Abstract
Cyclic adenosine monophosphate (cAMP) is a generic signaling molecule that, through precise control of its signaling dynamics, exerts distinct cellular effects. Consequently, aberrant cAMP signaling can have detrimental effects. Phosphodiesterase 4 (PDE4) enzymes profoundly control cAMP signaling and comprise different isoform types wherein enzymatic activity is modulated by differential feedback mechanisms. Because these feedback dynamics are non-linear and occur coincidentally, their effects are difficult to examine experimentally but can be well simulated computationally. Through understanding the role of PDE4 isoform types in regulating cAMP signaling, PDE4-targeted therapeutic strategies can be better specified. Here, we established a computational model to study how feedback mechanisms on different PDE4 isoform types lead to dynamic, isoform-specific control of cAMP signaling. Ordinary differential equations describing cAMP dynamics were implemented in the VirtualCell environment. Simulations indicated that long PDE4 isoforms exert the most profound control on oscillatory cAMP signaling, as opposed to the PDE4-mediated control of single cAMP input pulses. Moreover, elevating cAMP levels or decreasing PDE4 levels revealed different effects on downstream signaling. Together these results underline that cAMP signaling is distinctly regulated by different PDE4 isoform types and that this isoform specificity should be considered in both computational and experimental follow-up studies to better define PDE4 enzymes as therapeutic targets in diseases in which cAMP signaling is aberrant.
Significance
Cellular functioning relies on well-orchestrated intracellular signaling cascades. For example, by controlling the amplitude, duration, and localization of cyclic adenosine monophosphate (cAMP) signaling, distinct messages can be relayed using the same signaling system. cAMP signaling is extensively controlled by phosphodiesterase 4 (PDE4) enzymes, which exist as different isoform types that control cAMP levels in similar but distinct manners, as their enzymatic activity depends on dynamic feedback mechanisms. Here, by developing and applying a computational model, we show that the so-called long PDE4 isoforms predominantly regulate cAMP signaling. These findings represent an important step toward more specific targeting of long PDE4 types to achieve higher PDE4-mediated treatment efficacy in diseases in which cAMP signaling is aberrant.
Introduction
Sensing the environment and responding in an adaptive manner is crucial to cell survival and proper cell functioning. Relaying extracellular signals intracellularly to elicit an adaptive response therefore has to be tightly regulated in a dynamic, spatiotemporal manner. The pivotal intracellular signaling molecule cyclic adenosine monophosphate (cAMP) is synthesized by conversion of adenosine triphosphate (ATP) by both transmembrane and soluble adenylyl cyclases (ACs). Although AC activity is regulated by a wide variety of receptors, it is striking that these receptors, responding to different extracellular cues, all lead to the production of the generic signaling molecule cAMP, which subsequently can bind different effector proteins.
Among the cAMP effector proteins, protein kinase A (PKA) and exchange protein directly activated by cAMP (Epac) are the most well studied. PKA is a heterotetrametric protein complex which, upon binding cAMP, releases its catalytic subunits that can phosphorylate numerous target proteins (1,2). Binding of cAMP to Epac releases the autoinhibitory conformation of Epac, which can initiate the activation of Rap1, a small GTPase of the Ras superfamily (3,4). Subsequently, Rap1 can influence a variety of cellular processes through modulation of various downstream proteins (5). Moreover, by binding Popeye domain containing proteins and cyclic nucleotide-gated channels, cAMP signaling can modulate other biological functions (e.g., cell-cell adhesion and regulation of membrane potentials; reviewed in (6,7)). Although these different effector proteins all respond to cAMP, through regulation of localization and the dynamics of the cAMP signal (e.g., its amplitude and duration), cAMP can distinctly influence different intracellular processes (8).
Intriguingly, the only way by which cAMP is enzymatically degraded is via hydrolysis by phosphodiesterase (PDE) enzymes. In humans 11 PDE gene families exist (PDE1–11), which can be classified based on their substrate selectivity; some PDE types degrade cAMP selectively (PDE4, -7, -8) while others are selective to cyclic guanosine monophosphate (cGMP; PDE5, -6, -9). The remaining PDE types are dual-specific and degrade both cAMP and cGMP (PDE1, -2, -3, -10, -11). The different PDEs are heterogeneously distributed in a tissue- and cell-type-specific manner (9,10). The PDE4 gene family comprises the largest amount of cAMP-specific PDEs and plays a critical role in shaping the dynamics and spatiotemporal control of cAMP signaling in many tissues and cell types (9,10). As pivotal regulators of cAMP signaling, PDE4 enzymes provide interesting pharmacological targets to modulate cAMP levels in a wide variety of disorders (11, 12, 13). Consequently, PDE4 inhibition has been investigated as a therapeutic strategy to stimulate cAMP signaling. Although several PDE4 inhibitors are being used clinically, PDE4 inhibition may give rise to severe adverse effects (e.g., diarrhea, nausea, and emesis) with increasing doses. PDE4-mediated side effects are hypothesized to result, at least in part, from PDE4 inhibition in brainstem areas, and therefore more specific PDE4 inhibition is particularly required when, for the disease of interest, the therapeutic actions of PDE4 inhibition should occur in the brain (13). For example, in neurodegenerative disorders such as Alzheimer’s disease and multiple sclerosis, PDE4 inhibitors have to enter the central nervous system to exert a therapeutic effect, which makes these interventions also more prone to brainstem-mediated side effects (11,13,14). Therefore, a better understanding of how PDE4 mediates cAMP degradation is crucial to the optimization of PDE4 inhibition as a therapeutic strategy.
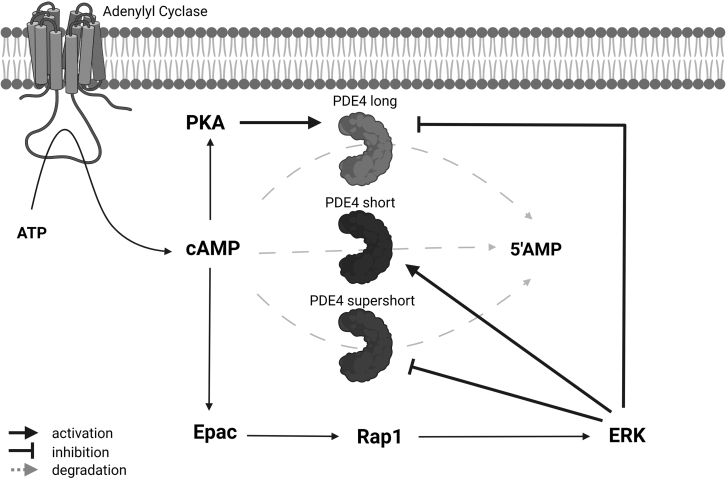
Human PDE4 enzymes are encoded by four genes (PDE4A, -B, -C, -D) that each generate multiple isoforms (e.g., PDE4D1–9) through the use of alternative promoters and alternative splicing. Although the different isoforms are protein products with the same main biological function (i.e., cAMP hydrolysis), their protein sequence differences allow for isoform-specific localization and regulation of enzymatic activity (13,15). Specifically, PDE4 isoforms can be categorized as long, short, and supershort based on the presence of regulatory domains. These regulatory domains influence the functional effect of phosphorylation by different kinases including PKA and extracellular signal-regulated kinase (ERK) on PDE4 enzymatic activity (13). Long PDE4 isoforms are activated when phosphorylated by PKA (16, 17, 18, 19, 20) whereas they are inhibited when phosphorylated by ERK (21, 22, 23, 24). In the case of phosphorylation by both PKA and ERK, the ERK-mediated inhibition of long PDE4 forms will be relieved by concurrent phosphorylation by PKA (22). Lacking the necessary regulatory domain, short PDE4 isoforms cannot be modulated by PKA but can be phosphorylated by ERK, resulting in enzyme activation. Similarly, supershort PDE4 isoforms can only be modulated by ERK but, in contrast to short isoforms, phosphorylation of ERK results in inhibition on supershort forms (21). Importantly, while ERK may phosphorylate all PDE4 subtypes, this phosphorylation has effects on the enzymatic activity of only PDE4B, PDE4C, and PDE4D, but not PDE4A, forms due to the differences in amino acid sequence (13,21,25). Interestingly, since PKA and ERK are activated downstream from cAMP signaling, their effects on PDE4 isoform activity act as feedback mechanisms on cAMP signaling (Fig. 1). PKA is activated directly upon binding cAMP, while ERK is activated by intermediate signaling molecules downstream from cAMP. In fact, cAMP and ERK signaling are intricately linked in cell-type-specific and cell-context-dependent manners (reviewed in (26)). Upon being activated by cAMP-bound Epac, Rap1 can initiate B-raf signaling, which eventually activates ERK in specific cell types (5,27,28).
Figure 1.
Schematic representation of cAMP signaling cascades that influence PDE4-mediated cAMP degradation. cAMP is created by conversion of ATP by adenylyl cyclases and is degraded by PDE4 enzymes to 5′-adenosine monophosphate (5′AMP). PDE4 enzymes comprise different isoform categories (i.e., long, short, and supershort), which are generated via the use of alternative promoters and alternative splicing from the four human PDE4 genes. The kinases PKA and ERK are activated downstream of cAMP and can influence the enzyme activity of PDE4 in an isoform-specific manner; long forms are activated upon phosphorylation by PKA, while phosphorylation by ERK causes inhibition of long and supershort PDE4 and activation of short PDE4. These feedback mechanisms contribute to the PDE4-mediated dynamic control of cAMP signaling. This figure was created with BioRender.com.
The fact that PDE4 isoforms show specific intracellular distribution patterns and their activity is dynamically and isoform-specifically regulated makes PDE4 isoforms crucial spatiotemporal regulators of cAMP signaling. Understanding the role of the different PDE4 isoform categories on this dynamic cAMP regulation will aid in determining which isoform type to inhibit to elicit the desired physiological effect. Consequently, inhibition of specific PDE4 isoforms may modulate cAMP signaling more effectively and may be therapeutically safer by inducing fewer or less severe side effects.
Signal termination, mediated by the different PDE4 isoform types, is critical in cAMP signaling and occurs very rapidly. This rapid termination makes experimental investigation of the role of specific PDE4 isoforms in spatiotemporal control of cAMP signaling difficult (29). Computational modeling of complex, dynamic biological mechanisms such as cAMP signaling can overcome several experimental limitations while providing pivotal insights into the importance of specific molecules by considering crosstalk and feedback mechanisms. Various mechanistic computational models have been developed previously to understand cAMP signaling pathway dynamics. These models have investigated, among others, the diffusion and stochastic effects on the information flow through PKA signaling (30), the dynamics of calcium-induced cAMP signaling (31, 32, 33, 34), the influence of receptor protein kinase and G-protein-coupled receptor crosstalk (35), and the localization of cAMP signaling in subcellular domains (36, 37, 38, 39). Despite their important role in inactivating cAMP, cAMP-degrading PDE enzymes and their isoforms, which respond differently to feedback mechanisms, are not always included in the computational modeling efforts. Some models have studied the influence of PDE4 (40), PDE4 and PDE1 (31,32,39,41), and PDE1 and PDE10 (38). Others have investigated theoretically how a single PDE or PDE complexes can create cAMP nanocompartments and how these depend on the cAMP degradation rate, cAMP diffusion rate, and geometrical and topological parameters (42). Only the framework of Oliveira et al. specifically modeled two different subtypes of PDE4, i.e., PDE4B (located in the submembrane region) and PDE4D (located in the cytosol), the most prevalent isozymes in HEK293 cells (43). Their simulation results demonstrated that the generation of a cAMP microdomain required a pool of PDE4D anchored in the cytosol as well as a PKA-mediated increase of PDE4D activity. Interestingly, cAMP microdomains did not require impeded diffusion of cAMP (43). To our knowledge, the influence of feedback mechanisms on general, but not isoform-specific, PDE4 activity has only been described by Song et al. (44).
In this study, we model the dynamics of cAMP signaling and investigate the involvement of specific PDE4 isoform types, i.e., long, short, and supershort, and the PKA/ERK feedback thereon, in the modulation of cAMP signaling dynamics. Using a computational approach, we intend to better understand and conceptualize the signaling feedback mechanisms that differentially modulate the PDE4 isoform activity to determine which PDE4 isoform type may provide a more efficacious target in diseases in which PDE4 inhibition shows therapeutic potential.
Materials and methods
Mathematical model development
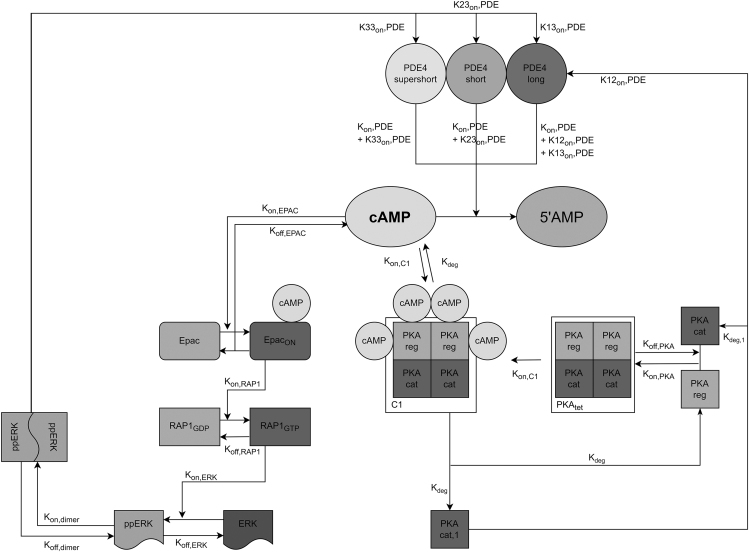
To simulate isoform-specific PDE4-mediated control of cAMP dynamics, a mathematical model was developed and implemented in the virtual cell environment VirtualCell (VCell; http://vcell.org) (45). Fifteen ordinary differential equations were established, which describe the dynamics of cAMP, PDE4, PKA, Epac, and RAP-1/ERK signaling in time. The following sections describe the mathematical framework of the model, the reactions, and corresponding kinematic parameters. Fig. 2 shows a schematic of the variables included in the model and their interactions.
Figure 2.
Schematic representation of the molecular reactions incorporated in the computational model. cAMP is modeled as an initial condition and is degraded to 5′AMP by different PDE4 isoforms (long, short, and supershort) that each can exhibit different degradation rates (e.g., K12onPDE). If not degraded, cAMP can bind and activate the tetramer PKA (PKAtet) to form C1, which causes PKAcat subunits to be released (PKAcat,1). Similarly, cAMP can bind and activate Epac to form Epacon, which can convert RAP1GDP into RAP1GTP to eventually elicit ERK phosphorylation and dimerization (ppERK). PKAcat,1 and ppERK can modulate PDE4 isoform activity and thereby provide feedback mechanisms of cAMP signaling.
Parameter values and initial conditions
Considering the high diffusion capability of free, unbound cAMP (46,47), we modeled the system as well-mixed using ordinary differential equations, which are listed, including initial concentrations, in Table 1. We also assume that all downstream, inactivated components are not present at the start of the simulation. All reactions and corresponding parameters are given in Tables 2 and 3. All reactions are assumed to be reversible, except the irreversible degradation of cAMP and phosphorylation-based actions on PDE4 isoforms.
Table 1.
Overview of reaction species, ordinary differential equations, and initial concentrations
| No. | Species | Description | Ordinary differential equation | Initial concentration (μM) | Reference for the used initial concentration |
|---|---|---|---|---|---|
| 1 | [cAMP] | cAMP concentration | = – (kon,PDE + k12on,PDE + k13on,PDE) × cAMP ×PDE4long– (kon,PDE + k22on,PDE + k23on,PDE) × cAMP × PDE4short– (kon,PDE + k32on,PDE + k33on,PDE) × cAMP × PDE4supershort– 4 × + 4 × kdeg × C1– + koff_Epac × Epacon | 0.3 (standard) or cyclic input (see text) | (48,49) |
| 2 | [AMP] | AMP concentration | = (kon,PDE + k12on,PDE + k13on,PDE) ×cAMP × PDE4long +(kon,PDE + k22on,PDE + k23on,PDE) × cAMP ×PDE4short + (kon,PDE + k32on,PDE + k33on,PDE) × cAMP × PDE4supershort | 0 | (50) |
| 3 | [PDE4] | PDE4 concentration | constant | 1 (standard) (hippocampal proportions, PDE4long: 0.7; PDE4short: 0.11; PDE4supershort: 0.19) | (50); supporting material |
| 4 | [PKAcat] | PKA catalytic subunits | = −2 kon,PKA ×PKAcat2 × PKAreg2 + kdeg,1 × PKAcat,1 + 2koff,PKA × PKAtet | 0.023 | (43) |
| 5 | [PKAreg] | PKA regulatory subunits | = –2kon,PKA × PKAreg2 × PKAcat2 + 2koff,PKA × PKAtet + 2kdeg × C1 | 0.048 | (43) |
| 6 | [PKAtet] | PKA tetramer | = kon_PKA × PKAcat2 × PKAreg2– koff_PKA × PKAtet | 0.173 | (32,50) |
| 7 | [C1] | cAMP-bound PKA tetramer | – kdeg × C1 | 0 | model assumption |
| 8 | [PKAcat1] | active PKA catalytic subunit | = 2kdeg × C1– kdeg,1 × PKAcat,1 | 0 | (43) |
| 9 | [EPAC] | inactivated Epac | + koff_Epac × Epacon | 0.488 | (49,51) |
| 10 | [EPACon] | cAMP-bound, activated Epac | =– koff_Epac × Epacon– kon,RAP1 × Epacon × RAP1_GDP + koff,RAP1 × RAP1GTP | 0 | (49) |
| 11 | [RAP1GDP] | inactive Rap1 | = –kon,RAP1 ×Epacon × RAP1GDP + koff,RAP1 × RAP1GTP | 0.2 | (52) |
| 12 | [RAP1GTP] | active Rap1 | = kon,RAP1 × Epac × RAP1GDP– koff,RAP1 × RAP1GTP– kon,ERK × RAP1GTP × ERK + koff × ppERK | 0 | model assumption |
| 13 | [ERK] | ERK concentration | = –kon,ERK × ERK × RAP1GTP + koff,ERK × ppERK | 0.8 | (53) |
| 14 | [ppERK] | activated ERK | = kon,ERK × ERK × RAP1GTP– koff,ERK × ppERK + 2 × koff,dimer × ERKdimer– 2kon,dimer × ppERK2 | 0 | model assumption |
| 15 | [ERKdimer] | dimerized ERK | = kon,dimer × ppERK2– koff dimer × ERKdimer | 0 | model assumption |
Table 2.
Kinematic parameters used to model activity of PDE4 isoforms upon phosphorylation by PKA and ERK
| Parameter | Description | KM (μM) | Vmax (μM−1 s−1) | % Vmax | References |
|---|---|---|---|---|---|
| kon,PDE | baseline PDE4-mediated cAMP degradation rate | / | 0.15 (mass action kinetics) | (50) | |
| k12on,PDE | PDE4 long PKA-phosphorylated | 1.3 | 21.76 | 272 | (22) |
| k13on,PDEa | PDE4 long ERK-phosphorylated | 1.3 | −2 | 25 | (22) |
| k22on,PDE | PDE4 short PKA-phosphorylated | 0 | 0 | ||
| k23on,PDE | PDE4 short ERK-phosphorylated | 1.3 | 10.4 | 130 | (24) |
| k32on,PDE | PDE4 supershort PKA-phosphorylated | 0 | 0 | ||
| k33on,PDEa | PDE4 supershort ERK-phosphorylated | 1.3 | −6.8 | 85 | (24) |
k13on,PDE and k33on,PDE cannot become larger than kon,PDE (0.15 μM s−1), the baseline PDE4-mediated cAMP degradation rate (kon,PDE).
Table 3.
Reactions and kinetic parameters
| # | Reaction | Rate constants | Value | References |
|---|---|---|---|---|
| 1 | cAMP AMP | kon,PDE | 0.15 μM−1 s−1 | (54) |
| 2 | 2PKAcat + 2PKAreg PKA | kon,PKA | 10 μM−3 s−1 | estimated |
| koff,PKA | 6 × 10−4 s−1 | |||
| 3 | PKA + 4cAMP C1 | kon,C1 | 0.0261 s−1 | (49,55) |
| km,PKA | 5.2 μM | |||
| 4 | C1 2PKAcat,1 + 2PKAreg + 4cAMP | kdeg | 0.21 s−1 | (32) |
| 5 | PKA cat,1 PKAcat | kdeg,1 | 0.0051 s−1 | (49) |
| 6 | Epac + cAMP Epacon | kon,Epac | 0.031 s−1 (estimated) | (49,56) |
| koff, Epac | 0.00651 s−1 | |||
| km,Epac | 30 μM | |||
| 7 | Epacon + RAP1GDP RAP1GTP | kon,RAP1 | 0.05 μM−1 s−1 (estimated) | (52) |
| koff,RAP1 | 1.166 10−4 s−1 | |||
| 8 | RAP1GTP + ERK ppERK | kon,ERK | 0.88 μM−1 s−1 | (57,58) |
| koff,ERK | 0.088 s−1 | |||
| 9 | ppERK + ppERK ERKdimer | kon,dimer | 0.2 μM−1 s−1 | estimated based on Kd = 7.5 nM (59) |
| koff, dimer | 0.0015 s−1 |
cAMP and PDE4 dynamics
cAMP signaling is initiated by the synthesis of cAMP by activated ACs. Since the primary focus of this study is the role of different PDE4 isoforms on cAMP signaling, we investigate three types of initial cAMP conditions: 1) an initial pulse of 0.1, 0.3, 1, or 3 μM; 2) a continuous, cyclic input of cAMP specified as follows:
which reflects the cAMP oscillations reported by Ohadi et al. (32); and 3) a ramp function of cAMP input defined as follows:
resulting in a gradual, time-dependent increase over 300 s (60) until a constant input of 0.3 μM, mimicking a sustained plateau of adenylyl cyclase activation. Note that we do not model adenylyl activity explicitly here.
We simulate in the computational model the PDE4-mediated enzymatic inactivation of cAMP (Table 1, Eqs. 1 and 2). Moreover, we model the total initial amount of all PDE4 isoforms combined as constant and equal to 1 μM (Table 1, Eq. 3) (50). The relative distribution of the PDE4 isoforms is modeled as follows: 70% long PDE4 isoform, 11% short PDE4 isoform, and 19% supershort PDE4 isoform. These distributions are based on in-house measurements of rat hippocampal tissue to exemplify tissue-specific isoform proportions (see supporting material). Since long PDE4 isoforms comprise 70% of the total in this tissue, the effect of long PDE4 isoforms on cAMP signaling may be biased by the fact that these long forms are most abundantly present. As such, we control for this potential bias by also investigating a scenario in which the isoforms have equal proportions. More specifically, one isoform is set to zero and the other isoforms are both present at 0.5 μM concentration in order to keep the total PDE4 concentration constant at 1 μM (Fig. 10). In the computational model, cAMP degradation is modeled with mass action kinetics, and has three contributions: 1) baseline degradation by PDE4 that is independent of isoform type; 2) isoform-specific modulation of the baseline degradation by the activated catalytic subunit of PKA (PKAcat,1); and 3) isoform-specific modulation of the baseline degradation by activated ERK (ERKdimer) (Table 1, Eqs. 1 and 2).
Figure 10.
Effects of isoform type specific PDE4 inhibition on cAMP and downstream signaling. The effect of isoform-specific PDE4 inhibition on cAMP signaling was tested while considering the relative proportions of isoform types measured in the hippocampus (A, C, E, and G) or considering equal expression proportions of the different PDE4 isoform types (B, D, F, and H) for a cyclic input of cAMP. Inhibition of long PDE4 isoforms (light-gray lines) produced the largest elevations in cAMP (A and B), PKA (C and D), Epac (E and F), and ERK levels (G and H) irrespective of the proportion of isoform types. Inhibition of short forms (black triangles) led to higher levels of cAMP and downstream molecules than inhibition of supershort forms (black diamonds) when considering equal isoform type proportions (B, D, F, H) but not in the case of hippocampal proportions (A, C, E, G), indicating that relative proportions determine the functional importance of isoform types. PKA, Epac, and ERK reflect the species PKAcat,1, Epacon, and ERKdimer, respectively. Simulations were run for 1500 s with oscillatory cAMP input (0.3 μM).
The activated catalytic subunit of PKA (PKAcat,1) and activated ERK (ERKdimer) are modeled to influence cAMP degradation rate by activating or inhibiting PDE4, depending on the PDE4 isoform involved (13). Therefore, k12on,PDE, k13on,PDE, k22on,PDE, k23on,PDE, k32on,PDE, and k33on,PDE are not constant but depend on PKAcat,1 or ERKdimer, which can have an inhibitory (Vmax is negative) or stimulating (Vmax is positive) effect depending on the affected PDE4 isoform. These phosphorylation-based activity effects were modeled with a Michaelis-Menten function as
| (1) |
Phosphorylation by PKA or ERK changes the rate of cAMP hydrolysis by PDE4 by changing Vmax without affecting KM (16,22). Moreover, it has been reported that the KM values for different PDE4 forms are similar (61, 62, 63). It has been reported that the measured basal Vmax (8 s−1) is increased to 272% due to phosphorylation by PKA and reduced by 75% due to phosphorylation by ERK in the case of the long isoform (16,22). The basal activity of the short isoform and supershort isoforms are increased to 130% and reduced to 85%, respectively due to phosphorylation by ERK (21). We would like to highlight that some literature reports mention a percent reduction in the Vmax value and others an overall activity reduction with respect to the baseline value. Here we have chosen to adapt the basal Vmax value (taken as 8 s−1 based on (50,64, 65, 66)). For example, a 272% increase in the Vmax of long isoform mutated to mimic PKA phosphorylation has been reported (22), which we captured via a Vmax of 21.76 (i.e., k12on,PDE). A similar reasoning was made for the other Vmax values (22,24). Parameters used for each isoform are given in Table 2. We also implemented that k13on,PDE and k33on,PDE cannot become larger than the baseline PDE4-mediated cAMP degradation rate (kon,PDE: 0.15 μM s−1), as this would result in “negative degradation” and therefore “production” of cAMP. The degradation of cAMP into AMP is assumed to be irreversible, as cAMP can only be synthesized by conversion of ATP via ACs.
PKA dynamics
PKA activation by cAMP is modeled as a multistep process using mass action kinetics. The inactive tetramer PKA (PKAtet) is formed by the association of two catalytic (PKAcat) and two regulatory (PKAreg) subunits with constant kon,PKA. We assume that four cAMP molecules can bind cooperatively to the tetramer with a Hill coefficient of 1.6 (2,67), to form a complex (C1). When C1 dissociates (with constant kdeg), the active catalytic subunits (PKAcat,1) are released. Inactivation of the active catalytic subunits (PKAcat,1) occurs at a constant rate kdeg,1, after which the catalytic subunits can reassociate with the regulatory subunits to form the PKA tetramer, PKAtet (Fig. 2 and Table 1, Eqs. 4–8).
At basal cAMP levels PKA activity has been found to be absent (55). We therefore assumed that cAMP was only able to significantly bind PKA when cAMP levels were higher than 5.2 μM (55), which we modeled by putting kmPKA to 5.2 μM (Table 1, Eq. 7).
Epac-RAP1-ERK dynamics
Epac is a guanine nucleotide exchange factor that aids in the activation of Rap1 (3, 4, 5). In the model, the binding of cAMP to Epac with rate constant kon,Epac releases Epac from its autoinhibitory conformation (EPAC), producing EPACon (Table 1, Eqs. 9 and 10). We assumed that Epac could only be significantly activated at cAMP concentrations higher than 30 μM, which we captured by setting kmEPAC to 30 μM (Table 1, Eq. 9) (56). Subsequently, EPACon is able to activate Rap1, which is mathematically represented in Table 1, Eqs. 11 and 12. Rap1 can, through complex, crosstalking signaling cascades, modulate ERK activation (5,26,28). Since PDE4 inhibitors that should exert therapeutic actions in the brain are also prone to induce PDE4-mediated side effects by actions in the brainstem, we sought to better understand PDE4-mediated cAMP degradation in neurons specifically. In neurons, cAMP increases ERK activity in an Rap1/B-raf dependent manner (28). Here, we assume that Rap1 directly activates ERK, as intermediate signaling via B-raf and MEK consists of linear reactions. ERK is activated after its dual phosphorylation and subsequent dimerization (Fig. 2 and Table 1, Eqs. 13–15). ERK dimerization is crucial for extranuclear/cytosolic actions, and PDE4 was found to associate with ERK2 dimers and not with monomers (68,69). Subsequently, the ERKdimer is able to phosphorylate PDE4 isoforms and stimulate or inhibit their rate isoform specifically (Eq. 1 and Table 2) (70).
Simulation settings
Simulations were run in VCell for a duration of 500 or 1500 s with a combined stiff solver (IDA/CVODE). The absolute and relative tolerance was set to 1 × 10−9. The models for a single pulse and continuous input can be accessed on the VCell public model repository https://vcell.org/vcell-published-models. The names of the models are as follows: for a single pulse, Carlier_cAMP_isoforms_v2; and for continuous cyclic input, Carlier_cAMP_isoforms_cyclic_v2.
Details on running a model in VCell can be found in the quick start guide on the VCell website, https://vcell.org/support. Data were analyzed using GraphPad Prism V9.1.0 (GraphPad, San Diego, CA).
Sensitivity analysis
A sensitivity analysis was performed on the standard model where the Vmax parameter values were altered to investigate which Vmax was most influential on the average and maximal cAMP values over 1500 s for cyclic input (see Fig. S3). The sensitivity was calculated as follows:
where cAMP(k) is the maximal or average cAMP concentration over 1500 s for cyclic input using the standard model settings, cAMP (k + Δk) is the maximal or average cAMP concentration over 1500 s for cyclic input at ±10% of the standard Vmax model parameter values, Δk is the varied parameter, and k is the standard model parameter value.
Results
Degradation dynamics of a single cAMP pulse
Different initial cAMP concentrations (0.1–3.0 μM) were used in the simulations to explore potential concentration-dependent effects on cAMP degradation (Fig. 3). Irrespective of the initial cAMP concentration, single cAMP pulses were completely degraded within 25 s. Comparison of the degradation rates revealed that initial cAMP concentrations of 1.0 and 3.0 μM induced a higher degradation rate compared with lower initial cAMP concentrations (Fig. 3 A, insert). Note that increases in initial cAMP concentrations are not proportional to the amount of cAMP present over time (reflected by the areas under the curve (AUC), Fig. 3 B). For example, for a pulse of 3.0 μM cAMP, a fold change of 10 would be expected while a fold change of 8.17 is observed, which indicates that higher initial cAMP concentrations elicit additional effects to facilitate its own degradation.
Figure 3.
Degradation dynamics of single cAMP pulses of different concentration predicted by the computational model. (A) Single cAMP pulses in a concentration range of 0.1–3.0 μM are quickly degraded for all simulated concentrations. In the inset, degradation rates are compared by log-transformation of the y axis showing equal degradation rates for cAMP concentration of 0.1–1.0 μM and profound increased degradation of cAMP in the case of an initial concentration of 3 μM cAMP. (B) Overview of plot characteristics per initial cAMP concentration. Fold changes in initial cAMP concentration, time to peak, peak value, and area under the curve (AUC) are shown compared with the 0.3 μM cAMP condition. These values indicate that the AUC does not change proportionally to the change in initial cAMP concentration. Simulations were run for 500 s considering estimated hippocampal PDE4 isoform proportions and a total initial (constant) amount of all PDE4 isoforms combined equal to 1 μM.
Effects of different cAMP concentrations on PKA, Epac, and ERK dynamics
Although distinct cAMP pulse concentrations were all found to be quickly degraded based on our initial simulations, we next sought to explore how downstream signaling cascades are affected by these different initial cAMP concentrations. In our computational model, we have focused on the downstream cAMP-PKA and cAMP-Epac-ERK pathways, as PKA and ERK affect cAMP signaling using feedback mechanisms through the modulation of PDE4 enzyme activity. In the following sections, the mention of PKA, Epac, and ERK reflects concentrations of the species PKAcat,1, Epacon, and ERKdimer, respectively.
Simulations using single cAMP pulses of different concentrations revealed that PKA and Epac are differentially activated depending on the cAMP concentration. For example, cAMP pulses of 0.3 and 1.0 μM lead to slightly higher peak activation of Epac compared with PKA, while a cAMP pulse of 3.0 μM induces profoundly higher peaks of PKA compared with Epac (Fig. 4, A, B, D, and E). Moreover, a distinction in PKA and Epac dynamics can be observed regarding their concentrations over time. Irrespective of the concentration cAMP, PKA levels subside more slowly compared with Epac (Fig. 3, A and B). ERK signaling is only activated if Epac is sufficiently activated (Figs. 4 C and 2). Higher initial cAMP levels resulted in profoundly higher ERKdimer concentrations until the endpoint of the simulation (500 s) (Fig. 4, C and F).
Figure 4.
Activation dynamics of downstream signaling by different cAMP concentrations predicted by the computational model. (A–F) Single cAMP pulses in a concentration range of 0.1–3.0 μM induced dose-dependent increases in PKA activation (A), Epac activation (B), and ERK activation (C). cAMP concentrations differentially affect PKA and Epac signaling as shown by differences in proportional peak height, time to peak, and AUC differences for the various cAMP concentrations compared with the default model using 0.3 μM cAMP as initial concentration (D and E). ERK activation did not reach a plateau during the 500 s simulation, but shows non-linear increases as higher initial cAMP concentrations were simulated (C and F). PKA, Epac, and ERK reflect the model species PKAcat,1, Epacon, and ERKdimer, respectively. Simulations were run for 500 s considering estimated hippocampal PDE4 isoform proportions.
Effects of PDE4 concentrations and isoform type on single-pulse cAMP signaling
Corresponding to literature, cAMP is rapidly degraded in the computational model after a single cAMP pulse (Fig. 3). PDE4 enzymes play a pivotal role the regulation of cAMP signaling. To explore the effect of PDE4-mediated cAMP regulation, simulations were run using different concentrations of total PDE4 enzyme. Total PDE4 enzyme concentrations ranged from 0.1 to 10 μM, while taking into account the relative proportions of PDE4 isoform types as measured in the rat hippocampus as an example of organ-specific PDE4 isoform type expression (see supporting material). Relative proportions of long (0.70), short (0.11), and supershort (0.19) isoforms were kept constant for all total PDE4 concentrations. As expected, higher total PDE4 concentrations resulted in a more rapid degradation of a single 0.3 μM cAMP pulse (Fig. 5 A). Dynamics of downstream PKA, Epac, and ERK signaling were highly non-linear for different PDE4 concentrations (Fig. 5, B–D). For example, compared with the default total PDE4 concentration of 1 μM, higher PDE4 concentrations (3–10 μM) lead to 3- and 10-fold lower AUC values for cAMP, PKA, and Epac. In contrast, lower PDE4 concentrations cause non-proportional increases in peak and AUC values for cAMP, PKA, and Epac (Fig. 5, B, C, and F). For example, a 10-fold higher initial cAMP pulse (3.0 μM) causes a 28.56- and 8.29-fold increase in the peak value for PKA and Epac, respectively. Regarding ERK activation, non-linear effects are observed for both lower and higher total PDE4 concentrations, implying that ERK levels are particularly sensitive to the amount of PDE4 present (Fig. 5, D and F).
Figure 5.
The influence of PDE4 concentration and isoform type on single-pulse cAMP signaling as predicted by the computational model. (A) Degradation of a single 0.3 μM cAMP pulse is dependent on the concentration of total PDE4 present. Relative proportions of long, short, and supershort PDE4 isoforms were kept constant. (B–D) Lower total PDE4 concentrations induced higher peaks, increased time to peak, and sustained activation of PKA (B), Epac (C), and ERK (D). (E) Simulations in which total PDE4 consists of only a specific PDE4 isoform type indicated that all PDE4 isoform types degrade a single 0.3 μM cAMP pulse identically. (F) Fold changes in time to peak, peak, and area under the curve (AUC) are listed for the different PDE4 concentration conditions, compared with the 1.0 μM PDE4 condition, for cAMP and for the downstream signaling molecules PKA, Epac, and ERK. PKA, Epac, and ERK reflect the species PKAcat,1, Epacon, and ERKdimer, respectively. Simulations were run for 500 s considering, except for (E), estimated hippocampal PDE4 isoform proportions.
Interestingly, downstream signaling cascades appear to respond differently to increases in initial cAMP pulses versus reductions in the amount of PDE4 present when comparing with the default model in which an initial cAMP pulse of 0.3 μM is simulated with 1 μM PDE4 present. Specifically, a 10-fold increase in initial cAMP concentration causes a larger fold change in AUC value and peak value for PKA (PKA peak 28.56 and AUC 28.31; Fig. 4 D) compared with a 10-fold decrease in PDE4 concentration (PKA peak 6.74 and AUC 8.66; Fig. 5 F). In contrast, a 10-fold increase in initial cAMP concentration causes a smaller fold change in AUC value for Epac (Epac AUC 7.79; Fig. 4 E) compared with a 10-fold decrease in PDE4 concentration (Epac AUC 8.89; Fig. 5 F). Moreover, changing the PDE4 concentration resulted in a more profound effect on the fold changes in time to peak for PKA and Epac (i.e., fold changes ranging from 0.18 to 3.72; Fig. 5 F) compared with changes in cAMP input concentrations (i.e., fold changes ranging from 0.81 to 1.04; Fig. 4, D and E). These findings indicate that changes in cAMP input or PDE4-mediated cAMP degradation differentially affect the amplitude and timing of activation of downstream signaling.
Because the enzyme activity of PDE4 isoform types is differentially altered by PKA and ERK feedback phosphorylation, we subsequently investigated whether the presence of a single PDE4 isoform type at a concentration of 1.0 μM, rather than a combination of them in specific proportions, resulted in isoform-specific dynamics of 0.3 μM cAMP pulse degradation. Unexpectedly, cAMP degradation was found to be identical irrespective of the PDE4 isoform type present (Fig. 5 E). These results suggest that, for a single pulse, the rate of cAMP degradation is determined by the total isoform concentration rather than the isoform type present or the relative proportion of multiple types.
Dynamics of oscillatory cAMP signaling and isoform-specific control by PDE4
Following up on the unexpected finding that different PDE4 isoform types show identical dynamics of single cAMP pulse degradation when present at the same concentration, we hypothesized that single cAMP pulses may not be sufficient to elicit the PKA- and ERK-based feedback mechanisms on PDE4 activity as observed in cell-based experiments. Accordingly, under physiological conditions cAMP synthesis occurs in a prolonged, oscillatory manner rather than as the production of single cAMP pulses (71,72). Previous studies have shown that calcium oscillates spontaneously and that these oscillations influence the cAMP/PKA dynamics (32,73). In particular, computational work has shown that cAMP/PKA is a leaky integrator of calcium dynamics, meaning that cAMP/PKA senses the lower frequency of the calcium dynamics. Here, we wanted to explore how different PDE4 concentrations and isoform types regulate downstream signaling dynamics of oscillatory cAMP signaling.
Similar to what is seen for a single cAMP pulse, the concentration of total PDE4 regulates oscillatory cAMP signaling and activation of downstream effectors (Figs. 5 and 6). Higher (3.0 μM) and lower (0.3 μM) PDE4 concentrations resulted, respectively, in a faster and slower degradation of oscillatory cAMP (Fig. 6 A). Accordingly, activation of the effectors PKA (PKAcat,1), Epac (Epacon), and ERK (ERKdimer) was similarly changed and these effects were highly non-linear (Fig. 6, B–D). Interestingly, and in contrast to a single pulse of cAMP, the isoform type or the relative proportion greatly influenced the rate of cAMP degradation in the simulated settings. More specifically, when keeping the total PDE4 concentration equal at 1 μM but varying the type of isoform, Fig. 7 shows that the long and short isoforms maintain an oscillatory cAMP profile (albeit with a higher initial peak for the short isoform with respect to the long isoform, Fig. 7 A) while the supershort isoform is not able to degrade the cyclic cAMP input at a sufficient rate, leading to a fast accumulation of cAMP (Fig. 7 A, inset). Also, downstream signaling is distinct when only a specific PDE4 isoform type is present. For the short and long isoform, the steady-state levels of activated PKA and Epac oscillate around 0.05 μM and 0.025 μM, respectively, whereas for the supershort isoform the steady-state Epac and PKA levels are almost 10- and 5-fold higher (0.24 μM and 0.27 μM, Fig. 7, B and C). A gradual increase in cAMP concentration followed by a sustained input resulted in findings similar to those of the cyclic cAMP input, i.e., the long and short isoforms maintain a stable cAMP profile while the supershort isoform is not able to degrade the sustained cAMP input at a sufficient rate, leading to a fast accumulation of cAMP (see Fig. S2). As such, these results nicely correspond to the findings of the Conti laboratory whereby sustained adenylyl cyclase activation has been shown to result in a transient increase in the intracellular cAMP concentration, after which the intracellular cAMP concentrations reach a steady-state level (for long isoform activation through PKA) (74). These simulations indicate that cAMP signaling is not effectively controlled in the presence of supershort PDE4 isoforms alone, suggesting that additional biological mechanisms would have to be employed to prevent cAMP levels from rising uncontrollably.
Figure 6.
The influence of PDE4 concentration on oscillatory cAMP signaling as predicted by the computational model. (A) Dynamic control of oscillatory cAMP pulses is dependent on the concentration of total PDE4 present. Relative proportions of long, short, and supershort PDE4 isoforms were kept constant. (B–D) Lower total PDE4 concentrations induced higher levels of activated PKA, Epac, and ERK. PKA, Epac, and ERK reflect the species PKAcat,1, Epacon, and ERKdimer, respectively. Simulations were run for 1500 s considering estimated hippocampal PDE4 isoform proportions.
Figure 7.
PDE4 isoform types differentially regulate oscillatory cAMP signaling. The effect of PDE4 isoform type on oscillatory cAMP signaling was simulated by including a single PDE4 isoform type only at a concentration of 1 μM. (A) Dynamic control of oscillatory cAMP signaling is distinct for different PDE4 isoform types. Presence of supershort PDE4 isoforms only leads to profound accumulation of cAMP (inset). When only short or long PDE4 isoforms are present, cAMP levels can be stabilized in a concentration range after an initial peak. (B and C) Similar to the effect on oscillatory cAMP control, PKA and Epac are most profoundly activated when only supershort PDE4 isoforms are present. In the case of only short PDE4 isoforms being present, an initial increase can be observed after which activation levels stabilize. (D) Similar to PKA and Epac activation, ERK activation increased mainly when only supershort isoforms were present. Presence of only short PDE4 isoforms led to a higher ERK activation compared with long PDE4 isoforms only. PKA, Epac, and ERK reflect the species PKAcat,1, Epacon, and ERKdimer, respectively. Simulations were run for 1500 s with oscillatory cAMP input (0.3 μM).
PDE4 activity changes over time in an isoform-specific manner
To understand why the type of PDE4 isoform only has an important impact for cyclic cAMP pulses and not for a single cAMP pulse in the computational model, we looked in detail at Eq. 1 in Table 1 (see materials and methods). The activity of the PDE4 isoform types is differentially affected upon phosphorylation events by activated PKA and ERK. As PKA and ERK themselves are dynamically regulated over time (Fig 7, B and D), we next sought to investigate how the degradation rates for the different PDE4 isoform types change over time as a response to oscillatory cAMP signaling.
For a single cAMP pulse, the degradation rates overlap for all isoform types (Fig. 5 E), which can be explained by the fact that the elevations in activated PKA and ERK concentrations are small (Fig. 5, B–D) resulting in no or negligible differences between the enzymatic activity of different PDE4 isoforms. In contrast, for cyclic cAMP pulses there is a continuous generation of activated ERK and PKA, leading to higher overall degradation rates by means of biological feedback loops impacting upon PDE4 activity (Fig 6 A). For the long isoform, whose activity is regulated by both PKA and ERK, the activation rate is higher than the inhibition rate, which reflects the Vmax settings (i.e., 21.76 for activation, −2 for inhibition), resulting in net increased activation (Figs. 8 A and S1). In other words, the activation of the long isoform by PKA keeps the PKA concentration under control, since a higher degradation rate of cAMP results in less PKA formation (a negative feedback loop), resulting in a compensatory k12on,PDE rate. The short isoform is activated by ERK. Since the activation of ERK is slower than that of PKA (Fig. 6, B and D), the increase in kon short (i.e., kon,PDE + k23on,PDE) is slower than the increase of kon long (i.e., kon,PDE + k12on,PDE + k13on,PDE), and overall a lower activation rate is reached for short PDE4 compared with long PDE4. Consequently, the cAMP has a larger initial peak when only the short isoform is present in comparison with when all (Fig. 6 A), or only the long isoform (Fig. 7 A) is present.
Figure 8.
Degradation rates of cAMP by PDE4 long, short, and supershort isoforms change differentially over time. The degradation rates of long (Kon long), short (Kon short), and supershort (Kon supershort) forms are shown in light gray, black, and dark gray, respectively. Degradation rates per PDE4 isoform are dependent on the baseline degradation rate and modulation by PKA and/or ERK and are calculated based on Tables 1 and 2 (Kon long = kon,PDE + k12on,PDE + k13on,PDE; Kon short = kon,PDE + k22on,PDE + k23on,PDE; Kon supershort = kon, PDE + k32on, PDE + k33on, PDE). (A) Changes in absolute degradation rates per PDE4 isoform type are plotted over time. (B) The contribution of different PDE4 isoform types to the total degradation changes over time. Changes in degradation rate correspond to changes in PKA and ERK levels as shown by the dashed line (PKA) and solid line (ERK). PKA and ERK reflect the species PKAcat,1 and ERKdimer, respectively. Simulations were run for 1500 s considering estimated hippocampal PDE4 isoform proportions (total 1 μM) and oscillatory cAMP input (0.3 μM).
The supershort isoform is inhibited by ERK, which represents a positive feedback loop. More specifically, a high ERK concentration inhibits the degradation of cAMP by long and supershort PDE4 forms, leading to downstream activation of ERK. Consequently, in the simulations of cAMP input with only the supershort isoform present, the kon supershort (i.e., kon,PDE + k33on,PDE) rate already flattens out after 500 s (Fig. 8, A and B), implying that there is no degradation of cAMP any longer (all PDE4 supershort isoforms are inhibited), resulting in a massive buildup of the cAMP concentration (Fig. 7 A). These simulations show that, in cases where only the supershort isoform is present, other mechanisms should be activated to limit the cAMP concentration increase, and in particular mechanisms that limit the activation of the ERK pathway or increase the cAMP degradation in a PDE4-independent mechanism.
The degradation rates of long and supershort PDE4 isoforms start to go down once ERK becomes activated while short forms are activated (Fig. 8, A and B). This implies that, in the case where ERK is already activated and PDE4 long and supershort forms are thus initially inhibited before cAMP synthesis is started, PDE4-mediated cAMP degradation could be diminished. This means that preceding ERK activation, by inhibition of long and supershort PDE4, could have “permissive and facilitating” actions on cAMP signaling.
To investigate the influence of Vmax parameter values on the model outcome, we performed a sensitivity analysis in which we varied the Vmax values by ±10% and looked at the average and maximum cAMP value over 1500 s (see Fig. S3). As can be appreciated from these results, the sensitivity of the model is similar for all Vmax values, with the highest for k12on,PDE, which captures the effect of the long isoform phosphorylation by PKA. In this respect, the sensitivity analysis is in line with the other reported results that highlight the important effect of the long isoform. Importantly, we highlight here that due to the scarcity of quantitative data, the baseline Vmax values are based on measures in different cell types using different assays.
Initial PKA and ERK concentrations influence PDE4-mediated cAMP degradation
Despite the large influence of PKA and ERK on PDE4-mediated cAMP degradation, our prior simulations did not take into account the initial presence of PKA and/or ERK and may therefore not fully represent the biological situation in which these species may already modulate PDE4 activity before cAMP is synthesized. Therefore, we also explored the influence of the activated PKA and ERK concentration on PDE4-mediated cAMP degradation by using non-zero initial concentrations (0.05 and 0.066 μM, respectively) based on their final values in the simulations shown in Fig. 6, B and D. The following simulations were run using a single pulse only, since cyclic pulses lead to continuous degradation and production, which overrules the effect of different initial PKA and ERK conditions.
Fig. 9 shows the degradation dynamics of a single 0.3 μM cAMP pulse for different initial concentrations of PKA and ERK. Similar to the aforementioned, these results indicate that the initial PKA concentration has an important influence on cAMP degradation by increasing the activity of long PDE4 isoforms and overruling a potential influence of initial ERK concentrations (i.e., note that the curves of the gray diamonds and black triangles overlap in Fig. 9). In the case of no PKA being initially present, non-zero initial concentrations of ERK also influence cAMP degradation by inhibiting long and supershort PDE4 isoforms while activating short PDE4 isoforms. Depending on the intracellular distribution of the different PDE4 isoform types and their relative proportions, these initial PKA and ERK concentrations may give rise to specific intracellular cAMP gradients by distinctly influencing PDE4 activity.
Figure 9.
The initial concentrations of PKA and ERK influence degradation of a single cAMP pulse. Elevated initial PKA levels lead to quicker degradation of a single 0.3 μM cAMP pulse (black triangles), while increased initial ERK levels diminish degradation (light-gray triangles) compared with the default simulation in which PKA and ERK are initially absent (black squares). When both PKA and ERK are initially present, PKA overrules the effect of ERK indicating that PKA, by activating long PDE4 isoforms, has a major influence on PDE4-mediated cAMP degradation (gray diamonds). PKA and ERK reflect the species PKAcat,1 and ERKdimer, respectively. Simulations were run considering estimated hippocampal PDE4 isoform proportions (total 1 μM) with a single cAMP pulse input (0.3 μM).
Inhibition of PDE4 long isoforms has the most profound effect on cAMP signaling
Based on the observations that cAMP signaling is best kept under control when long PDE4 isoforms are present (Fig. 7), long isoforms contribute largely to the total PDE4-mediated cAMP degradation (Fig. 8), and the dominant effect of PKA over ERK in modulating PDE4 activity by acting on long PDE4 forms specifically (Fig. 9), we sought to investigate whether inhibition of long PDE4 isoforms specifically impact cAMP signaling most profoundly compared with inhibition of the other isoform types.
Inhibition of single PDE4 isoform types was simulated by setting this isoform to zero while leaving the other two types unaffected. In prior simulations we have used the relative proportions of long, short, and supershort PDE4 isoforms based on those measured in the hippocampus as an example of tissue-specific expression, i.e., 0.7 long, 0.11 short, and 0.19 supershort. Since long PDE4 isoforms comprised 70% of the total in this tissue, the observations that long PDE4 isoforms exert a large control on cAMP signaling may have been biased by the fact that these long forms are most abundantly present. To control for this potential bias, we also simulated the inhibition of an isoform type while keeping the remaining forms in equal proportions, i.e., one isoform is set to zero and the other isoforms are both present at 0.5 μM concentration in order to keep the total PDE4 concentration constant at 1 μM.
Fig. 10 visualizes the effect of PDE4 isoform type inhibition on cAMP signaling considering both the hippocampal proportion of isoform types and equal proportions. Irrespective of the proportions considered, inhibition of long PDE4 forms led, compared with inhibition of short or supershort isoforms, to higher levels of cAMP (light-gray curve in Fig. 10, A and B), PKA (light-gray curve in Fig. 10, C and D), Epac (light-gray curve in Fig. 10, E and F), and ERK signaling (light-gray curve in Fig. 10, G and H). Inhibition of short isoforms differed from the inhibition of supershort forms only when considering equal proportion of isoform types (Fig. 10, B, D, F, and H), which demonstrates that unequal abundance of PDE4 isoform types (e.g., in the case of the hippocampus) influences the functional importance of a particular isoform type.
Discussion
Various intra- and extracellular stimuli all induce the synthesis of cAMP but eventually evoke distinct cellular effects. This “repurposing” of the same signaling machinery by controlling its dynamics is beneficial from an evolutionary perspective compared with developing separate pathways for each stimulus, receptor, or response (75). Accordingly, by means of compartmentalization and multiple (dynamic) feedback mechanisms, cAMP can convey signals from multiple different sources to induce distinct responses. By degrading cAMP, PDE4 enzymes exert profound control over cAMP signaling dynamics. Specifically, PDE4 enzymes consist of multiple isoform types, the enzyme activity of which is dynamically regulated in a feedback-based manner in response to downstream cAMP signaling. Here, we constructed a computational model to explore the role of PDE4 and its different isoforms in the control of cAMP dynamics.
Based on our model, we explored the effects of different concentrations cAMP and PDE4 on activation of the downstream cAMP-PKA and cAMP-Epac pathways. It was identified that PKA activation was mainly influenced by the concentration of cAMP while Epac activation was more sensitive to the amount of PDE4 present. Moreover, simulations using different total PDE4 concentrations caused substantial changes in PKA and Epac activation dynamics by changing time to peak values while simulation using different cAMP concentrations did not. Changes in PDE4 concentration led to non-linear changes in the dynamics of cAMP and downstream effectors, which provides an estimation of the magnitude of effect of experimentally observed changes in PDE4 expression. For example, the 1.5- to 4.0-fold increases in PDE4 at the mRNA, protein, and enzyme activity level that have been reported in physiological and disease-associated conditions do affect cAMP and downstream signaling based on our model (76, 77, 78).
In light of investigating the effect of PKA- and ERK-based feedback on PDE4 isoform activity, our simulations indicated that oscillatory cAMP signaling, as opposed to single cAMP input pulses, is necessary to induce downstream effector activation that can influence PDE4 isoform activity. As the change in enzyme activity upon phosphorylation by PKA and/or ERK is PDE4 isoform specific, we examined how oscillatory cAMP signaling affects the activity of these PDE4 isoform types. The results indicated that long PDE4 isoforms exert the largest control on dynamics of cAMP and downstream effector signaling (Fig. 7) and that long PDE4 isoforms contribute most to total PDE4-mediated cAMP degradation (Fig. 8). These long-dominant effects may have been biased by the fact that we considered PDE4 isoform proportions based on measurements in rat hippocampal tissue, in which long forms were predominantly (i.e., 70%) present. However, in simulations in which each of the PDE4 isoform type were present in equal amount and were separately inhibited, long forms also exhibited the largest impact on cAMP and downstream signaling dynamics (Fig. 10). This seeming importance of long PDE4 forms is supported by the fact that the four PDE4 genes (PDE4A, -B, -C, -D) encode more long isoforms than short and supershort isoforms (13), which may imply that long forms are involved in a broader array of cellular functions. Indeed, these various long PDE4 isoforms are known to localize to specific intracellular compartments owing to their unique N-terminus amino acid stretches to engage in specific protein-protein interactions (13,15). As such, long PDE4 isoforms can control cAMP signaling in a precisely located and efficient manner, as their activity can be quickly increased upon phosphorylation by PKA.
In contrast to long forms, supershort PDE4 forms were found to inadequately control oscillatory cAMP signaling (Fig. 7). These isoforms cannot be phosphorylated, and activated, by PKA, but are only affected by ERK phosphorylation. However, phosphorylation by ERK actually decreases the enzyme activity of supershort PDE4 while modestly increasing the activity of short PDE4 forms. Thus, phosphorylation-based feedback mechanisms appear insufficient in increasing supershort PDE4 isoform activity. This may imply that cAMP levels could become uncontrollable in compartments where only supershort PDE4 forms are present. However, early studies have reported the transcription-based upregulation of (super)short PDE4 in response to cAMP signaling activation as an alternative feedback mechanism (79, 80, 81). Moreover, (super)short forms have been shown to localize throughout the cytosol where they may control cAMP signaling globally compared with cAMP control by long PDE4 forms in specific locations (64,82,83).
The results of this study, which indicate the effects of PDE4 isoform types on cAMP signaling dynamics, should be interpreted in the light of the following assumptions and limitations. Firstly and most importantly, the model simulations predict that all cAMP is degraded in approximately 15 s (single cAMP pulse), which differs from the reported cAMP measurements in live cells (i.e., 100–300 s (84)). Although this discrepancy may be cell dependent, we hypothesize that this might be due to the ordinary differential equation formalism we are using, which as such do not fully account for the spatial regulation of cAMP signaling via cAMP compartmentalization and local subcellular cAMP gradients. More specifically, precise subcellular localization of PDE isoforms is proposed to be important for shaping cAMP gradients (85, 86, 87). Here, we provide a very rough approximation of how different isoforms of a certain PDE4 subtype may be proportionally expressed in (parts of) an organ. As such, our approximation of the ratio of different PDE4 isoform categories by means of a western blot of just the PDE4D subtype does not reflect or provide information on the expression of all PDE4 subtypes and isoforms in specific cell types and their localization within these cells. Interestingly, of the 21 reported human PDE4 isoforms, 15 are long (70%), 2 are short (10%), and 4 are supershort isoforms (20%), which reflects very similar proportions as reported for PDE4D isoforms here (13). As soon as isoform-specific intracellular expression patterns are determined, these details can be included in future models. In addition, local production by ACs, cAMP buffering (by e.g., PKA), physical barriers (i.e., the cytoskeleton), export by multidrug resistance proteins, and cell shape are also believed to contribute to cAMP compartmentalization (88). Importantly, recent insights indicated that cAMP is primarily buffered by PKA regulatory subunit condensates and that PDEs effectively reduce cAMP signaling in highly localized, nanometer-sized compartments (46,89,90). Also, A-kinase anchoring proteins (AKAPs) have the ability to anchor PKA, Epac, and PDE4 to specific subcellular locations to form local signaling complexes with high signaling specificity and efficacy (91,92). By tethering PKA to specific subcellular locations, it can specifically activate effector proteins in its vicinity. Besides, AKAPs can also directly bind effector proteins to spatially and temporally influence the signal transduction (91,92). AKAPs are thus important players in establishing compartmentalized cAMP signaling by contributing to the subcellular localization of signaling components, but it is not yet completely understood how the intracellular positioning of cAMP effector proteins (i.e., PDE4, Epac, PKA) by AKAPs shapes cellular cAMP signaling.
Secondly, considering the short time scales that we model, we assume a constant amount of protein, thus ignoring potential production and degradation processes as well as transcriptional or epigenetic regulation (13,78,93). For example, the transcriptional upregulation of (super)short PDE4 as feedback mechanism may take longer than 1500 s as simulated here, but could be implicated in future models. Earlier studies reported large increases in (super)short transcripts, but these were observed after a period of multiple hours (80,94). Because of this difference in time frames and the difficulty of translating cAMP increases to downstream transcriptional upregulation, we opted to not include transcription-regulated feedback in this model as yet. Thirdly, this study focuses on PDE4, whereas other cAMP-degrading PDEs also play an important role in the spatiotemporal dynamics of cAMP signaling. Similarly, the PDE4 activity can be modulated through a wide variety of post-translational modifications and interactions with partner proteins (13), which are not all captured in the current model. For example, the effect of phosphatases, which would remove phosphorylation of PDE4 or undo the effects of PKA in Raf-1-expressing cells on phosphorylation of Raf1 and its downstream signaling to ERK (26), providing yet other cell-specific routes for cAMP and ERK signaling pathways to interact, could be incorporated in future work. Finally, the parameter values of the model (e.g., proportional/relative concentrations of included signaling molecules and the way they influence each other) are highly cell-type-, context-, and compartment-specific (26,28,95). Future work should focus on acquiring these cell-type- and context-specific experimental data (e.g., using next-generation cAMP-sensing techniques (46)) in order to better calibrate the computational models (including, if necessary, stochastic simulation techniques such as reported in (49)) and simulate these dedicated scenarios.
The current study pointed out that different PDE4 isoforms distinctly regulate cAMP and downstream signaling dynamics and that these isoform-specific differences should be considered in future computational and experimental work on PDE4/cAMP signaling. Computational follow-up studies could focus on PDE4/cAMP signaling in specific cellular compartments or cell types by adapting the model presented here. Moreover, this model can provide insights into PDE4 drug design by simulating how PDE4 inhibitors, with different affinities to the different isoform types, impact overall cAMP signaling. Experimentally, future computational work should validate the influence of specific PDE4 isoforms on cell-type-specific cAMP-regulated processes by using, for example, RNA silencing or (epi)genetic editing.
Author contributions
D.P., S.H., and A.C. developed the computational model. D.P., S.H., and A.C. analyzed its results. D.P., T.V., J.P., and A.C. contributed to the design and conception of the study. D.P., S.H., and A.C. drafted the manuscript. D.P., S.H., D.v.d.H., T.V., and J.P. contributed to manuscript revision, and read and approved the submitted version.
Acknowledgments
We thank Zeynep Karagöz for independently confirming our simulation results. A.C. gratefully acknowledges the financial support of the Dutch province of Limburg in the LINK (FCL67723) (“Limburg INvesteert in haar Kenniseconomie”) knowledge economy project and a VENI grant (number 15075).This work was supported by Alzheimer Nederland (grant WE.03-2016-07).
Declaration of interests
T.V. and J.P. have a proprietary interest in selective PDE4D inhibitors for the treatment of demyelinating disorders. J.P. has a proprietary interest in the PDE4 inhibitor roflumilast for the treatment of cognitive impairment as well as PDE4D inhibitors for the treatment of Alzheimer’s disease.
Editor: Alexandr Kornev.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.06.019.
Supporting material
References
- 1.Herberg F.W., Taylor S.S., Dostmann W.R.G. Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase. Biochemistry. 1996;35:2934–2942. doi: 10.1021/bi951647c. [DOI] [PubMed] [Google Scholar]
- 2.Kim C., Cheng C.Y., et al. Taylor S.S. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt M., Dekker F.J., Maarsingh H. Exchange protein directly activated by cAMP (epac): a multidomain cAMP mediator in the regulation of diverse biological functions. Pharmacol. Rev. 2013;65:670–709. doi: 10.1124/pr.110.003707. [DOI] [PubMed] [Google Scholar]
- 4.de Rooij J., Zwartkruis F.J.T., et al. Wittinghofer A. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 5.Bos J.L., de Rooij J., Reedquist K.A. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 6.Schindler R.F.R., Brand T. The Popeye domain containing protein family--A novel class of cAMP effectors with important functions in multiple tissues. Prog. Biophys. Mol. Biol. 2016;120:28–36. doi: 10.1016/j.pbiomolbio.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaupp U.B., Seifert R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 8.Musheshe N., Schmidt M., Zaccolo M. cAMP: from long-range second messenger to nanodomain signalling. Trends Pharmacol. Sci. 2018;39:209–222. doi: 10.1016/j.tips.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Baillie G.S., Tejeda G.S., Kelly M.P. Therapeutic targeting of 3′, 5′-cyclic nucleotide phosphodiesterases: inhibition and beyond. Nat. Rev. Drug Discov. 2019;18:770–796. doi: 10.1038/s41573-019-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakics V., Karran E.H., Boess F.G. Quantitative comparison of phosphodiesterase mRNA distribution in human brain and peripheral tissues. Neuropharmacology. 2010;59:367–374. doi: 10.1016/j.neuropharm.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Blokland A., Heckman P., et al. Prickaerts J. Phosphodiesterase type 4 inhibition in CNS diseases. Trends Pharmacol. Sci. 2019;40:971–985. doi: 10.1016/j.tips.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Peng T., Qi B., et al. Shi J. Advances in the development of phosphodiesterase-4 inhibitors. J. Med. Chem. 2020;63:10594–10617. doi: 10.1021/acs.jmedchem.9b02170. [DOI] [PubMed] [Google Scholar]
- 13.Paes D., Schepers M., et al. Prickaerts J. The molecular biology of phosphodiesterase 4 enzymes as pharmacological targets: an interplay of isoforms, conformational states, and inhibitors. Pharmacol. Rev. 2021;73:1016–1049. doi: 10.1124/pharmrev.120.000273. [DOI] [PubMed] [Google Scholar]
- 14.Schepers M., Tiane A., et al. Vanmierlo T. Targeting phosphodiesterases-towards a tailor-made approach in multiple sclerosis treatment. Front. Immunol. 2019;10:1727. doi: 10.3389/fimmu.2019.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houslay M.D. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 2010;35:91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann R., Wilkinson I.R., et al. Houslay M.D. cAMP-specific phosphodiesterase HSPDE4D3 mutants which mimic activation and changes in rolipram inhibition triggered by protein kinase A phosphorylation of Ser-54: generation of a molecular model. Biochem. J. 1998;333:139–149. doi: 10.1042/bj3330139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez R., Sette C., et al. Conti M. Activation and selective inhibition of a cyclic AMP-specific phosphodiesterase, PDE-4D3. Mol. Pharmacol. 1995;48:616–622. [PubMed] [Google Scholar]
- 18.Sette C., Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J. Biol. Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 19.Sette C., Vicini E., Conti M. The ratPDE3/IVd phosphodiesterase gene codes for multiple proteins differentially activated by cAMP-dependent protein kinase. J. Biol. Chem. 1994;269:18271–18274. [PubMed] [Google Scholar]
- 20.MacKenzie S.J., Baillie G.S., et al. Houslay M.D. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1) Br. J. Pharmacol. 2002;136:421–433. doi: 10.1038/sj.bjp.0704743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baillie G.S., MacKenzie S.J., et al. Houslay M.D. Sub-family selective actions in the ability of Erk2 MAP kinase to phosphorylate and regulate the activity of PDE4 cyclic AMP-specific phosphodiesterases. Br. J. Pharmacol. 2000;131:811–819. doi: 10.1038/sj.bjp.0703636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann R., Baillie G.S., et al. Houslay M.D. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenhard J.M., Kassel D.B., et al. Luther M. Phosphorylation of a cAMP-specific phosphodiesterase (HSPDE4B2B) by mitogen-activated protein kinase. Biochem. J. 1996;316:751–758. doi: 10.1042/bj3160751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKenzie S.J., Baillie G.S., et al. Houslay M.D. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J. Biol. Chem. 2000;275:16609–16617. doi: 10.1074/jbc.275.22.16609. [DOI] [PubMed] [Google Scholar]
- 25.Lario P.I., Bobechko B., et al. Vrielink A. Purification and characterization of the human PDE4A catalytic domain (PDE4A330-723) expressed in Sf9 cells. Arch. Biochem. Biophys. 2001;394:54–60. doi: 10.1006/abbi.2001.2513. [DOI] [PubMed] [Google Scholar]
- 26.Stork P.J.S., Schmitt J.M. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–266. doi: 10.1016/s0962-8924(02)02294-8. [DOI] [PubMed] [Google Scholar]
- 27.Hoy J.J., Salinas Parra N., et al. Iglesias-Bartolome R. Protein kinase A inhibitor proteins (PKIs) divert GPCR-Gαs-cAMP signaling toward EPAC and ERK activation and are involved in tumor growth. FASEB J. 2020;34:13900–13917. doi: 10.1096/fj.202001515R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dugan L.L., Kim J.S., et al. Gutmann D.H. Differential effects of cAMP in neurons and astrocytes: role of B-RAF. J. Biol. Chem. 1999;274:25842–25848. doi: 10.1074/jbc.274.36.25842. [DOI] [PubMed] [Google Scholar]
- 29.Beavo J.A., Brunton L.L. Cyclic nucleotide research—still expanding after half a century. Nat. Rev. Mol. Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 30.Bhalla U.S. Signaling in small subcellular volumes. I. Stochastic and diffusion effects on individual pathways. Biophys. J. 2004;87:733–744. doi: 10.1529/biophysj.104.040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohadi D., Rangamani P. Geometric control of frequency modulation of cAMP oscillations due to calcium in dendritic spines. Biophys. J. 2019;117:1981–1994. doi: 10.1016/j.bpj.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohadi D., Schmitt D.L., et al. Rangamani P. Computational modeling reveals frequency modulation of calcium-cAMP/PKA pathway in dendritic spines. Biophys. J. 2019;117:1963–1980. doi: 10.1016/j.bpj.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jędrzejewska-Szmek J., Damodaran S., et al. Blackwell K.T. Calcium dynamics predict direction of synaptic plasticity in striatal spiny projection neurons. Eur. J. Neurosci. 2017;45:1044–1056. doi: 10.1111/ejn.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shumilov A.V., Gotovtsev P.M. Modeling the activity of the dopamine signaling pathway by combination of analog electrical circuit and mathematical approaches. Heliyon. 2021;7:e05879. doi: 10.1016/j.heliyon.2020.e05879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Getz M., Swanson L., et al. Rangamani P. A predictive computational model reveals that GIV/girdin serves as a tunable valve for EGFR-stimulated cyclic AMP signals. Mol. Biol. Cell. 2019;30:1621–1633. doi: 10.1091/mbc.E18-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenner B., Getz M., et al. Zhang J. Spatially compartmentalized phase regulation of a Ca2+-cAMP-PKA oscillatory circuit. Elife. 2020;9:e55013. doi: 10.7554/elife.55013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone N., Shettlesworth S., et al. Phan A.V. A two-dimensional finite element model of cyclic adenosine monophosphate (cAMP) intracellular signaling. SN Appl. Sci. 2019;1:1713. doi: 10.1007/s42452-019-1757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira R.F., Kim M., Blackwell K.T. Subcellular location of PKA controls striatal plasticity: stochastic simulations in spiny dendrites. PLoS Comput. Biol. 2012;8:e1002383. doi: 10.1371/journal.pcbi.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M., Park A.J., et al. Blackwell K.T. Colocalization of protein kinase A with adenylyl cyclase enhances protein kinase A activity during induction of long-lasting long-term-potentiation. PLoS Comput. Biol. 2011;7:e1002084. doi: 10.1371/journal.pcbi.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chay A., Zamparo I., et al. Blackwell K.T. Control of βAR- and N-methyl-D-aspartate (NMDA) receptor-dependent cAMP dynamics in hippocampal neurons. PLoS Comput. Biol. 2016;12:e1004735. doi: 10.1371/journal.pcbi.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindskog M., Kim M., et al. Kotaleski J.H. Transient calcium and dopamine increase PKA activity and DARPP-32 phosphorylation. PLoS Comput. Biol. 2006;2:e119. doi: 10.1371/journal.pcbi.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohse C., Bock A., et al. Bauer W.R. Experimental and mathematical analysis of cAMP nanodomains. PLoS One. 2017;12:e0174856. doi: 10.1371/journal.pone.0174856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveira R.F., Terrin A., et al. Blackwell K.T. The role of type 4 phosphodiesterases in generating microdomains of cAMP: large scale stochastic simulations. PLoS One. 2010;5:e11725. doi: 10.1371/journal.pone.0011725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song R.S., Massenburg B., et al. Neves S.R. ERK regulation of phosphodiesterase 4 enhances dopamine-stimulated AMPA receptor membrane insertion. Proc. Natl. Acad. Sci. USA. 2013;110:15437–15442. doi: 10.1073/pnas.1311783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slepchenko B.M., Loew L.M. Use of virtual cell in studies of cellular dynamics. Int Rev Cell Mol Biol. 2010;283:1–56. doi: 10.1016/S1937-6448(10)83001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock A., Annibale P., et al. Lohse M.J. Optical mapping of cAMP signaling at the nanometer scale. Cell. 2020;182:1519. doi: 10.1016/j.cell.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang P.-C., Boras B.W., et al. Clancy C.E. A computational modeling and simulation approach to investigate mechanisms of subcellular cAMP compartmentation. PLoS Comput. Biol. 2016;12:e1005005. doi: 10.1371/journal.pcbi.1005005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal S.R., Clancy C.E., Harvey R.D. Mechanisms restricting diffusion of intracellular cAMP. Sci. Rep. 2016;6:19577. doi: 10.1038/srep19577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jȩdrzejewska-Szmek J., Luczak V., et al. Blackwell K.T. β-adrenergic signaling broadly contributes to LTP induction. PLoS Comput. Biol. 2017;13:e1005657. doi: 10.1371/journal.pcbi.1005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neves S.R., Tsokas P., et al. Iyengar R. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salonikidis P.S., Zeug A., et al. Richter D.W. Quantitative measurement of cAMP concentration using an exchange protein directly activated by a cAMP-based FRET-sensor. Biophys. J. 2008;95:5412–5423. doi: 10.1529/biophysj.107.125666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasagawa S., Ozaki Y.-i., et al. Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Liu R.-Y., et al. Byrne J.H. Computational design of enhanced learning protocols. Nat. Neurosci. 2011;15:294–297. doi: 10.1038/nn.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin W., Tran T.M., et al. Rich T.C. Roles of GRK and PDE4 activities in the regulation of beta2 adrenergic signaling. J. Gen. Physiol. 2008;131:349–364. doi: 10.1085/jgp.200709881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koschinski A., Zaccolo M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: implications for compartmentalization of cAMP signalling. Sci. Rep. 2017;7:14090. doi: 10.1038/s41598-017-13021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purves G.I., Kamishima T., et al. Dart C. Exchange protein activated by cAMP (Epac) mediates cAMP-dependent but protein kinase A-insensitive modulation of vascular ATP-sensitive potassium channels. J. Physiol. 2009;587:3639–3650. doi: 10.1113/jphysiol.2009.173534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fujioka A., Terai K., et al. Matsuda M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 58.Radhakrishnan K., Edwards J.S., et al. Oliver J.M. Sensitivity analysis predicts that the ERK-pMEK interaction regulates ERK nuclear translocation. IET Syst. Biol. 2009;3:329–341. doi: 10.1049/iet-syb.2009.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khokhlatchev A.V., Canagarajah B., et al. Cobb M.H. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 60.Calebiro D., Nikolaev V.O., et al. Lohse M.J. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009;7:e1000172. doi: 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saldou N., Obernolte R., et al. Huber A. Comparison of recombinant human PDE4 isoforms: interaction with substrate and inhibitors. Cell. Signal. 1998;10:427–440. doi: 10.1016/s0898-6568(97)00169-1. [DOI] [PubMed] [Google Scholar]
- 62.Bolger G.B., McPhee I., Houslay M.D. Alternative splicing of cAMP-specific phosphodiesterase mRNA transcripts. Characterization of a novel tissue-specific isoform, RNPDE4A8. J. Biol. Chem. 1996;271:1065–1071. doi: 10.1074/jbc.271.2.1065. [DOI] [PubMed] [Google Scholar]
- 63.Jin S.L., Swinnen J.V., Conti M. Characterization of the structure of a low Km, rolipram-sensitive cAMP phosphodiesterase. Mapping of the catalytic domain. J. Biol. Chem. 1992;267:18929–18939. [PubMed] [Google Scholar]
- 64.Bolger B.G., Erdogan S., et al. Houslay D.M. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem. J. 1997;328:539–548. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim J., Pahlke G., Conti M. Activation of the cAMP-specific phosphodiesterase PDE4D3 by phosphorylation. Identification and function of an inhibitory domain. J. Biol. Chem. 1999;274:19677–19685. doi: 10.1074/jbc.274.28.19677. [DOI] [PubMed] [Google Scholar]
- 66.Reeves M.L., Leigh B.K., England P.J. The identification of a new cyclic nucleotide phosphodiesterase activity in human and Guinea-pig cardiac ventricle. Implications for the mechanism of action of selective phosphodiesterase inhibitors. Biochem. J. 1987;241:535–541. doi: 10.1042/bj2410535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang P., Kornev A.P., et al. Taylor S.S. Discovery of allostery in PKA signaling. Biophys Rev. 2015;7:227–238. doi: 10.1007/s12551-015-0170-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrero A., Pinto A., et al. Crespo P. Small molecule inhibition of ERK dimerization prevents tumorigenesis by RAS-ERK pathway oncogenes. Cancer Cell. 2015;28:170–182. doi: 10.1016/j.ccell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Casar B., Pinto A., Crespo P. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol. Cell. 2008;31:708–721. doi: 10.1016/j.molcel.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 70.Houslay M., Baillie G. Portland Press Ltd.; 2003. The Role of ERK2 Docking and Phosphorylation of PDE4 cAMP Phosphodiesterase Isoforms in Mediating Cross-Talk between the cAMP and ERK Signalling Pathways. [DOI] [PubMed] [Google Scholar]
- 71.Dyachok O., Isakov Y., et al. Tengholm A. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 2006;439:349–352. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- 72.Huff T.C., Camarena V., et al. Wang G. Oscillatory cAMP signaling rapidly alters H3K4 methylation. Life science alliance. 2020;3 doi: 10.26508/lsa.201900529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Rochefort N., et al. Konnerth A. Reactivation of the same synapses during spontaneous up states and sensory stimuli. Cell Rep. 2013;4:31–39. doi: 10.1016/j.celrep.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 74.Rich T.C., Xin W., et al. Conti M. Cellular mechanisms underlying prostaglandin-induced transient cAMP signals near the plasma membrane of HEK-293 cells. Am. J. Physiol. Cell Physiol. 2007;292:C319–C331. doi: 10.1152/ajpcell.00121.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Purvis J.E., Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levallet G., Levallet J., et al. Bonnamy P.-J. Expression of the cAMP-phosphodiesterase PDE4D isoforms and age-related changes in follicle-stimulating hormone-stimulated PDE4 activities in immature rat sertoli cells. Biol. Reprod. 2007;76:794–803. doi: 10.1095/biolreprod.106.055343. [DOI] [PubMed] [Google Scholar]
- 77.Peter D., Jin S.L.C., et al. Zitt C. Differential expression and function of phosphodiesterase 4 (PDE4) subtypes in human primary CD4+ T cells: predominant role of PDE4D. J. Immunol. 2007;178:4820. doi: 10.4049/jimmunol.178.8.4820. [DOI] [PubMed] [Google Scholar]
- 78.Paes D., Lardenoije R., et al. Prickaerts J. Increased isoform-specific phosphodiesterase 4D expression is associated with pathology and cognitive impairment in Alzheimer's disease. Neurobiol. Aging. 2021;97:56–64. doi: 10.1016/j.neurobiolaging.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Conti M. Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol. Endocrinol. 2000;14:1317–1327. doi: 10.1210/mend.14.9.0534. [DOI] [PubMed] [Google Scholar]
- 80.Swinnen J.V., Tsikalas K.E., Conti M. Properties and hormonal regulation of two structurally related cAMP phosphodiesterases from the rat Sertoli cell. J. Biol. Chem. 1991;266:18370–18377. [PubMed] [Google Scholar]
- 81.Liu H., Palmer D., et al. Maurice D.H. Expression of phosphodiesterase 4D (PDE4D) is regulated by both the cyclic AMP-dependent protein kinase and mitogen-activated protein kinase signaling pathways: a potential mechanism allowing for the coordinated regulation OFPDE4D activity and expression in cells. J. Biol. Chem. 2000;275:26615–26624. doi: 10.1074/jbc.M001634200. [DOI] [PubMed] [Google Scholar]
- 82.Huston E., Lumb S., et al. Russell A. Molecular cloning and transient expression in COS7 cells of a novel human PDE4B cAMP-specific phosphodiesterase, HSPDE4B3. Biochem J. 1997;328:549–558. doi: 10.1042/bj3280549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cheung Y.F., Kan Z., et al. Castle J.C. PDE4B5, a novel, super-short, brain-specific cAMP phosphodiesterase-4 variant whose isoform-specifying N-terminal region is identical to that of cAMP phosphodiesterase-4D6 (PDE4D6) J. Pharmacol. Exp. Therap. 2007;322:600–609. doi: 10.1124/jpet.107.122218. [DOI] [PubMed] [Google Scholar]
- 84.Violin J.D., DiPilato L.M., et al. Lefkowitz R.J. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J. Biol. Chem. 2008;283:2949–2961. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 85.Feinstein W.P., Zhu B., et al. Rich T.C. Assessment of cellular mechanisms contributing to cAMP compartmentalization in pulmonary microvascular endothelial cells. Am. J. Physiol. Cell Physiol. 2012;302:C839–C852. doi: 10.1152/ajpcell.00361.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barnes A.P., Livera G., et al. Milgram S.L. Phosphodiesterase 4D forms a cAMP diffusion barrier at the apical membrane of the airway epithelium. J. Biol. Chem. 2005;280:7997–8003. doi: 10.1074/jbc.M407521200. [DOI] [PubMed] [Google Scholar]
- 87.Xin W., Feinstein W.P., et al. Rich T.C. Estimating the magnitude of near-membrane PDE4 activity in living cells. Am. J. Physiol. Cell Physiol. 2015;309:C415–C424. doi: 10.1152/ajpcell.00090.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saucerman J.J., Greenwald E.C., Polanowska-Grabowska R. Mechanisms of cyclic AMP compartmentation revealed by computational models. J. Gen. Physiol. 2014;143:39–48. doi: 10.1085/jgp.201311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang J.Z., Lu T.-W., et al. Zhang J. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell. 2020;182:1531–1544.e15. doi: 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jackson P.K. cAMP signaling in nanodomains. Cell. 2020;182:1379–1381. doi: 10.1016/j.cell.2020.08.041. [DOI] [PubMed] [Google Scholar]
- 91.Omar M.H., Scott J.D. AKAP signaling islands: venues for precision pharmacology. Trends Pharmacol. Sci. 2020;41:933–946. doi: 10.1016/j.tips.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dema A., Perets E., et al. Klussmann E. Pharmacological targeting of AKAP-directed compartmentalized cAMP signalling. Cell. Signal. 2015;27:2474–2487. doi: 10.1016/j.cellsig.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 93.Tilley D.G., Maurice D.H. Vascular smooth muscle cell phenotype-dependent phosphodiesterase 4D short form expression: role of differential histone acetylation on cAMP-regulated function. Mol. Pharmacol. 2005;68:596. doi: 10.1124/mol.105.014126. [DOI] [PubMed] [Google Scholar]
- 94.Swinnen J.V., Joseph D.R., Conti M. The mRNA encoding a high-affinity cAMP phosphodiesterase is regulated by hormones and cAMP. Proc. Natl. Acad. Sci. USA. 1989;86:8197–8201. doi: 10.1073/pnas.86.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dodge-Kafka K.L., Soughayer J., et al. Scott J.D. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.