Abstract
Competition assays are a simple phenotyping strategy that confront two bacterial strains to evaluate their relative fitness. Because they are more accurate than single-strain growth assays, competition assays can be used to highlight slight differences that would not otherwise be detectable. In the frame of host-pathogens interactions, they can be very useful to study the contribution of individual bacterial genes to bacterial fitness and lead to the identification of new adaptive traits. Here, we describe how to perform such competition assays by taking the example of the model phytopathogenic bacterium Xanthomonas campestris pv. campestris during infection of the mesophyll of its cauliflower host. This phenotypic assay is based on the use of a Competitive Index (CI) that compares the relative abundance of co-inoculated strains before and after inoculation. Since multiplication is a direct proxy for bacterial fitness, the evolution of the ratio between both strains in the mixed population is a direct way to assess differences in fitness in a given environment. In this protocol, we exploit the blue staining of GUS-expressing bacteria to count blue vs. white colonies on plates and estimate the competitiveness of the strains of interest in plant mesophyll.
Keywords: Competitive Index, Competition assay, Fitness, Mesophyll, Xanthomonas, Cauliflower, GUS, Brassica oleracea
Background
Over the years, topics of ecology and evolution have been increasingly important when studying host-microbe interactions. In addition to simply assessing the virulence of bacterial pathogens, matters of fitness and adaptation to the environment are now being addressed. Recently, large-scale high-throughput screening tools, such as RB-TnSeq (Randomly-Barcoded Transposon insertion site Sequencing), have been developed to efficiently identify the genetic determinants of bacterial fitness. RB-TnSeq relies on the co-inoculation of a library of mutagenized bacteria, all carrying a unique barcode, to simultaneously evaluate the contribution of each gene to bacterial survival and growth ( Wetmore et al., 2015 ). Such studies result in the identification of genes putatively implicated in bacterial adaptation to the host that then need to be confirmed by reverse genetics using simpler fitness assays.
Early on, it was proposed that competition assays could be used as an answer to this need. Typically, such competition assays are performed by co-inoculating two strains in a 1:1 ratio into a given environment and following the evolution of this ratio over time. If one of the strains grows more than the other, it is considered as more fit (in this specific environment) than its counterpart. The main limiting factor is then to be able to count separately the two mixed populations. A quite straightforward solution here is to use discriminating markers, such as antibiotic resistance, to count the abundance of each strain after plating on selective medium. Differential antibiotic resistances have been widely used in co-inoculation assays ( Macho et al., 2007 and 2010; Feng et al., 2012 ), but it is known that such resistances often carry a strong cost of fitness that could interfere in studies that involve fine fitness mechanisms. Also, they require plating the samples twice, once on each selective medium, which can introduce a bias in the evaluation of the strains relative frequencies. Fluorescent markers provide an alternative to antibiotics but require costly imaging equipment ( Perrier et al., 2019 ). Here, we describe a visual blue/white screening using the enzymatic properties of the beta-glucuronidase (GUS) enzyme. The assay is then performed by co-inoculating a GUS-expressing strain in competition with a non-tagged strain: after plating on medium containing X-gluc (the substrate of the GUS enzyme), colonies of tagged bacteria will turn blue while the others will remain white. A similar Bio-protocol had been proposed before using LacZ as a marker in the Vibrio cholerae animal pathogen (Fu and Mekalanos, 2014). However, even though the concept is similar, we feel that a detailed protocol for plant inoculations is very much needed.
Here, we chose the bacterial phytopathogen Xanthomonas campestris pv. campestris (Xcc) as a model to illustrate this protocol. Xcc is particularly interesting because it has the specificity to infect multiple organs and tissues of its cauliflower host (Brassica oleracea pv. botrytis cv. Clovis). After entry through the hydathodes, Xcc spreads through the xylem sap-conducting vessels and colonizes the leaf mesophyll of its host, where it causes the typical symptoms of the Black rot disease (Vicente and Holub, 2013; An et al., 2019 ). We recently performed an RB-TnSeq screening on Xcc during cauliflower infection ( Luneau et al., 2022 ), which ultimately led us to set up a competition assay. In this protocol, we describe how this competition assay can be performed in the plant leaf mesophyll. We take the example of the well-described hrpX (transcriptional activator of the Type III Secretion System), hrcV (Type III Secretion System component), and xopAC (Type III Effector) mutants that display contrasted fitness phenotypes when confronted with the Xcc 8004 wild-type strain in planta. This protocol provides a basis that can be adapted for other plant compartments, such as hydathodes or xylem, as well as in vitro studies. Availability of such protocol is particularly relevant given the number of plant pathogens that enter through and thrive in their host leaves.
Materials and Reagents
2-mL microcentrifuge tubes (Sarstedt, catalog number: 72.695.400)
5-mL tubes (Sarstedt, catalog number: 72.701)
Petri dishes 92 × 16 mm (Sarstedt, catalog number: 82.1473)
1-mL Blunt syringe (SOFT-JECT HSW, catalog number: 5010.200V0)
Sterile 4-mm diameter glass beads (Dutscher, catalog number: 068502)
Sterile 96-well V-bottom clear polystyrene microplates (Greiner Bio-One, catalog number: 651101)
Sterile pipette tips
Compost (Proveen, catalog number: 14926)
-
Cauliflower: Brassica oleracea var. botrytis cv. Clovis F1 (Vilmorin)
Note: Plants are grown under greenhouse conditions and used at 4 weeks after sowing. All inoculations were performed using the second true leaf (second leaf above the cotyledons). After inoculation, plants are kept in classical growth chamber conditions: 9 h light, 22°C, and 70% relative humidity.
-
Xanthomonas campestris pv. campestris (Xcc) strain 8004::GUS-GFP (WT hereafter; Cerutti et al., 2017 ) and variants: Xcc 8004::GUS*-GFP* (WT* hereafter, carrying the point mutations Y468A and Y66A inactivating the catalytic sites of the β-glucuronidase and GFP proteins respectively); Xcc 8004::GUS-GFP ΔhrpX; Xcc 8004::GUS-GFP ΔhrcV; Xcc 8004::GUS-GFP ΔxopAC
Note: With concerns to the cost of fitness related to the constitutive expression of the GUS-GFP cassette, we decided to create the non-functional version of the cassette for our reference strain. With this system, the two challenged strains carry and express the GUS-GFP cassette, but only one of them will actually appear as tagged. We thus remove the possibility of a fitness cost associated with the cassette while still being able to differentiate the two mixed strains.
Spectrophotometer cells (Standard Polystyrene Semi-micro Cuvettes, Sarstedt, catalog number: 67.742)
Sterile distilled water
Magnesium chloride (MgCl2) (Merck, catalog number: 105832)
Yeast extract (Sigma-Aldrich, catalog number: Y1625)
Casamino acids (BD, BD Biosciences, catalog number: 223050)
Potassium phosphate dibasic (K2HPO4) (Merck, catalog number: 105101)
Agar (BD, BD Biosciences, catalog number: 214010)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Merck, catalog number: 105886)
X-Gluc (5-Bromo-4-chloro-3-indoxyl-beta-D-glucuronic acid, cyclohexylammonium salt monohydrate) (BIOSYNTH, catalog number: B-7300)
DMF (N,N Dimethylformamide) (Sigma-Aldrich, catalog number: D4551)
Rifampicin (Euromedex, catalog number: 1059)
Methanol (VWR, catalog number: 20847.320)
Pimaricin (DSM, DEVLOCID Instant, catalog number: 00226)
MOKA medium (see Recipes)
1 mM MgCl2 solution (see Recipes)
X-gluc (see Recipes)
Rifampicin (see Recipes)
Pimaricin (see Recipes)
Equipment
6-mm diameter Hollow punch (Harris, Uni-Core, Electron Microscopy Sciences, catalog number: 69039-60)
Hand-held colony counter (Heathrow Scientific, catalog number: HS120000)
28°C bacteria incubator
Centrifuge (Eppendorf, model: 5415D)
Beads-assisted grinder (Retsch, catalog number: MM400)
Spectrophotometer (Amersham Biosciences, model: Ultrospec 2100 pro)
Sterile environment (bunsen burner or microbiology hood)
Multi-channel pipette (10–100 µL)
Mono-channel pipettes
Procedure
-
Preparation of bacterial cells for the inoculation
Bacterial cells grown on MOKA plates with appropriate antibiotics (i.e., Rifampicin 50 µg/mL here) are used to inoculate 5 mL of liquid MOKA medium (with same antibiotics) and grown overnight at 28°C under agitation at 200 rpm.
Centrifuge 2 mL of bacterial preculture for 5 min at 6,000 rpm (i.e., 3.3 × g) and wash twice in 1 mM MgCl2. Resuspend in 1 mM MgCl2 and measure OD600nm.
Prepare 1 mL of bacterial suspension in a 2-mL tube so that, for each strain, final OD600nm = 0.1 (i.e., 1 × 108 cfu/mL; note: this estimation has to be verified when working with other species by plating on solid medium) in 1 mM MgCl2.
Dilute each suspension to 1/100 in a 5-mL tube to obtain a final inoculum at OD600nm = 0.001 (i.e., 1 × 106 cfu/mL) in 5 mL of 1 mM MgCl2. For the WT* reference strain, prepare 10 mL of suspension at OD600nm = 0.001 as it will be used in all mixes.
Mix 2 mL of WT* suspension with 2 mL of each of the strains to test in another 5-mL tube (so that each mixed strain is at 5 × 105 cfu/mL, but the final concentration of the inoculum is 1 × 106 cfu/mL).
-
Inoculation
Using a blunt syringe filled with one milliliter of bacterial suspension, infiltrate the mesophyll of the 2nd true leaf of cauliflower (for pictures of the procedure, see Cohn et al., 2015 ). A large enough area should be infiltrated in order to later be able to sample a 6-mm leaf disc without taking any of the tissues damaged by the syringe. Typically, up to seven separate spots can be infiltrated on the same cauliflower leaf, thus allowing the inoculation of multiple strains or mixes on the same leaf. In this case, be careful to well separate infiltration areas to exclude cross-contamination.
Circle the infiltrated area on the lower side of the leaf to mark the infected tissue for later sampling.
Place the plants in the growth chamber and water them appropriately.
-
Plating of the inoculation suspensions and counting of the initial relative frequencies in mixed populations
Transfer 100 µL of inoculation suspension into a 96-well plate with conical bottom.
Perform serial 1/10 dilutions (10 µL in 90 µL of 1 mM MgCl2) using a multichannel pipette.
Take 100 µL of dilution 10-3 and spread out them on round 92-mm MOKA plates containing the appropriate antibiotics (i.e., Rifampicin 50 µg/mL here) and fungicide (i.e., Pimaricin 30 µg/mL here) as well as X-Gluc 200 µM. In our case, spreading is performed by shaking using 4-mm glass beads until homogeneous repartition of the suspension.
Incubate 4–5 days at 28°C to allow bacterial growth and blue staining of GUS-expressing colonies.
Count blue and white colonies, e.g., using a hand-held colony counter, to determine the ratio of mixed strains at T0. This is necessary for proper normalization during later calculation of the CI (see Data analysis section).
-
Sampling, plating, and counting of relative frequencies in mixed populations after incubation
Three days after inoculation, sample mesophyll tissues by punching out a 6-mm leaf disc in the infiltrated area defined by the marker label and placing it in 200 µL of 1 mM MgCl2.
Grind leaf samples using 4-mm glass beads in 2-mL tubes (2 beads per tube) using the beads-assisted grinder for 2 min at 30 Hz.
Centrifuge shortly (5 s) the tubes containing the ground material to gather it at the bottom.
Transfer 100 µL of ground tissue to a 96-well plate with conical bottom.
Proceed as described in Steps C2 to C5. However, because each strain displays a different phenotype, and to ensure the presence of colonies from both strains, plate 50 µL of 10-2 and 10-3 dilutions.
Note: Classically, we advise collecting bacterial titers by counting the plates displaying between 30 and 300 colonies. However, because competition assays require to finely determine the number of CFU for each of the mixed strains, we recommend counting plates where there are at least 100 colonies for mix-inoculated samples.
Data analysis
For reproducibility purposes, experiments should be performed in at least four biological replicates. A biological replicate is defined as a replication of a complete experimental procedure with fresh buffers, fresh bacterial cells suspension prepared from cells grown on new solid medium, and new batches of plants. Results of competition assays are displayed as a Competitive Index (CI), which is defined as the ratio of the relative abundance of mutant over WT* strain after incubation normalized by the relative abundance of mutant over WT* strain in the inoculum ( Taylor et al., 1987 ; Macho et al., 2007 ):
N: number of colonies
dpi: days post inoculation
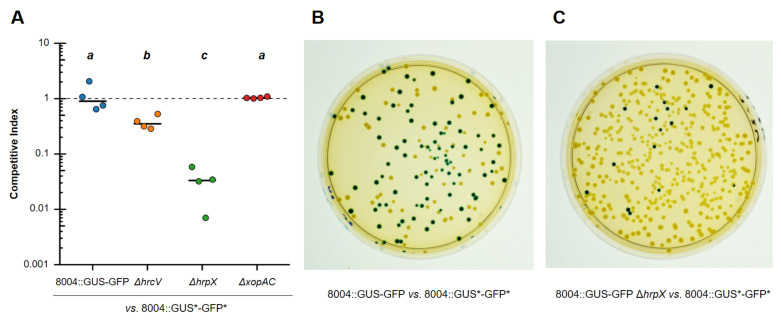
Here, we used the Xcc 8004::GUS-GFP ΔhrpX, ΔhrcV, and ΔxopAC mutants as an example for this protocol. These three strains are Type III Secretion System-related mutants that exhibit contrasted fitness in planta ( Feng et al., 2012 ; Guy et al., 2013 ; Wang et al., 2015 ). Competitive indices of these three strains, when co-inoculated with the WT* strain, are shown in Figure 1. Xcc 8004::GUS-GFP is a negative control whose fitness should be the same as the WT* reference strain (i.e., CI ~1). Strains with CI < 1 are considered less fit than the WT strain in the mesophyll environment.
Figure 1. Competition assay of Xcc 8004::GUS-GFP strain and related mutants in cauliflower mesophyll.
All competition assays were performed by co-inoculating the Xcc 8004::GUS*-GFP* reference strain with Xcc 8004::GUS-GFP derivatives in a 1:1 ratio. Mesophyll discs were sampled at 3 days post-inoculation. A. Graphical representation of competition assays results. Strains showing CI ~1 are considered as fit as the WT strain, while strains with CI < 1 are considered less fit than the WT strain in the mesophyll environment. Horizontal bars show the median for four independent biological replicates of one plant sample each. Statistical significance of fitness differences was evaluated by a Kruskal-Wallis test. Groups identified as significantly different (P < 0.05) are displayed using the letters ‘a’, ‘b’, and ‘c’. Results showing the same letter belong to the same statistical group. B. Photo of a plate displaying colonies from the WT vs. WT* sample at 3 dpi in the mesophyll. Visually, the two populations are roughly present in a 1:1 ratio, illustrating the expected equivalent fitness of both strains. The same 1:1 ratio is obtained after plating any of the inoculation suspensions. C. Photo of a plate displaying colonies from the ΔhrpX vs. WT* sample at 3 dpi in the mesophyll. Blue-stained colonies of the ΔhrpX strain are strongly under-represented, clearly exposing the large deficit of fitness of this strain.
We used four independent biological replicates of one plant sample each. The plot was generated using the ggplot2 package of the R software (Wickham, 2016; https://github.com/tidyverse/ggplot2). We purposely set a log10-based scale to stretch out low CI values that would be hardly differentiable otherwise. Statistical treatment of the data was also achieved with R using the Kruskal-Wallis test. Groups identified as significantly different (P < 0.05) are displayed using the letters ‘a’, ‘b’, and ‘c’ in Figure 1A.
Recipes
-
MOKA medium
Mix 4 g of yeast extract, 8 g of casamino acids, 2 g of K2HPO4, and 0.3 g of MgSO4·7H2O.
Add distilled water to 1 L.
Mix until complete dissolution of the reagents.
Sterilize 250-mL aliquots of the mixture by autoclaving at 120°C for 20 min.
Store at room temperature.
Solid MOKA medium for plates is prepared identically and complemented with 3.75 g of agar added to each individual 250-mL aliquot before autoclaving.
-
1 mM MgCl2 solution
Prepare a stock solution of 1 M MgCl2 by dissolving 2.033 g of MgCl2·6H2O in 10 mL of dH2O.
Sterilize by autoclaving at 120°C for 20 min.
Prepare 1 mM MgCl2 by diluting the stock solution 1/1,000 in sterile dH2O.
Both solutions are stored at room temperature.
-
X-gluc
Prepare X-gluc 500x stock solution by dissolving 0.54 g of X-gluc in 10 mL of DMF.
Store at -20°C.
Add 500 µL of X-gluc stock solution to 250 mL of MOKA + Agar solution to get a final concentration of 200 µM of X-gluc in plates used for CI measurement.
-
Rifampicin
Dissolve rifampicin powder in methanol to obtain a stock solution at 10 mg/mL.
Store at -20°C.
Dilute rifampicin stock solution to 1/200 in MOKA media to obtain a final concentration of 50 µg/mL.
-
Pimaricin
Resuspend pimaricin powder in water to obtain a stock solution at 30 mg/mL.
Store at 4°C.
Dilute pimaricin stock solution to 1/1,000 in MOKA media to obtain a final concentration of 30 µg/mL.
Acknowledgments
This work was supported by a Ph.D. grant from the French Ministry of National Education and Research to JSL. JSL, LDN, EL, and AB are supported by the ANR (ANR-19-CE20-0014-01). LIPM is part of the French Laboratory of Excellence project (TULIP ANR-10-LABX-41; ANR-11-IDEX-0002-02).
Competing interests
There are no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. An S. Q., Potnis N., Dow M., Vorhölter F. J., He Y. Q., Becker A., Teper D., Li Y., Wang N., Bleris L. and Tang J. L.(2019). Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas . FEMS Microbiol Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cerutti A., Jauneau A., Auriac M. C., Lauber E., Martinez Y., Chiarenza S., Leonhardt N., Berthome R. and Noel L. D.(2017). Immunity at cauliflower hydathodes controls systemic Infection by Xanthomonas campestris pv campestris . Plant Physiol 174(2): 700-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohn M., Shybut M., Dahlbeck D., and Staskawicz B.(2015). Assays to Assess Virulence of Xanthoonas axonopodis pv. manihotis on Cassava . Bio-protocol 5(13): 1522. [Google Scholar]
- 4. Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C. and Zhou J. M.(2012). A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases . Nature 485(7396): 114-118. [DOI] [PubMed] [Google Scholar]
- 5. Fu Y. and Mekalanos J. J.(2014). Infant rabbit colonization competition assays. Bio-protocol 4(11): e1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guy E., Lautier M., Chabannes M., Roux B., Lauber E., Arlat M. and Noel L. D.(2013). xopAC-triggered immunity against Xanthomonas depends on Arabidopsis receptor-like cytoplasmic kinase genes PBL2 and RIPK . PLoS One 8(8): e73469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luneau, J. S., Baudin, M., Quiroz Monnens, T., Carrère, S., Bouchez, O., Jardinaud, M. F., Gris, C., François, J., Ray, J., Torralba, B., Arlat, M., Lewis, J. D., Lauber, E., Deutschbauer, A. M., Noël, L. D., Boulanger, A. Genome-wide identification of fitness determinants in the Xanthomonas campestris bacterial pathogen during early stages of plant infection. New Phytol. 2022 Jun 15. [DOI] [PMC free article] [PubMed]
- 8. Macho A. P., Guidot A., Barberis P., Beuzón C. R. and Genin S.(2010). A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants . Mol Plant Microbe Interact 23(9): 1197-1205. [DOI] [PubMed] [Google Scholar]
- 9. Macho A. P., Zumaquero A., Ortiz-Martin I. and Beuzón C. R.(2007). Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions . Mol Plant Pathol 8(4): 437-450. [DOI] [PubMed] [Google Scholar]
- 10. Perrier A., Barlet X., Rengel D., Prior P., Poussier S., Genin S. and Guidot A.(2019). Spontaneous mutations in a regulatory gene induce phenotypic heterogeneity and adaptation of Ralstonia solanacearum to changing environments . Environ Microbiol 21(8): 3140-3152. [DOI] [PubMed] [Google Scholar]
- 11. Taylor R. K., Miller V. L., Furlong D. B. and Mekalanos J. J.(1987). Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84(9): 2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vicente J. G. and Holub E. B.(2013). Xanthomonas campestris pv. campestris(cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops . Mol Plant Pathol 14(1): 2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang G., Roux B., Feng F., Guy E., Li L., Li N., Zhang X., Lautier M., Jardinaud M. F., Chabannes M., et al.(2015). The decoy Substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-Self recognition and immunity in plants. Cell Host Microbe 18(3): 285-295. [DOI] [PubMed] [Google Scholar]
- 14. Wetmore K. M., Price M. N., Waters R. J., Lamson J. S., He J., Hoover C. A., Blow M. J., Bristow J., Butland G., Arkin A. P., et al.(2015). Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6(3): e00306-00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wickham H.(2016). ggplot2: elegant graphics for data analysis. In: Wickham, H.(Ed.). ggplot2. Springer-Verlag New York. ISBN: 978-3-319-24277-4. [Google Scholar]