Abstract
Objective
Deep learning models for clinical event forecasting (CEF) based on a patient’s medical history have improved significantly over the past decade. However, their transition into practice has been limited, particularly for diseases with very low prevalence. In this paper, we introduce CEF-CL, a novel method based on contrastive learning to forecast in the face of a limited number of positive training instances.
Materials and Methods
CEF-CL consists of two primary components: (1) unsupervised contrastive learning for patient representation and (2) supervised transfer learning over the derived representation. We evaluate the new method along with state-of-the-art model architectures trained in a supervised manner with electronic health records data from Vanderbilt University Medical Center and the All of Us Research Program, covering 48 000 and 16 000 patients, respectively. We assess forecasting for over 100 diagnosis codes with respect to their area under the receiver operator characteristic curve (AUROC) and area under the precision-recall curve (AUPRC). We investigate the correlation between forecasting performance improvement and code prevalence via a Wald Test.
Results
CEF-CL achieved an average AUROC and AUPRC performance improvement over the state-of-the-art of 8.0%–9.3% and 11.7%–32.0%, respectively. The improvement in AUROC was negatively correlated with the number of positive training instances (P < .001).
Conclusion
This investigation indicates that clinical event forecasting can be improved significantly through contrastive representation learning, especially when the number of positive training instances is small.
Keywords: clinical event forecasting, unsupervised representation learning, contrastive learning, electronic health records
INTRODUCTION
Electronic health records (EHRs) provide a historical accounting of a patient’s medical status. When deployed in large healthcare organizations, the depth and breadth of the data stored in EHR systems can provide a substantial quantity of training data for modern machine learning frameworks and artificial intelligence applications. In particular, deep learning has the potential to profoundly impact many problems in healthcare,1–3 including the forecasting of a patient’s future risk of health problems.4–8 However, one of the greatest concerns over the application of deep learning is that the resulting models tend to adapt to, and thus perform inference over, the noise (ie, spurious feature-label correlation) inherent in training data,9 neglecting the intuitive causal relationships that clinicians typically recognize. As a consequence, deep learning is, at times, insufficiently reliable to transition from theory to practice.10
Recently, representation learning,11,12 a branch of machine learning that trains large and deep models in an unsupervised manner, has emerged as a potential solution to this problem. Through representation learning, a forecasting problem is resolved through a simple classifier built on top of the learned representation that can be informative without drawing upon the assistance of ground-truth labels. Among the various approaches for representation learning, contrastive learning has, perhaps, demonstrated the greatest potential.13,14 However, the evidence to date has been generated in the domains of computer vision and natural language processing.15–18 To the best of our knowledge, this approach has not been adapted to clinical event data, which is a composition of semantically heterogenous domains (eg, diagnoses, medications, or demographics).
In this article, we focus on deep learning-based diagnosis forecasting applied to a large population. We specifically aim to support more accurate forecasts of which patients should be prioritized for resources in resource-constrained environments. One example of such a situation is to identify patients who might need prospective genotyping.19 There is evidence, for instance, that patient morbidity can be significantly reduced when the right drug is provided at the right time with the right dosage. As such, patients can be genotyped prior to an event that requires use of the pharmacogenetics. However, the resources available for genotyping are, at the present moment in time, limited due to the fact that insurers do not reimburse for prospective genotyping, such that healthcare organizations (or patients) are left with deciding whether or not to pay for the generation of relevant genetic data. Thus, to balance costs against clinical outcomes, a medical center could provide risk forecasting for its population to identify the patients who would benefit the most from prospective genotyping.
We introduce CEF-CL, a novel method for individual-level Clinical Event Forecasting through Contrastive Learning. We illustrate the potential for this method with data derived from two clinical data sets. The first, corresponds to data from the NIH-sponsored All of Us Research Program,20 a publicly available resource that enables reproducibility of this investigation. The second is an EHR data set from Vanderbilt University Medical Center.21 We conduct experiments with five different types of sequential models trained with a purely supervised paradigm (as a baseline) and CEF-CL across over 100 diagnosis code forecasting tasks. We show that the performance improvement achieved by CEF-CL over the baseline is inversely proportional to the prevalence of the diagnoses. We use a large number of forecasting tasks to illustrate that this new learning method has potential for a wide range of applications, but also highlight several clinical phenomena for which it appears to achieve notable performance.
MATERIALS AND METHODS
Challenges for few-shot learning
Few-shot learning (FSL)22 represents a set of machine learning problems where only a limited number of training instances with supervised information (eg, known class labels) are available to the model training process. With respect to health data, a clinical event forecasting model is typically learned from a case group (ie, patients with the target diagnosis), which is assigned positive labels before the training process. In many scenarios, the case group may be quite small due to the rarity of the target disease. As such, the general clinical event forecasting problem falls within the scope of FSL.
In an FSL setting, the training instances in the case group are an insufficient representation of the corresponding population’s data distribution, which leads to several problems. First, as shown by Prabhu and colleagues,23 diseases in the long tail of the prevalence distribution tend to exhibit large intraclass variability (ie, the training instances that are members of the same class exhibit very different patterns). Second, the low volume of case instances hinders regularization during a parameter-learning process supervised by the case/control labels. As a consequence, it is difficult to train an effective and generalizable classifier.
As neural networks grow in their depth and number of parameters, their ability to learn complex patterns improves.24–27 This flexibility makes it seem as though deep learning can model the complex processes that govern disease development—particularly when they are influenced by multiple contributing factors and exhibit complicated trajectories over time.28–31 However, when model architectures based on modern deep learning techniques, which we refer to as backbone models, are deployed in an FSL setting, there is an imbalance in the quantity of parameters that are to support modeling power and the amount of supervised information available to guide the optimization of the parameters (ie, too many parameters but too little supervised information). Consequently, optimizing for high modeling power could produce undesirable generalization behaviors, such as: (1) memorization, where the model fits a transformation from features to labels in the training set (even when the label is arbitrarily assigned), which perpetuates spurious relationships32; (2) overconfidence: the model generates an output with high confidence for test instances that are outside of the training distribution33; (3) instability: small perturbations = to the input induce large changes in the model’s output.34 It is possible that a multitask learning strategy35 could resolve these problems (ie, the model is trained for multiple tasks simultaneously). For instance, Choi and colleagues36 showed that a joint-training strategy can induce a strong regularization effect on the learning process for healthcare related tasks. However, this is usually not feasible in practice because: (1) it requires the incorporation of extra knowledge that is not readily available (eg, labels for auxiliary tasks) and (2) many labeled cohorts are created for a single learning task only.
Representation learning as a basis
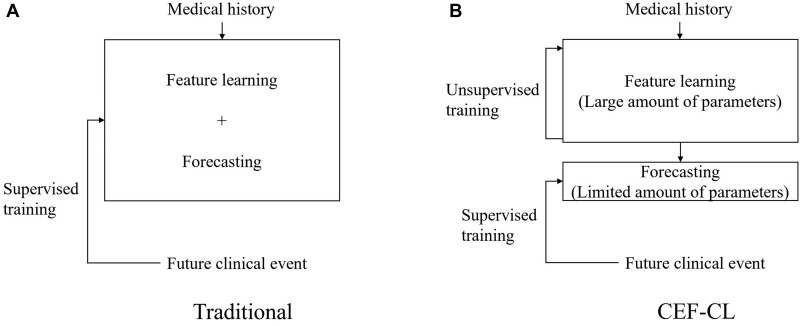
We build on the work of Ma and colleagues37 and alter the learning schema from the traditional end-to-end supervised paradigm. In doing so, we combine unsupervised representation learning (URL) and supervised transfer learning (STL). This method separates the model into two components. The first component, which possesses almost all of the parameters in the model, is trained to learn informative patient representations in an unsupervised manner. The second component, which is composed of a limited number of parameters, is trained to fit the derived patient representation with ground-truth labels. Figure 1 illustrates the relationship between traditional end-to-end supervised paradigm and the proposed hybrid method. The hybrid method is notable for several reasons. First, it reduces the risk of overfitting the model to the training data by encouraging the training process to reduce its reliance on training labels. Second, when the minority class is composed of a substantially smaller number of instances than the majority class (eg, as is the case in rare disorders), the relationships between the features for the minority class can be sufficiently learned by leveraging instances from the majority class that share the same feature space.
Figure 1.
An illustration of (A) a traditional end-to-end supervised training paradigm, where the feature learning and forecasting processes are integrated into one model trained under the supervision of class labels; and (B) the new paradigm based on unsupervised-representation learning and supervised-transfer learning, where the two processes are processed by separated models and only the one for forecasting is trained under the supervision of class labels.
To realize such a learning paradigm, we introduce contrastive learning into the representation modeling of EHR data. Contrastive learning is a discriminative approach for learning the latent representation of data by: (1) maximizing the semantic similarity between instances in the same predefined latent class and (2) minimizing the semantic similarity between instances in different predefined latent classes.38 The essential component of contrastive learning is to acquire an instance’s latent class as an auxiliary input to the learning algorithm. It is straightforward to obtain the latent class via data augmentation for certain tasks (eg, in computer vision tasks, images and their crops/rotations are in the same latent class). Yet it is not evident how the latent class for EHR data in the form of temporally organized clinical events (eg, the assignment of series of diagnosis codes) should be defined. Thus, we introduce a data augmentation strategy to adapt contrastive learning to the needs of EHR modeling.
Learning method framework
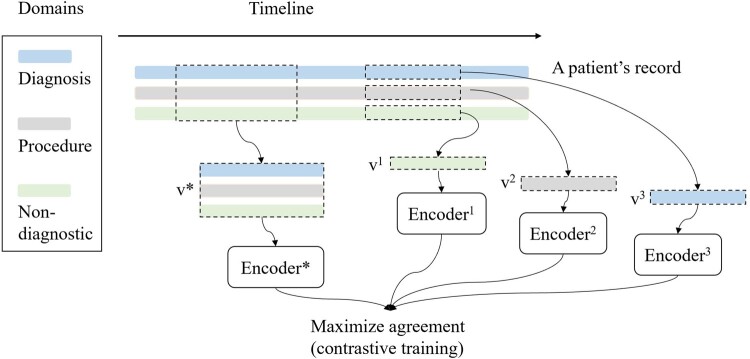
We introduce a learning method for clinical event forecasting with longitudinal data. In this setting, each patient is associated with a medical history represented as a sequence of episodes—inclusive of inpatient stays and outpatient visits. Each episode includes information for three semantically disparate domains: diagnoses, procedures, and nondiagnostic information in the form of patient demographics, the episode length, and time between episodes. Note that this method is not restricted to these domains and can be applied on data with additional domains, such as prescribed medications or laboratory test results. This framework consists of two stages: (1) URL and (2) STL. In the URL stage, we apply a contrastive method and design the corresponding data augmentation strategy for EHR data. The data augmentation and contrastive pretraining subsections as follows collectively describe the URL stage. Figure 2 provides a summary of this stage of the process.
Figure 2.
An illustration of data augmentation and contrastive pretraining for data derived from EHRs. v1, v2, v3, and v* correspond to the type of partial views.
Data augmentation
Data augmentation produces latent classes by generating partial views for each patient. Specifically, each patient in the data set corresponds to a latent class, including the patient’s record and the record’s partial views For each patient, we generate a pair of views in each training epoch through a two-step process. First, following the augmentation method introduced by Giorgi and colleagues,39 we select two subsequences of consecutive episodes from each patient’s record. Next, we remove all information except for an arbitrarily selected medical concept type (eg, diagnoses) from one subsequence. We represent the partial views as for the subsequence with all domains and for the subsequence with the domain .
Contrastive pretraining
The objective for contrastive pretraining is to learn latent representations for the diagnosis and procedure event history of patients that can be effectively leveraged for a downstream forecasting task.
We use multiple encoders with the same architecture, each of which corresponds to a type of augmented partial view. Specifically, and each are mapped into the latent space through encoders denoted as and , respectively. Note that the encoder can be used with the architecture of any existing backbone model.
The optimization objective for contrastive pretraining is defined as:
| (1) |
where and denotes the th and th patients in the data set, respectively, and
where τ is a positive-valued temperature hyperparameter. This objective is referred to as the NT-Xent by Chen and colleagues40 and is equivalent to the infoNCE41 loss, which approximates the mutual information between different views of the same record.
Transfer learning
In this step, the objective is to obtain a classifier for forecasting. We first use the trained to derive a latent representation for each patient. We then perform transfer learning42 by training a logistic regression classifier with the representations and labels.
Materials
To assess the potential for contrastive learning, we performed an empirical analysis with EHR data from two distinct resources. The first corresponds to deidentified EHR data from Vanderbilt University Medical Center (VUMC). The second corresponds to the publicly available Registered Tier data from the NIH-sponsored All of Us Research Program.
We refined the data sets for this study in the following manner. First, we selected data to cover similar time periods of length that sufficiently characterize changes in clinical status for patients that evolve over time. Specifically, the VUMC data covers January 1, 2005 to December 31, 2011, while the All of Us data covers July 1, 2011 to June 30, 2018, as the observation period. Note, for deidentification purposes, in each resource each patient record was independently date-shifted between −1 and −365 days. We acknowledge that these two resources cover different time periods, but we do not believe this influences our results, as we are not combining these resources for analytic purposes.
Second, we reduced the data set to focus on patients with a sufficient number of observations to support machine learning. Specifically, we retained patients with at least 25 episodes within the observation period defined by this study. At the same time, for computational efficiency, we restricted the total number of episodes per patient to the final 200 during the observation period.
Third, we limited our analysis to the set of patients whose medical history is relatively completely recorded by the data source. Specifically, we follow criteria that is similar to that introduced by Schildcrout et al,43 retaining patients who experienced at least five episodes in the last two years of the observation period.
Finally, we assess the performance of the forecasting models using the 6-month period (ie, prediction window) after a patient’s final episode in the observation period (ie, index date), which we refer to as the forecasting period. Specifically, the data from the observation and forecasting periods are used for the URL and the STL stages, respectively. We define a future diagnosis of a patient as one that was only seen in the forecasting period (ie, there was no indication in the observation period). The patients with and without the future diagnosis are labeled as positive and negative, respectively. As such, to ensure we utilize data that reflects the relatively authentic clinical status for those in this study, we removed patients who lacked a visit in the forecasted (ie, future) timeframe.
We mapped all diagnosis and procedure codes into their Clinical Classifications Software (CCS) form (https://www.hcup-us.ahrq.gov/tools_software.jsp, last accessed May 31, 2021). Table 1 provides summary statistics about the resulting data sets. It should be noted that the information for age was calculated based on the age at the final episode in the patients’ observation period, which is at most 6 months before the end of the forecasting period.
Table 1.
A summary of the data sets used in this study
| Data set | Patients (episodes) | Episodes per patient (mean; median) | Age (min, median, max) | Gender (male, female) | CCS diagnosis codes | CCS procedure codes |
|---|---|---|---|---|---|---|
| VUMC | 48 547 (2 725 373) | 56; 45 | 0, 55, 90 | 41%, 59% | 262 | 244 |
| All of Us | 16 123 (1 305 589) | 81; 64 | 17, 62, 87 | 40%, 60% | 282 | 244 |
EXPERIMENTS AND RESULTS
The forecasting tasks correspond to the CCS diagnosis codes with more than 50 patients in each respective data set. There are 181 and 120 tasks for the VUMC and All of Us data sets, respectively. Each data set is partitioned into a training, validation, and testing set according to a 1:1:1 ratio. We perform experiments with various backbone models trained in both an end-to-end supervised paradigm (as baselines) and CEF-CL to predict future diagnoses in the forecasting period based on the episodes in the observation period. In the experiments, the models are trained for each unique task. We evaluate forecasting performance using two measures for each task: (1) the area under the receiver operating characteristic (AUROC) curve and (2) the area under the precision-recall (AUPRC) curve. We observed that, in the FSL setting, the forecasting performance of a model trained in a supervised manner highly depends on the initialization of its parameter. Therefore, to obtain a robust performance estimation, we repeat the training process three times per baseline per task.
Backbone models
We use five backbone models in our experiments. The first two correspond to state-of-the-art model architectures, used by COmplementary patterN AugmentatioN (CONAN)28 and Long-term dependencies and Short-term correlations with the utilization of a hierarchical Attention Network (LSAN).30 These models incorporate an attention mechanism44 both within and beyond the episode level. We also use naïve backbones with the simple structure of recurrent neural network (RNN), long short-term memory (LSTM),45 and Gated Recurrent Unit (GRU). We provide details for the model architecture in Supplementary Appendix A.
Forecasting performance improvement with the proposed method
Tables 2 and 3 report the average AUROC and AUPRC across the forecasting tasks for each ⟨backbone model, training method⟩ pair. Specifically, the averaging is performed on the min, max, and median performance of the outcomes for the three independent runs on all tasks. It can be seen that contrastive pretraining improves upon the AUROC of the backbone models by 5.5%–13.5% and 6.1%–35.1%, on an average, for the VUMC and All of Us data, respectively. The AUPRC improvement is 3.1%–31.9% and 23.8%–88.1%, for VUMC and All of Us data, respectively.
Table 2.
A summary of the AUROC for the forecasting tasks
| Backbone model | End-to-end supervised training | CEF-CL | Improvement |
|---|---|---|---|
| VUMC (181 tasks) | |||
| RNN | 0.607 (0.584–0.629) | 0.689 (0.677–0.702) | 13.5% |
| GRU | 0.637 (0.624–0.650) | 0.680 (0.668–0.693) | 6.8% |
| LSTM | 0.638 (0.617–0.657) | 0.684 (0.670–0.697) | 7.2% |
| CONAN | 0.633 (0.620–0.647) | 0.668 (0.652–0.679) | 5.5% |
| LSAN | 0.618 (0.597–0.637) | 0.662 (0.649–0.677) | 7.1% |
| All of Us (120 tasks) | |||
| RNN | 0.572 (0.544–0.609) | 0.773 (0.758–0.791) | 35.1% |
| GRU | 0.683 (0.660–0.701) | 0.772 (0.753–0.788) | 13.0% |
| LSTM | 0.690 (0.656–0.719) | 0.784 (0.767–0.798) | 13.6% |
| CONAN | 0.717 (0.700–0.735) | 0.761 (0.742–0.779) | 6.1% |
| LSAN | 0.670 (0.628–0.702) | 0.764 (0.746–0.780) | 14.0% |
Note: In this table, the results are depicted as a (b−c), where a represents the average performance score calculated of three independent runs, while b and c represent the minimum score and the maximum score.
Table 3.
A summary of the AUPRC for the forecasting tasks
| Backbone model | End-to-end supervised training | CEF-CL | Improvement |
|---|---|---|---|
| VUMC (181 tasks) | |||
| RNN | 2.41 (2.12–2.83)×0−2 | 2.86 (2.62–3.30)×10−2 | 18.7% |
| GRU | 2.56 (2.32–2.78)×10−2 | 2.64 (2.45–3.00)×10−2 | 3.1% |
| LSTM | 2.24 (2.00–2.49)×10−2 | 2.72 (2.46–3.07)×10−2 | 21.4% |
| CONAN | 2.10 (1.96–2.36)×10−2 | 2.43 (2.21–3.02)×10−2 | 15.7% |
| LSAN | 1.85 (1.66–2.06)×10−2 | 2.44 (2.19–2.79)×10−2 | 31.9% |
| All of Us (120 tasks) | |||
| RNN | 1.93 (1.64–2.30)×10−2 | 3.63 (3.24–4.19)×10−2 | 88.1% |
| GRU | 2.71 (2.46–3.09)×10−2 | 3.64 (3.21–4.32)×10−2 | 34.3% |
| LSTM | 2.80 (2.42–3.22)×10−2 | 3.71 (3.35–4.36)×10−2 | 32.5% |
| CONAN | 2.81 (2.57–3.09)×10−2 | 3.48 (3.06–4.08)×10−2 | 23.8% |
| LSAN | 2.57 (2.20–3.00)×10−2 | 3.51 (3.10–4.21)×10−2 | 36.6% |
Note: In this table, the results are depicted as a (b−c), where a represents the average performance score calculated of three independent runs, while b and c represent the minimum score and the maximum score.
In addition, we also compare the performance of the best backbone models between the training methods (eg, for the VUMC data, the best performing model, in terms of AUROC, is LSTM for end-to-end supervised learning and RNN for CEF-CL). The result indicates an AUROC and AUPRC improvement of 8.0% and 11.7% for the VUMC data and 9.3% and 32.0%, respectively, for the All of Us data. In addition, CEF-CL achieves better AUROC performance than the baseline in 167 of the 181 (92.2%) tasks for VUMC data and 113 of the 120 (94.2%) tasks for the All of Us data. Additionally, CEF-CL achieves better AUPRC in 154 of the 181 (85.0%) tasks for VUMC data and 106 of the 120 (88.3%) tasks for the All of Us data. We refer the reader to Supplementary Appendix D for the forecasting performance for each task.
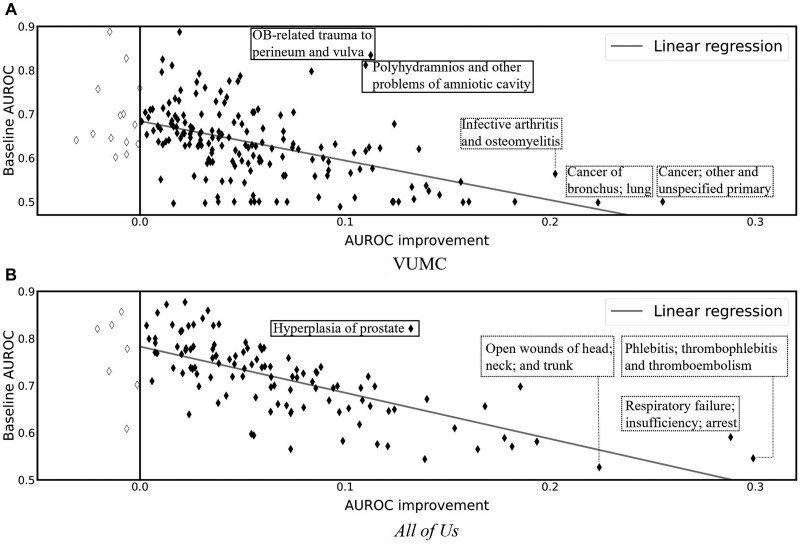
Next, we analyzed the relationship between the improvement brought about by CEF-CL and the performance of the baseline with the best-performing backbone over the set of CCS codes in Figure 3, where x-axis represents the AUROC gap between the baseline and CEF-CL, while the y axis represents the baseline AUROC performance. It can be seen that the improvement achieved through CEF-CL is inversely correlated with the AUROC achieved by the baseline. This is not surprising because the forecasting tasks with weak baseline results clearly have larger room for improvement. However, it should be recognized that the tasks that benefited the most from CEF-CL in the VUMC and All of Us data (as shown in the dashed boxes in Figure 3) differed. Notably, in the VUMC data, the tasks that benefited the most were Cancer; other and unspecified primary (AUROC improvement, baseline AUROC: 0.255, 0.755), Cancer of bronchus; lung (0.223, 0.498), and Infective arthritis and osteomyelitis (0.202, 0.564). By contrast, the tasks that benefited the most in the All of Us data were Phlebitis; thrombophlebitis and thromboembolism (0.300, 0.489), Respiratory failure; insufficiency; arrest (0.299, 0.546), and Open wounds of head; neck; and trunk (0.288, 0.591).
Figure 3.
AUROC improvement achieved through CEF-CL versus AUROC of the baseline with the best performing backbone over the set of CCS codes. Each marker corresponds to a unique task defined by a CCS diagnosis code (the significantly enhanced forecasts with AUROC improvement greater than 0.2 and 0.1 are highlighted with dashed boxes for VUMC and All of Us data, respectively). The gray line is a linear regression line of the observations (VUMC: Slope = −0.90, r2=0.23; All of Us: Slope = −0.98, r2=0.51) as an indication of the correlation between AUROC improvement and the baseline AUROC.
At the same time, we acknowledge that the forecasting performance of CEF-CL, in terms of AUROC, for some of these tasks remain is sufficiently high to support real world deployment. Therefore, we also highlight the tasks that are seemingly well-forecasted with the baseline and still moderate performance improvement through CEF-CL (with solid boxes in Figure 3). Specifically, examples of these tasks are OB-related trauma to perineum and vulva (0.113, 0.834) and Polyhydramnios and other problems of amniotic cavity (0.11, 0.812) for the VUMC data; Hyperplasia of prostate (0.132, 0.821) for the All of Us data.
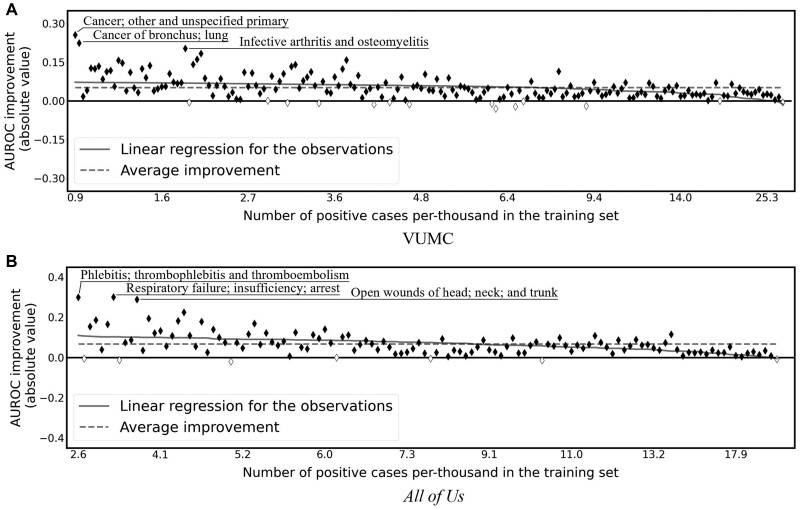
To further investigate how CEF-CL improves forecasting for tasks for which the baseline performed poorly, we analyze the performance improvement achieved by CEF-CL with respect to the frequency of each task’s target concept. Figure 4 compares the best-performing backbone for CEF-CL and end-to-end supervised learning in terms of forecasting performance improvement over the set of target CCS diagnosis codes. Specifically, the x-axis represents the prevalence of positive cases in the data set and the y-axis represents the absolute improvement in terms of AUROC. It should be noted that the relative and absolute improvement exhibited highly similar patterns, such that we present only the absolute improvement here and defer the relative improvement to the Supplementary Material. We focus on AUROC because it is insensitive to label imbalance. We performed a linear regression between the x and y values for the tasks. It is evident that the CEF-CL outperforms the best baseline—particularly on the tasks with more imbalanced training data (ie, few-shot tasks). We statistically confirmed this finding by running a Wald Test with the null hypothesis that the slope is zero. It was found that P = 1.2 × 10−8 and 3.7 × 10−8 for the VUMC and All of Us data sets, respectively. As such, the evidence suggests that the proposed framework is particularly more adept at few-shot learning tasks than the baseline.
Figure 4.
Absolute AUROC differences in CCS code forecasting tasks with training data under different levels of the number of positive cases. The x-axis corresponds to the number of positive cases per 1000 instances in the training set. Each marker corresponds to a unique task, where solid (hollow) markers indicate positive (negative) improvement. The forecasts with AUROC improvement greater than 0.2 are highlighted with dashed boxes. The solid line is the linear regression line. The dashed horizontal line is a baseline of average improvement of all tasks.
DISCUSSION
Though these findings are notable, there are several open issues that require further consideration. First, we believe that the performance gain occurs because the end-to-end supervised training paradigm is incapable of: (1) handling high intraclass variance for the positive class and (2) model discriminative patterns in the samples with the same label. By comparison, the contrastive pretraining resolves both of these problems, which leads to a greater improvement in the face of imbalanced training data. Second, the performance improvements differ between tasks. Although we demonstrated that the improvement was correlated with the baseline performance and the frequency of the task’s target concept, other factors could contribute to the differences, such as the complexity of the forecasting tasks or the amount of data necessary to achieve stability over the tasks. To account for potential performance variation and stability issues of the tasks, we recommend future research incorporate known confounders (eg, detailed demographic information) in the models and to conduct experiment on larger data sets. Third, it was observed that there are differences in the tasks that achieve the greatest forecasting performance improvement achieved through CEF-CL with respect to the data sets studied. There are numerous potential reasons for the difference, such as the fact that All of Us is composed of data from a wide variety of organizations’ EHRs whereas the VUMC data are drawn from a single organization. However, determining the driving factors for such differences is beyond the scope of this investigation.
It should be recognized that this investigation has implications for longitudinal EHR modeling more generally. This is particularly because EHR modeling relies on a mechanism that is capable of deriving meaningful representations of a patient at the current moment and historically. As such, we believe that the contrastive representation learning framework introduced in this study can be used to facilitate a broad scope of research including both of discriminative (eg, predictive modeling) and generative tasks (eg, synthetic data generation46).
While our work sets a foundation for learning in low prevalence environments, there are opportunities to further extend our methodology, particularly with respect to its scalability, and clinical viability, in several ways. First, the backbone models in our experiments used the feature and outcome spaces built on CCS codes. However, CEF-CL can use other types of models based on data from other clinical coding systems, such as the International Classification of Diseases (ICD), as well as various semantic types of clinical concept (eg, medications, vital signs, or laboratory test results). In addition, CEF-CL leverages the sequential property only between healthcare episodes. Thus, this approach can be adapted to the scenario of outcome prediction for a single medical encounter (eg, an ICU stay) by using backbone models designed for modeling the temporal trajectory of patient health status recorded in a finer granularity (eg, with 1 hour as a unit). Second, in the URL step, we directly utilize the multidomain nature of EHR data, using medical concepts from each domain as a separate augmentation. This step leverages insights gained from the principle of compositional generalization47—an intelligent system’s capability of generalizing learned knowledge to new tasks and situations comes partly from learning the knowledge in a compositional manner (eg, learning syntax and semantics of language separately48).9 However, we anticipate CEF-CL can be further improved by incorporating colearning,49 where each group of separated features is treated as being conditionally independent. Therefore, a strategy considering the feature independence in the URL step might lead to even better downstream performance.
Finally, we acknowledge that in our experiments, repeating each training process only three times might lead to relatively higher variance in the performance estimation for each task. However, each experiment requires a nontrivial amount of computation (eg, on an NVIDIA 2080Ti GPU, the model training process was ∼1 hour per task, which implies running all 181 tasks on all baselines takes more than 500 GPU hours). Still, since we aim to compare the performance of the models in an overall manner, the variance for each individual task is amortized in the average AUROC and AUPRC across all tasks, such that the results can be compared in a reliable manner.
CONCLUSION
This paper introduced a framework to enhance clinical event forecasting through a 2-stage process of URL followed by transfer learning. We specifically illustrated how to adapt contrastive representation learning and a corresponding data augmentation strategy for to EHR data organized in a longitudinal manner. This investigation is notable in that the new approach significantly outperforms the traditional end-to-end supervised training paradigm, especially for FSL tasks. The findings of this study indicated that not all forecasting tasks could be improved upon using the new framework. Further research can be conducted to investigate the reason of lack of improvement for certain tasks.
FUNDING
The research is sponsored, in part, by NIH grants U2COD023196 and UL1TR002243.
AUTHOR CONTRIBUTIONS
ZZ, CY, XZ, and BAM contributed to the formulation of the study. ZZ designed the methods and carried out the experiments. ZZ and SLN performed the data collection. ZZ drafted the paper. ZZ, CY, and BAM interpreted the results. CY, XZ, SLN, and BAM contributed to editing, reviewing, and approving the final manuscript.
ETHICS APPROVAL
The Institutional Review Board at Vanderbilt University Medical Center approved the study under IRB#061099. The All of Us Research Program approved the usage of All of Us data in this study.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
Supplementary Material
Contributor Information
Ziqi Zhang, Department of Computer Science, Vanderbilt University, Nashville, Tennessee, USA.
Chao Yan, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Xinmeng Zhang, Department of Computer Science, Vanderbilt University, Nashville, Tennessee, USA.
Steve L Nyemba, Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Bradley A Malin, Department of Computer Science, Vanderbilt University, Nashville, Tennessee, USA; Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Department of Biostatistics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
DATA AVAILABILITY STATEMENT
The VUMC data analyzed in this study are not publicly available due to the potential identifying nature of the records, but a deidentified version of the data is available from the authors upon reasonable request to the VUMC IRB. The All of Us data used in this study are publicly available at https://allofus.nih.gov/.
REFERENCES
- 1. Xiao C, Choi E, Sun J.. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc 2018; 25(10): 1419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ching T, Himmelstein DS, Beaulieu-Jones BK, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 2018; 15(141): 20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang F, Casalino LP, Khullar D.. Deep learning in medicine—promise, progress, and challenges. JAMA Intern Med 2019; 179(3): 293–4. [DOI] [PubMed] [Google Scholar]
- 4. Choi E, Bahadori MT, Schuetz A, et al. Doctor AI: predicting clinical events via recurrent neural networks. J Mach Learn Res Workshop Conf Proc 2016; 56:301–18. [PMC free article] [PubMed] [Google Scholar]
- 5. Miotto R, Li L, Kidd BA, et al. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pham T, Tran T, Phung D, et al. DeepCare: a deep dynamic memory model for predictive medicine. In: Proceedings of the Pacific-Asia Conference on Knowledge and Discovery in Databases; 2016: 30–41.
- 7. Cheng Y, Wang F, Zhang P, et al. Risk prediction with electronic health records: a deep learning approach. In: Proceedings of the SIAM International Conference on Data Mining; 2016: 432–40.
- 8. Pham T, Tran T, Phung D, et al. Predicting healthcare trajectories from medical records: a deep learning approach. J Biomed Inform 2017; 69: 218–29. [DOI] [PubMed] [Google Scholar]
- 9. Lake BM, Ullman TD, Tenenbaum JB, et al. Building machines that learn and think like people. Behav Brain Sci 2017; 40: e253. [DOI] [PubMed] [Google Scholar]
- 10. Ghassemi M, Naumann T, Schulam P, et al. A review of challenges and opportunities in machine learning for health. AMIA Jt Summits Transl Sci Proc 2020; 2020: 191–200. [PMC free article] [PubMed] [Google Scholar]
- 11. Jing L, Tian Y.. Self-supervised visual feature learning with deep neural networks: a survey. IEEE Trans Pattern Anal Mach Intell 2020; 43(11), 4037–4058. [DOI] [PubMed] [Google Scholar]
- 12. Bengio Y, Courville A, Vincent P.. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 2013; 35(8): 1798–828. [DOI] [PubMed] [Google Scholar]
- 13. Le-Khac PH, Healy G, Smeaton AF.. Contrastive representation learning: a framework and review. IEEE Access 2020; 8: 193907–34. [Google Scholar]
- 14. Jaiswal A, Babu AR, Zadeh MZ, et al. A survey on contrastive self-supervised learning. Technologies (Basel) 2020; 9(1): 2. [Google Scholar]
- 15. Li J, Zhou P, Xiong C, et al. Prototypical contrastive learning of unsupervised representations. In: Conference Track Proceedings of the 9th International Conference on Learning Representations; 2021.
- 16. Kim Y, Shin J, Yang E, et al. Few-shot visual reasoning with meta-analogical contrastive learning. In: Advances in Neural Information Processing Systems; La Jolla, CA: Neural information processing systems foundation; 2020.
- 17. O’Neill J, Buitelaar P. Few shot transfer learning between word relatedness and similarity tasks using a gated recurrent siamese network. In: Proceedings of the AAAI Conference on Artificial Intelligence; 2018: 5342–9.
- 18. Liang PP, Wu P, Ziyin L, et al. Cross-modal generalization: learning in low resource modalities via meta-alignment. In: Proceedings of the 29th ACM International Conference on Multimedia; New York, NY: Association for Computing Machinery, Inc.; 2021: 2680–9.
- 19. Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012; 92(1): 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.All of Us Research Program Investigators. The “All of Us” research program. N Engl J Med 2019; 381(7): 668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roden DM, Pulley JM, Basford MA, et al. Development of a large‐scale de‐identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84(3): 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Yao Q, Kwok JT, et al. Generalizing from a few examples: a survey on few-shot learning. ACM Comput Surv 2021; 53(3): 1–34. [Google Scholar]
- 23. Prabhu V, Kannan A, Ravuri M, et al. Few-shot learning for dermatological disease diagnosis. Proc Mach Learn Res 2019; 105: 532–52. [Google Scholar]
- 24. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: Conference Track Proceedings of the 3rd International Conference on Learning Representations; 2015.
- 25. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Los Alamitos, CA: IEEE Computer Society; 2015: 1–9.
- 26. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016: 770–78.
- 27. Srivastava RK, Greff K, Schmidhuber J. Training very deep networks. In: Advances in Neural Information Processing Systems; La Jolla, CA: Neural information processing systems foundation; 2015: 2377–85.
- 28. Cui L, Biswal S, Glass LM, et al. CONAN: complementary pattern augmentation for rare disease detection. Proc AAAI Conf Artif Intell 2020; 34(01): 614–21. [Google Scholar]
- 29. Choi E, Bahadori MT, Kulas JA, et al. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. In: Advances in Neural Information Processing Systems; La Jolla, CA: Neural information processing systems foundation; 2015: 3504–12.
- 30. Ye M, Luo J, Xiao C, et al. LSAN: modeling long-term dependencies and short-term correlations with hierarchical attention for risk prediction. In: Proceedings of the ACM SIGKDD Conference on Knowledge Discovery and Data Mining; New York, NY: Association for Computing Machinery; 2020: 1753–62.
- 31. Ma F, Chitta R, Zhou J, et al. Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In: Proceedings of the ACM SIGKDD Conference on Knowledge Discovery and Data Mining; New York, NY: Association for Computing Machinery; 2017: 1903–11.
- 32. Zhang C, Bengio S, Hardt M, Recht B, Vinyals O.. Understanding deep learning requires rethinking generalization. Commun ACM 2021; 64(3): 107–15. [Google Scholar]
- 33. Gal Y, Ghahramani Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: The 33rd International Conference on Machine Learning; International Machine Learning Society; 2016: 1050–9.
- 34. Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks. In: Conference Track Proceedings of the 2nd International Conference on Learning Representations; 2014.
- 35. Caruana R. Multitask learning. Mach Learn 1997; 28(1): 41–75. [Google Scholar]
- 36. Choi E, Xiao C, Stewart WF, et al. Mime: multilevel medical embedding of electronic health records for predictive healthcare. In: Advances in Neural Information Processing Systems; La Jolla, CA: Neural information processing systems foundation; 2018: 4552–62.
- 37. Ma T, Zhang A.. Affinitynet: semi-supervised few-shot learning for disease type prediction. Proc AAAI Conf Artif Intell 2019; 33(01): 1069–76. [Google Scholar]
- 38. Saunshi N, Plevrakis O, Arora S, et al. A theoretical analysis of contrastive unsupervised representation learning. In: The 36th International Conference on Machine Learning; International Machine Learning Society; 2019: 5628–37.
- 39. Giorgi JM, Nitski O, Bader GD, et al. Declutr: deep contrastive learning for unsupervised textual representations. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing; Stroudsburg, PA: Association for Computational Linguistics; 2021: 879–95. [Google Scholar]
- 40. Chen T, Kornblith S, Norouzi M, et al. A simple framework for contrastive learning of visual representations. In: The 37th International Conference on Machine Learning; International Machine Learning Society; 2020: 1597–1607.
- 41. Oord AV, Li Y, Vinyals O. Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748. July 10, 2018.
- 42. Pan SJ, Yang Q.. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010; 22(10): 1345–59. [Google Scholar]
- 43. Schildcrout JS, Denny JC, Bowton E, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther 2012; 92(2): 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: Advances in Neural Information Processing Systems; La Jolla, CA: Neural information processing systems foundation; 2017: 5998–6008.
- 45. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9(8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 46. Zhang Z, Yan C, Lasko TA, et al. SynTEG: a framework for temporal structured electronic health data simulation. J Am Med Inform Assoc 2021; 28(3): 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y, Zhao L, Wang J, et al. Compositional generalization for primitive substitutions. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and 9th International Joint Conference on Natural Language Processing; Stroudsburg, PA: Association for Computational Linguistics; 2019: 4293–302. [Google Scholar]
- 48. Russin J, Jo J, O’Reilly RC, et al. Compositional generalization by factorizing alignment and translation. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: Student Research Workshop; Stroudsburg, PA: Association for Computational Linguistics; 2020: 313–27. [Google Scholar]
- 49. Blum A, Mitchell T. Combining labeled and unlabeled data with co-training. In: Proceedings of the Annual Conference on Computational Learning Theory; 1998: 92–100.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The VUMC data analyzed in this study are not publicly available due to the potential identifying nature of the records, but a deidentified version of the data is available from the authors upon reasonable request to the VUMC IRB. The All of Us data used in this study are publicly available at https://allofus.nih.gov/.