Abstract
We have evaluated the use of [1,2-13C2]propionate for the analysis of propionic acid metabolism, based on the ability to distinguish between the methylcitrate and methylmalonate pathways. Studies using propionate-adapted Escherichia coli MG1655 cells were performed. Preservation of the 13C-13C-12C carbon skeleton in labeled alanine and alanine-containing peptides involved in cell wall recycling is indicative of the direct formation of pyruvate from propionate via the methylcitrate cycle, the enzymes of which have recently been demonstrated in E. coli. Additionally, formation of 13C-labeled formate from pyruvate by the action of pyruvate-formate lyase is also consistent with the labeling of pyruvate C-1. Carboxylation of the labeled pyruvate leads to formation of [1,2-13C2]oxaloacetate and to multiply labeled glutamate and succinate isotopomers, also consistent with the flux through the methylcitrate pathway, followed by the tricarboxylic acid (TCA) cycle. Additional labeling of TCA intermediates arises due to the formation of [1-13C]acetyl coenzyme A from the labeled pyruvate, formed via pyruvate-formate lyase. Labeling patterns in trehalose and glycine are also interpreted in terms of the above pathways. The information derived from the [1,2-13C2]propionate label is contrasted with information which can be derived from singly or triply labeled propionate and shown to be more useful for distinguishing the different propionate utilization pathways via nuclear magnetic resonance analysis.
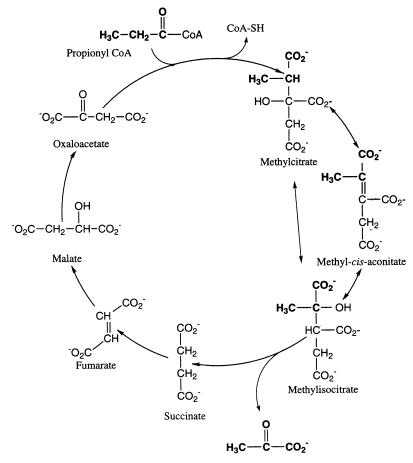
The metabolism of propionic acid by various organisms has been the object of extensive analysis which has elucidated at least six different pathways (10, 34). In most vertebrates, propionyl coenzyme A (propionyl-CoA), the obligate intermediate, is carboxylated to d-methylmalonyl-CoA, isomerized to l-methylmalonyl-CoA, and rearranged to yield succinyl-CoA via a vitamin B12-dependent enzyme (14, 19). Alternatively, plants, microorganisms, and insects make use of a variety of alternate pathways which include reduction to acryloyl-CoA followed by α or β oxidation (11, 15, 16, 27), reductive carboxylation to yield α-ketobutyrate (5, 26), and Claisen condensations with either glyoxylate or oxaloacetate (OA) (25, 32–34, 36, 37). In yeast and Escherichia coli, condensation with OA leads to the methylcitrate cycle (Fig. 1), whereby methylcitrate is isomerized to methylisocitrate, which is cleaved to yield pyruvate and succinate, with subsequent regeneration of OA from the latter via the usual tricarboxylic acid (TCA) cycle transformations. In view of the variety of metabolic pathways for propionate, the interpretation of propionate metabolism has often been difficult. Recent proposals that metabolite labeling patterns observed in E. coli grown on 13C-labeled propionate can provide evidence for metabolite channeling (7, 8) make it important to fully elucidate the pathway(s) involved in propionate metabolism.
FIG. 1.
Methylcitrate cycle for the oxidation of propionate to pyruvate.
Although the introduction of carbon-13 isotopic labeling greatly facilitates metabolic analysis due to the ease with which the labeled position(s) can be determined, the unique feature of this technique is the ability to follow the fate of more complex structural units which are identified on the basis of scalar coupling interactions. Thus, the use of correlated labeling patterns provides a capability beyond that of analogous 14C labeling studies. In the present study, [1,2-13C2]propionate was used to distinguish metabolism via the methylmalonyl-CoA pathway and via the methylcitrate pathway in E. coli. The labeling patterns derived from [1,2-13C2]propionate provide a basis for this analysis due to the very different labeling which will result from flux through these two pathways. In general, this approach will be optimal when sufficient levels of alanine are produced to allow determination of the label distribution in the pyruvate precursor. In E. coli extracts, both alanine and alanine-containing peptides involved in cell wall turnover can provide this information. Other metabolic intermediates, particularly succinate and peptidyl-glycine, provide less direct but nevertheless useful information regarding the metabolic pathways for propionate.
MATERIALS AND METHODS
[1,2-13C2]sodium propionate was obtained from Isotec, Inc. (Miamisburg, Ohio). All other chemicals were from Sigma or Mallinckrodt. E. coli MG1655 (F− λ−; ATCC 47076) was a gift from Roel Schaaper.
Bacteria and culture conditions.
The cells were grown on a minimal medium (growth medium) containing 60 mM K2HPO4, 33 mM KH2PO4, 76 mM (NH4)2SO4, 2 mM trisodium citrate, 0.1% trace elements solution SL10, 1 mM MgSO4, and 20 mM sodium propionate, as described by Textor et al. (34).
The cultures were grown in a shaking incubator (37°C, 250 rpm) over a period of several days. This adaptation period was required in order to obtain the high yields of cells necessary to perform these experiments (34). Once the E. coli cells had adapted to the medium, the growth period could be reduced to 1 to 2 days.
Initially, the adaptation process involved growing the cells overnight in a 3-ml aliquot of LB medium. These cells were then resuspended in 100 ml of the growth medium and allowed to incubate for at least 48 h. (Cell growth was monitored by measuring the optical density [OD] at 578 nm.) When the OD of the 100-ml culture was >0.5, 10 ml was removed and used to inoculate 500 ml of growth medium in a 2-liter flask. The large culture grew to an OD of at least 0.6 before the labeling reactions were performed. This growth period usually lasted 3 or more days.
Labeling reactions.
The 500-ml cultures were harvested by centrifugation for 20 min at 4,000 rpm in sterile, weighed, 1-liter centrifuge bottles. The supernatant was removed, and the cells were weighed. The cell pellet was then washed in approximately 25 ml of labeling medium. The cells were resuspended in enough labeling buffer to yield approximately 1 g of cells/15 ml of buffer. This mixture was then transferred to sterile 100-ml beakers, and the cells were resuspended in a medium containing a 20 mM concentration of the labeled sodium propionate and 2 mM unlabeled citrate as the primary carbon sources. In a second series of studies, the labeled sodium propionate was replaced by a mixture of [1,2-13C2]propionate plus unlabeled propionate, so that the total added propionate remained set at 20 mM. The [1,2-13C2]propionate/unlabeled propionate ratios used were 2/18, 4/16, 10/10, and 20/0. The labeling mixture was then placed in a shaking incubator (37°C, 225 rpm) for 1.75 to 3.5 h. The labeling reactions were stopped by adding 60% perchloric acid (PCA) to a final concentration of 4% and freezing the cells at −70°C.
Preparation of PCA extracts.
The thawed PCA-treated cells were placed on ice and sonicated for 15 min. The sonicator was set to cycle on/off such that it was on 60% of the time and off 40% of the time to control heating. The cell debris was removed by centrifuging the sample at 6,000 rpm for 20 min at 4°C. The supernatant was collected and brought to a neutral pH with 10 M KOH. The sample was then centrifuged at 10,000 rpm for 20 min at 4°C. The remaining supernatant was collected and lyophilized. For the peptide hydrolysis experiment, the PCA extract was treated with 1.5 ml of hydrochloric acid (final concentration, 6 N HCl) and refluxed for 48 h at 115°C. The sample was then neutralized to pH 7 with potassium hydroxide, lyophilized, and prepared for nuclear magnetic resonance (NMR) analysis by adding 0.8 ml of D2O, centrifuging and collecting the supernatant.
NMR studies.
The NMR samples were prepared by dissolving approximately 220 mg of lyophilized sample into 0.6 ml of D2O, and the pH (uncorrected for isotope effects) was adjusted to 7.1 with DCl and NaOD. Any undissolved material was removed by centrifugation prior to loading the sample into a 5-mm NMR tube. In addition, 20 mM sodium azide was added to the sample to limit bacterial growth. NMR studies were performed at a 13C frequency of 125.892 MHz on a Varian Unity 500 NMR spectrometer (Varian Associates, Inc., Palo Alto, Calif.) using a 5-mm broadband probe. For proton-decoupled 13C experiments, a WALTZ-16 decoupling scheme (28) was used. With the labeling strategy used, there is little formation of [2-13C]acetyl-CoA and hence little labeling of the aliphatic carbons of TCA cycle-derived metabolites. Thus, significantly longer acquisition times were required in comparison with studies using labeled precursors leading to the formation of [2-13C]acetyl-CoA. Proton-decoupled 13C spectra of extracts typically ran for ∼100,000 pulses, using a recycle time of 1.3 s and a pulse width of 30°. Carbon-13 chemical shifts were referenced to the shifts of internal glutamate, using the values given in the appendix of reference 17. Two-dimensional 1H-13C heteronuclear single quantum coherence (HSQC) (2, 4) and HSQC-total correlation spectroscopy (TOCSY) (21) experiments were performed with an inverse probe in order to correlate the proton and carbon resonances. The TOCSY portion of the HSQC-TOCSY experiment was implemented by using an MLEV-17 spinlock sequence (1).
RESULTS
Formation of pyruvate and formate.
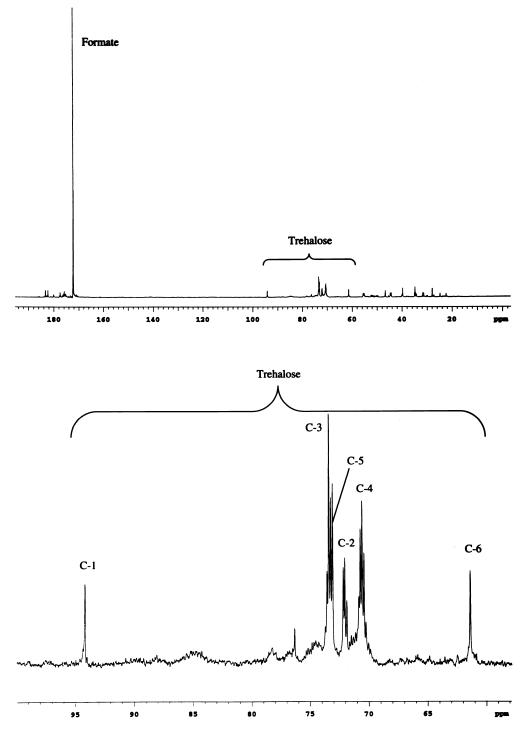
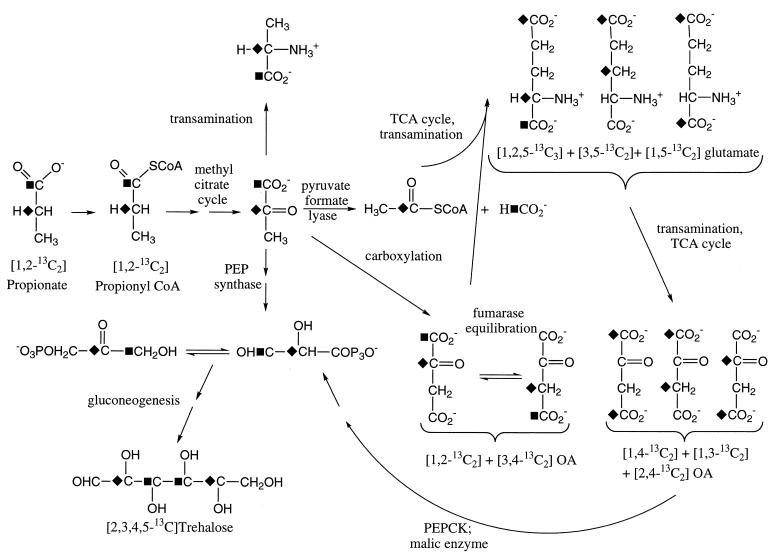
For these studies, we used a prototrophic strain of E. coli, MG1655, which appeared to grow most successfully on the high-propionate medium relative to several strains evaluated. The proton-decoupled spectrum of [1,2-13C2]propionate shows the expected doublet pattern for the C-1 (δ = 186.2 ppm) and C-2 (δ = 31.9 ppm) positions with J12 = 51.2 Hz. The proton-decoupled 13C spectrum corresponding to a PCA extract of the propionate-adapted MG1655 cells, which had been labeled in medium containing 20 mM [1,2-13C2]propionate and 2 mM citrate as the primary carbon sources, is shown in Fig. 2. The presence of a large formate resonance is most readily explained by the conversion of [1,2-13C2]propionate to [1,2-13C2]pyruvate via the enzymes of the methylcitrate pathway (34), followed by formation of [1-13C]acetyl-CoA plus [13C]formate as a result of the action of pyruvate-formate lyase (Fig. 3). Thus, despite the preservation of the three-carbon structural unit of propionate as it is converted into pyruvate, further coupling interactions are lost due to the scission of the C-1–C-2 bond by pyruvate-formate lyase. Consequently, the observation of scalar coupling interactions in these studies arises either as a result of the direct incorporation of doubly labeled pyruvate or as a fortuitous consequence of bond formation between 13C nuclei derived from different, labeled precursors.
FIG. 2.
Proton-decoupled 13C spectrum derived from an extract of E. coli MG1655 cells adapted to growth on propionate and subjected to a 3.5-h period of labeling with 20 mM [1,2-13C2]propionate plus 2 mM citrate (upper trace). An expansion of the region of the spectrum containing the trehalose resonances is shown in the lower trace. The spectrum represents 100,000 transients with a pulse recycle time of 1.3 s, a flip angle of 35°, and a 3-Hz exponential multiplication.
FIG. 3.
Principal metabolic pathways involved in the uptake and metabolism of propionate and the labeling of observed metabolic intermediates.
Labeling of amino acid α-carbons.
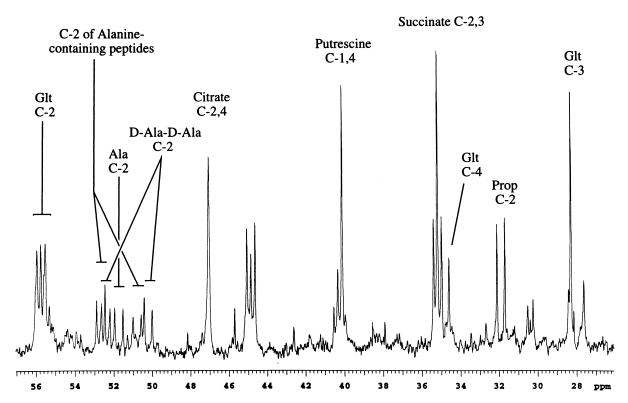
To obtain more detailed insight into the metabolism of the labeled propionate and to observe the less concentrated metabolites, longer periods of signal averaging were used. The spectral region containing the C-2 resonances of the amino acids exhibits a number of multiplets (Fig. 4). A set of resonances, δ = 55.8 ppm corresponding to singlet plus doublet with J = 54.2 Hz, is in reasonable agreement with the shift and 1JCo-Cα value of 53.4 Hz reported for glutamate at pH 7 (18). A more detailed analysis of the labeling of glutamate carbons is given below.
FIG. 4.
Aliphatic region of the 13C-{1H} spectrum corresponding to the sample in Fig. 2, using the assignments indicated. Glt, glutamate; prop, propionate.
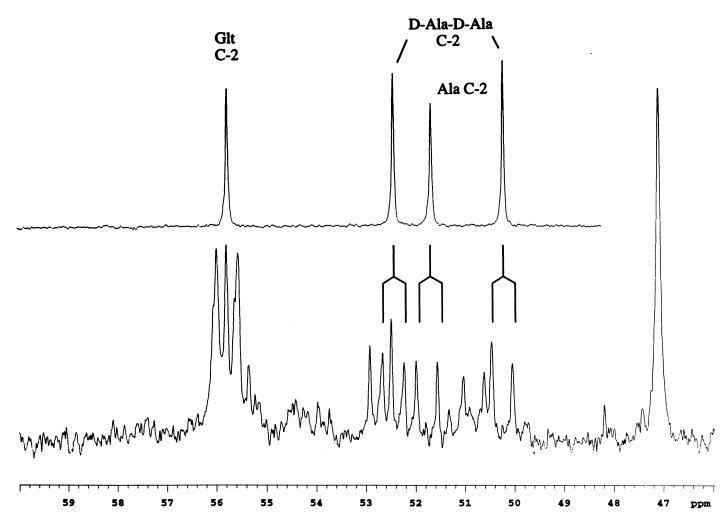
In addition to the glutamate C-2 resonances, the spectra exhibit a group of at least five doublets in the chemical shift range from 50 to 53 ppm. The coupling constants range from 52.6 to 54.1 Hz, consistent with values of 54.1 and 52.7 Hz reported for alanine 1JCo-Cα at pH values of 6.4 and 11.1, respectively (35). The latter value might be expected to correspond more closely to C-terminal alanine. Comparison with standards indicates that three of these doublets can be assigned to free alanine and to d-Ala-d-Ala dipeptide (Fig. 5). Most significantly, there is little measurable singlet intensity for these resonances, indicating that the set of doublets corresponds to [1,2-13C2]alanine and to peptides containing this labeled amino acid. There is no evidence for the production of other labeled species such as [2,3-13C2]alanine [1,2,3-13C3]alanine that might arise due to possible metabolite interconversions. This labeling pattern is consistent with the metabolic scheme outlined in Fig. 3, showing that the series of conversions from propionate to pyruvate to alanine proceeds with preservation of the carbon skeleton. It is known that the peptide components of the bacterial cell wall undergo significant turnover and that a number of alanine-containing cell wall precursors and breakdown products can be observed in the cytosol (6, 12, 22). A comparison of the spectral region containing the amino acid C-2 resonances with a standard consisting of glutamate, l-alanine, and d-alanyl-d-alanine indicates that two of the other doublets can be assigned to the dipeptide (Fig. 5). Hence, the detection of both alanine and a number of alanine-containing peptides provides further evidence for the labeling of the pyruvate pool in these cells. The acid hydrolysis of the sample results in the disappearance of the group of doublets attributed to the alanine-containing peptides and the appearance of a single set of doublet peaks with a shift and coupling constant expected for [1,2-13C2]alanine (Fig. 6A and B). There is a weak resonance in the center which may be part of a small doublet (Fig. 6A) or may correspond to [2-13C]alanine arising from the naturally abundant 13C in nominally unlabeled alanine in the sample or possibly indicate some reverse flux involving synthesis of pyruvate from OA.
FIG. 5.
Comparison of the region of the extract spectrum containing the amino acid α-carbon resonances with a standard containing glutamate (Glt), l-alanine, and d-alanyl-d-alanine (D-Ala-D-Ala).
FIG. 6.
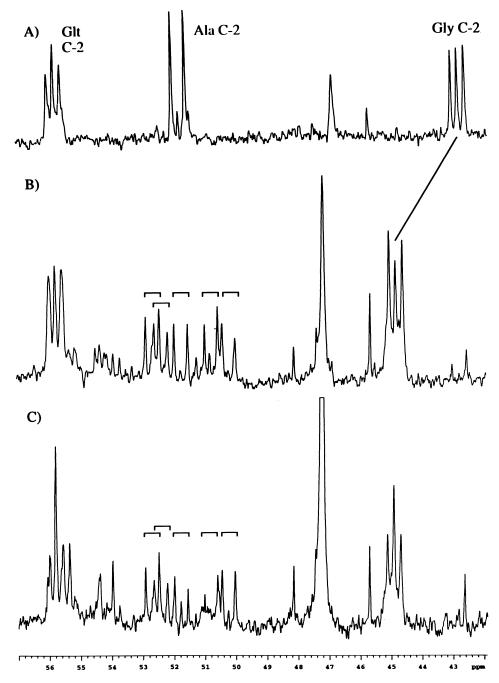
Comparison of the 42- to 57-ppm region of the 13C NMR spectra of PCA extracts corresponding to the sample in Fig. 4 after hydrolysis in 6 N HCl to effect peptide hydrolysis (A), the corresponding region of the sample prior to hydrolysis (B), and the corresponding region of a PCA extract derived from cells provided 4 mM [1,2-13C2]propionate, 16 mM unlabeled propionate, and 2 mM citrate during the labeling period. Glt, glutamate.
Condensation of labeled 13CO2 with pyruvate to form OA represents an alternative labeling pathway for metabolites derived from TCA intermediates. If each of the carbons in 20 mM propionate–2 mM citrate is considered to be metabolized to CO2 to the same extent, the fraction of 13CO2 would be 55%. In general, the enrichment of the CO2 pool is limited by the catabolism of endogenous, unlabeled compounds and by atmospheric CO2. Thus, the CO2 enrichment is probably significantly lower than 55%. To experimentally determine the significance of 13CO2 incorporation on the labeling of the alanine pool, we performed a series of studies in which a mixture of unlabeled and [1,2-13C2]propionate was added during the labeling period, such that the total added propionate remained fixed at 20 mM. [1,2-13C2]propionate/unlabeled propionate concentration ratios (millimolar values) used were 2/18, 4/16, 10/10, and 20/0. Proton-decoupled 13C spectra obtained for the lower ratios of propionate labeling were similar to those shown in Fig. 4 except that the multiplet/singlet ratios were reduced. This result is illustrated in a spectrum of the region of the 13C NMR spectrum containing the amino acid C-2 resonances derived from the 4/16 labeling experiment (Fig. 6C). Although the use of the lower enrichment significantly reduces the spectral signal/noise ratio, it is apparent that labeling of the alanine resonances is largely unaffected by the large variation in propionate labeling. The set of doublets arising from [1,2-13C2]alanine and [1,2-13C2]alanine-containing peptides observed near 52 ppm is closely analogous to that observed with 20 mM [1,2-13C2]propionate (Fig. 6B). Thus, the multiply labeled alanine does not arise primarily from the formation of bonds between labeled carbon nuclei but rather is due to retention of the coupling interactions derived from the doubly labeled propionate. A small increase in the alanine C-2 singlet resonances observed in Fig. 6C most probably arises from the reduced amount of labeled alanine relative to the natural abundance background.
Resonances corresponding to the C-2 carbons of other amino acids such as glutamine, aspartate, or asparagine were not present at sufficient concentration to allow further analysis. Close inspection of the glutamate C-2 resonances indicates the presence of small shoulders suggesting the presence of a closely related species such as a γ-glutamyl-linked peptide, e.g., glutathione (see below). Finally, as discussed below, the multiplet group centered at δ = 44.9 ppm probably arises from a C-terminal glycine residue in a peptide.
Labeling of glutamate and TCA cycle flux.
Glutamate labeling by [1-13C]acetyl-CoA formed as shown in Fig. 3, in combination with unlabeled OA, will result in a 50:50 mixture of [5-13C]glutamate and [1,5-13C2]glutamate isotopomers (17). Inclusion of flux through the glyoxylate shunt will also introduce label at the C-1 position of glutamate. The aliphatic region of the glutamate spectrum will appear similar to that obtained previously for the glutamate-producing organism Brevibacterium flavum (38) grown on [1-13C]acetate, with a high doublet/singlet ratio for C-4, a lower doublet/singlet ratio for C-2, and a C-3 singlet. The glutamate labeling pattern shown in Fig. 4 is not consistent with labeling derived from [1-13C]acetyl-CoA only. Comparison of resonance intensities indicates significantly greater labeling at C-2 and C-3 than at C-4 and relatively little doublet intensity for C-3 or C-4. The labeling observed for C-2 and C-3 is readily explained in terms of a significant anaplerotic formation of labeled OA from [1,2-13C2]pyruvate, formed as shown in Fig. 3. Thus, pyruvate carboxylation will result in [1,2-13C2]OA which may equilibrate via fumarase to [3,4-13C2]OA. These precursors will lead to formation of glutamate labeled in the 1 and 2 positions or the 3 position. At steady state, the glutamate isotopomers produced from a combination of [1,2-13C2]OA and [1-13C]acetyl-CoA are [1,2,5-13C3]-, [3,5-13C2]-, [1,3,5-13C3]-, [2,5-13C2]-, and [5-13C]glutamate. In particular, the 1,2,5-13C3- and 2,5-13C2-labeled isotopomers will produce doublet and singlet patterns, respectively, for glutamate C-2, while 3,5-13C2- and 1,3,5-13C3-labeled isotopomers will correspond to the glutamate C-3 singlet. In contrast, the low doublet/singlet ratio for glutamate C-4 indicates that there are additional, unlabeled sources of acetyl-CoA, in addition to the [1-13C]acetyl-CoA derived from propionate as shown in Fig. 3. The glutamate C-5 carboxyl is predicted to be a singlet, and the C-1 carboxyl is predicted to be a singlet plus doublet, as observed (spectrum not shown). Thus, under the conditions of the study, the labeling of glutamate provides additional support for the flux through the methylcitrate cycle by retention of coupling information in the glutamate C-1–C-2–C-3 fragment derived from the labeled pyruvate and ultimately from propionate.
The 13C spectra derived from the 4/16 propionate labeling experiment show evidence of label dilution in the form of reduced doublet/singlet ratios for all of the multiply labeled metabolites observed in Fig. 4, including glutamate C-2. This is apparent from a comparison of the glutamate C-2 resonances in Fig. 6B and C. In this case, the labeling information derived from pyruvate is further reduced by the presence of unlabeled pyruvate derived from unlabeled propionate. The increased ratio of singlet/doublet resonance intensities can arise from (i) the naturally abundant 13C nuclei in the metabolites and (ii) a very low degree of labeling via formation of small amounts of [2-13C]acetyl-CoA which arises due to secondary pathways not depicted in Fig. 3. A low flux through the latter pathway can contribute significantly to the singlet intensities of glutamate C-2, C-3, and C-4. Additionally, the reduced [1-13C]acetyl-CoA/unlabeled acetyl-CoA ratio in the 4/16 experiment will reduce the labeling of glutamate C-1 and C-5, and thus the fractional C-2 and C-4 doublet intensities, even in the case of many turns of the TCA cycle.
Gluconeogenesis.
The observation of significant trehalose in the cells is consistent with some degree of osmotic stress that results from labeling at higher cell density. The glucose carbons of trehalose can become labeled either via phosphorylation of pyruvate followed by conversion to triose phosphates, or via formation of phosphoenolpyruvate (PEP) through the action of malic enzyme or PEP carboxykinase (PEPCK). In the first case, labeling of the trehalose C-1/6, C-2/5, and C-3/4 carbons will parallel the labeling of pyruvate and pyruvate-derived metabolites such as alanine, resulting in equal labeling of carbons 2, 3, 4, and 5 and no labeling of carbons 1 and 6. In the second scenario, the labeling of the trehalose positions will be more analogous to the labeling of TCA metabolites derived from malate. Carbons 1/6, 2/5, and 3/4 will have multiplet structure similar to glutamate C-3, C-2, and C-1, respectively, except for the obvious difference that C-3 and C-4 will be coupled to each other. The trehalose spectrum reveals the highest labeling level at positions C-3 and C-4 and lesser but significant labeling at C-2 and C-5. The trehalose C-1 and C-6 resonances appear predominantly as singlets, and the resolved C-2 resonance appears as a singlet plus doublet, with 1JCC = 36.5 Hz, which would correspond to coupling with C-3 rather than C-1 (39). Based on the trehalose spectrum, it is apparent that under the conditions of the present studies, the triosephosphate pool is derived primarily from C-4 precursors via the action of malic enzymes and/or PEPCK. The spectra differ significantly from predictions based on direct phosphorylation of pyruvate, although the latter pathway may contribute to a very limited extent. In particular, it is noted that for the resolved trehalose C-2 resonance, the singlet/doublet ratio is 0.61, compared with an expected value near zero if pyruvate were the direct precursor.
Methylmalonate pathway.
A principal rationale for the use of [1,2-13C2]propionate is to distinguish pathways preserving the propionate carbon skeleton from the methylmalonate pathway, in which the coupling pattern will be lost initially. Entry of [1,2-13C2]propionate via this pathway will result in [1,3-13C2]succinyl-CoA and a mixture of [1,3-13C2]- and [2,4-13C2]OA. Initially, condensation with (unlabeled) acetyl-CoA will yield [1,3-13C2]- and [2-13C]α-ketoglutarate and glutamate, in contrast with the 1,2-13C2 and 3-13C labeling pattern observed (Fig. 4). Input of label through acetyl-CoA not considered, further turns of the citric acid cycle will label α-ketoglutarate and ultimately glutamate at carbons 1, 2, and 3 but without simultaneously labeling directly bonded carbons. The labeled OA can also lead to labeling of pyruvate (1,3-13C2 plus 2-13C) and, via pyruvate dehydrogenase or pyruvate-formate lyase, to labeled acetyl-CoA (1-13C plus 2-13C). The [1-13C]acetyl-CoA will label glutamate C-1 and C-5, again with no directly bonded carbons labeled. The [2-13C]acetyl-CoA provides a precursor leading to multiply labeled glutamate; however, in this case, the glutamate labeling will be very different, with a highly labeled C-4 position, in contrast with the observed spectrum (Fig. 4). The [1,3-13C2]- and [2-13C]pyruvate formed will be transaminated to yield [1,3-13C2]- and [2-13C]alanine, in contrast with the observation of [1,2-13C2]alanine. Thus, under these growth conditions, the labeling observed in alanine, and in the TCA-derived metabolites, provides no evidence for flux through the methylmalonate pathway.
Entry of labeled propionate via the methylmalonyl-CoA pathway involving condensation with catabolically produced 13CO2 can result in the production of [1,3,4-13C3]succinyl-CoA and thus [1,2,4-13C3]OA and [1,3,4-13C3]OA isotopomers. These can be converted to a mixture of 1,2-13C2- and 1,3-13C2-labeled pyruvate and alanine. However, as discussed above, lowering the [1,2-13C2]propionate/unlabeled propionate ratio from 20/0 to 4/16 reduced the maximum theoretical enrichment of the CO2 pool from 55 to 11% (ignoring endogenous and atmospheric sources of unlabeled CO2), but the 13C alanine spectrum obtained was nearly unaffected. A small fractional increase in the intensity of alanine C-2 singlets is observed, most probably indicative of the relatively more significant natural abundance background arising from unlabeled alanine.
Other metabolites.
Multiplet resonances corresponding to succinate C-2/3 are observed at 35.2 ppm. The pattern observed is consistent with a mixture of [2/3-13C]- and [1,2-13C2]/[3,4-13C2]succinate, as expected to arise from labeled OA produced as outlined in Fig. 3. In contrast, entry of the [1,2-13C2]propionate via the methylmalonate pathway would be expected to yield 1,3-13C2- and 2,4-13C2-labeled isotopomers, so that multiplet intensity due to 1J12 coupling would not be observed. Hence, the succinate labeling pattern also is indicative of propionate metabolism via the methylcitrate pathway. The large flux through pyruvate-formate lyase and the significant accumulation of succinate observed in this study are more typical of anaerobic metabolism. Although the cells were aerobically grown, the aerobic metabolism of the microorganism may be compromised under the conditions of growth on propionate.
The identification of a resonance due to putrescine at δ = 40.1 ppm is based on comparison with a standard sample, as well as the known biosynthetic pathway for this metabolite (20). The carbon skeleton of putrescine is derived via formation of ornithine from glutamate, followed by decarboxylation. Hence, the labeled C-2 and C-5 resonances of glutamate contribute to the observed C-1,4 singlet, explaining its large intensity. The 1J12 coupling interaction that is observed for glutamate C-2 is lost as a result of decarboxylation. Previous NMR studies of putrescine in E. coli have involved direct addition of the labeled compound to cell suspensions (9).
A multiplet observed at δ = 44.9 ppm, which appears to consist of a singlet plus doublet with 1J12 = 54.2 Hz, characteristic of coupling to a carboxyl carbon, was prominent in the 13C spectra. Based on the chemical shift, two types of structural units, the methylene group of citrate, malate, or potentially 2-methylcitrate or the methylene group of a C-terminal glycine, are the most likely assignments. Predictions of the shifts for these metabolites based on substituent effects (29) and on expected symmetries of labeling eliminated the first group of possibilities, leaving C-terminal glycine as the most likely candidate. Consistent with this conclusion, a 1H-13C HSQC spectrum of the extract indicates that the carbon multiplet is coupled to a closely spaced AB pattern in the proton spectrum with a mean shift of 3.6 ppm. Such a pattern would be consistent with a C-terminal glycine residue, with the proton inequivalence arising from a chirality elsewhere in the molecule. Furthermore, the AB spin system exhibits no additional proton-proton coupling in the 1H-13C HSQC-TOCSY spectrum, providing further support for this assignment. Further confirmation of this assignment is obtained by analysis of the acid hydrolyzed sample (Fig. 6A) in which the multiplet was found to shift upfield to 42.7 ppm, as expected for free glycine.
Based on the assignment given above, the multiplet structure of this resonance indicates the existence of a substantial pool of doubly labeled glycine. For growth on glucose, glycine is typically derived from 3-phosphoserine, derived in turn from 3-phosphoglycerate. The 3-phosphoglycerate can be produced by oxidation of glyceraldehyde-3-phosphate. Hence, the glycine C-1 and C-2 carbons would be expected to have a labeling pattern similar to that of C-1 and C-2 of the 3-phosphoglycerate. If the 3-phosphoglycerate is derived from glyceraldehyde-3-phosphate, the labeling of glycine C-1 and C-2 should be similar to the labeling of C-3 and C-2 of trehalose. Alternatively, for growth on other carbon sources, glycine can be derived from threonine, which is derived via aspartate from OA. Hence, in either case, the glycine labeling pattern is expected to be derived ultimately from the OA pool, which contains particularly [1,2-13C2]OA. This is consistent with the high doublet/singlet ratio observed for this resonance, which appears somewhat greater than the corresponding ratio in glutamate (Fig. 4). If this C-terminal glycine arises primarily from glutathione, a substantial fraction of the observed glutathione C-2 resonance intensity must also correspond to the γ-glutamyl group in glutathione. In fact, weak resonances can be observed at 27.6 and 32.7 ppm. These resonances most probably correspond to the γ-glutamyl C-3 and C-4 resonances of glutathione (24). However, the greater intensities of the free glutamate resonances indicate that either the C-terminal glycyl residue of glutathione becomes more heavily labeled than the γ-glutamyl residue or the C-terminal glycine corresponds to additional peptide(s).
DISCUSSION
The large range of propionate metabolic pathways represents a considerable analytical challenge. Perhaps as a result of this complexity, the existence of the methylcitrate pathway in E. coli was not demonstrated until 1997 (34). The extent to which several different pathways may contribute in various organisms is still not clear. The present study was motivated by the question of whether an approach which leads to qualitative, rather than merely quantitative, metabolite labeling differences which are dependent on the metabolic pathways involved could be developed. The results demonstrate the feasibility of using [1,2-13C2]propionate to unequivocally determine retention of the 13C-13C-12C carbon skeleton in pyruvate and alanine under the conditions of the study involving propionate-adapted E. coli. In this case, retention of the carbon skeleton of the propionate is consistent with flux via the methylcitrate pathway but not with flux through the methylmalonyl pathway. The methylmalonate pathway, which represents the primary alternate route for propionate uptake in some species, would in contrast result in the loss of coupling interaction of the precursor after formation of succinyl-CoA.
This and analogous approaches are limited by the complexity of the metabolic pathways, and by the possibility of synthetic coupling of a pair of labeled nuclei which can reintroduce the coupling interaction. Probably the most significant possibility in the present case relates to the incorporation of carbon-13 via 13CO2 fixation. However, these routes will in almost all cases be characterized by significant label dilution, so that the particular isotopomers will be diluted by the presence of other, less labeled isotopic forms of the molecule. In the present study, the pattern of labeling of alanine is largely retained, even if the [1,2-13C2]propionate precursor is substantially diluted with unlabeled propionate.
In contrast with the use of [1,2-13C2]propionate described here, the use of singly or triply labeled propionate precursors results in labeling patterns that are more ambiguous to analyze in terms of the metabolic flux. Previous NMR studies using [2-13C]propionate yielded glutamate labeled strongly at C-5, indicating formation of [1-13C]acetyl-CoA, as well as some enrichment at C-2 and C-3 (8, 34). Labeling at these latter positions can be interpreted to indicate anaplerotic flux via methylmalonyl-CoA formed by carboxylation of propionyl-CoA (8). However, with the [2-13C]propionate precursor, flux via the methylcitrate pathway can lead to [2-13C]pyruvate and to [2-13C]OA, which can result in an analogous labeling of glutamate C-2 or C-3.
[1,2,3-13C3]propionate has also been used to elucidate propionate metabolism (7, 13, 40). Metabolism via the methylmalonate pathway will lead to [1,2,3-13C3]succinyl-CoA and ultimately to [1,2,3-13C3]OA and [2,3,4-13C3]OA. Alternatively, metabolic conversion via the methylcitrate pathway will lead to [1,2,3-13C3]pyruvate and, after carboxylation, to [1,2,3-13C3]OA. Hence, although there will be some differences in the predicted label distribution of some metabolites, the [1,2,3-13C3]propionate label is considerably less able to distinguish between these alternative pathways.
The labeling pattern of pyruvate, as reflected most directly in the labeling of alanine and alanine-containing peptides, provides unequivocal confirmation of the functioning of the methylcitrate pathway. Consistent with previous studies (8), alanine levels in E. coli are very low, so that often analysis of the labeling of the pyruvate pool is based on the observations of metabolites derived more indirectly from this precursor. Thus, Evans et al. (8) observed no significant alanine C-2 resonance in E. coli labeled with [2-13C]propionate. As shown here, this observation arises from the low level of alanine in the cells rather than from a low level of labeling of the alanine. The structural cell wall or murein sacculus of E. coli contains the unit N-acetylglucosaminyl-β-1,4-N-acetylmuramic acid (MurNAc) with l-alanyl-d-glutamyl-mesodiaminopimelic acid attached to the muramic acid carboxyl group and with zero, one, or two d-alanine residues linked to the l-anomer of the diaminopimelic acid (6, 12, 22). The peptide components of the cell wall undergo rapid turnover, and intracellular pools of d-alanyl-d-alanine, diaminopimelic acid, d-glutamic acid, and various UDP-MurNAc peptides have all been identified. In the present study, we observed at least five doublets near 52 ppm which correspond to alanine and alanine-containing peptides. Although NMR has often been used to study the components of the bacterial cell wall, this is to our knowledge the first identification of intracellular peptides that are involved in cell wall turnover. This finding raises the possibility of more extensive studies of cell wall turnover using NMR in combination with stable isotope labeling of cell wall precursors such as alanine.
In addition to basic questions relating to the metabolic flux of propionate, the metabolism of propionate has been of interest recently due to its application to the determination of channeling of cellular metabolites (7, 8, 30). The latter phenomena can be studied by comparing experimental isotopic label distributions to those predicted to result from complete equilibration of label between symmetric sites on intermediates such as succinate or fumarate (3, 30). To perform such analyses, it is necessary to determine the metabolic pathways involved (23). As is apparent from the above discussion as well as the recent studies of Textor et al. (34), it is not necessary to invoke flux through the methylmalonyl pathway to explain the isotopic labeling patterns of metabolites derived from 13C-labeled propionate under the conditions of these studies. The use of [1,2-13C2]propionate should be valuable for determining whether such a pathway contributes to the utilization of propionate under other conditions.
ACKNOWLEDGMENT
We are grateful to Roel Schaaper for the suggestion and gift of E. coli MG1655 cells used in these studies.
REFERENCES
- 1.Bax A, Davis D G. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 2.Bax A, Ikura M, Kay L E, Torchia D A, Tschudin R. Comparison of different modes of 2-dimensional reverse-correlation NMR for the study of proteins. J Magn Reson. 1990;86:304–318. [Google Scholar]
- 3.Bernhard S A, Tompa P. The mechanism of succinate or fumarate transfer in the tricarboxylic acid cycle allows molecular rotation of the intermediate. Arch Biochem Biophys. 1990;276:191–198. doi: 10.1016/0003-9861(90)90026-u. [DOI] [PubMed] [Google Scholar]
- 4.Bodenhausen G, Ruben D J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 5.Buchanan B B. Role of ferredoxin in the synthesis of α-ketobutyrate from propionyl coenzyme-A and carbon dioxide by enzymes from photosynthetic and non-photosynthetic bacteria. J Biol Chem. 1969;244:4218–4223. [PubMed] [Google Scholar]
- 6.de Roubin M R, Mengin-Lecreulx D, van Heijenoort J. Peptidoglycan biosynthesis in Escherichia coli: variations in the metabolism of alanine and d-alanyl-d-alanine. J Gen Microbiol. 1992;138:1751–1757. doi: 10.1099/00221287-138-8-1751. [DOI] [PubMed] [Google Scholar]
- 7.Evans C T. Metabolic engineering of a non-allosteric citrate synthase in an Escherichia coli citrate synthase mutant. J Mol Recognit. 1995;8:327–333. doi: 10.1002/jmr.300080602. [DOI] [PubMed] [Google Scholar]
- 8.Evans C T, Sumegi B, Srere P A, Sherry A D, Malloy C R. [13C]propionate oxidation in wild-type and citrate synthase mutant Escherichia coli: evidence for multiple pathways of propionate utilization. Biochem J. 1993;291:927–932. doi: 10.1042/bj2910927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frydman B, Frydman R B, de los Santos C, Garrido D A, Goldemberg S H, Algranati I D. Putrescine distribution in Escherichia coli studied in vivo by 13C nuclear magnetic resonance. Biochim Biophys Acta. 1984;805:337–344. doi: 10.1016/0167-4889(84)90016-8. [DOI] [PubMed] [Google Scholar]
- 10.Halarnkar P P, Blomquist G J. Comparative aspects of propionate metabolism. Comp Biochem Physiol. 1989;92B:227–231. doi: 10.1016/0305-0491(89)90270-8. [DOI] [PubMed] [Google Scholar]
- 11.Hofmeister A E M, Buckel W. (R)-lactyl-CoA dehydratase from Clostridium propionicum: stereochemistry of the dehydration of (R)-2-hydroxybutyryl-CoA to crotonyl-CoA. Eur J Biochem. 1992;206:547–552. doi: 10.1111/j.1432-1033.1992.tb16958.x. [DOI] [PubMed] [Google Scholar]
- 12.Holtje J-V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 13.Jones J G, Naidoo R, Sherry A D, Jeffrey F M, Cottam G L, Malloy C R. Measurement of gluconeogenesis and pyruvate recycling in the rat liver: a simple analysis of glucose and glutamate isotopomers during metabolism of [1,2,3-13C3]propionate. FEBS Lett. 1997;412:131–137. doi: 10.1016/s0014-5793(97)00764-3. [DOI] [PubMed] [Google Scholar]
- 14.Kaziro Y, Leone E, Ochoa S. Biotin and propionyl carboxylase. Proc Natl Acad Sci USA. 1960;46:1319–1327. doi: 10.1073/pnas.46.10.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchta R D, Abeles R H. Lactate reduction in Clostridium propionicum. J Biol Chem. 1985;260:13181–13189. [PubMed] [Google Scholar]
- 16.Ladd J N, Walker D J. The fermentation of lactate and acrylate by the rumen micro-organism LC. Biochem J. 1959;71:364–373. doi: 10.1042/bj0710364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.London R E. 13C labeling in studies of metabolic regulation. Prog NMR Spectrosc. 1988;20:317–383. [Google Scholar]
- 18.London R E, Walker T E, Kollman V H, Matwiyoff N A. Studies of the pH dependence of 13C shifts and carbon-carbon coupling constants of [U-13C]aspartic and -glutamic acids. J Am Chem Soc. 1978;100:3723–3729. [Google Scholar]
- 19.Mazumder R, Sajakawa T, Ochoa S. Metabolism of propionic acid in animal tissues. X. Methylmalonyl coenzyme A mutase holoenzyme. J Biol Chem. 1963;238:50–53. [PubMed] [Google Scholar]
- 20.Morris D R, Pardee A B. Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem. 1966;241:3129–3135. [PubMed] [Google Scholar]
- 21.Norwood T J, Boyd J, Heritage J E, Soffe N, Campbell I D. Comparison of techniques for H-1 detected heteronuclear H-1-N-15 spectroscopy. J Magn Reson. 1990;87:488–501. [Google Scholar]
- 22.Park J T. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 23.Pronk J T, van der Linden-Beuman A, Verduyn C, Scheffers W A, van Dijken J P. Propionate metabolism in Saccharomyces cerevisiae: implications for the metabolon hypothesis. Microbiology. 1994;140:717–722. doi: 10.1099/00221287-140-4-717. [DOI] [PubMed] [Google Scholar]
- 24.Rabenstein D L, Keire D A. Nuclear magnetic resonance spectroscopy of glutathione. In: Dolphin D, Poulson R, Avramovic O, editors. Glutathione: chemical, biochemical, and medical aspects, part A. New York, N.Y: Wiley-Interscience; 1989. pp. 67–101. [Google Scholar]
- 25.Reeves H C, Ajl S C. α-Hydroxyglutaric acid synthetase. J Bacteriol. 1962;84:186–187. doi: 10.1128/jb.84.1.186-187.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer F D, Erfle J D, Mahadevan S. Amino acid biosynthesis in mixed rumen cultures. Biochem J. 1975;150:357–372. doi: 10.1042/bj1500357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweiger G, Buckel W. Studies on the dehydration of (R)-2-hydroxyglutarate in Acidaminococcus fermentans. A radical mechanism? Arch Microbiol. 1984;137:302–307. [Google Scholar]
- 28.Shaka A J, Keeler J, Freeman R. Evaluation of a new broadband decoupling sequence: WALTZ-16. J Magn Reson. 1983;53:313–340. [Google Scholar]
- 29.Stothers J B. Carbon-13 NMR spectroscopy. New York, N.Y: Academic Press; 1972. [Google Scholar]
- 30.Sumegi B, Sherry A D, Malloy C R. Channeling of TCA cycle intermediates in cultured Saccharomyces cerevisiae. Biochemistry. 1990;29:9106–9110. doi: 10.1021/bi00491a002. [DOI] [PubMed] [Google Scholar]
- 31.Sumegi B, Sherry A D, Malloy C R, Srere P A. Evidence for orientation-conserved transfer in the TCA cycle in Saccharomyces cerevisiae: 13C NMR studies. Biochemistry. 1993;32:12725–12729. doi: 10.1021/bi00210a022. [DOI] [PubMed] [Google Scholar]
- 32.Tabuchi T, Serizawa N, Uchiyama H. A novel pathway for the partial oxidation of propionyl-CoA to pyruvate via seven-carbon tricarboxylic acids in yeasts. Agric Biol Chem. 1974;38:2571–2572. [Google Scholar]
- 33.Tabuchi T, Uchiyama H. Methylcitrate condensing and methylisocitrate cleaving enzymes: evidence for the pathway of oxidation of propionyl-CoA to pyruvate via C7-tricarboxylic acids. Agric Biol Chem. 1975;39:2035–2042. [Google Scholar]
- 34.Textor S, Wendisch V F, De Graaf A A, Muller U, Linder M L, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168:428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- 35.Tran-Dinh S, Fermandjian S, Sala E, Mermet-Bouvier R, Cohen M, Fromageot P. 13C nuclear magnetic resonance studies of 85% 13C-enriched amino acids. Chemical shifts, coupling constants JC-C and conformation. J Am Chem Soc. 1974;96:1484–1493. doi: 10.1021/ja00812a035. [DOI] [PubMed] [Google Scholar]
- 36.Uchiyama H, Ando M, Toyonaka Y, Tabuchi T. Subcellular localization of the methylcitric-acid-cycle enzymes in propionate metabolism of Yarrowia lipolytica. Eur J Biochem. 1982;125:523–527. doi: 10.1111/j.1432-1033.1982.tb06713.x. [DOI] [PubMed] [Google Scholar]
- 37.Uchiyama H, Tabuchi T. Properties of methylcitrate synthase from Candida lipolytica. Agric Biol Chem. 1976;40:1411–1418. [Google Scholar]
- 38.Walker T E, Han C H, Kollman V H, London R E, Matwiyoff N A. 13C nuclear magnetic resonance studies of the biosynthesis by Microbacterium ammoniaphilum of l-glutamate selectively enriched with carbon-13. J Biol Chem. 1982;257:1189–1195. [PubMed] [Google Scholar]
- 39.Walker T E, London R E, Whaley T W, Barker R, Matwiyoff N A. Carbon-13 nuclear magnetic resonance spectroscopy of [1-13C] enriched monosaccharides. Signal assignments and orientational dependence of geminal and vicinal carbon-carbon and carbon-hydrogen spin-spin coupling constants. J Am Chem Soc. 1976;98:5807–5813. [Google Scholar]
- 40.Winter S M, Weber G L, Gooley P R, MacKenzie N E, Sipes I G. Identification and comparison of the urinary metabolites of [1,2,3-13C3]acrylic acid and [1,2,3-13C3]propionic acid in the rat by homonuclear 13C nuclear magnetic resonance spectroscopy. Drug Metab Dispos. 1992;20:665–672. [PubMed] [Google Scholar]