shown in blue.
In Fig. 3, we show how a variation in these two parameters gives rise to different structural phases with varying degrees of nematic ordering. In the - phase diagram in Fig. 3, we define all structures with S < 0.4 as disordered assemblies. S > 0.6 were defined as ordered assemblies while those with intermediate values of were labeled as partially ordered structures. The S values were averaged across 100 different equilibrium configurations and across 100 polymer chains. When both homo- and heterodomain interactions are weak ( and < 0.5 kT), we encounter structures which are disordered and exhibit spherical geometry. As we increase the strength of homodomain interactions, in the limit of weak heterodomain interactions ( < 0.5 kT), we encounter ordered structures with the PLD domains displaying a high degree of orientational order with “fibril-like” geometry. When both interactions are competitively strong ( and > 0.5 kT), we observe structures with partial order. Therefore, in the regime where homotypic interactions between PLD domains dominate in Fig. 2 A), the self-assembled state has a high propensity to organize into ordered structures stabilized by PLD-PLD contacts. An interplay between these homodomain and heterodomain interactions can therefore result in structures with varying morphologies.
Phosphorylation of the PLD is a barrier to the formation of ordered structures
Our results so far show that even within a narrow range of interaction strengths, in the thermally relevant limit, the interplay between homo- and heterodomain interactions can result in ordered structures. Phase-separated structures in the cell could localize proteins at concentrations as high as 50–100 times that of their average bulk concentrations (37,38). Such a large local density of PLDs in these condensates makes these structures prone to forming filamentous structures. Previous experimental studies suggest that post-translational modifications could potentially act as a barrier to liquid-solid transitions in cellular condensates (25,39).
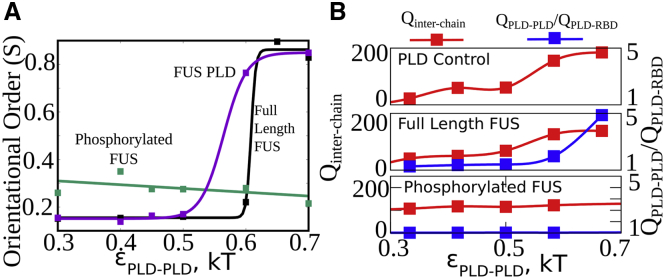
In this context, we modeled a key post-translational modification that is typically associated with the FUS PLD, i.e., phosphorylation. The FUS PLD in primates harbors 31 potential phosphosites (26,40). Here, we model phosphorylation by introducing a single negative charge per phosphosite in our coarse-grained PLD beads. The net negative charge on each phosphorylation PLD bead corresponds to the number of phosphosites (26) residing in a particular 8-residue sequence onto which the coarse-grained bead maps (see Table S2). The net charge on the phosphorylated variant of the PLD is −31. In Fig. 4 A, we compare the propensity of three variants of FUS—the PLD domain only, full-length FUS, and phosphorylated FUS—to form ordered structures. The PLD control simulations reveal that the PLD domain can undergo a disorder-order transition on its own in the limit of > 0.5 kT. Similarly, the full-length FUS, for = 0.5 kT, also shows a -dependent disorder-order transition, with lower values than the PLD-only control simulations. The phosphorylated FUS, on the other hand, shows no such propensity to form highly ordered structures (S > 0.6) even at approaching 1 kT. This result suggests that the high local density of negative charges in the PLD of phosphorylated FUS results in an abrogation of order formation in these structures.
Figure 4.
Phosphorylation abrogates order. (A) Orientational order as a function of εPLD-PLD for the PLD alone, full-length FUS, and phosphorylated FUS. Phosphorylation results in disordered structures even at strong εPLD-PLD. (B) Mean interchain contacts per FUS chain () and the contact ratio () for PLD alone (top), full-length FUS (middle), and phosphorylated FUS (bottom) as a function of εPLD-PLD. Phosphorylation results in lower values of contact ratios. The solid lines are a guide to the eye.
We further probe whether this loss of ordering in phosphorylated version of FUS is a result of overall loss of intermolecular contacts within the FUS cluster. As evident from the versus curve for the phosphorylated FUS (Fig. 4 B, bottom), the modification to the PLD results in number of intermolecular contacts () comparable with the unmodified variant. However, unlike the other two scenarios, the phosphorylated FUS shows no increase in contact ratio () even at strong interactions. However, the phosphorylated FUS clusters are stabilized by interactions between the negatively charged phosphorylated PLD and the positively charged RBD. Phosphorylated structures, therefore, show higher ordering than their unmodified counterparts in the limit of weak PLD-PLD interactions (Fig. 4 A, < 0.5 kT). This ordering, however, is driven by interdomain electrostatic interactions, and order remains unaltered upon an increase in PLD-PLD interaction strength (Fig. 4 A, green curve). In the corresponding limit of strong , the full-length FUS displays contact ratio values approaching 5. This shows that phosphorylation results in an upregulation of the heterodomain interactions over homodomain interactions, and thereby results in a more disordered condensed phase.
Disorder-order transitions are characterized by sharp density transitions
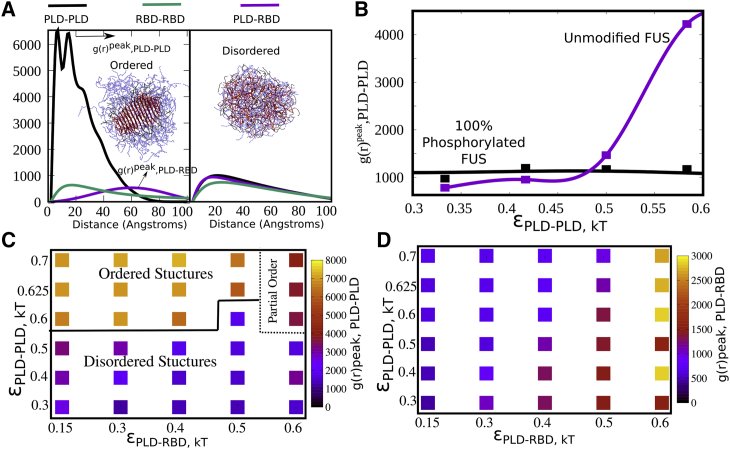
The transition from droplet-like morphology to a more fibrillar morphology in FUS protein assembly is also accompanied by a loss of dynamicity of the condensate (18). We therefore probe how tuning and alters local densities of PLD and RBDs within the clusters. In Fig. 5 A, we plot the pair correlation function, g(r), for different domains in the condensed phase. The g(r) plotted in Fig. 5 A is the mean number of atom pairs of any type at a given radial distance, normalized by a pair of ideal gas particles for the same bulk density. The g(r) values in Fig. 5 refer to the average number of atom pairs of any type (PLD-PLD, PLD-RBD, RBD-RBD) found at any radial distance (between r and r + dr), normalized by the equivalent values for a system of ideal gas particles of the same bulk density. As the peak value of g(r) 1, the system exhibits gas-like organization. The higher this peak in g(r), ( in Fig. 5 A), the system exhibits more solid-like organization. Also, the higher the g(r) peak, the denser the packing and potentially less dynamic the condensed phase.
Figure 5.
Local density of domains. (A) Pair correlation function, g(r), for PLD-PLD (black), RBD-RBD (green), and PLD-RBD (purple) contacts within the largest cluster, for ordered and disordered structures. The g(r) here is essentially the probability of finding any two particle types (PLD-PLD, PLD-RBD, RBD-RBD) within a certain distance of each other normalized by corresponding value for a pair of ideal gas particles at the same bulk density. For ordered structures, the g(r) for PLD-PLD interactions peaks severalfold higher than the other two curves, suggesting a high local density of PLD domains within the cluster. For disordered structures the three curves peak at similar values, indicating a more homogeneous density throughout the cluster. (B) The peak values of g(r) plotted as a function of εPLD-PLD. For full-length FUS, the g(r) peak for PLD-PLD contacts shows a sharp sigmoidal transition with increasing εPLD-PLD, suggesting a sharp transition from a low to high density phase. For phosphorylated variants of FUS, such a transition was not observed. (C and D) Phase diagrams showing the density of PLD-PLD and PLD-RBD packing, respectively. An increase in PLD-PLD interactions results in a high packing density for PLD domains within the cluster, as evident from the g(r) peak values for PLD-PLD interactions. In contrast, little variation in PLD-RBD g(r) peaks was observed throughout the parameter space explored.
In Fig. 5 A, we show representative g(r) curves for ordered and disordered structures. For ordered structures, the g(r) for PLD-PLD interactions shows peak values severalfold higher than the corresponding curves for PLD-RBD and RBD-RBD interactions. This suggests that the ordered structures have a dense PLD core and a less dense RBD shell, as also evident from the structures shown in Fig. 3. The two peaks in the g(r) profile are indicative of the morphology of the ordered structures. The first peak (∼10 Å) in the pair correlation function corresponds to the spacing between adjacent PLD chains (ith and (i + 1)th neighbors in Fig. S6) in the ordered assembly while the second peak (∼10 Å) is indicative of the spacing with the next nearest (ith and (i + 2)th neighbor in Fig. S6). The PLDs get packed within a 60 Å radius core within the FUS cluster, and the g(r) curve starts decaying around length scales, suggesting that the outer shell of these structures is composed primarily of the RBD and linker regions. The significantly higher peaks in the g(r) profile for PLD-PLD in Fig. 5 A is indicative of the difference in packing densities for the demixed, PLD-rich core and the RBD-rich outer shell. For disordered structures formed in the limit of weak interactions ( and < 0.5 kT), the g(r) curves for all three pairwise interactions are comparable, with peak values severalfold lower than the peak g(r) values for PLD-PLD packing in the ordered structures. This result suggests that intracluster densities of polymer domains in the disordered structure are more uniform compared with the ordered structures, which exhibit a dense core and a less dense shell.
We further probe how phosphorylation influences the peak densities of PLD domains within the FUS clusters by plotting the for PLD-PLD contacts as a function of . Strikingly, even at higher , the phosphorylated variant shows no change in peak densities within the condensed phase. The full-length FUS, on the other hand, shows a discontinuous transition from uniform, less dense structures to the more dense core-shell structures that typify order. We further plot the peak densities for the PLD-PLD and PLD-RBD pairwise contacts as a function of varying and . Interestingly, the regime of interaction strengths that corresponds to a high contact ratio (QPLD-PLD >> QPLD-RBD in Fig. 2 A), and high orientational order (Fig. 3) also corresponds to large peaks in g(r) for PLD-PLD (Fig. 5 C). However, the peak values of g(r) for PLD-RBD interactions (Fig. 5 D) are significantly lower even for strong PLD-RBD interactions.
The g(r) peak values for PLD-RBD show a five-fold increase as we increase from 0.15 to 0.7 kT. However, the g(r) peak values for PLD-RBD interactions are about two-fold lower than those of PLD-PLD interactions, even at strong (Fig. 5 D). This is consistent with an absence of ordering through strong PLD-RBD interactions. The physical barrier preventing dense packing via PLD-RBD interactions is the high density of charges in the RBD of FUS. While the strength of RBD-RBD interactions remains unaltered across the phase space, an increase in has an indirect effect on the peak densities of RBD-RBD interactions (Fig. S7). As we increase strong from 0.15 to 0.6 kT, we observe a three-fold increase in peak heights for RBD-RBD interactions. Therefore, the local densities required for ordered structures cannot be achieved through PLD-RBD interactions owing to the concomitant increase in contact probability of charged RBD-RBD interactions for this packing. Therefore, even for strong PLD-RBD interactions, we observe partially ordered structures instead of the fully ordered structures that are observed at low PLD-RBD and strong PLD-PLD interactions. Thus, the most feasible mechanism of forming ordered FUS structures is via demixing of the PLD and RBD domains within the condensed phase, enabling high packing densities reminiscent of the solid-like state.
Assemblies can switch from fully mixed to multi-phase architecture on switching interaction networks
Multi-component mixtures (composed of several protein types) can phase separate into multi-phase assemblies with subphases that localize different components based on interaction preferences. Diversity in interactions and interfacial tensions has been known to be a key driver in segregation of components between different subphases. Due to the multi-domain architecture of RBPs and the ability to form clusters stabilized by different interaction networks (Fig. 2 A), we probed whether multi-phase assemblies could result from single-component mixtures of FUS alone. In Fig. 5 we show how the interplay between the homodomain and heterodomain interactions can result in hugely altered local densities of the PLD and RBD domains within the condensates. Given how density differences are a vital signature of multi-phase condensates, we looked at how different domains are packed within the single-component FUS assemblies stabilized by different interaction networks. In Fig. 6, we show how FUS condensates could exhibit different subcluster organization for different combinations of interaction strengths. For weak interactions ( and < 0.5 kT), when PLD-PLD interactions dominate ( = 0.4 kT and = 0.2 kT), we observe spherical clusters with a multi-phase structure. Here, PLD segments form the core of the condensates while the RBD domains organize themselves in the outer shell. This is evident from the distributions of the radius of gyration of PLD domains which peak at a much lower value (∼35 Å) compared with that of RBD domains (∼70 Å), showing that the PLD domains form a condensed core. However, these core-shell structures formed by PLD, in the limit of weak interactions, do not exhibit nematic order. As the strengths of the two interactions become comparable, we observe single-phase clusters where the two domains are fully mixed (Fig. 6 B), with the radius of gyration distributions for PLD and RBD segments showing significant overlap. In the strong interaction regime where PLD-PLD interactions dominate, we again observe core-shell structures. However, unlike the spherical disordered core-shell structures observed when the two interactions are less than 0.5 kT, the PLD core (for ) here shows fibril-like nematic ordering. Differential interaction strengths can thereby result in core-shell structures, even for assemblies that are made up of only one type of protein.
Figure 6.
Different architectures of the FUS assemblies. (A) For the limit of weak interactions when εPLD-PLD > εPLD-RBD, we observe disordered structures with the PLDs (red beads) phase separating from the RBDs (blue beads). (B) As εPLD-PLD and εPLD-RBD become comparable, we observe a disordered assembly where the PLD and RBD domains are fully mixed. (C) Above a critical interaction strength, when εPLD-PLD > εPLD-RBD, we observe core-shell structures with distinct PLD and RBD phases within the assembly. Unlike the disordered core-shell structures for weak interactions, the PLD core here shows a high degree of orientational order. The black and purple curves in (A), (B), and (C) show the distribution of radius of gyration of PLD and RBD domains, respectively. Small values of Rg indicate a collapsed core while large values indicate that the domains are spread out. When the two distributions do not overlap a core-shell structure is indicated, while overlapping distributions suggest a fully mixed cluster. (D) The ratio of radius of gyration of the PLD (RgPLD) and RBD (RgRBD) domains in the cluster. Vaues approaching 1 indicate a fully mixed, single-phase cluster. Low values of this ratio indicate a dense PLD core and an RBD shell.
Dramatic slowdown of intracluster dynamics upon disorder-to-order structural transition
Our simulations reveal that the interplay between homodomain and heterodomain interactions can result in self-assembled structures with diverse architectures (single versus multi-phase, Fig. 6) and varying degrees of orientational order. However, aging of droplets and condensates in vivo and in vitro is not only characterized by structural transitions but also by a slowdown in intradroplet dynamics. In this context, it becomes critical to ascertain whether the sharp disorder-order transitions in structural order and local densities that we observe above a critical εPLD-PLD are also accompanied by altered intracluster dynamics. To probe the microenvironment within the FUS clusters, we run long 10-μs-long simulations with the self-assembled clusters (localizing ∼100 FUS chains) shown in Fig. 3. Using these trajectories, we first plotted the mean square displacements (MSD) of the center of masses of PLD and RBD domains within the cluster (Figs. S8 and S9). Here,
| (8) |
The angular brackets in the equation for MSD indicate an ensemble average across all molecules within a cluster as well as different time origins along the trajectory.
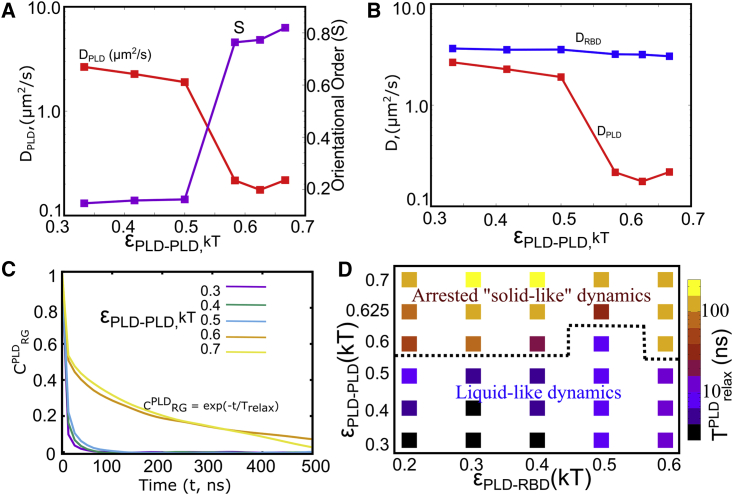
We compute the diffusion coefficients (DPLD and DRBD) from these MSD curves (by fitting the curves to MSD = 6 Dt). In Fig. S8, we show the MSD profiles for PLD domains in various scenarios. As evident from Figs. S8 and S9, the diffusion coefficient for monomeric FUS in solution (∼11 μm2/s) shows a fivefold reduction upon self-assembly into disordered FUS-rich clusters (∼2.5 μm2/s). However, while the disordered structures still exhibit diffusive behavior, structures with orientational order (Fig. S8, red curve) exhibit flat MSD profiles with two orders of magnitude decrease in DPLD. In Fig. 7A, we plot DPLD (intracluster diffusion coefficient of PLD domains) as a function of homodomain interaction strengths for a fixed εPLD-RBD of 0.15 kT. We observed two distinct dynamics regime and a sharp transition between them at εPLD-PLD > 0.5 kT. As we increase the homodomain interaction strength from ∼0.5 to ∼0.6 kT, we observe a dramatic—two orders of magnitude—decrease in diffusion coefficients (red curve in Figs. 7 A and S8), suggesting that the PLD domains exhibit solid-like dynamics in this regime (also evident from the flat MSD profiles in Figs. S8 and S9). Interestingly, this sharp transition in diffusion coefficients is concomitant with a sharp increase in structural order (Fig. 7 A, purple curve), suggesting that solid-like intradroplet dynamics emerges because of structural ordering. While the disorder-order structural transitions result in an order-of-magnitude reduction in DPLD, the RBD regions show a comparatively negligible drop in diffusion coefficients even in highly ordered structures, suggesting that these domains continue to exhibit liquid-like dynamics within ordered structures. The spatial heterogeneity in local densities that characterizes PLD-driven assemblies also results in spatially dependent dynamics within the cluster, with a solid ordered core and a liquid-like shell. In Fig. S10, we plot detailed phase diagrams that show how the diffusion coefficients of the two domains vary as a function of the homo- and heterodomain interaction strengths.
Figure 7.
Intracluster dynamics. (A) Diffusion coefficients of the PLD domain of polymer chains within the self-assembled cluster, as a function of εPLD-PLD (red curve, left-hand-side y-axis). As we increase the interaction strength for homodomain interactions above 0.5 kT, we observe a sharp slowdown in intradroplet dynamics, with an order-of-magnitude decrease in DPLD. This drop in DPLD coincides with a sharp increase in orientational order of PLD domains within the cluster (purple curve, right-hand-side y-axis). (B) Diffusion coefficients for PLD (red) and RBD (blue) domains within the cluster as a function of εPLD-PLD. At higher values of homodomain interactions, the diffusion coefficient of PLD domains shows an order-of-magnitude decrease. There is negligible reduction in DRBD, in comparison. (C) Normalized autocorrelation function (CPLDRG) for the radius of gyration of PLD domains within FUS chains as a function of time. For less viscous intracluster environment, the ACF curves decay rapidly to zero, suggesting that the system loses its memory at short timescales. On the other hand, the ACF profiles decay much slower at stronger values of εPLD-PLD where we encounter ordered structures. CPLDRG = exp(−t/Trelax) (D) Measuring polymer dynamics as a function of εPLD-PLD and εPLD-RBD. Trelax is the timescale that defines the rate at which autocorrelation function decays (CPLDRG = exp(−t/Trelax)). The higher this Trelax, the slower the dynamics of polymers within the protein-rich cluster and the more viscous the intracluster environment. Low values of Trelax, on the other hand, indicate that the system loses its memory rapidly and signify a relatively less viscous intradroplet environment.
We further computed the autocorrelation function, ACF (computed from time traces of radius of gyration of FUS chains within the clusters), to study the characteristic timescales at which the system loses its memory (41,42). The normalized autocorrelation function of the radius of gyration of FUS chains, CRG, can be used to describe the dynamics of polymers within the dense polymer-rich clusters.
| (9) |
Here, the < > denotes an ensemble average over all the polymer chains that are part of the FUS assembly as well as using different timesteps as starting point along the trajectory.
As shown in Fig. 7 C, the ACF profiles decay with time, with faster rate of decay suggesting more liquid-like dynamics while a slower rate of decay is symbolic of more viscous intracluster environments. To quantify relaxation dynamics, we define an order parameter Trelax, the timescale that defines the rate at which autocorrelation function decays (CPLDRG = exp(−t/Trelax)). Trelax allows us to compare the relaxation dynamics of polymer chains within various self-assembled clusters and thereby offers us window into intradroplet dynamics. The higher this Trelax, the slower the dynamics of polymers within the protein-rich cluster and more viscous the intracluster environment. Low values of Trelax, on the other hand, indicate that the system loses its memory rapidly and signifies a relatively less viscous intradroplet environment. In Fig. 7 D, we show a phase diagram of polymer relaxation times as a function of the two interaction strengths. Consistent with our MSD profiles, we observed a sharp slowdown in polymer relaxation dynamics at strong εPLD-PLD, suggesting that the regime at which we observe ordered structures is inherently less dynamic. Overall, the different structural phases that we encounter upon tuning the two interaction parameters correspond to distinct dynamic regimes, suggesting that subtle changes in interaction strengths and alterations in interaction networks can result in arrested droplet dynamics.
Slowdown in intracluster polymer dynamics results in altered intercluster coalescence behavior
Disorder-order structural transitions result in altered intracluster dynamics of polymer chains, with a marked slowdown in polymer relaxation times and diffusion coefficients in the regime corresponding to the ordered phase (Fig. 7). Does this altered intracluster environment result in altered coalescence of large assemblies? This question is highly relevant in the context of biomolecular assemblies where the multi-droplet state has often been observed to be long living (at biologically relevant timescales). Here, we employ metadynamics simulations to probe whether assemblies with varying structural order, formed under different regimes of εPLD-PLD and εPLD-RBD, have altered coalescence behavior. To address this question, we start off with two large droplets of 100 chains each and run metadynamics simulations with a biasing potential along a collective variable that captures coalescence: DCOM. Here, the collective variable DCOM refers to the distance between the center of masses of the two clusters. Previous simulations with system sizes of 50–200 chains have shown that these droplet sizes can recapitulate equilibrium and dynamic quantities in FUS droplets (19). Large values of DCOM (>80 Å) correspond to a state where the clusters are not in contact with each other, while short distances (<50 Å) suggest that the clusters have undergone complete coalescence. Intermediate values of DCOM indicate that the clusters are in contact, without undergoing complete coalescence.
The free-energy profiles along this coalescence reaction coordinate—DCOM—suggest that disordered, single-phase homogeneous condensates (Fig. 8 A) do not encounter large barriers for coalescence, with a distinct minimum at ∼20 Å showing that the two clusters undergo complete mixing to form a single large cluster. Similarly, even core-shell structures formed in the limit of weak interactions show complete coalescence (Fig. S11). Strikingly, the partially ordered structures formed in the regime of strong interactions (εPLD-PLD = 0.7 kT and εPLD-RBD = 0.6 kT) do not show a free-energy minimum corresponding to the coalesced states (Fig. 8 B). Conversely, due to the strong involvement of both domains in interchain contacts, these clusters are uniformly dense (Fig. 5, C and D; Fig. S7). A relatively higher packing density (Fig. 5 D) of charged domains in this regime (Fig. S7) results in an electrostatically driven emulsification of these droplets. This is consistent with previous studies suggesting that surface charge density can result in an enhanced emulsion stability in biomolecular condensates (22). The droplets with an ordered PLD core and a less dense, liquid-like RBD shell (Fig. 8 C) display a free-energy minimum for intermediate values of DCOM. In this regime, the two droplets combine with the ordered “fibril-like” cores joining end to end. However, unlike the partially ordered structures (Fig. 8 B), in this regime (εPLD-PLD = 0.6 kT and εPLD-RBD = 0.2 kT), these structures have a sixfold lower packing local density (Fig. 5 D) of charged RBDs, thereby allowing the liquid-like shells of the clusters to penetrate each other. However, the solid-like packing of PLD cores in these structures act as a barrier against complete coalescence. Overall, the altered intracluster dynamics of polymers in the various interaction regimes (Fig. 7) results in significantly altered coalescence behavior for large clusters. These results suggest that phase transitions that result in a slowdown of intracluster dynamics could result in arrested coalescence behavior of biomolecular condensates.
Figure 8.
Free-energy profiles for coalescence of large clusters. Free-energy profiles for coalescence of two large protein assemblies with different structures, formed at different regimes of εPLD-PLD and εPLD-RBD. (A) Homogeneous, disordered droplets (εPLD-PLD and εPLD-RBD = 0.2 kT) undergo complete coalescence, with a minimum that corresponds to the fully coalesced state. (B) Partially ordered structures formed in the limit of strong PLD-PLD and PLD-RBD interactions (εPLD-PLD = 0.7 kT and εPLD-RBD = 0.6 kT) do not coalesce into one large cluster, and remain as individual droplets. (C) Assemblies with an ordered PLD core and a disordered RBD shell (εPLD-PLD = 0.2 kT and εPLD-RBD = 0.6 kT) assemble into one large assembly where the solid cores join end to end without complete coalescence. The order parameter used to track coalescence is the distance between the center of mass of all chains that are members of the first droplet (initially) with those that are part of the second droplet (initially).
Ordered structures can be seeded by strong nonspecific interactions with other scaffold molecules
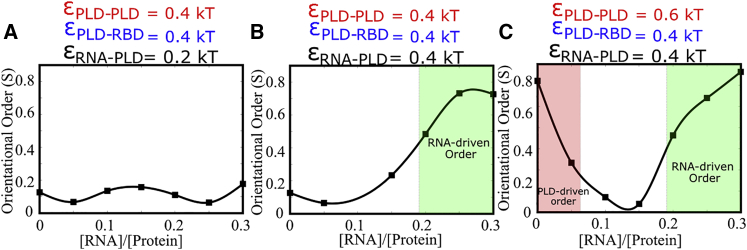
The contact ratios (Fig. 2 A), orientational order (Figs. 2 B and 3), and peak intracluster densities (Fig. 5) suggest that disordered-ordered transition in FUS clusters is mediated primarily by PLD-driven interactions. Further, the PLD switches from collapsed configuration to an extended ordered configuration above a critical PLD-PLD interaction strength. Therefore, could interactions with domains or regions from other biopolymers also influence ordering within FUS proteins? To address this question, we performed simulations with a 200-bead-long, homopolymer mimicking RNA molecules that localize within RNP condensates. We first fix the interaction strengths for RNA and RBD beads () to 0.7 kT and then systematically vary the strength of promiscuous RNA binding to the PLD (). The strength of and was set to 0.5 kT, a scenario that results in partially ordered structures in the absence of RNA. The ratio of protein to RNA molecules was initially set at 3:1. In Fig. 9, we show how RNA-PLD interactions can influence the extent of order in the RNA-protein condensates. In the limit of weak RNA-PLD interactions, the RNA interactions are extremely specific, binding to the RBD alone, and the system displays partial ordering in PLD domains, akin to the scenario with no RNA molecules. As we increase the strength of nonspecific RNA binding to the PLD from 0.1 to 0.3 kT, we see an initial decrease in order because RNA chains compete with PLD domains from other chains for PLD binding (reduction in contact ratio, Fig. 9, green curve). However, a further increase in results in an increase in orientational order within these structures, with the PLD and the positively charged RBD showing alignment along the negatively charged RNA backbone.
Figure 9.
Ordering of FUS-RNA condensates in response to varying strengths of RNA-PLD binding. An initial increase in εPLD-RNA results in a decrease in orientational order and therefore disordered FUR-RNA assemblies (black curve). A further increase in εPLD-RNA, however, results in an increase in orientational order, with the PLD domains aligning themselves along the RNA spline. The intermolecular contact ratios (green curve) confirm that the order within the clusters is a result of RNA-PLD contacts and not PLD-PLD contacts.
We further probed the effect of RNA on structural order in a broad range of RNA concentrations, for different regimes of protein-protein ( and ) and protein-RNA interactions (). When protein-protein interactions are weak (Fig. 10 A, and = 0.4 kT), the structures that form in the absence of RNA (RNA/protein → 0 in Fig. 10, A and B) are disordered. In this regime, when RNA-protein interactions are weak (, Fig. 10 A) the RNA-protein assemblies remain disordered for all RNA concentrations under study. However, as RNA binding becomes stronger (, Fig. 10 B), we observe ordered structures for large RNA concentrations (Fig. 10 B) in the regime of weak protein-protein interactions. Therefore, for weak protein-protein and strong RNA-protein interactions, RNA molecules can scaffold ordered structures.
Figure 10.
Ordering in FUS-RNA condensates, at varying RNA concentrations. (A) When both protein-protein and protein-RNA interactions are weak, εPLD-PLD and εPLD-RBD < 0.5 kT and εRNA-PLD = 0.2 kT, the ensuing structures remain disordered throughout the concentration range. (B) As we increase RNA-protein interaction, εRNA-PLD = 0.4 kT, we see an RNA-driven ordering of clusters at high [RNA/protein] ratios. (C) When PLD-PLD interactions are strong, the structures that form in the absence of RNA are ordered. As we introduce RNA to the mixture, in this regime of interactions there is an initial decrease in order. A further increase in RNA concentrations results in ordered RNA-protein clusters, with nematic order as a result of strong RNA-protein interactions. The solid lines are a guide to the eye.
When PLD-PLD interactions are strong (Fig. 10 C), the structures observed in the absence of RNA display PLD-driven nematic order (Fig. 10 C, RNA/protein → 0; Fig. S12 A). For an initial increase in RNA/protein ratio, we see a decrease in nematic order due to a competition between RNA and PLD domains for PLD-PLD binding. A further increase in RNA/protein ratio results in an increase in nematic order (Fig. 10 C). However, the ordering in this regime is a result of RNA-protein interactions, with the RNA forming a backbone around which the ordered structures become stabilized (Figs. 10 C and S12 B).
Therefore, the RNP clusters exhibit liquid-like, disordered structures only when the strength of RNA-protein interactions is within a narrow range. On the other hand, interactions between the PLD domains of different, non-FUS proteins (stronger than a critical ) could also result in ordered structure formation, making this an accessible self-assembled geometry for RBPs with IDRs.
Discussion
The role of PLDs as key drivers of phase separation can be ascertained by a strong positive correlation between the number of tyrosine residues in the PLD of FUS family proteins and their measured saturation concentrations for LLPS (16). While the low-complexity, PLD within FUS can phase separate on its own, the threshold concentrations for LLPS of PLD alone are higher than that of the full-length protein, suggesting that a hierarchy of interactions could drive self-assembly (13). PLD domains on their own can either exist in a disordered droplet-like state or an ordered, cross--rich amyloid-fibrillar state. This gives rise to an interesting paradox—while the low-complexity domains are crucial drivers of phase separation, a high density of PLDs within the condensed phase makes these structures prone to fibril formation. Indeed, liquid-solid transitions have been widely reported in vitro and upon prolonged stress in vivo (18,25,43). While the phenomenon of droplet aging and liquid-solid transition is ubiquitous in biological LLPS, its molecular underpinnings are largely unclear. In this study, we examine the structural transitions in FUS assemblies within a biologically relevant range of interaction strengths. Are ordered and disordered structural phases characterized by distinct interaction networks? Are morphological transitions accompanied by large changes in intradroplet densities? Can post-translational modifications act as a barrier to ordered structure formation? To address these questions, we employ a coarse-grained, minimal representation of the multi-domain FUS protein with few interaction parameters in order to understand how the interplay between homo- and heterodomain interactions could shape the structure of the FUS-rich clusters.
Disorder-order transition occurs within a biologically relevant regime of interaction parameters and is sensitive to mutations and modifications
Interestingly, the minimal model can effectively capture ordered structures in the narrow range of thermally relevant interaction strengths ( and in the range of 0.1–0.7 kT). Our simulations reveal that the disorder-order transition is accompanied by a switch in the predominant interaction network that stabilizes the self-assembled structures. While homo- and heterodomain interactions are both equally likely in single-phase, disordered assemblies (Figs. 3 and 6), the ordered structures are significantly enriched in PLD-PLD interactions (Figs. 2 A and 3). Crucially, we observe that this transition from the disordered to ordered self-assembled state has the hallmarks of a highly cooperative first-order phase transition (Fig. 2 B). A small change in homodomain interaction strengths (≈0.1 kT per bead, Figs. 2 B, C and 3) could result in dramatically altered structures. Mutations of single or small numbers of PLD residues, resulting in altered strength of interactions and/or interaction network could therefore cause disorder-order transitions in FUS protein droplets. Further evidence that ordered structures are dependent on PLD-PLD interactions comes from simulations with phosphorylated PLDs that localize a high density of negative charges. Consistent with experimental findings (25), phosphorylation resulted in downregulation of PLD-PLD contacts resulting in structures with low nematic order. Phosphorylation not only resulted in disordered structures (Fig. 4) but also lower local densities (Fig. 5 B) of FUS PLD compared with assemblies composed of unmodified FUS. Also, ordered structures exhibited severalfold higher peak local densities compared with disordered phases, suggesting that these structures could also have vastly different material properties.
The origins of structural order in the semiflexible polymer model
The disorder-order transition in our simulations is reminiscent of the isotropic-nematic transitions in liquid crystals that occur at low temperatures or high density. However, unlike liquid crystals, the disorder-order transition in our model occurs in the semiflexible limit ( and ) as opposed to the rod-like limit, indicating that the ordered phases are formed to maximize favorable PLD-PLD interactions at the expense of competing, less-favorable PLD-RBD and RBD-RBD interactions. Interaction-driven ordering of polymer self-assemblies has been previously reported for homopolymer chains (30). This mechanism is also observed in polymer crystallization where higher density lamellae consisting of ordered chains intertwine with disordered regions (44,45). Diblock copolymers have previously been known to assemble into structures with liquid-crystalline order within a condensed phase (46). In this context, the multi-domain architecture of FUS, with the uncharged PLD and the positively charged RBD, is analogous to a diblock polymer. Interchain interactions between biopolymers, like proteins, can therefore play the role of density modulators within the condensed state. When the strength of these interactions exceeds a critical limit it results in sharp density transitions within the self-assembled phase (Fig. 5 C) reminiscent of an Onsager-like isotropic-nematic transition (47). Unlike the Onsager-like model where the liquid crystals are rod-like molecules that exclude volume, the polymer chains in our simulations are self-associative and semiflexible. Above a critical strength of interactions, the loss of entropy (loss of flexibility) from ordering is compensated for by the gain of enthalpy by strong interchain interactions in the densely packed ordered state where resulting sheet-like ordering provides higher local density and fewer defects. While structural order could be a result of either homotypic (PLD-PLD) or heterotypic interactions (with RNA or with RBD) stronger than a critical limit, the high density of positive charges on the RBD of FUS does not allow local densities large enough to promote ordering (Figs. 5 and S7). Demixing of PLDs and RBDs within the condensed phase in order to maximize PLD-PLD interactions and significantly increase local packing density is therefore the most plausible mechanism for ordering within diblock copolymer condensates such as FUS. Given that subtle changes to interaction strengths result in dramatically altered order, such transitions could be a response to even slight changes in interaction parameters such as point mutations or solvent conditions.
Disorder-order transitions are liquid-solid transitions
Strikingly, the sharp density (and structural) transitions resulting from tuning the interaction parameters are also accompanied by dramatically altered intradroplet dynamics, as shown by the intradroplet diffusion coefficients and polymer relaxation times (Figs. 7 and S8–S10). These results reveal that different structural phases also exhibit distinct material properties ranging from liquid-like (fast relaxation dynamics) to irreversible, trapped, solid-like (slow relaxation dynamics). While liquid-like disordered structures have diffusion coefficient 5-times lower than the dilute phase, the ordered structures show a 50-fold reduction in diffusion coefficients (Figs. S8–S10). This vastly altered dynamics is highly relevant to biological condensates where an irreversible phase transition to the solid-like state is a signature of aberrant behavior (48). Indeed, FUS condensates have been identified to span several different material states, with ALS mutations accelerating their transition to the irreversible, solid-like state that results in impaired RNP-granule functions (18,48). Our results therefore provide a minimal, mechanistic framework that links structural properties of the protein assembly to its material state.
Testable predictions
A key feature of the FUS protein (and other RBPs of the FUS family) is that the two domains—the PLD and the RBD—have distinct amino acid compositions. The PLD is rich in tyrosine residues while the disordered regions of the RBD localize glycine and arginine residues. Previous studies have shown that the number of tyrosines in the PLD and arginines in the RBD contribute to the multi-valency of FUS (17). Our findings emphasize that the structural and material fate of the FUS assembly is decided by the interaction network that stabilizes the condensed phase. The interaction parameters in our simulations, and , can be tuned by sequence modification (mutations, phosphorylation of PLD, methylation of RBD) or by varying solvent conditions (salt concentration, pH, addition of RNA). Here, we propose some experiments that could be used to test the key findings from our simulations.
-
1)
Strengthening PLD-PLD interactions and weakening PLD-RBD interactions results in an increased propensity to self-assemble into ordered aggregates stabilized by PLD-PLD interactions. In this context, experiments that selectively weaken pi-cation interactions between tyrosine and arginine residues of the PLD and RBD domains could result in a PLD-dominant condensed phase. Therefore, a systematic experimental study of FUS assembly at increasing salt concentrations would be extremely relevant for testing the predictions from our model. Our results suggest that at low salt concentrations, the heterodomain pi-cation interactions between tyrosine and arginine residues would dominate, whereas at high salt one would expect condensates that are stabilized by homodomain pi-pi interactions between tyrosine residues of the PLD. It must, however, be noted that the threshold concentrations for LLPS could be different at different salt concentrations.
-
2)
Similarly, one could also setup experiments wherein hydrophobic interactions between the tyrosine residues of PLDs can be selectively disrupted in order to test our predictions of PLD-driven ordering. 1,6-Hexanediol, an aliphatic alcohol, has previously been employed to probe material properties of phase-separated droplets in response to selective disruption of hydrophobic interactions (49). Our simulations predict that assemblies that form at increased concentrations of such a hydrophobic disruptor (which weakens PLD-PLD interactions) would show a lower likelihood of transitioning to an ordered nematic-like assembly.
-
3)
Our simulations also suggest that subtle changes to the interaction strengths (of the order of 1 kT) could result in dramatically altered ordering within the assembly, confirmed by abrogation of order within the phosphorylated variants. In this context, studying sequence variants wherein the arginine residues of the RBD are substituted to reduce their net interaction propensity with the tyrosine residues of the PLD could also result in a switch in the interaction network. Similarly, we also predict that one could also reduce the propensity to form ordered structures by studying variants with tyrosine substitutions in the PLD.
-
4)
One of the most striking predictions from our model is that the ordered states that are characterized by PLD-PLD interactions are also dynamically solid-like. In this context, studying the material properties of condensates (using fluorescence recovery after photobleaching studies) formed under different experimental conditions described above (varying salt, 1,6-hexanediol concentrations) could be truly relevant to establishing the relationship between structure (determined by the key stabilizing interaction network) and material properties of the droplets.
Conclusions
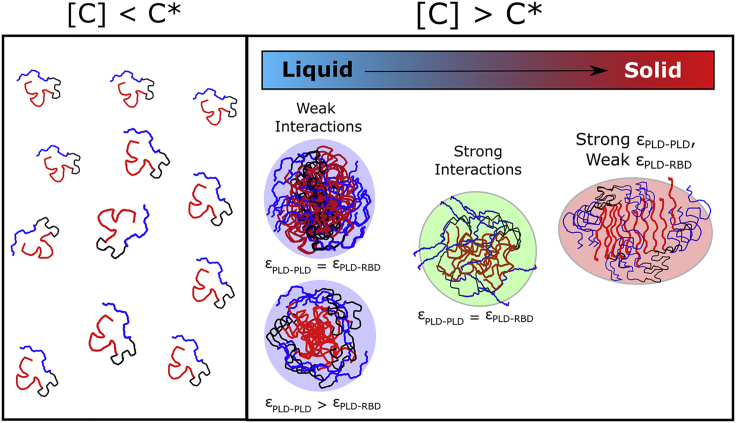
The fact that a minimal model lacking residue-level resolution can capture diverse structural phases associated with protein condensates—single-phase, core-shell disordered, and nematically ordered structures—suggests that the assembly of these structures could follow some generic physical mechanisms. While the current study employed a coarse-grained version of the FUS protein, the general principles for forming ordered, nematic-like structural phases could be applicable to other, related multi-domain RBPs that harbor intrinsically disordered domains (Fig. 11). The observation that a rich diversity of structural phases could exist for a narrow range of interaction strengths suggests that these transitions are subject to tight regulatory control. Any deviation from this tight regulation could result in aberrant phase transitions that are an outcome of subtle changes to the interaction networks.
Figure 11.
Graphical summary of the findings. Below a critical concentration C∗, the multi-domain RBPs remain in their soluble, fully-mixed state. However, above the critical concentration, these structures could exist in different structural and material phases that are an outcome of the interplay between homodomain (PLD-PLD) and heterodomain (PLD-RBD) interactions.
Author contributions
S.R. was involved in conceptualization of the work, analysis. and writing the paper. E.S. conceptualized the work, analyzed the results, and wrote the paper. S.R. performed the simulations.
Acknowledgments
The study was supported by the following grants: NIH-5RO1EY030444-03 & NIH-5R35GM139571-02.
Declaration of interests
The authors declare no competing interests.
Editor: Robert Best.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.06.013.
Supporting material
References
- 1.Banani S.F., Lee H.O., et al. Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi J.M., Holehouse A.S., Pappu R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Standart N., Weil D.P.-B. P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet. 2018;34:612–626. doi: 10.1016/j.tig.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Gonçalves A.C., Towers E.R., et al. Gale J.E. Drug-induced stress granule formation protects sensory hair cells in mouse cochlear explants during ototoxicity. Sci. Rep. 2019;9:12501. doi: 10.1038/s41598-019-48393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchan J.R., Yoon J.-H., Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Protter D.S.W., Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- 8.Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillén-Boixet J., Kopach A., et al. Franzmann T.M. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361.e17. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feric M., Vaidya N., et al. Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molliex A., Temirov J., et al. Taylor J. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maharana S., Wang J., et al. Alberti S. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Choi J.M., et al. Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmon T.S., Holehouse A.S., et al. Pappu R.V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife. 2017;6:e30294. doi: 10.7554/elife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo F., Gui X., et al. Liu C. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 2018;25:341–346. doi: 10.1038/s41594-018-0050-8. [DOI] [PubMed] [Google Scholar]
- 16.Burke K.A., Janke A.M., et al. Fawzi N.L. Residue-by-Residue view of In Vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L., Shorter J. It’s raining liquids: RNA tunes viscoelasticity and dynamics of membraneless organelles. Mol. Cell. 2015;60:189–192. doi: 10.1016/j.molcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel A., Lee H., et al. Alberti S. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 19.Benayad Z., von Bülow S., et al. Hummer G. Simulation of FUS protein condensates with an adapted coarse-grained model. J. Chem. Theor. Comput. 2021;17:525–537. doi: 10.1021/acs.jctc.0c01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kar M., Posey A.E., et al. Pappu R.V. Glycine-rich peptides from FUS have an intrinsic ability to self-assemble into fibers and networked fibrils. Biochemistry. 2021;60:3213–3222. doi: 10.1021/acs.biochem.1c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignon G.L., Zheng W., et al. Mittal J. Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput. Biol. 2018;14:e1005941. doi: 10.1371/journal.pcbi.1005941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsh T.J., Krainer G., et al. Knowles T.P. Surface electrostatics govern the emulsion stability of biomolecular condensates. bioRxiv. 2020 doi: 10.1101/2020.04.20.047910. Preprint at. [DOI] [PubMed] [Google Scholar]
- 23.Garaizar A., Espinosa J.R., et al. Collepardo-Guevara R. Intermolecular reorganisation of single-component condensates during ageing promotes multiphase architectures. bioRxiv. 2021 doi: 10.1101/2021.10.09.463670. Preprint at. [DOI] [Google Scholar]
- 24.Qamar S., Wang G., et al. St George-Hyslop P. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e15. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monahan Z., Ryan V.H., et al. Fawzi N.L. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–2967. doi: 10.15252/embj.201696394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoads S.N., Monahan Z.T., et al. Shewmaker F.P. The role of post-translational modifications on prion-like aggregation and liquid-phase separation of FUS. Int. J. Mol. Sci. 2018;19:886. doi: 10.3390/ijms19030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loughlin F.E., Lukavsky P.J., et al. Allain F.H.T. The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell. 2019;73:490–504.e6. doi: 10.1016/j.molcel.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Kang J., Lim L., et al. Song J. A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 2019;17:e3000327. doi: 10.1371/journal.pbio.3000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellesia G., Shea J.-E. Effect of β-sheet propensity on peptide aggregation. J. Chem. Phys. 2009;130:145103. doi: 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]
- 30.Ranganathan S., Maji S.K., Padinhateeri R. Defining a physical basis for diversity in protein self-assemblies using a minimal model. J. Am. Chem. Soc. 2016;138:13911–13922. doi: 10.1021/jacs.6b06433. [DOI] [PubMed] [Google Scholar]
- 31.Dignon G.L., Zheng W., et al. Mittal J. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U S A. 2018;115:9929–9934. doi: 10.1073/pnas.1804177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. doi: 10.1006/jcph.1995.1039. [DOI] [Google Scholar]
- 33.Laio A., Gervasio F.L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601. doi: 10.1088/0034-4885/71/12/126601. [DOI] [Google Scholar]
- 34.Selinger J.V. Springer; 2016. Introduction to the Theory of Soft Matter. [DOI] [Google Scholar]
- 35.Seeber M., Cecchini M., et al. Caflisch A. Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara S., Sato T. Molecular dynamics simulation of structural formation of short polymer chains. Phys. Rev. Lett. 1998;80:991–994. doi: 10.1103/physrevlett.80.991. [DOI] [Google Scholar]
- 37.Mitrea D.M., Kriwacki R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing W., Muhlrad D., et al. Rosen M.K. A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. Elife. 2020;9:e56525. doi: 10.7554/eLife.56525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofweber M., Dormann D. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019;294:7137–7150. doi: 10.1074/jbc.tm118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasmeh P., Wagner A. Natural selection on the phase-separation properties of FUS during 160 My of mammalian evolution. Mol. Biol. Evol. 2021;38:940–951. doi: 10.1093/molbev/msaa258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestryaev E.M. Comparison of various correlation times in polymer melts by molecular dynamics simulation. J. Phys. Conf. 2011;324:012031. doi: 10.1088/1742-6596/324/1/012031. [DOI] [Google Scholar]
- 42.Kalathi J.T., Kumar S.K., et al. Grest G.S. Rouse mode analysis of chain relaxation in polymer nanocomposites. Soft Matter. 2015;11:4123–4132. doi: 10.1039/c5sm00754b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q., Teplow D., et al. Abelson J. Domain structure in yeast tRNA ligase. Biochemistry. 1990;29:6132–6138. doi: 10.1021/bi00478a004. [DOI] [PubMed] [Google Scholar]
- 44.Olmsted P.D., Poon W.C.K., et al. Ryan A. Spinodal-assisted crystallization in polymer melts. Phys. Rev. Lett. 1998;81:373–376. doi: 10.1103/physrevlett.81.373. [DOI] [Google Scholar]
- 45.Strobl G.R. Springer; 1996. The Physics of Polymers. [DOI] [Google Scholar]
- 46.Muthukumar M., Ober C.K., Thomas E.L. Competing interactions and levels of ordering in self-organizing polymeric materials. Science. 1997;277:1225–1232. doi: 10.1126/science.277.5330.1225. [DOI] [Google Scholar]
- 47.Onsager L. The effects of shape on the interaction of colloidal particles. Ann. N Y Acad. Sci. 1949;51:627–659. doi: 10.1111/j.1749-6632.1949.tb27296.x. [DOI] [Google Scholar]
- 48.Murakami T., Qamar S., et al. St George-Hyslop P. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krainer G., Welsh T.J., et al. Knowles T.P.J. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 2021;12:1085. doi: 10.1038/s41467-021-21181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.