Abstract
Objectives
The HL7® fast healthcare interoperability resources (FHIR®) specification has emerged as the leading interoperability standard for the exchange of healthcare data. We conducted a scoping review to identify trends and gaps in the use of FHIR for clinical research.
Materials and methods
We reviewed published literature, federally funded project databases, application websites, and other sources to discover FHIR-based papers, projects, and tools (collectively, “FHIR projects”) available to support clinical research activities.
Results
Our search identified 203 different FHIR projects applicable to clinical research. Most were associated with preparations to conduct research, such as data mapping to and from FHIR formats (n = 66, 32.5%) and managing ontologies with FHIR (n = 30, 14.8%), or post-study data activities, such as sharing data using repositories or registries (n = 24, 11.8%), general research data sharing (n = 23, 11.3%), and management of genomic data (n = 21, 10.3%). With the exception of phenotyping (n = 19, 9.4%), fewer FHIR-based projects focused on needs within the clinical research process itself.
Discussion
Funding and usage of FHIR-enabled solutions for research are expanding, but most projects appear focused on establishing data pipelines and linking clinical systems such as electronic health records, patient-facing data systems, and registries, possibly due to the relative newness of FHIR and the incentives for FHIR integration in health information systems. Fewer FHIR projects were associated with research-only activities.
Conclusion
The FHIR standard is becoming an essential component of the clinical research enterprise. To develop FHIR’s full potential for clinical research, funding and operational stakeholders should address gaps in FHIR-based research tools and methods.
Keywords: fast healthcare interoperability resources (FHIR), health information interoperability, data management, health information management, electronic health records
BACKGROUND AND SIGNIFICANCE
The Health Level Seven International® (HL7®) Fast Healthcare Interoperability Resources® (FHIR®)1 specification has been rapidly adopted in healthcare to enable clinical data exchange. FHIR is a set of data models and aligned technologies that define data formats, data elements, and application programming interface (API) protocols to enable the exchange of healthcare-related information.1 FHIR is built on modern computing standards (eg, JavaScript Object Notation [JSON], Secure HTTP [https]) and has data elements that are organized into data models called Resources. Data exchange rules, including additional data elements and constraints on the data model, are specified in Profiles. Implementation Guides serve as “recipes” or standard operating procedures for consistent use of FHIR Resources and APIs to support workflows in specified domains, thereby standardizing processes in addition to data.2 Together, these core components of rules and procedures enable data exchange among a growing number of computer applications in healthcare. Integral to the adoption and development of FHIR has been the Substitutable Medical Applications and Reusable Technologies (SMART) API standard, which allows applications or “apps” to be used across a variety of health information systems without modification. A SMART on FHIR application implements standardized authorization and authentication protocols with the data interoperability specifications of FHIR.3
FHIR was first proposed in 20114 as a new specification from the HL7 standards development organization, based on emerging industry approaches and work accomplished in previous versions of HL7 standards development. Since then, federal agencies and insurers have begun promoting its use, particularly in response to the US 21st Century Cures Act of 2016 (Cures Act), which calls for simplified access, exchange, and use of healthcare information, via APIs, to support increased interoperability.5 In March 2020, the Office of the National Coordinator for Health Information Technology (ONC) released a rule6 by which interoperability provisions of the Cures Act5 are to be implemented, providing a clearer path for health data interoperability. The National Institutes of Health (NIH) Strategic Plan for Data Science7 and the National Library of Medicine (NLM) 10-year strategic plan8 to ensure that research data are Findable, Accessible, Interoperable, and Reusable (FAIR)9 have helped prioritize the development of standardized data exchange for research.10,11 The current challenge is to implement modern data exchange standards for research in an industry that, so far, has focused on providers, payors, and patients.
In July 2019, NIH issued a notice (NOT-OD-19-122) to encourage investigators to explore applications of FHIR to “capture, integrate, and exchange clinical data for research purposes.”12 Vanderbilt University Medical Center was awarded a contract to pursue these goals with NLM support.13 Because FHIR usage for research purposes is relatively new and growing at a rapid pace, no compilation currently exists of such projects. For this reason, we conducted a scoping review of FHIR-based papers, projects, and tools (collectively, “FHIR projects”) available or in development that address the use of FHIR in support of clinical research. Our review differs from previous a review of FHIR use in clinical data exchange14 as our review is focused on FHIR applications and projects in clinical research. This article presents a compilation and evaluation of our findings.
METHODS
Protocol
The Preferred Reporting Items of Systematic Reviews and Meta-Analysis (PRISMA) methodology extension for scoping reviews (PRISMA-ScR) was used to develop our review protocol, which is summarized below. The objective of this review was to systematically identify and categorize FHIR projects that self-identify as being designed for, useful for, or relevant to clinical research. The review was designed to address the following questions:
What FHIR projects currently exist or are being developed for clinical research preparation, planning, recruitment, management, or conduct, or for sharing and analysis of clinical research data?
What gaps are there in the landscape of FHIR projects for clinical research that have yet to be filled?
Eligibility criteria
Items eligible for inclusion in the review described a project, study, resource, method, tool, or application that uses or proposes to use FHIR to design, implement, or test tools, resources, and applications to advance clinical research. Publication years were restricted to 2015–2021. We excluded items unrelated to FHIR and ones that did not fit the conceptual framework of our review, including those describing clinical uses of FHIR with no research applications, those focusing on technology or computer science aspects, those using only features of the SMART on FHIR protocol that were not part of the HL7 FHIR specification (eg, user authentication), and general descriptive articles that did not reference practical methods or tools. We also excluded projects not written in the English language.
Information sources and search strategy
The following online bibliographic databases were searched for peer-reviewed publications: PubMed, ScienceDirect, and SpringerLink. For the search for federally funded projects, we queried NIH RePORTER15 and the websites of the National Science Foundation (NSF),16 the Agency for Healthcare Research and Quality (AHRQ),17 and the Office of the National Coordinator for Health Information Technology (ONC).18 For the tool search, we searched the app stores of leading electronic health record (EHR) vendors (Allscripts App Expo, Cerner App Gallery, Epic Apple Orchard, SMART App Gallery). In addition, we hand-searched the reference lists of relevant publications, reviewed project implementations listed on the FHIR website (“FHIR Applications Registry”). We also conducted a web search to locate any relevant gray literature on FHIR use for research, such as review papers, white papers, expert opinions, internet reports, government websites, and book chapters.
A research information specialist (N.K.) and informatics researcher (S.D.) developed the search strategies and conducted the searches. The proprietary online search engine provided by each database or website was employed when available. In most cases, the terms “FHIR” and “Fast Healthcare Interoperability Resources” were the terms searched. The full list of search strategies is presented in Table 1. We included papers published from January 1, 2015 (to avoid early concept papers irrelevant to our review), through September 15, 2021. Our search for funded projects and applications was conducted on September 15, 2021. We restricted our applications search to those apps that explicitly stated they were designed for research or could be used to facilitate research activities.
Table 1.
Data sources and search strategies
| Source | URL | Search strategy |
|---|---|---|
| PubMed | https://pubmed.ncbi.nlm.nih.gov/ | (FHIR) OR (Fast Healthcare Interoperability Resources): filtered by 01/01/2015-09/15/2021 |
| SpringerLink | https://link.springer.com/ | FHIR OR “Fast Healthcare Interoperability Resources”: filtered by 2015-2021 and English language |
| ScienceDirect | https://www.sciencedirect.com/ | (FHIR OR “Fast Healthcare Interoperability Resources”): filtered by 2015-2021 |
| NIH RePORTER | https://reporter.nih.gov/ | FHIR |
| AHRQ | https://digital.ahrq.gov/ahrq-funded-projects/search | FHIR OR “Fast Healthcare Interoperability Resources” |
| NSF | https://www.nsf.gov/awardsearch/ | FHIR OR “Fast Healthcare Interoperability Resources” |
| ONC | https://www.healthit.gov/topic/scientific-initiatives | All |
| Tool-Allscripts | https://storealpha.allscripts.com/ | Category=FHIR Apps |
| Tool-Cerner | https://code.cerner.com/apps | FHIR |
| Tool-Epic | https://apporchard.epic.com/Gallery | FHIR & Categories=Research |
| Tool-SMART App Gallery | https://apps.smarthealthit.org/apps | Category=FHIR Tools |
| Website-FHIR | https://www.fhir.org/implementations/registry/ | All |
| https://www.google.com/search?q=%2Bfhir+%2Bresearch | “+fhir +research” |
Selection of sources
All candidate records were deposited first into a reference manager software program and then compiled into a single Microsoft Excel spreadsheet. If a publication, funding award, and/or application and website referenced the same project, they were merged into a single project listing that included all relevant citations. The items extracted for each record included, as applicable: title, first author or funding award recipient, journal, URL, publication or award year (where specified), abstract, and source. After excluding duplicates, the first authors (S.D. and N.K.) independently screened each record’s title, abstract, description, or summary, as applicable, depending on whether the project was a paper, funded award, or application. We excluded those records not related to FHIR and those for which the full text could not be retrieved. For the remaining records, the full text was reviewed. Records were excluded that were not related to a specific FHIR clinical research use such as those with only a technology focus, only about the potential promise of FHIR, or only about SMART on FHIR. Disagreements regarding inclusion or exclusion were resolved through discussion between the two adjudicators (S.D. and N.K.), who together approved the final list of included projects.
Data items and charting
The same authors (S.D. and N.K.) independently reviewed all included projects and labeled them according to how FHIR was being proposed or used to support research (eg, to extract data from an EHR, map data between formats, or standardize genomic data formats). Each project could receive more than one label.
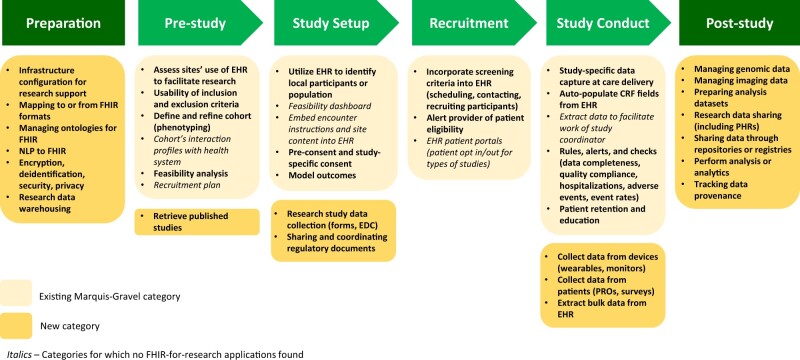
To support consistent labeling, we used a flowchart defined by Marquis-Gravel et al19 that outlines opportunities for leveraging EHRs for clinical trials. This trial-oriented framework, applicable to clinical research studies in general, was also useful as a foundation for organizing and synthesizing many of the FHIR projects discovered in our search, as FHIR is a principal means of extracting and repurposing EHR data. The Marquis-Gravel framework includes the major EHR-based elements capable of supporting a clinical trial, such as cohort identification, consent procedures, recruitment and retention, study management, and data collection. Within the established framework, we added categories for collecting data from patients (eg, surveys, patient-reported outcomes [PROs]), collecting data from devices such as wearables and monitors, and sharing and coordinating regulatory documentation. Given that research uses for FHIR extend beyond the conduct of a single study, we extended this framework to include a clinical research preparation stage that encompasses such organization-level activities as establishing data pipelines and infrastructure, or mapping between FHIR and other data formats (eg, Observational Medical Outcomes Partnership20 Common Data Model to FHIR, FHIR to Clinical Data Interchange Standards Consortium21 formats, custom datasets to FHIR Resources). We also extended the framework beyond study conduct, to include post-study activities such as preparation of analysis datasets, research data sharing, and depositing data in registries or repositories. These added categories were determined through prior reviews of the literature and discussion with all authors. If the reviewers determined a need for additional categories during the labeling process, they were discussed with the other authors and added to the categorization system.
Synthesis of results
The lead authors (S.D. and N.K.) independently assigned each of the FHIR projects identified in the search to one or more of these research use categories and then compared and harmonized the selection of labels for each FHIR project based on discussion. Each project could receive multiple labels to describe its use of FHIR for research. Categorization differences were resolved through a joint re-review of the materials and discussion. Any persisting conflicts in labeling were to be resolved by a third author (P.H.). We calculated the counts and frequencies of each label, as well as the number of projects with labels in each study phase (eg, Preparation, Recruitment, Study Conduct).
RESULTS
Selection and characteristics of sources of evidence
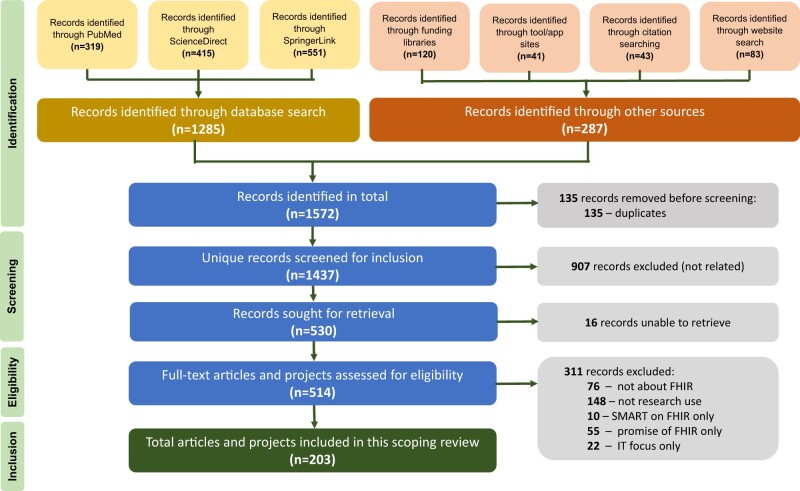
Our searches identified 1572 candidate FHIR projects through searches of publication databases (n = 1285), funding libraries (n = 120), tool/app stores (n = 41), citation searches (n = 43), and other websites and search engines (n = 83). We dropped 135 duplicate records and screened out an additional 907 records after a review of the title, abstract, or other preview material. Of the 530 records sought for retrieval, 16 could not be obtained, primarily due to expired web links that could not be located elsewhere. A total of 530 candidate FHIR projects were reviewed in depth, of which 311 were dropped after review, which were mostly projects not addressing a research use for FHIR (n = 148) or papers that were not actually about FHIR, often mentioning FHIR only once in a background text or citation (n = 76). Of the original 1572 candidate projects, 203 (12.9%) were selected for inclusion in the scoping review (Figure 1). Note that our search found 16 projects that included more than one source, such as a paper and tool, funding award and tool, website and tool, or paper and website. For these 16 cases, we classified the search source as the one through which we originally found the project. The final 203 projects are thus represented by 125 articles from publication databases (plus their associated tools and websites),11,22–155 29 projects from funding award libraries (plus their associated tools),156–187 12 standalone tools from app stores,188–199 and 37 items from website and citation searches (plus their associated tools or websites).200–238
Figure 1.
PRISMA flowchart identification of FHIR projects related to clinical research. Figure acronyms: FHIR, fast health interoperability resources; IT, information technology; SMART, Substitutable Medical Applications and Reusable Technologies.
Thirty-eight different labels were available using the expanded Marquis-Gravel categories to categorize how each project used FHIR to contribute to research activities (Figure 2). Reviewer 1 used an average of 1.64 labels per item and reviewer 2 an average of 1.66 labels. Approximately, one-third of FHIR projects were labeled identically between reviewers (n = 73, 36%) and 40 did not match (19.7%), with the remainder overlapping with some or most categories. All categorization differences were resolved through joint re-review of the materials and discussion and did not require intervention of the third reviewer. After the harmonization of the labels, projects had a mean of 1.8 labels, with 89 projects receiving only one label and two projects with five labels.
Figure 2.
Expanded Marquis-Gravel categorization19—major components of clinical research facilitated by FHIR capabilities. CRF, case report form; EDC, electronic data collection [system]; EHR, electronic health record; NLP, natural language processing; PHR, personal health record; PRO, patient-reported outcomes.
Characteristics of sources and synthesis of results
Two-thirds of FHIR projects were funded or published in the most recent 3 years, with 41 in 2019 (20%), 44 in 2020 (22%), and 43 projects through mid-September of 2021 (21%). The remaining third of the projects were older, with 26 in 2018 (13%), 23 in 2017 (11%), 13 in 2015–2016 (6%), and 13 with no date specified (6%).
Across the trajectory of clinical research activities, most projects focused on general research preparation including infrastructure and development of data pipelines (n = 152, 74.9%). The second major category for research-related FHIR projects was post-study activities (n = 93, 45.8%), including analyzing data, managing specific types of collected data, and sharing data, followed by study conduct (n = 52, 25.6%). The individual categories with the most research-related FHIR projects involved mapping data to and from FHIR formats (n = 66, 32.5% of projects), managing ontologies for FHIR (n = 30, 14.8%), sharing data through FHIR-enabled repositories or registries (n = 24, 11.8%), research data sharing including personal health records (n = 23, 11.3%), managing genomic data (n = 21, 10.3%), and cohort phenotyping (n = 19, 9.4%). FHIR projects appeared less common among prestudy feasibility assessment activities (n = 27, 13.3%), study setup (n = 29, 14.3%), and recruitment (n = 5, 2.5%) (Table 2). All FHIR projects and their labels are listed in Supplementary Table S1.
Table 2.
Synthesis of results—research-related FHIR projects by category
| Categories | Count | % projects | Category total |
|---|---|---|---|
| Preparation | 152 (74.9%) | ||
| Mapping to or from FHIR research formats | 66 | 32.5 | |
| Managing ontologies for FHIR | 30 | 14.8 | |
| Infrastructure configuration for research support | 17 | 8.4 | |
| NLP to FHIR | 16 | 7.9 | |
| Research data warehousing | 14 | 6.9 | |
| Encryption, deidentification, security, privacy | 9 | 4.4 | |
| Prestudy | 27 (13.3%) | ||
| Define and refine cohort (phenotyping) | 19 | 9.4 | |
| Assess sites’ use of EHR to facilitate research | 3 | 1.5 | |
| Usability of inclusion and exclusion criteria | 2 | 1.0 | |
| Retrieve published studies | 2 | 1.0 | |
| Feasibility analysis | 1 | 0.5 | |
| Study setup | 29 (14.3%) | ||
| Research study data collection (forms, EDC) | 12 | 5.9 | |
| Preconsent and study-specific consent | 7 | 3.4 | |
| Utilize EHR to identify local participants or population | 6 | 3.0 | |
| Sharing and coordinating regulatory documents | 3 | 1.5 | |
| Model outcomes | 1 | 0.5 | |
| Recruitment | 5 (2.5%) | ||
| Incorporate screening criteria into EHR (scheduling, contacting, recruiting participants) | 4 | 2.0 | |
| Alert provider of patient eligibility | 1 | 0.5 | |
| Study conduct | 52 (25.6%) | ||
| Collect data from patients (PROs, surveys) | 14 | 6.9 | |
| Collect data from devices (wearables, monitors) | 13 | 6.4 | |
| Autopopulate CRF fields from EHR | 12 | 5.9 | |
| Extract bulk data from EHR | 8 | 3.9 | |
| Rules, alerts, and checks | 2 | 1.0 | |
| Study-specific data capture at care delivery | 2 | 1.0 | |
| Patient retention and education | 1 | 0.5 | |
| Post-study | 93 (45.8%) | ||
| Sharing data through repositories or registries | 24 | 11.8 | |
| Research data sharing (including PHRs) | 23 | 11.3 | |
| Managing genomic data | 21 | 10.3 | |
| Preparing analysis datasets | 12 | 5.9 | |
| Managing imaging data | 5 | 2.5 | |
| Perform analysis or analytics | 5 | 2.5 | |
| Tracking data provenance | 3 | 1.5 | |
| Total labels | 358 (for 203 projects) | ||
CRF, case report form; EDC, electronic data collection [system]; EHR, electronic health record; NLP, natural language processing; PHR, personal health record; PRO, patient-reported outcomes.
Unused labels from the Marquis-Gravel categorization included two prestudy activities (cohort’s interaction profiles with the health system, recruitment plan), two study setup activities (feasibility dashboard, embed study instructions in EHR), one recruitment task (EHR health portals with patient opt in/opt out), and one study activity (extracting data to facilitate the work of the study coordinator.)
DISCUSSION
Summary of evidence
Our landscape assessment revealed a growing number of FHIR-related projects, but limited penetration of FHIR in current research operations, especially compared to its more robust use in clinical applications for direct patient care.14 Most FHIR projects to date appear focused on the two ends of our clinical research trajectory: the development of FHIR-based data infrastructure and pipelines or the storage, analysis, or sharing of data generated from the study. This may reflect the relative newness of the FHIR specification; researchers and software developers are still building the foundations and working on transmitting data in FHIR. Moreover, many of the remaining research- and FHIR-related projects we did find were linked to clinical systems such as EHRs, patient-facing data systems, and registries, a situation possibly driven by patient, provider, or payor needs. We found relatively few projects associated with prestudy research activities, such as participant recruitment, consenting, and management of study documents.
Among the published papers that describe FHIR-based research application projects, a fair number of examples fall into the realm of demonstration projects, single-purpose applications, or concept ideas for FHIR tool development.89,93,112,128,225 Similarly, while various funded projects are developing FHIR infrastructure and tools to achieve their research aims, few are actively engaged in the development of FHIR-based research tools as their primary objective.
Limited adoption to date of FHIR for research may also be due to gaps in the FHIR specification. Essential enhancements to FHIR and its accompanying Implementation Guides are needed, including additional research-related FHIR Resources and protocols for study document management and participant consent. Many of these projects are in formative phases, and the ever-expanding FHIR implementer community is continually working to evaluate proposed additions to the specification and broaden the range of research use cases for FHIR. Connectathon events, organized regularly by HL7, engage participants from industry and academia in hands-on development and testing of new FHIR-based software solutions, including solutions benefiting research.239 The HL7 FHIR Accelerator program comprises a growing list of defined user communities across the spectrum of healthcare to improve data interoperability, including CodeX (data exchange for cancer research), the Da Vinci Project (payor–provider data interoperability), and the Gravity Project (social determinants of health).240 Of particular interest is HL7’s Vulcan FHIR Accelerator, a recently formed, multistakeholder program to advance the development, refinement, and use of the FHIR standard to bridge gaps between healthcare and clinical research by fostering collaborations, maximizing shared resources, and developing FHIR Research Resources.241
Despite gaps in project coverage and the FHIR specification itself, FHIR usage in the research context is evolving quickly. The body of literature reporting on FHIR-related projects for clinical research is growing steadily; most manuscripts included in this review were published between 2019 and 2021, demonstrating growing interest in this topic among researchers. Funding by NIH for developing FHIR tools to support research is also increasing.13 Notably, a priority of the recently released Policy and Development Agenda on National Health IT Priorities for Research promulgated by ONC is to improve the interoperability of healthcare data and the underlying documentation to enable investigators to more productively exploit FHIR-based APIs for research.242,243 The ONC Cures Act Final Rule also confirms the adoption of the FHIR Bulk Data Access implementation specification, providing a mandate for prioritizing further development and use of this technology.6 This specification may be transformative in enabling clinical research data retrieval to be both timely and efficient. In addition, a December 31, 2022, compliance deadline for the Cures Act components in the ONC Health IT Certification Program244 requires FHIR R4.0.1, US Core profiles, and SMART on FHIR. This will likely lead to further adoption of FHIR in EHRs, especially at academic medical centers, thereby accelerating the network effect and allowing researchers to develop FHIR applications and benefit from the data interoperability.
Limitations
Our scoping review has several limitations. FHIR usage in the research domain is changing rapidly and our review was conducted within a finite timeframe. Our search of the gray literature was not exhaustive and may have missed research-related work presented in other forums. Indeed, we may have missed FHIR-related tools for “the middle” of the research trajectory (prestudy, study setup, recruitment) if such tools are only presented in closed EHR vendor conferences. We also excluded FHIR projects designed strictly for clinical use, although we recognize such tools may be repurposed for research in appropriate circumstances.
The Marquis-Gravel schema provided an essential framework for our labeling of projects, but the categories were designed to describe EHR use in clinical trials and did not reflect all the possible uses of FHIR in a clinical research context. We attempted to remedy this by the development of additional categories. Our assignment of categories was also subjective; although we had two authors independently reviewing the citations, discussion and additional review of the material was necessary for almost two-thirds of the projects. While useful for this scoping review, we recognize this organizational framework is an approximation of the clinical research process, that not all research studies require all categories, and that some research activities may span individual process steps. Nevertheless, our review revealed gaps and opportunities in the application of FHIR for research that may inform future development and implementation efforts.
Conclusions
Despite significant interest in FHIR among investigators and the potential of the FHIR standard to transform the clinical research landscape, relatively few FHIR projects that address research needs are fully operational. Moreover, FHIR specifications for research operations, while developing at a fast pace, are not yet mature. Although more FHIR-enabled apps for research are entering the marketplace, a scattershot approach is unlikely to create a truly interoperable research ecosystem. Promoting and investing in the further development and use of FHIR Implementation Guides to support research through programs such as Vulcan will encourage the broad and substantial base needed to ensure that interoperability is accessible and attainable for all researchers.
FUNDING
This work was funded in part through the U.S. National Library of Medicine contract 75N97019P00279 with Vanderbilt University Medical Center and the U.S. National Library of Medicine at the National Institutes of Health.
AUTHOR CONTRIBUTIONS
S.D. and N.K. contributed equally to the manuscript. P.H., T.Z.-C., N.K., D.C., and S.D. conceived the study. N.K. and S.D. conducted the searches, reviewed and classified the findings, and drafted the manuscript. All authors revised the manuscript and approved the final submission.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Supplementary Material
Contributor Information
Stephany N Duda, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Department of Biomedical Informatics, Vanderbilt University School of Medicine, Nashville, Tennessee, USA.
Nan Kennedy, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Douglas Conway, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Alex C Cheng, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Department of Biomedical Informatics, Vanderbilt University School of Medicine, Nashville, Tennessee, USA.
Viet Nguyen, Stratametrics LLC, Salt Lake City, Utah, USA; HL7 Da Vinci Project, Ann Arbor, Michigan, USA.
Teresa Zayas-Cabán, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
Paul A Harris, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Department of Biomedical Informatics, Vanderbilt University School of Medicine, Nashville, Tennessee, USA.
Data Availability
No new data were generated or analyzed in support of this research.
REFERENCES
- 1.FHIR. HL7 FHIR Release 4. https://www.hl7.org/fhir/ Accessed August 26, 2020.
- 2.FHIR Implementation Guide Registry. HL7 FHIR Release 4. http://fhir.org/guides/registry/ Accessed September 10, 2020.
- 3. Mandel JC, Kreda DA, Mandl KD, et al. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc 2016; 23 (5): 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hatt N. A Brief History of FHIR and Its Impact on Connectivity. MedCity News. 2017.https://medcitynews.com/2017/11/fhir-and-its-impact-on-connectivity/ Accessed July 6, 2020. [Google Scholar]
- 5.21st Century Cures Act, Pub. L. No. 114-225, 130 Stat. 1034. 2016.
- 6.21st Century Cures Act: Interoperability, Information Blocking, and the ONC Health IT Certification Program, 45 C.F.R. § 170 and 171. Federal Register. 2020.
- 7.NIH Strategic Plan for Data Science | Data Science at NIH. https://datascience.nih.gov/strategicplan Accessed January 6, 2020.
- 8.A Platform for Biomedical Discovery and Data-Powered Health. https://www.nlm.nih.gov/pubs/plan/lrp17/NLM_StrategicReport2017_2027.html Accessed April 7, 2021.
- 9. Wilkinson MD, Dumontier M, Aalbersberg I, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016; 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kush RD, Warzel D, Kush MA, et al. FAIR data sharing: the roles of common data elements and harmonization. J Biomed Inform 2020; 107: 103421. [DOI] [PubMed] [Google Scholar]
- 11. Sinaci AA, Núñez-Benjumea FJ, Gencturk M, et al. From raw data to FAIR data: the FAIRification workflow for health research. Methods Inf Med 2020; 59 (S 01): e21–32. [DOI] [PubMed] [Google Scholar]
- 12.NOT-OD-19-122: Fast Healthcare Interoperability Resources (FHIR) Standard. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-122.html Accessed January 6, 2020.
- 13.NIH Awards Contracts to Increase Availability of High-Quality Data Using FHIR Standard | Data Science at NIH. https://datascience.nih.gov/news/fhir-awards-announcement-high-quality-data Accessed January 6, 2020.
- 14. Ayaz M, Pasha MF, Alzahrani MY, et al. The fast health interoperability resources (FHIR) standard: systematic literature review of implementations, applications, challenges and opportunities. JMIR Med Inform 2021; 9 (7): e21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NIH RePORTER. https://reporter.nih.gov/ Accessed May 2, 2022.
- 16.NSF - National Science Foundation. https://www.nsf.gov/ Accessed May 2, 2022.
- 17.Agency for Healthcare Research and Quality. https://www.ahrq.gov/ Accessed May 2, 2022. [DOI] [PubMed]
- 18.Scientific Initiatives | HealthIT.gov. Office of the National Coordinator for Health Information Technology (ONC). https://www.healthit.gov/topic/scientific-initiatives Accessed May 2, 2022.
- 19. Marquis-Gravel G, Roe MT, Turakhia MP, et al. Technology-enabled clinical trials. Circulation 2019; 140 (17): 1426–36. [DOI] [PubMed] [Google Scholar]
- 20.OMOP Common Data Model – OHDSI. https://www.ohdsi.org/data-standardization/the-common-data-model/ Accessed May 19, 2022.
- 21.CDISC | Clear Data. Clear Impact. https://www.cdisc.org/ Accessed May 19, 2022.
- 22. Gruendner J, Gulden C, Kampf M, et al. A framework for criteria-based selection and processing of fast healthcare interoperability resources (FHIR) data for statistical analysis: design and implementation study. JMIR Med Inform 2021; 9 (4): e25645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones J, Gottlieb D, Mandel JC, et al. A landscape survey of planned SMART/HL7 bulk FHIR data access API implementations and tools. J Am Med Inform Assoc 2021; 28 (6): 1284–7. doi: 10.1093/jamia/ocab028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wesley DB, Blumenthal J, Shah S, et al. A novel application of SMART on FHIR architecture for interoperable and scalable integration of patient-reported outcome data with electronic health records. J Am Med Inform Assoc 2021; 28 (10): 2220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartschke A, Börner Y, Thun S.. Accessing the ECG data of the apple watch and accomplishing interoperability through FHIR. Stud Health Technol Inform2021; 278: 245–50. [DOI] [PubMed] [Google Scholar]
- 26. De A, Huang M, Feng T, et al. Analyzing patient secure messages using a fast health care interoperability resources (FIHR)-based data model: development and topic modeling study. J Med Internet Res 2021; 23 (7): e26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banach A, Ulrich H, Kroll B, et al. APERITIF – automatic patient recruiting for clinical trials based on HL7 FHIR. Stud Health Technol Inform2021; 281: 58–62. [DOI] [PubMed] [Google Scholar]
- 28. Lenert LA, Ilatovskiy AV, Agnew J, et al. Automated production of research data marts from a canonical fast healthcare interoperability resource data repository: applications to COVID-19 research. J Am Med Inform Assoc 2021; 28 (8): 1605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riepenhausen S, Mertens C, Dugas M.. Comparing SDTM and FHIR for real world data from electronic health records for clinical trial submissions. Stud Health Technol Inform2021; 281: 585–9. [DOI] [PubMed] [Google Scholar]
- 30. Wen A, Rasmussen LV, Stone D, et al. CQL4NLP: development and integration of FHIR NLP extensions in clinical quality language for EHR-driven phenotyping. AMIA Jt Summits Transl Sci Proc 2021; 2021: 624–33. [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao D, Song C, Nakamura N, et al. Development of an application concerning fast healthcare interoperability resources based on standardized structured medical information exchange version 2 data. Comput Methods Programs Biomed 2021; 208: 106232. [DOI] [PubMed] [Google Scholar]
- 32. Madrigal E, Le LP.. Digital media archive for gross pathology images based on open-source tools and fast healthcare interoperability resources (FHIR). Mod Pathol 2021; 34 (9): 1686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gencturk M, Teoman A, Alvarez-Romero C, et al. End user evaluation of the FAIR4Health data curation tool. Stud Health Technol Inform 2021; 281: 8–12. [DOI] [PubMed] [Google Scholar]
- 34. Garza MY, Rutherford MW, Adagarla B, et al. Evaluating site-level implementations of the HL7 FHIR standard to support eSource data exchange in clinical research. Stud Health Technol Inform 2021; 281: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garza MY, Rutherford M, Myneni S, et al. Evaluating the coverage of the HL7® FHIR® standard to support eSource data exchange implementations for use in multi-site clinical research studies. AMIA Annu Symp Proc 2021 2020; 2020: 472–81. [PMC free article] [PubMed] [Google Scholar]
- 36. Hund H, Wettstein R, Heidt CM, et al. Executing distributed healthcare and research processes – the HiGHmed data sharing framework. Stud Health Technol Inform2021; 278: 126–33. [DOI] [PubMed] [Google Scholar]
- 37. Schmidt CO, Darms J, Shutsko A, et al. Facilitating study and item level browsing for clinical and epidemiological COVID-19 studies. Stud Health Technol Inform 2021; 281: 794–8. [DOI] [PubMed] [Google Scholar]
- 38. Sass J, Zabka S, Essenwanger A, et al. Fast healthcare interoperability resources (FHIR) representation of medication data derived from German procedure classification codes (OPS) Using identification of medicinal products (IDMP) compliant terminology. Stud Health Technol Inform2021; 278: 231–6. [DOI] [PubMed] [Google Scholar]
- 39. Wettstein R, Hund H, Kobylinski I, et al. Feasibility queries in distributed architectures – concept and implementation in HiGHmed. Stud Health Technol Inform2021; 278: 134–41. [DOI] [PubMed] [Google Scholar]
- 40. Oehm J, Storck M, Fechner M, et al. FhirExtinguisher: a FHIR resource flattening tool using FHIRPath. Stud Health Technol Inform2021; 281: 1112–3. doi: 10.3233/SHTI210369. [DOI] [PubMed] [Google Scholar]
- 41. Oehm J. FhirExtinguisher 2022. https://github.com/JohannesOehm/FhirExtinguisher Accessed April 28, 2022.
- 42. Rinaldi E, Thun S.. From OpenEHR to FHIR and OMOP data model for microbiology findings. Stud Health Technol Inform 2021; 281: 402–6. doi: 10.3233/SHTI210189. [DOI] [PubMed] [Google Scholar]
- 43. Murugan M, Babb LJ, Taylor CO, et al. Genomic considerations for FHIR®; eMERGE implementation lessons. J Biomed Inform 2021; 118: 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Majeed RW, Stöhr MR, Günther A.. HIStream-import: a generic ETL framework for processing arbitrary patient data collections or hospital information systems into HL7 FHIR bundles. Stud Health Technol Inform2021; 278: 75–9. [DOI] [PubMed] [Google Scholar]
- 45. Liu S, Luo Y, Stone D, et al. Integration of NLP2FHIR representation with deep learning models for EHR phenotyping: a pilot study on obesity datasets. AMIA Jt Summits Transl Sci Proc 2021; 2021: 410–9. [PMC free article] [PubMed] [Google Scholar]
- 46. Bauer DC, Metke‐Jimenez A, Maurer‐Stroh S, et al. Interoperable medical data: the missing link for understanding COVID‐19. Transbound Emerg Dis 2021; 68 (4): 1753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zong N, Ngo V, Stone DJ, et al. Leveraging genetic reports and electronic health records for the prediction of primary cancers: algorithm development and validation study. JMIR Med Inform 2021; 9 (5): e23586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alper BS, Dehnbostel J, Afzal M, COVID-19 Knowledge Accelerator (COKA) Initiative, et al. Making science computable: developing code systems for statistics, study design, and risk of bias. J Biomed Informatics 2021; 115: 103685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zong N, Stone DJ, Sharma DK, et al. Modeling cancer clinical trials using HL7 FHIR to support downstream applications: a case study with colorectal cancer data. Int J Med Inform 2021; 145: 104308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lambarki M, Kern J, Croft D, et al. Oncology on FHIR: a data model for distributed cancer research. Stud Health Technol Inform 2021; 278: 203–10. [DOI] [PubMed] [Google Scholar]
- 51. Guérin J, Laizet Y, Texier VL, et al. OSIRIS: a minimum data set for data sharing and interoperability in oncology. JCO Clin Cancer Informatics 2021; 5: 256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gulden C, Blasini R, Nassirian A, et al. Prototypical clinical trial registry based on fast healthcare interoperability resources (FHIR): design and implementation study. JMIR Med Inform 2021; 9 (1): e20470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng AC, Duda SN, Taylor R, et al. REDCap on FHIR: clinical data interoperability services. J Biomed Inform 2021; 121: 103871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burkhardt HA, Brandt PS, Lee JR, et al. StayHome: a FHIR-native mobile COVID-19 symptom tracker and public health reporting tool. Online J Public Health Inform 2021; 13 (1): e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rinaldi E, Saas J, Thun S.. Use of LOINC and SNOMED CT with FHIR for microbiology data. Stud Health Technol Inform 2021; 278: 156–62. [DOI] [PubMed] [Google Scholar]
- 56. Dolin RH, Gothi SR, Boxwala A, et al. vcf2fhir: a utility to convert VCF files into HL7 FHIR format for genomics-EHR integration. BMC Bioinformatics 2021; 22 (1): 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weber M, Griessbach A, Grossmann R, et al. A FHIR-based eConsent app for the digital hospital. Stud Health Technol Inform 2020; 270: 3–7. [DOI] [PubMed] [Google Scholar]
- 58. Kiourtis A, Mavrogiorgou A, Kyriazis D.. A semantic similarity evaluation for healthcare ontologies matching to HL7 FHIR resources. Stud Health Technol Inform 2020; 270: 13–7. [DOI] [PubMed] [Google Scholar]
- 59. Ulrich H, Germer S, Kock-Schoppenhauer A-K, et al. A smart mapping editor for standardised data transformation. Stud Health Technol Inform 2020; 270: 1185–6. [DOI] [PubMed] [Google Scholar]
- 60. Lenivtceva I, Kashina M, Kopanitsa G.. Category of allergy identification from free-text medical records for data interoperability. Stud Health Technol Inform 2020; 273: 170–5. [DOI] [PubMed] [Google Scholar]
- 61. Ryu B, Shin S-Y, Baek R-M, et al. Clinical genomic sequencing reports in electronic health record systems based on international standards: implementation study. J Med Internet Res 2020; 22 (8): e15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fischer P, Stöhr MR, Gall H, et al. Data integration into OMOP CDM for heterogeneous clinical data collections via HL7 FHIR bundles and XSLT. Stud Health Technol Inform 2020; 270: 138–42. [DOI] [PubMed] [Google Scholar]
- 63. Margheri A, Masi M, Miladi A, et al. Decentralised provenance for healthcare data. Int J Med Inform 2020; 141: 104197. [DOI] [PubMed] [Google Scholar]
- 64. Reinecke I, Gulden C, Kümmel M, et al. Design for a modular clinical trial recruitment support system based on FHIR and OMOP. Stud Health Technol Inform 2020; 270: 158–62. [DOI] [PubMed] [Google Scholar]
- 65. Zong N, Sharma DK, Yu Y, et al. Developing a FHIR-based framework for phenome wide association studies: a case study with a pan-cancer cohort. AMIA Jt Summits Transl Sci Proc 2020; 2020: 750–9. [PMC free article] [PubMed] [Google Scholar]
- 66. Zong N, Wen A, Stone DJ,. et al. Developing an FHIR-Based Computational Pipeline for Automatic Population of Case Report Forms for Colorectal Cancer Clinical Trials Using Electronic Health Records. JCO Clin Cancer Inform 2020; 4: 201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Alterovitz G, Heale B, Jones J, et al. FHIR genomics: enabling standardization for precision medicine use cases. NPJ Genom Med 2020; 5 (1): 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xu H, Cox S, Stillwell L, et al. FHIR PIT: an open software application for spatiotemporal integration of clinical data and environmental exposures data. BMC Med Inform Decis Mak 2020; 20 (1): 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. FHIR PIT. NCATS Data Translator Project - Tangerine Team 2021. https://github.com/NCATS-Tangerine/FHIR-PIT Accessed April 28, 2022.
- 70.FAIR4Health. GitHub. https://github.com/fair4health Accessed April 28, 2022.
- 71. Lee H-A, Kung H-H, Lee Y-J, et al. Global infectious disease surveillance and case tracking system for COVID-19: development study. JMIR Med Inform 2020; 8 (12): e20567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu D, Sahu R, Ignatov V, et al. High performance computing on flat FHIR files created with the new SMART/HL7 bulk data access standard. AMIA Annu Symp Proc 2019; 2019: 592–6. [PMC free article] [PubMed] [Google Scholar]
- 73.HL7.FHIR.UV.BULKDATA\Bulk Data IG Home Page - FHIR v4.0.1. https://hl7.org/fhir/uv/bulkdata/ Accessed 28 Apr 2022.
- 74. Tanaka K, Yamamoto R.. Implementation of a secured cross-institutional data collection infrastructure by applying HL7 FHIR on an existing distributed EMR storages. Stud Health Technol Inform 2020; 272: 155–8. [DOI] [PubMed] [Google Scholar]
- 75. Osterman TJ, Terry M, Miller RS.. Improving cancer data interoperability: the promise of the minimal common oncology data elements (mCODE) initiative. JCO Clin Cancer Inform 2020; 4: doi:10.1200/CCI.20.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ploner N, Prokosch H-U.. Integrating a secure and generic mobile app for patient reported outcome acquisition into an EHR infrastructure based on FHIR resources. Stud Health Technol Inform 2020; 270: 991–5. [DOI] [PubMed] [Google Scholar]
- 77. Gruendner J, Wolf N, Tögel L, et al. Integrating genomics and clinical data for statistical analysis by using GEnome MINIng (GEMINI) and fast healthcare interoperability resources (FHIR): system design and implementation. J Med Internet Res 2020; 22 (10): e19879. doi: 10.2196/19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gründner J. genomics_data_service. 2020. https://github.com/juliangruendner/genomics_data_service Accessed April 28, 2022.
- 79. Persons KR, Nagels J, Carr C, et al. Interoperability and considerations for standards-based exchange of medical images: HIMSS-SIIM Collaborative White Paper. J Digit Imaging 2019; 33 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mandl KD, Gottlieb D, Mandel JC, et al. Push button population health: the SMART/HL7 FHIR bulk data access application programming interface. NPJ Digit Med 2020; 3 (1): 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sayeed R, Gottlieb D, Mandl KD.. SMART markers: collecting patient-generated health data as a standardized property of health information technology. NPJ Digit Med 2020; 3: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Garcia SJ, Zayas-Cabán T, Freimuth RR.. Sync for genes: making clinical genomics available for precision medicine at the point-of-care. Appl Clin Inform 2020; 11 (2): 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sass J, Bartschke A, Lehne M, et al. The German Corona Consensus Dataset (GECCO): a standardized dataset for COVID-19 research in university medicine and beyond. BMC Med Inform Decis Mak 2020; 20 (1): 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brandt PS, Kiefer RC, Pacheco JA, et al. Toward cross-platform electronic health record-driven phenotyping using clinical quality language. Learn Health Syst 2020; 4 (4): e10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bild R, Bialke M, Buckow K, et al. Towards a comprehensive and interoperable representation of consent-based data usage permissions in the German medical informatics initiative. BMC Med Inform Decis Mak 2020; 20 (1): 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Khvastova M, Witt M, Krefting D.. towards interoperability in clinical research: enabling FHIR on the open source research platform XNAT. Stud Health Technol Inform 2019; 258: 3–5. [PubMed] [Google Scholar]
- 87. XNAT FHIR Plugin. Somnonetz 2022. https://github.com/somnonetz/xnat-fhir-plugin Accessed April 28, 2022.
- 88. Watkins M, Viernes B, Nguyen V, et al. Translating social determinants of health into standardized clinical entities. Stud Health Technol Inform 2020; 270: 474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hylock RH, Zeng X.. A blockchain framework for patient-centered health records and exchange (HealthChain): evaluation and proof-of-concept study. J Med Internet Res 2019; 21 (8): e13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vaidyam A, Halamka J, Torous J.. Actionable digital phenotyping: a framework for the delivery of just-in-time and longitudinal interventions in clinical healthcare. Mhealth 2019; 5: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kiourtis A, Nifakos S, Mavrogiorgou A, et al. Aggregating the syntactic and semantic similarity of healthcare data towards their transformation to HL7 FHIR through ontology matching. Int J Med Inform 2019; 132: 104002. [DOI] [PubMed] [Google Scholar]
- 92. Hong N, Wang K, Wu S, et al. An interactive visualization tool for HL7 FHIR specification browsing and profiling. J Healthc Inform Res 2019; 3 (3): 329–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dullabh P, Hovey L, Heaney-Huls K, et al. Application programming interfaces (APIs) in health care: findings from a current-state assessment. Stud Health Technol Inform 2019; 265: 201–6. [DOI] [PubMed] [Google Scholar]
- 94. Zohner J, Marquardt K, Schneider H, et al. Challenges and opportunities in changing data structures of clinical document archives from HL7-V2 to FHIR-based archive solutions. Stud Health Technol Inform 2019; 264: 492–5. [DOI] [PubMed] [Google Scholar]
- 95. Daumke P, Heitmann KU, Heckmann S, et al. Clinical text mining on FHIR. Stud Health Technol Inform 2019; 264: 83–7. [DOI] [PubMed] [Google Scholar]
- 96. Hong N, Wen A, Stone DJ, et al. Developing a FHIR-based EHR phenotyping framework: a case study for identification of patients with obesity and multiple comorbidities from discharge summaries. J Biomed Inform 2019; 99: 103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. nlp2fhir-obesity-phenotyping. BD2KOnFHIR 2019. https://github.com/BD2KOnFHIR/nlp2fhir-obesity-phenotyping Accessed April 28, 2022.
- 98. Hong N, Wen A, Shen F, et al. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. JAMIA Open 2019; 2 (4): 570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pfaff ER, Champion J, Bradford RL, et al. Fast healthcare interoperability resources (FHIR) as a meta model to integrate common data models: development of a tool and quantitative validation study. JMIR Med Inform 2019; 7 (4): e15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Metke-Jimenez A, Hansen D.. FHIRCap: transforming REDCap forms into FHIR resources. AMIA Jt Summits Transl Sci Proc 2019; 2019: 54–63. [PMC free article] [PubMed] [Google Scholar]
- 101. Eapen BR, Costa A, Archer N, et al. FHIRForm: an open-source framework for the management of electronic forms in healthcare. Stud Health Technol Inform 2019; 257: 80–5. [PubMed] [Google Scholar]
- 102. Braunstein ML. Health care in the age of interoperability part 6: the future of FHIR. IEEE Pulse 2019; 10 (4): 25–7. [DOI] [PubMed] [Google Scholar]
- 103.FHIRPath (Normative Release). http://hl7.org/fhirpath/ Accessed September 21, 2020.
- 104. Tahar K, Müller C, Dürschmid A, et al. Integrating heterogeneous data sources for cross-institutional data sharing: requirements elicitation and management in SMITH. Stud Health Technol Inform 2019; 264: 1785–6. [DOI] [PubMed] [Google Scholar]
- 105. Matney SA, Heale B, Hasley S, et al. Lessons learned in creating interoperable fast healthcare interoperability resources profiles for large-scale public health programs. Appl Clin Inform 2019; 10 (1): 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Deppenwiese N, Duhm-Harbeck P, Ingenerf J, et al. MDRCupid: a configurable metadata matching toolbox. Stud Health Technol Inform 2019; 264: 88–92. [DOI] [PubMed] [Google Scholar]
- 107. Zhang XA, Yates A, Vasilevsky N, et al. Semantic integration of clinical laboratory tests from electronic health records for deep phenotyping and biomarker discovery. NPJ Digit Med 2019; 2: doi:10.1038/s41746-019-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kiourtis A, Mavrogiorgou A, Menychtas A, et al. Structurally mapping healthcare data to HL7 FHIR through ontology alignment. J Med Syst 2019; 43 (3): 62. [DOI] [PubMed] [Google Scholar]
- 109. Kilintzis V, Chouvarda I, Beredimas N, et al. Supporting integrated care with a flexible data management framework built upon linked data, HL7 FHIR and ontologies. J Biomed Inform 2019; 94: 103179. [DOI] [PubMed] [Google Scholar]
- 110. Saripalle R, Runyan C, Russell M.. Using HL7 FHIR to achieve interoperability in patient health record. J Biomed Inform 2019; 94: 103188. [DOI] [PubMed] [Google Scholar]
- 111. Solbrig HR, Hong N, Murphy SN, et al. Automated population of an i2b2 clinical data warehouse using FHIR. AMIA Annu Symp Proc 2018; 2018: 979–88. [PMC free article] [PubMed] [Google Scholar]
- 112. Prasser F, Kohlbacher O, Mansmann U, et al. Data integration for future medicine (DIFUTURE). Methods Inf Med 2018; 57 (S 01): e57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang P, White J, Schmidt DC, et al. FHIRChain: applying blockchain to securely and scalably share clinical data. Comput Struct Biotechnol J 2018; 16: 267–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Paris N, Mendis M, Daniel C, et al. i2b2 implemented over SMART-on-FHIR. AMIA Jt Summits Transl Sci Proc 2018; 2018: 369–78. [PMC free article] [PubMed] [Google Scholar]
- 115. Blumenthal S. Improving interoperability between registries and EHRs. AMIA Jt Summits Transl Sci Proc 2018; 2018: 20–5. [PMC free article] [PubMed] [Google Scholar]
- 116. Hong N, Wen A, Shen F, et al. Integrating structured and unstructured EHR data using an FHIR-based type system: a case study with medication data. AMIA Jt Summits Transl Sci Proc 2018; 2018: 74–83. [PMC free article] [PubMed] [Google Scholar]
- 117. Gulden C, Mate S, Prokosch H-U, et al. Investigating the capabilities of FHIR search for clinical trial phenotyping. Stud Health Technol Inform 2018; 253: 3–7. [PubMed] [Google Scholar]
- 118. Bialke M, Bahls T, Geidel L, et al. MAGIC: once upon a time in consent management—a FHIR® tale. J Transl Med 2018; 16 (1): 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Metke-Jimenez A, Steel J, Hansen D, et al. Ontoserver: a syndicated terminology server. J Biomed Semantics 2018; 9 (1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.FHIR ValueSet $expand Comparison Tool. https://ontoserver.csiro.au/vstool/ Accessed April 28, 2022.
- 121. Crump JK, Del Fiol G, Williams MS, et al. Prototype of a standards-based EHR and genetic test reporting tool coupled with HL7-compliant infobuttons. AMIA Jt Summits Transl Sci Proc 2018; 2017: 330–9. [PMC free article] [PubMed] [Google Scholar]
- 122. Warner JL, Prasad I, Bennett M, et al. SMART cancer navigator: a framework for implementing ASCO workshop recommendations to enable precision cancer medicine. JCO Precis Oncol 2018; (2): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hong N, Wen A, Mojarad MR, et al. Standardizing heterogeneous annotation corpora using HL7 FHIR for facilitating their reuse and integration in clinical NLP. AMIA Annu Symp Proc 2018; 2018: 574–83. [PMC free article] [PubMed] [Google Scholar]
- 124. Walonoski J, Kramer M, Nichols J, et al. Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J Am Med Inform Assoc 2018; 25 (3): 230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Aerts J. Towards a single data exchange standard for use in healthcare and in clinical research. Stud Health Technol Inform 2018; 248: 55–63. [PubMed] [Google Scholar]
- 126. Jiang G, Kiefer RC, Sharma DK, et al. A consensus-based approach for harmonizing the OHDSI common data model with HL7 FHIR. Stud Health Technol Inform 2017; 245: 887–91. [PMC free article] [PubMed] [Google Scholar]
- 127. Boussadi A, Zapletal E.. A fast healthcare interoperability resources (FHIR) layer implemented over i2b2. BMC Med Inform Decis Mak 2017; 17 (1): 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jiang G, Kiefer R, Prud'hommeaux E, et al. Building interoperable FHIR-based vocabulary mapping services: a case study of OHDSI vocabularies and mappings. Stud Health Technol Inform 2017; 245 (1327): 1327. [PMC free article] [PubMed] [Google Scholar]
- 129. Swaminathan R, Huang Y, Astbury C, et al. Clinical exome sequencing reports: current informatics practice and future opportunities. J Am Med Inform Assoc 2017; 24 (6): 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim D-Y, Hwang S, Kim M-G, et al. Development of Parkinson patient generated data collection platform using FHIR and IoT devices. Stud Health Technol Inform 2017; 245: 141–5. [PubMed] [Google Scholar]
- 131. Wagholikar KB, Jain R, Oliveira E, et al. Evolving research data sharing networks to clinical app sharing networks. AMIA Jt Summits Transl Sci Proc 2017; 2017: 302–7. [PMC free article] [PubMed] [Google Scholar]
- 132. Solbrig HR, Prud'hommeaux E, Grieve G, et al. Modeling and validating HL7 FHIR profiles using semantic web Shape Expressions (ShEx). J Biomed Inform 2017; 67: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. FHIRShExTest. caCDE-QA 2017. https://github.com/caCDE-QA/FHIRShExTest Accessed April 28, 2022.
- 134. Lee J, Hulse NC, Wood GM, et al. Profiling fast healthcare interoperability resources (FHIR) of family health history based on the clinical element models. AMIA Annu Symp Proc 2016; 2016: 753–62. [PMC free article] [PubMed] [Google Scholar]
- 135. Hong N, Prodduturi N, Wang C, et al. Shiny FHIR: an integrated framework leveraging shiny R and HL7 FHIR to empower standards-based clinical data applications. Stud Health Technol Inform 2017; 245: 868–72. [PMC free article] [PubMed] [Google Scholar]
- 136. Wagholikar KB, Mandel JC, Klann JG, et al. SMART-on-FHIR implemented over i2b2. J Am Med Inform Assoc 2017; 24 (2): 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chute CG, Huff SM.. The pluripotent rendering of clinical data for precision medicine. Stud Health Technol Inform 2017; 245: 337–40. [PubMed] [Google Scholar]
- 138. Geßner S, Neuhaus P, Varghese J, et al. The portal of medical data models: where have we been and where are we going? Stud Health Technol Inform 2017; 245: 858–62. [PubMed] [Google Scholar]
- 139. Leroux H, Metke-Jimenez A, Lawley MJ.. Towards achieving semantic interoperability of clinical study data with FHIR. J Biomed Semantics 2017; 8 (1): 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hochheiser H, Castine M, Harris D, et al. An information model for computable cancer phenotypes. BMC Med Inform Decis Mak 2016; 16 (1): 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Pfiffner PB, Pinyol I, Natter MD, et al. C3-PRO: Connecting ResearchKit to the health system using i2b2 and FHIR. PLoS One 2016; 11 (3): e0152722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Doods J, Neuhaus P, Dugas M.. Converting ODM metadata to FHIR questionnaire resources. Stud Health Technol Inform 2016; 228: 456–60. [PubMed] [Google Scholar]
- 143. Jiang G, Kiefer RC, Rasmussen LV, et al. Developing a data element repository to support EHR-driven phenotype algorithm authoring and execution. J Biomed Inform 2016; 62: 232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ulrich H, Kock A-K, Duhm-Harbeck P, et al. Metadata repository for improved data sharing and reuse based on HL7 FHIR. Stud Health Technol Inform 2016; 228: 162–6. [PubMed] [Google Scholar]
- 145. Clunie DA, Dennison DK, Cram D, et al. Technical challenges of enterprise imaging: HIMSS-SIIM collaborative white paper. J Digit Imaging 2016; 29 (5): 583–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Jiang G, Solbrig HR, Kiefer R, et al. A standards-based semantic metadata repository to support EHR-driven phenotype authoring and execution. Stud Health Technol Inform 2015; 216 (1098): 1098. [PMC free article] [PubMed] [Google Scholar]
- 147. Alterovitz G, Warner J, Zhang P, et al. SMART on FHIR Genomics: facilitating standardized clinico-genomic apps. J Am Med Inform Assoc 2015; 22 (6): 1173–8. [DOI] [PubMed] [Google Scholar]
- 148. Lenivtceva I, Slasten E, Kashina M, et al. Applicability of Machine Learning Methods to Multi-label Medical Text Classification In Krzhizhanovskaya VV, Závodszky G, Lees MH, et al. , eds. Computational Science – ICCS 2020. Cham, Switzerland: Springer International Publishing; 2020. 509–22. [Google Scholar]
- 149. Uciteli A, Beger C, Kirsten T, et al. Ontological representation, classification and data-driven computing of phenotypes. J Biomed Semantics 2020; 11 (1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kiourtis A, Mavrogiorgou A, Nifakos S, et al. A string similarity evaluation for healthcare ontologies alignment to HL7 FHIR resources. In: Arai K, Bhatia R, Kapoor S, eds. Intelligent Computing. Cham, Switzerland: Springer International Publishing; 2019: 956–70. [Google Scholar]
- 151. Aberdeen J, Bayer S, Clark C, et al. An annotation and modeling schema for prescription regimens. J Biomed Semantics 2019; 10 (1): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Stenzhorn H. A simple tool to enrich clinical trial data with multiontology-based conceptual tags. In: Da Silveira M, Pruski C, Schneider R, eds. Data Integration in the Life Sciences. Cham, Switzerland: Springer International Publishing; 2017: 17–21. [Google Scholar]
- 153. Helmer TT, Lewis AA, McEver M, et al. Creating and implementing a COVID-19 recruitment data mart. J Biomed Inform 2021; 117: 103765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Eccher C, Gios L, Zanutto A, et al. TreC platform. An integrated and evolving care model for patients’ empowerment and data repository. J Biomed Inform 2020; 102: 103359. [DOI] [PubMed] [Google Scholar]
- 155. Campbell WS, Carter AB, Cushman-Vokoun AM, et al. A model information management plan for molecular pathology sequence data using standards: from sequencer to electronic health record. J Mol Diagn 2019; 21 (3): 408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ondersma S. Accelerating Collaborative, Cumulative, and Open Intervention Science with an E-Intervention Authoring Platform. https://projectreporter.nih.gov/project_info_description.cfm?aid=9882671&icde=47497059&ddparam=&ddvalue=&ddsub=&cr=13&csb=default&cs=ASC&pball= Accessed July 24, 2020.
- 157.CIAS Mobile. App Store. https://apps.apple.com/us/app/cias-mobile/id1129075949 Accessed April 28, 2022.
- 158. Osborne JP. An Interoperable HL7 FHIR-based Medical Device Data System (MDDS) For Accessing And Integrating Live Point-Of-Care Data From High-Acuity Bedside Patient Monitoring EquipmentNIH Reporter Project Information. 2021. https://reporter.nih.gov/project-details/10353084 Accessed April 22, 2022. [Google Scholar]
- 159. Buse JB. CAMP FHIR: Lightweight, Open-Source FHIR Conversion Software to Support EHR Data Harmonization and Research. NIH Reporter Project Information. 2021.https://reporter.nih.gov/project-details/10353084 Accessed April 22, 2022. [Google Scholar]
- 160. Jiang G. FHIRCAT: Enabling the Semantics of FHIR and Terminologies for Clinical and Translational Research. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=10005525&icde=47496777 Accessed August 5, 2020. [Google Scholar]
- 161.FHIRCat - Enabling the Semantics of FHIR and Terminologies for Clinical and Translational Research. FHIRCat 2021. https://github.com/fhircat/FHIRCat Accessed April 28, 2022.
- 162. Haendel MA. A Phenomics-First Resource for Interpretation of Variants. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9855880&icde=51222519&ddparam=&ddvalue=&ddsub=&cr=3&csb=default&cs=ASC&pball= Accessed August 12, 2020.
- 163. Manion F. CLAMP-CS: A Cloud-Based, Service-Oriented, High-Performance Natural Language Processing Platform for Healthcare. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=10011177&icde=51222519&ddparam=&ddvalue=&ddsub=&cr=30&csb=default&cs=ASC&pball= Accessed August 12, 2020.
- 164. Vreeman D. Transferring Harmonized Laboratory Data from Healthcare Institutions to Registries Using FHIR Protocol. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=10118277&icde=51222519&ddparam=&ddvalue=&ddsub=&cr=26&csb=default&cs=ASC&pball= Accessed 12 Aug 2020. [Google Scholar]
- 165. Mandl KD. Instrumenting the Delivery System for a Genomics Research Information Commons. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9818382&icde=46878221&ddparam=&ddvalue=&ddsub=&cr=8&csb=default&cs=ASC&pball= Accessed July 20, 2020. [Google Scholar]
- 166. McDonald CJ. NLM R&D Tools for Standardization of Electronic Medical Records and Clinical Genetic Test Reports for Care and Research. NIH Reporter Project Informationhttps://projectreporter.nih.gov/project_info_description.cfm?aid=9792197&icde=47497059&ddparam=&ddvalue=&ddsub=&cr=9&csb=default&cs=ASC&pball= Accessed July 20, 2020.
- 167.LHC FHIR Tools. https://lhcforms.nlm.nih.gov/ Accessed September 21, 2020.
- 168. Schatz M. Implementing the Genomic Data Science Analysis, Visualization, and Informatics Lab-Space (AnVIL). NIH Reporter Project Information. 2018.https://reporter.nih.gov/project-details/10220581 Accessed April 22, 2022. [Google Scholar]
- 169. Natter MD. Expanding HL7 FHIR to Support Post-Marketing Research and Surveillance within Multi-Source, Chronic Disease Registries. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9757693&icde=47497059&ddparam=&ddvalue=&ddsub=&cr=12&csb=default&cs=ASC&pball= Accessed August 5, 2020. [Google Scholar]
- 170. Hollenbach JA. Integrated Exchange and Storage of Current- and Future-Generation Immunogenomic Data. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9640391&icde=47496321 Accessed August 5, 2020. [Google Scholar]
- 171. Pathak J. National Infrastructure for Standardized and Portable EHR Phenotyping Algorithms. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9774075&icde=46878221&ddparam=&ddvalue=&ddsub=&cr=16&csb=default&cs=ASC&pball= Accessed August 5, 2020. [Google Scholar]
- 172. Sim I. Open mHealth: Community-Based Data and Metadata Standards for Mobile Health. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9698354&icde=51222519&ddparam=&ddvalue=&ddsub=&cr=22&csb=default&cs=ASC&pball= Accessed August 12, 2020. [Google Scholar]
- 173. Hastak S. Architectural Review of the BRIDG Model. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9277217&icde=0 Accessed August 5, 2020. [Google Scholar]
- 174. Starren J. Improving Patient Reported Outcome Data for Research Through Seamless Integration of the PROMIS Toolkit into EHR Workflows. https://projectreporter.nih.gov/project_info_details.cfm?aid=9527207 Accessed July 24, 2020.
- 175. Jiang G. Tools for Standardizing Clinical Research Metadata Using HL7 FHIR. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=9551055&icde=47495935 Accessed July 20, 2020. [Google Scholar]
- 176. Jiang G. CACDE-QA: A Quality Assurance Platform for Cancer Study Common Data Elements. NIH Reporter Project Information. https://projectreporter.nih.gov/project_info_description.cfm?aid=8913908&icde=26360245&ddparam=&ddvalue=&ddsub=&cr=1&csb=default&cs=ASC Accessed July 20, 2020. [Google Scholar]
- 177. Wesley D, Ratwani RM.. Advancing the Collection and Use of Patient-Reported Outcomes through Health Information Technology. AHRQ Digital Healthcare Research: Informing Improvement in Care Quality, Safety, and Efficiency. https://digital.ahrq.gov/ahrq-funded-projects/advancing-collection-and-use-patient-reported-outcomes-through-health Accessed August 12, 2020. [Google Scholar]
- 178. Clifford GD. Leveraging Heterogeneous Data across International Borders in a Privacy Preserving Manner for Clinical Deep Learning. NSF Award. https://www.nsf.gov/awardsearch/showAward?AWD_ID=1822378 Accessed August 5, 2020. [Google Scholar]
- 179.Advancing Standards for Precision Medicine | HealthIT.gov. https://www.healthit.gov/topic/advancing-standards-precision-medicine Accessed April 27, 2022.
- 180.Sync for Science | HealthIT.gov. https://www.healthit.gov/topic/sync-science Accessed April 27, 2022.
- 181.Cumulus: A Universal Research Sidecar for a SMART Learning Healthcare System | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/leap/cumulus-universal-research-sidecar Accessed April 27, 2022.
- 182.Development and Testing of Data Sharing Functionality for Health System Participating in National Cardiovascular Disease Registries | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/leap/data-sharing-functionality Accessed April 27, 2022.
- 183.FHIR Factories: An Evolving Digital Architecture to Scale Health Research | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/leap/fhir-factories Accessed April 27, 2022.
- 184.FHIRedApp: An API-based patient engagement platform for the 21st Century. Leading Edge Acceleration Projects (LEAP) in Health Information Technology. https://www.healthit.gov/topic/leading-edge-acceleration-projects-leap-health-information-technology-health-it Accessed July 24, 2020.
- 185.Common Data Model Harmonization | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/pcor/common-data-model-harmonization-cdm Accessed April 27, 2022.
- 186.Coordinated Registry Network for Women’s Health Technologies | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/pcor/coordinated-registry-network-womens-health-technologies-crn Accessed April 27, 2022.
- 187.Patient-Reported Outcomes through Health IT Project | HealthIT.gov. https://www.healthit.gov/topic/scientific-initiatives/pcor/patient-reported-outcomes-through-healthit-pro Accessed April 27, 2022.
- 188.EDETEK. https://www.edetek.com/ Accessed August 18, 2020.
- 189.1upHealth Clinical Research Connector. 1upHealth. https://apporchard.epic.com/Gallery?id=12515 Accessed April 29, 2022.
- 190.AppScript by IQVIA. Realizing the Value of digital health. 2018.https://www.iqvia.com/blogs/2018/11/realizing-the-value-of-digital-health Accessed August 24, 2020.
- 191.Fora – A Platform to support health conversations. https://fora.health/ Accessed August 19, 2020.
- 192.Atlas: Automated Abstraction. Carta Healthcare. https://www.carta.healthcare/atlas/ Accessed August 18, 2020.
- 193.Clinical & Patient Registry Software to Improve Healthcare Outcomes. ArborMetrix. https://www.arbormetrix.com/registryx Accessed April 29, 2022.
- 194.Archer. Epic App Orchard. https://apporchard.epic.com/Gallery?id=6270 Accessed April 29, 2022.
- 195.CIBMTR Reporting SMART-on-FHIR App. https://nmdp-bioinformatics.github.io/cibmtr-reporting-fhir/ Accessed August 5, 2020.
- 196.Clinical Pipe. https://www.clinicalpipe.com Accessed July 20, 2020.
- 197.MyLinks. https://www.mylinks.com/ Accessed July 20, 2020.
- 198.Clinical Trials Search. https://apps.smarthealthit.org/app/clinical-trials-search Accessed April 27, 2022.
- 199.Health Wizz. https://apps.smarthealthit.org/app/health-wizz Accessed April 27, 2022.
- 200. FHIR to HPO. OCTRI 2021. https://github.com/OCTRI/fhir2hpo Accessed April 22, 2022.
- 201. Börner Y. Einthoven 2022. https://github.com/ylboerner/Einthoven Accessed April 22, 2022.
- 202. Saripalle R, Sookhak M, Haghparast M.. An interoperable UMLS terminology service using FHIR. Future Internet 2020; 12 (11): 199. [Google Scholar]
- 203. Wang J, Mathews WC, Pham HA, et al. Opioid2FHIR: s system for extracting FHIR-compatible opioid prescriptions from clinical text. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Seoul, Korea (South); 1748–51. doi: 10.1109/BIBM49941.2020.9313258 [DOI] [Google Scholar]
- 204. Choudhury A, van Soest J, Nayak S, et al. Personal health train on FHIR: a privacy preserving federated approach for analyzing FAIR data in healthcare. In: Bhattacharjee A, Borgohain SK, Soni B, et al. , eds. Machine Learning, Image Processing, Network Security and Data Sciences. Singapore: Springer; 2020: 85–95. [Google Scholar]
- 205. Beattie Z, Miller LM, Almirola C, et al. The collaborative aging research using technology initiative: an open, sharable, technology-agnostic platform for the research community. Digit Biomark 2020; 4 (Suppl 1): 100–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. CORD-19-on-FHIR – Semantics for COVID-19 Discovery. FHIRCat 2022. https://github.com/fhircat/CORD-19-on-FHIR Accessed April 22, 2022.
- 207. FHIR Ontology External Module. The Australian e-Health Research Centre 2021. https://github.com/aehrc/redcap_fhir_ontology_provider Accessed April 22, 2022.
- 208.NLP2FHIR: A FHIR-Based Clinical Data Normalization Pipeline and Its Applications. 2020.https://github.com/BD2KOnFHIR/NLP2FHIR Accessed August 5, 2020.
- 209. Kasthurirathne SN, Cormer KF, Devadasan N, et al. Development of a FHIR Based Application Programming Interface for Aggregate-Level Social Determinants of Health. AMIA Informatics summit 2019 Conference Proceedings 2019. https://scholarworks.iupui.edu/handle/1805/19635 Accessed August 11, 2020.
- 210. Mavrogiorgou A, Kiourtis A, Touloupou M, et al. Internet of medical things (IoMT): acquiring and transforming data into HL7 FHIR through 5G network slicing. Emerg Sci J 2019; 3 (2): 64–77. [Google Scholar]
- 211. Abolafia J, Gopal P, Hume S. Use of FHIR in Clinical Research: From Electronic Medical Records to Analysis. In: Proceedings of PhUSE. Amsterdam, Netherlands: 2019. https://www.lexjansen.com/phuse/2019/rw/RW04.pdf Accessed September 20, 2020.
- 212.Data Capacity for Patient-Centered Outcomes Research through Creation of an Electronic Care Plan for People with Multiple Chronic Conditions. ASPE. https://aspe.hhs.gov/data-capacity-patient-centered-outcomes-research-through-creation-electronic-care-plan-people-0 Accessed April 22, 2022.
- 213.Creation of LOINC Equivalence Classes. ASPE. 2019.https://aspe.hhs.gov/creation-loinc-equivalence-classes Accessed August 5, 2020.
- 214.Award of the Imaging Data Commons: Bringing Multi-modal Imaging Data to the Cancer Research Community | CBIIT. https://datascience.cancer.gov/news-events/blog/award-imaging-data-commons-bringing-multi-modal-imaging-data-cancer-research Accessed July 21, 2020.
- 215.Phenopackets: Standardizing and Exchanging Patient Phenotypic Data. https://www.ga4gh.org/news/phenopackets-standardizing-and-exchanging-patient-phenotypic-data/ Accessed April 22, 2022.
- 216. Making Electronic Health Record (EHR) Data More Available for Research and Public Health. ASPE 2019; https://aspe.hhs.gov/making-electronic-health-record-ehr-data-more-available-research-and-public-health Accessed August 5, 2020. [Google Scholar]
- 217. Jafir M. Scalable Consent Framework for the Advancement of Interoperability with Fast Healthcare Interoperability Resources (FHIR®) based APIs. Leading Edge Acceleration Projects (LEAP) in Health Information Technology. https://www.healthit.gov/topic/leading-edge-acceleration-projects-leap-health-information-technology-health-it Accessed July 21, 2020.
- 218.Strengthening the Data Infrastructure for Outcomes Research on Mortality Associated with Opioid Poisonings. ASPE. 2019.https://aspe.hhs.gov/strengthening-data-infrastructure-outcomes-research-mortality-associated-opioid-poisonings Accessed August 5, 2020.
- 219. Lackerbauer AM, Lin AC, Krauss O, et al. A model for implementing an interoperable electronic consent form for medical treatment using HL7 FHIR. EJBI 2018; 14 (3): doi:10.24105/ejbi.2018.14.3.6 [Google Scholar]
- 220. Wu H, Toti G, Morley KI, et al. SemEHR: a general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research. J Am Med Inform Assoc 2018; 25 (5): 530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Hume S, Abolafia J, Low G. Use of HL7 FHIR as eSource to Pre-populate CDASH Case Report Forms using a CDISC ODM API. Frankfurt, Germany: 2018. https://www.lexjansen.com/phuse/2018/rw/RW02.pdf Accessed March 30, 2021.
- 222. Brodey B. Development and Validation of Software for an Electronic-Based DISC-5, the NETDISC-5https://projectreporter.nih.gov/project_info_description.cfm?aid=9678771&icde=46984723 Accessed July 24, 2020.
- 223. Sharma DK, Solbrig HR, Prud'hommeaux E, et al. D2Refine: a platform for clinical research study data element harmonization and standardization. AMIA Jt Summits Transl Sci Proc 2017; 2017: 259–67. [PMC free article] [PubMed] [Google Scholar]
- 224. Savova GK, Tseytlin E, Finan S, et al. DeepPhe: a natural language processing system for extracting cancer phenotypes from clinical records. Cancer Res 2017; 77 (21): e115–8–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Zopf R, Abolafia J, Reddy B. Use of Fast Healthcare Interoperability Resources (FHIR) in the Generation of Real World Evidence (RWE). In: Proceedings of PhUSE. Edinburgh, UK: 2017. https://www.cdisc.org/sites/default/files/resource/Use_of_Fast_Healthcare_Interoperability_Resources_in_the_Generation_of_Real_World_Evidence.pdf Accessed April 22, 2022.
- 226.XpertScreen. XpertDox. https://www.xpertdox.com/ Accessed August 20, 2020.
- 227. OMOPonFHIR SQLRender server deployment. OMOP on FHIR 2022. https://github.com/omoponfhir/omoponfhir-main-r4-sql Accessed April 22, 2022.
- 228.The FHIR Project at Georgia Tech. GT-FHIR2, the OMOP on FHIR Project. http://omoponfhir.org/ Accessed December 31, 2019.
- 229.Connecting Patients With FHIR® Based Clinical Data Registry — Darena Solutions. https://www.darenasolutions.com/blog/connecting-patients-with-fhir-based-clinical-data-registry Accessed September 11, 2020.
- 230.VTOC Research & Registries | VisionTree | Patient-Centered Outcomes & Healthcare Solutions. https://www.visiontree.com/solutions/research/patient-centered-outcomes/ Accessed July 20, 2020.
- 231.FHIR Bulk Data Access (Flat FHIR) :/. https://hl7.org/fhir/uv/bulkdata/ Accessed September 11, 2020.
- 232.HL7.FHIR.US.SIRB\Single IRB (sIRB) - FHIR v4.0.1. https://build.fhir.org/ig/HL7/fhir-sirb/ Accessed April 27, 2022.
- 233.ResearchStudy - FHIR v4.0.1. https://www.hl7.org/fhir/researchstudy.html Accessed July 24, 2020.
- 234.Consent - FHIR v4.0.1. https://www.hl7.org/fhir/consent.html Accessed July 9, 2020.
- 235.ResearchSubject - FHIR v4.0.1. https://www.hl7.org/fhir/researchsubject.html Accessed July 24, 2020.
- 236. Hoffman RA, Wu H, Venugopalan J, et al. Intelligent mortality reporting with FHIR. IEEE EMBS Int Conf Biomed Health Inform 2017; 2017: 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Braunstein ML. Death-on-FHIR: Analytics-Driven Mortality Reporting Using a SMART-on-FHIR Apphttps://fdocuments.net/document/death-on-fhir-analytics-driven-mortality-reporting-1-3a-informant-s-name-c.html?page=1 Accessed April 27, 2022.
- 238.Solutions - Precision Digital Health. https://precisiondigitalhealth.com/solutions/ Accessed April 27, 2022.
- 239.HL7 FHIR Connectathons | HL7 International. http://www.hl7.org/events/fhir-connectathon/index.cfm?ref=nav Accessed August 24, 2020.
- 240.HL7® FHIR® Accelerator Program | HL7 International. https://www.hl7.org/about/fhir-accelerator/ Accessed August 24, 2020.
- 241.Vulcan | HL7 International. https://www.hl7.org/vulcan/ Accessed August 24, 2020.
- 242. Zayas-Cabán T, Chaney KJ, Rucker DW.. National health information technology priorities for research: a policy and development agenda. J Am Med Inform Assoc 2020; 27 (4): 652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Zayas-Cabán T, Abernethy AP, Brennan PF, et al. Leveraging the health information technology infrastructure to advance federal research priorities. J Am Med Inform Assoc 2020; 27 (4): 647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.An Upcoming Milestone in Our Interoperability Journey. Health IT Buzz. 2022.https://www.healthit.gov/buzz-blog/healthit-certification/an-upcoming-milestone-in-our-interoperability-journey Accessed May 19, 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analyzed in support of this research.